Abstract
An interferon-stimulated response element (ISRE)/interferon regulatory element (IRE) spanning nucleotide coordinates 1091–1100 is present in the enhancer 1/X gene promoter region of the hepatitis B virus (HBV) genome. In the context of a minimal promoter element, the enhancer 1/X gene promoter ISRE/IRE was shown to be a functional regulatory site capable of mediating interferon α- (IFNα) and interferon-stimulated gene factor 3 (ISGF3)-specific transcriptional activation in transient transfection analysis. The enhancer 1/X gene promoter ISRE/IRE was also shown to mediate interferon regulatory factor (IRF) 1 and IRF7 activation of transcription from a minimal promoter construct. In contrast, IFNα and the IRFs had minimal effect on HBV transcription and replication in the context of the viral genome in cell culture.
INTRODUCTION
Hepatitis B virus (HBV) is an enveloped virus associated with significant morbidity and mortality (1,2). HBV causes both acute and chronic liver disease. Chronic HBV infection is associated with liver cirrhosis and hepatocellular carcinoma (3,4). It is estimated that there are ∼300 000 000 chronic HBV carriers in the world. Current therapeutic interventions often fail to resolve the viral infection although they can improve the prognosis of the patients (5–8). A widely used therapeutic intervention is treatment with interferon α (IFNα) (6–11). IFNα therapy can reduce viral replication and liver damage associated with viral infection. The mechanisms of action of IFNα responsible for the improvement in the patients’ condition are poorly defined.
The genome of HBV is a partially double-stranded circular 3.2 kb DNA molecule. The DNA genome of the virus is transcribed in the nucleus of the hepatocyte to produce the 3.5, 2.4, 2.1 and 0.7 kb viral transcripts (2,12). The level of these transcripts are regulated by the enhancer 2/core promoter, the large surface antigen promoter, the major surface antigen promoter and the enhancer 1/X gene promoter, respectively (13–15). The 3.5 kb pregenomic HBV transcript encodes the viral polymerase and the nucleocapsid or core polypeptide (16). The HBV polymerase binds to the packaging sequence (ɛ) in the pregenomic RNA and this complex is encapsidated by core polypeptides to form an immature viral capsid (17–19). The reverse transcriptase/DNA polymerase activity of the viral polymerase converts the pregenomic RNA into the partially double-stranded DNA genome present in the mature capsid (20). The mature capsid interacts with the surface antigen polypeptides and subsequently buds into the lumen of the endoplasmic reticulum as a virus particle (21–23). Virions pass through the endoplasmic reticulum and Golgi apparatus prior to release from hepatocytes (12).
As the pregenomic RNA is the substrate for the generation of viral DNA replication intermediates, it is apparent that regulation of HBV transcription can directly influence synthesis of the virus (24). The presence of a sequence element in the enhancer 1/X gene promoter region with homology to an interferon-stimulated response element (ISRE) and an interferon regulatory element (IRE) suggested that IFNα might directly regulate HBV transcription and, consequently, HBV replication (25–29). The relationship between IFNα and the activation of transcription mediated through ISRE/IRE sequences is complex. IFNα binds to the type 1 interferon receptor and activates the latent signal transducer and activator of transcription 1 (STAT1), STAT2 and interferon regulatory factor (IRF)9/p48/ISGF3γ, which form a trimeric complex, the interferon-stimulated gene factor 3 (ISGF3). ISGF3 translocates into the nucleus where it binds to ISRE sequences and directly activates transcription of interferon-stimulated genes (ISG) (30). One of the target genes for ISGF3 is IRF7 (31). In addition, IFNβ has been shown to stimulate IRF1 and IRF2 expression (32). Therefore, IFNα/β can indirectly activate transcription of genes possessing IRE sequences in their enhancers and promoters. However, the similarity in the consensus sequences for ISGF3 and IRF binding can make it difficult to establish the relative importance of these transcription factors for transcriptional activation from promoters containing ISRE/IRE sequences (26,28,29).
Initial analysis suggested that IRF9/p48/ISGF3γ was involved in the IFNα-induced suppression of HBV enhancer 1 activity (25). However, further studies suggested that IFNα suppressed viral replication by an indirect mechanism that did not involve the ISRE/IRE sequence in the HBV enhancer 1 region (27). In this study the functional properties of the HBV ISRE/IRE sequence in the enhancer 1 region were further characterized in cell culture. In the context of a minimal promoter, the HBV IRSE/IRE sequence was shown to mediate ISGF3- and IRF-mediated activation of transcription. In contrast, in the context of the viral genome, the HBV ISRE/IRE sequence was unable to mediate IFNα- or IRF-dependent modulation of transcription. This suggests that if HBV transcription and replication are directly regulated by IFNα and IRFs modulating enhancer 1 activity during a viral infection, this situation has not yet been reproduced in cell culture.
MATERIALS AND METHODS
Plasmid constructs
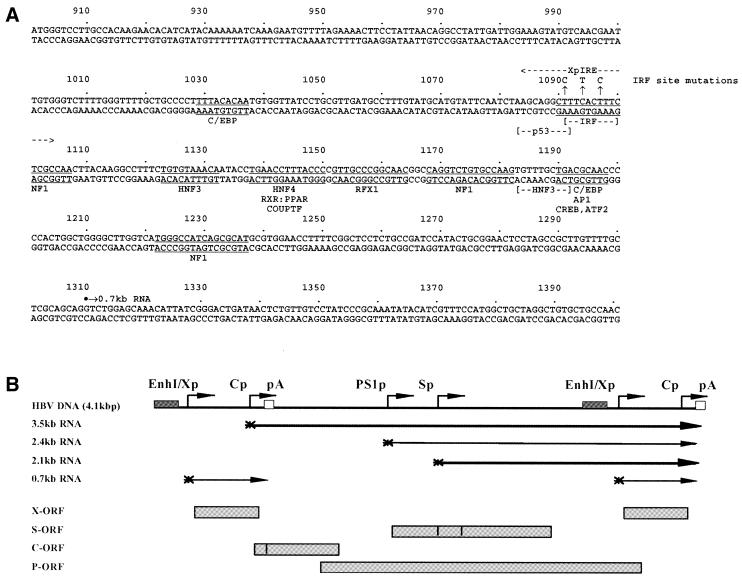
The steps in cloning of the plasmid constructs used in the transfection experiments were performed by standard techniques (33). HBV DNA sequences in these constructs were derived from plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (34). The firefly luciferase (LUC) reporter gene in these constructs was derived from plasmid p19DLUC (35). pHBVTATALUC was constructed by inserting a double-stranded oligonucleotide containing the large surface antigen promoter TATA-box element, produced by annealing the oligonucleotides CTATATTATATAAGAGAGAAGCT and TCTCTCTTATATAATATAGGTAC (spanning HBV coordinates 2773–2791), into the SacI and KpnI sites of p19DLUC in the same orientation as the TATA-box element occurs in the HBV genome (36). XpIRE(1)TATALUC, XpIRE(4)TATALUC, XpIREmut(4)TATALUC and ISG15-ISRE(3)TATALUC were made by inserting one to four copies (as indicated in the construct designation) of the XpIRE, XpIREmut and ISG15-ISRE double-stranded oligonucleotides into the unique SalI site of pHBVTATALUC. The oligonucleotide pairs used to generate the XpIRE, XpIREmut and ISG15-ISRE double-stranded oligonucleotides were TCGAGCAGGCTTTCACTTTCTCGC and TCGAGCGAGAAAGTGAAAGCCTGC (oligo XpIRE, HBV nucleotide coordinates 1084–1104), TCGAGCAGGCcTTtACcTTCTCGC and TCGAGCGAGAAgGTaAAgGCCTGC (oligo XpIREmut, mutated nucleotides in the HBV ISRE/IRE are indicated in lower case) and TCGAGCCTCGGGAAAGGGAAACCGAAACTGAA and TCGATTCAGTTTCGGTTTCCCTTTCCCGAGGC (oligo ISG15-ISRE, nucleotide sequence located between position –119 and –92 of the ISG15 promoter). The XpIRE double-stranded oligonucleotide spans the HBV enhancer 1/X gene promoter IRF-binding site (Fig. 1A; 25,27). The ISG15-ISRE double-stranded oligonucleotide spans the ISRE of interferon-stimulated gene 15 (ISG15) (37,38). The luciferase reporter gene constructs, ISG54LUC and p[(AAGTGA)4]5tkΔ(–39)lucter, have been described previously (39,40). The ISG54LUC construct contains the hamster interferon-stimulated gene 54 (ISG54) promoter spanning –429 and +31 and controlling the level of expression of the luciferase reporter gene (39). The p[(AAGTGA)4]5tkΔ(–39)lucter construct contains five copies of a synthetic IRE upstream of the herpes simplex virus minimal thymidine kinase promoter regulating the level of expression of the luciferase reporter gene (40).
Figure 1.
Sequence and structure of the HBV DNA constructs utilized in transient transfection analysis. (A) Sequence of the HBV enhancer 1/X gene promoter region (subtype ayw). The nucleotide coordinates are shown and an arrow indicates the approximate location of a putative HBV X gene transcription initiation site (58–60). The coordinates of the enhancer 1/X gene promoter region are derived from the GenBank genetic sequence data bank. The locations of the C/EBP (61), p53 (52), NF1 (50,51), IRF (25), HNF3 (62,63), HNF4 (64), RXR:PPAR (64–66), COUPTF (45,64), RFX1 (64,67,68), AP1 (51), CREB (69) and ATF2 (69) binding sites are shown. The location of the double-stranded oligonucleotide spanning the interferon regulatory element (XpIRE) and used in transient transfection analysis is also indicated. The 3 nt IRF site mutation is shown above the wild-type sequence. The nucleotide substitutions do not alter the HBV polymerase polypeptide sequence. (B) Structure of the HBV DNA (4.1 kb) construct used in transient transfection analysis. The 4.1 kb greater-than-genome length HBV DNA sequence in this construct spans coordinates 1072–3182 and 1–1990 of the HBV genome (subtype ayw). The locations of the HBV 3.5, 2.4, 2.1 and 0.7 kb transcripts are indicated. EnhI/Xp, enhancer 1/X gene promoter region; Cp, nucleocapsid or core promoter; pA, polyadenylation site; PS1p, presurface antigen promoter; Sp, major surface antigen promoter; X, X gene; S, surface antigen gene; C, core gene; P, polymerase gene; ORF, open reading frame.
The HBV DNA (4.1 kb) construct, containing 1.3 copies of the HBV genome, includes the viral sequence from nucleotide coordinates 1072–3182 plus 1–1990 (Fig. 1B). This plasmid was constructed by cloning the NsiI–BglII HBV DNA fragment (nucleotide coordinates 1072–1990) into pUC13, generating pHBV(1072–1990). Subsequently, a complete copy of the 3.2 kb viral genome linearized at the NcoI site (nucleotide coordinates 1375–3182 plus 1–1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1072–1990), generating the HBV DNA (4.1 kb) construct. The plasmid 4.1IREmut was derived by introducing clustered point mutations into the ISRE/IRE of the enhancer 1/X gene promoter in the HBV DNA (4.1 kb) construct using the Chameleon double-stranded, site-directed mutagenesis kit (Stratagene Cloning Systems, La Jolla, CA) according to the manufacturer’s instructions. The IREmut mutation converted the 10 nt ISRE/IRE sequence located between nucleotide coordinates 1091 and 1100 from GAAAGTGAAA to GAAgGTaAAg (Fig. 1A) but did not alter the HBV polymerase polypeptide sequence. The nucleotide substitutions introduced into the enhancer 1/X gene promoter construct were verified by dideoxynucleotide sequencing (41). Both enhancer 1/X gene promoter regions in the terminally redundant HBV DNA (4.1 kb) construct were mutated for this analysis.
The pCMVSTAT1α, pCMVSTAT2, pCMVISGF3γ p48, pCMVIRF1, pCMVIRF2, pBPSRT1IRF3 and pCMVIRF7 vectors express STAT1, STAT2, IRF9/p48/ISGF3γ, IRF1, IRF2, IRF3 and pIRF7 polypeptides from the human STAT1, human STAT2, human IRF9/p48/ISGF3γ, human IRF1, human IRF2, mouse IRF3 and mouse IRF7 cDNAs, respectively, using the cytomegalovirus immediate early promoter (pCMV) (31,39,42).
Cells and transfections
The human hepatoma cell line HepG2 was grown in RPMI-1640 medium and 10% fetal bovine serum at 37°C in 5% CO2/air. Transfections using luciferase reporter gene constructs were performed as previously described (43,44), except that 6-well plates, containing ∼3 × 105 cells, were used. The transfected DNA mixture comprised 5 µg LUC plasmid and 0.25 µg pCMVβ, which served as an internal control for transfection efficiency. pCMVβ directs expression of the Escherichia coli β-galactosidase (β-gal) gene using the cytomegalovirus (CMV) immediate early promoter (Clontech Laboratories, Palo Alto, CA). When appropriate, the DNA mixture also included 0.5 µg STAT1, STAT2, IRF9/p48/ISGF3γ, IRF1, IRF2, IRF3 and pIRF7 expression vectors, pCMVSTAT1α, pCMVSTAT2, pCMVISGF3γ p48, pCMVIRF1, pCMVIRF2, pBPSRT1IRF3 and pCMVIRF7, respectively, or the control expression vector, pCMV (45). The DNA was removed 4–6 h after transfection and washed with 2 ml of medium. Fresh RPMI medium containing recombinant human IFNα (PBL Biomedical Laboratories, New Brunswick, NJ) at a final concentration of 1000 U/ml replaced the standard medium as required 10 h prior to harvesting cells. Serum-free DMEM containing poly(I)·poly(C) (Fluka Chemical Corp., NY) and DEAE–dextran at final concentrations of 10 and 500 µg/ml, respectively, replaced the standard medium as required 10 h prior to harvesting cells. After 1 h the serum-free medium containing poly(I)·poly(C) was replaced with RPMI-1640 medium containing 10% fetal bovine serum. Cell extracts were prepared 40–48 h after transfection. Cells were lysed in 150 µl of lysis buffer (0.1 M potassium phosphate, pH 7.8, 0.2% v/v Triton X-100) and the cell debris was pelleted by centrifugation for 2 min at 13 000 r.p.m. in an Eppendorf 5417C microcentrifuge with an F45-30-11 rotor. The supernatant was assayed for luciferase activity essentially as previously described (46) and for β-galactosidase activity using a Galacto-Light kit (Tropix Inc.) as instructed by the manufacturer. The level of β-galactosidase activity observed was not specifically affected by any of the exogenously expressed transciption factors. The luciferase activities were normalized to the level of β-galactosidase activity in each transfection experiment.
Transfections for viral RNA and DNA analysis were performed as previously described (47) using 10 cm plates, containing ∼1 × 106 cells. DNA and RNA isolation was performed 3 days post-transfection. The transfected DNA mixture was composed of 10 µg HBV DNA (4.1 kb) plus 1.5 µg transcription factor expression vectors, pCMVIRF1, pCMVIRF2, pBPSRT1IRF3 and pCMVIRF7 (39,42). Controls were derived from cells transfected with HBV DNA (4.1 kb) and the pCMV expression vector lacking a transcription factor cDNA insert (45). Fresh RPMI medium containing recombinant human IFNα (PBL Biomedical Laboratories) at a final concentration of 1000 U/ml replaced the standard medium as required 10 h prior to harvesting cells.
Characterization of HBV transcripts and viral replication intermediates
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (48), with minor modifications. For RNA isolation (49) the cells were lysed in 1.8 ml of 25 mM sodium citrate, pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (v/v) sarcosyl, 0.1 M 2-mercaptoethanol. After addition of 0.18 ml of 2 M sodium acetate, pH 4.0, the lysate was extracted with 1.8 ml of water-saturated phenol plus 0.36 ml of chloroform/isoamyl alcohol (49:1). After centrifugation for 30 min at 3000 r.p.m. in a Sorval RT6000 rotor, the aqueous layer was precipitated with 1.8 ml of isopropanol. The precipitate was resuspended in 0.3 ml of 25 mM sodium citrate, pH 7.0, 4 M guanidinium isothiocyanate, 0.5% (v/v) sarcosyl, 0.1 M 2-mercaptoethanol and precipitated with 0.6 ml of ethanol. After centrifugation for 20 min at 14 000 r.p.m. in an Eppendorf 5417C microcentrifuge with an F45-30-11 rotor, the precipitate was resuspended in 0.3 ml of 10 mM Tris–HCl, pH 8.0, 5 mM EDTA, 0.1% (w/v) sodium lauryl sulfate and precipitated with 45 µl of 2 M sodium acetate plus 0.7 ml of ethanol.
For the isolation of viral DNA replication intermediates the cells were lysed in 0.4 ml of 100 mM Tris–HCl, pH 8.0, 0.2% (v/v) NP40. The lysate was centrifuged for 1 min at 14 000 r.p.m. in an Eppendorf 5417C microcentrifuge with an F45-30-11 rotor to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate plus 200 µg/ml DNase I and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl, 10 mM EDTA, 0.8% (w/v) sodium lauryl sulfate, 1.6 mg/ml pronase and incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 vol of ethanol and resuspended in 100 µl of 10 mM Tris–HCl, pH 8.0, 1 mM EDTA. RNA (northern) and DNA (Southern) filter hybridization analyses were performed using 10 µg total cellular RNA and 30 µl viral DNA replication intermediates, respectively, as described (33).
RESULTS
ISGF3 and IFNα activate transcription through the HBV enhancer 1 ISRE/IRE sequence in the context of a minimal promoter
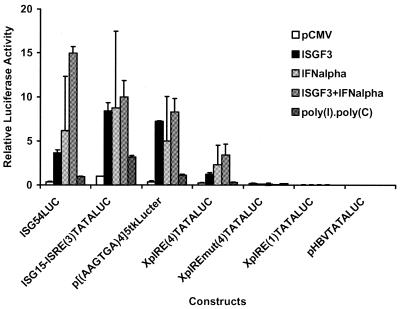
The enhancer 1/X gene promoter ISRE/IRE sequence (Fig. 1A) was examined in the context of a minimal TATA-box element located upstream of the luciferase open reading frame (Fig. 2). These synthetic promoter constructs were tested for their transcriptional activity in the HepG2 cell line in the absence or presence of ectopic expression of STAT1, STAT2 and IRF9/p48/ISGF3γ, which constitute the ISGF3 transcription factor. The level of transcription from the minimal promoter construct XpIRE(4)TATALUC, containing four copies of the enhancer 1/X gene promoter ISRE/IRE sequence, increased 4.7-fold in the presence of ectopic expression of ISGF3 (ratio of relative activities from Fig. 2, 1.21/0.26). Similarly, IFNα treatment, which activates endogenous ISGF3, increased the level of transcription from the XpIRE(4)TATALUC construct ∼9-fold (ratio of relative activities from Fig. 2, 2.27/0.26). The combined effect of ectopic expression of ISGF3 and IFNα treatment was approximately additive and resulted in a 13-fold increase in transcription (ratio of relative activities from Fig. 2, 3.42/0.26). Mutation of the enhancer 1/X gene promoter ISRE/IRE sequence in the construct XpIREmut(4)TATALUC completely inhibited activation of transcription from the minimal promoter by ectopic expression of ISGF3 or IFNα treatment (Fig. 2). Poly(I)·poly(C) failed to greatly modulate transcription from the XpIRE(4)TATALUC construct, suggesting that this treatment poorly activated transcription factors, including ISGF3 and IRFs, which mediate their effects on transcription through the ISRE/IRE sequence in these cells. The single copy of the enhancer 1/X gene promoter ISRE/IRE sequence present in the XpIRE(1)TATALUC construct failed to mediate ISGF3- or IFNα-dependent transcription, indicating that in the context of a minimal promoter multiple ISRE/IRE sequences are required for activation of transcription (Fig. 2). However, activation of transcription by ISGF3 or IFNα treatment demonstrates that the enhancer 1/X gene promoter ISRE/IRE sequence can act as an interferon-stimulated response element in the context of a minimal promoter element.
Figure 2.
Functional analysis of the HBV enhancer 1/X gene promoter ISRE/ IRE in the context of a minimal promoter. The XpIRE(1)TATALUC, XpIRE(4)TATALUC, XpIREmut(4)TATALUC and ISG15-ISRE(3)TATALUC constructs contain the XpIRE (enhancer 1/X gene promoter IRE), XpIREmut (enhancer 1/X gene promoter mutant IRE) and ISG15-ISRE (ISG15 promoter ISRE) double-stranded oligonucleotides cloned into the minimal promoter construct pHBVTATALUC, respectively. The number of copies of the oligonucleotide in the constructs is shown in parentheses in the construct name. The ISG54LUC construct contains the hamster ISG54 promoter spanning –429 to +31 controlling the level of expression of the luciferase reporter gene (39). The p[(AAGTGA)4]5tkΔ(–39)lucter construct contains five copies of a synthetic IRE upstream of the herpes simplex virus minimal thymidine kinase promoter that regulates the level of expression of the luciferase reporter gene (40). Relative activities of the constructs in HepG2 cells in the absence of treatment or ectopically expressed transcription factors (pCMV), in the presence of ectopically expressed interferon-stimulated gene factor 3 (ISGF3), after IFNα treatment (IFNalpha), in the presence of ectopically expressed ISGF3 with IFNα treatment (ISGF3+IFNalpha) and after poly(I)·poly(C) treatment are indicated. ISGF3 was expressed by co-transfecting the expression vectors pCMVSTAT1α, pCMVSTAT2 and pCMVISGF3γ p48. The transcriptional activities are reported relative to the ISG15-ISRE(3)LUC plasmid in the absence of any expression vector or treatment and is designated as having a relative activity of 1.0. The relative activities of the ISG54LUC construct under the various conditions were divided by 10 so that all constructs could be shown using the same scale. The standard deviation of the mean is indicated by an error bar. The internal control used to correct for transfection efficiencies was pCMVβ.
The ISG54LUC, ISG15-ISRE(3)TATALUC and p[(AAGTGA)4]5tkΔ(–39)lucter constructs demonstrated similar, but somewhat greater, responsiveness to ectopic expression of ISGF3 and IFNα treatment, confirming that the enhancer 1/X gene promoter ISRE/IRE sequence can act as an interferon-stimulated response element in a manner similar to previously characterized ISRE/IRE-containing promoters (37–40). These constructs also showed a 3-fold increase in transcription in response to poly(I)·poly(C) treatment, suggesting that this stimulus weakly activates transcription factors that can mediate their effects through ISRE/IRE sequences in HepG2 cells (Fig. 2). Similar results using these constructs were also obtained using Huh7 cells (data not shown).
IRF1 and IRF7 activate transcription through the HBV enhancer 1 ISRE/IRE sequence in the context of a minimal promoter
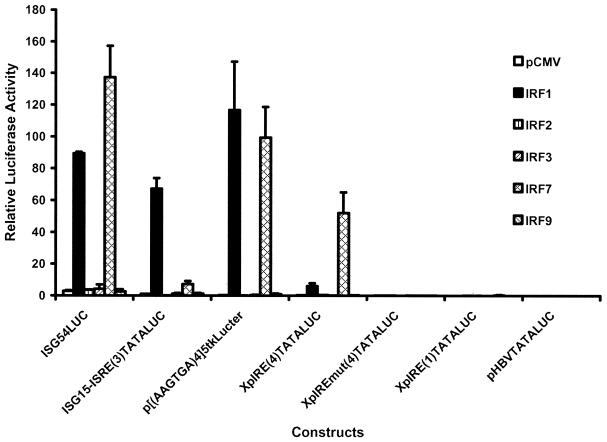
The enhancer 1/X gene promoter ISRE/IRE sequence (Fig. 1A) was examined in the context of a minimal TATA-box element located upstream of the luciferase open reading frame. These synthetic promoter constructs were tested for their transcriptional activities in the HepG2 cell line in the absence or presence of the ectopic expression of IRF1, IRF2, IRF3, IRF7 and IRF9/p48/ISGF3γ (Fig. 3). The level of transcription from the minimal promoter construct XpIRE(4)TATALUC, containing four copies of the enhancer 1/X gene promoter ISRE/IRE sequence, was increased ∼25- and 220-fold in the presence of ectopic expression of IRF1 and IRF7, respectively (ratio of relative activities from Fig. 3, 6.06/0.23 and 52.05/0.23, respectively). IRF2, IRF3 and IRF9/p48/ISGF3g failed to alter the level of transcription from the XpIRE(4)TATALUC construct. Mutation of the enhancer 1/X gene promoter ISRE/IRE sequence in the construct XpIREmut(4)TATALUC completely inhibited activation of transcription from the minimal promoter by ectopic expression of IRF1 and IRF7 (Fig. 3). IRF1 and IRF7 activated transcription from the XpIRE(1)TATALUC construct 2- and 6-fold, respectively, demonstrating that modest transcriptional activation by these transcription factors was possible from a minimal promoter containing a single copy of the enhancer 1/X gene promoter ISRE/IRE sequence (ratio of relative activities from Fig. 3, 0.08/0.04 and 0.24/0.04, respectively). These results demonstrate that the enhancer 1/X gene promoter ISRE/IRE sequence is capable of mediating IRF-dependent transcriptional activation from a minimal promoter construct. The ISG54LUC, ISG15-ISRE(3)TATALUC and p[(AAGTGA)4]5tkΔ(–39)lucter constructs demonstrated qualitatively similar responsiveness to ectopic expression of IRF1 and IRF7, confirming that the enhancer 1/X gene promoter ISRE/IRE sequence can act as an interferon regulatory element in a manner similar to previously characterized ISRE/IRE-containing promoters (37–40). Similar results using these constructs were also obtained using Huh7 cells (data not shown).
Figure 3.
Effect of interferon regulatory factors on the HBV enhancer 1/X gene promoter ISRE/IRE in the context of a minimal promoter. The constructs used are as described in Figure 2. Relative activities of the constructs in HepG2 cells in the absence (pCMV) or presence of ectopically expressed IRF1, IRF2, IRF3, IRF7 and IRF9 using the expression vectors pCMV, pCMVIRF1, pCMVIRF2, pBPSRT1IRF3, pCMVIRF7 and pCMVISGF3γ p48, respectively, are indicated. The transcriptional activities are reported relative to the ISG15-ISRE(3)LUC plasmid in the absence of expression vectors and is designated as having a relative activity of 1.0. The standard deviation of the mean is indicated by an error bar. The internal control used to correct for transfection efficiencies was pCMVβ.
The HBV enhancer 1 ISRE/IRE sequence does not influence HBV transcription and replication in the context of the viral genome
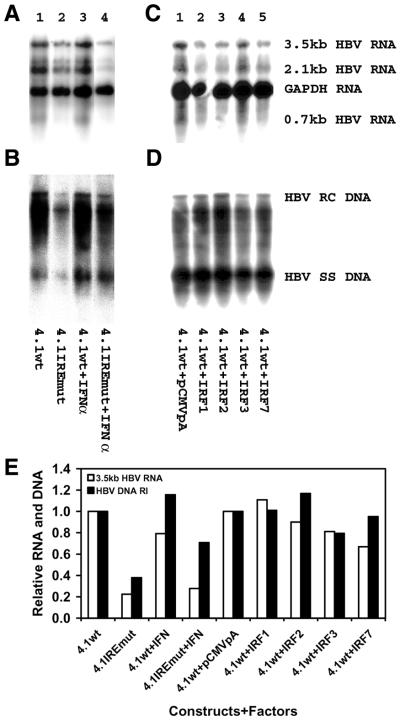
Using minimal promoter constructs, it was possible to demonstrate that the enhancer 1/X gene promoter ISRE/IRE sequence can mediate activation of transcription by IFNα and the transcription factors ISGF3, IRF1 and IRF7 (Figs 2 and 3). In contrast, reporter gene constructs containing the complete viral genome and utilizing the large surface antigen promoter (PS1pLUC), the major surface antigen promoter (SpLUC), the enhancer 1/X gene promoter (XpLUC) and the enhancer 2/core promoter (CpLUC) (35), respectively, failed to demonstrate any alteration in the level of transcription in response to treatment with IFNα or ectopic expression of IRF1 and IRF7 (data not shown). Similarly, viral transcription and replication derived from the HBV DNA (4.1 kb) construct (Fig. 1B) was not altered by treatment with IFNα or ectopic expression of IRF1 and IRF7 (Fig. 4). Mutation of the enhancer 1/X gene promoter ISRE/IRE sequence in the HBV DNA (4.1 kb) construct, generating the 4.1IREmut construct, reduced the level of the HBV 3.5 kb transcripts and viral replication intermediates 2– 4-fold (Fig. 4). This indicated that this regulatory element contributes to the level of pregenomic RNA expression. As this region of the enhancer 1/X gene promoter has been reported to bind p53 and NF1 in addition to the IRF transcription factors (25,27,50–52), it is reasonable that this element might contribute to the constitutive level of viral replication in HepG2 cells. However, it does not appear to be able to mediate ISGF3- or IRF-dependent alterations in viral transcription in the context of the viral genome (Fig. 4).
Figure 4.
The IRE in the enhancer 1/X gene promoter does not modulate HBV transcription and replication in HepG2 cells in response to IFN treatment or ectopic expression of IRF transcription factors. HepG2 cells were transiently transfected with the wild-type HBV DNA (4.1 kb) construct (4.1wt) or the HBV IREmut DNA (4.1 kb) construct (4.1IREmut) containing a 3 nt mutation (Fig. 1A) in the enhancer 1/X gene promoter ISRE. Both enhancer 1/X gene promoter regions in this terminally redundant HBV construct (Fig. 1B) were mutated for this analysis. (A and B) Cells were treated with IFNα (lanes 3 and 4) at 1000 U/ml. (C and D) The expression vectors, pCMV, pCMVIRF1, pCMVIRF2, pBPSRT1IRF3 and pCMVIRF7 (lanes 1–5), were included in the transient transfection analyses. (A and C) RNA (northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B and D) DNA (Southern) filter hybridization analysis of HBV replication intermediates. (E) Quantitative analysis of the 3.5 kb HBV RNA and HBV DNA replication intermediates. The levels of the 3.5 kb HBV RNA and HBV DNA replication intermediates are reported relative to the wild-type HBV DNA (4.1 kb) construct in the absence of IFNα treatment or ectopic transcription factor expression (lane 1), which are designated as having a relative activity of 1.0.
DISCUSSION
Chronic HBV infection is a serious health problem which is commonly treated with interferon therapy (7,8). IFN therapy suppresses viral replication and can relieve the clinical symptoms associated with HBV infection (7,8). However, IFN therapy does not efficiently resolve chronic HBV infections and the molecular mechanisms of IFN action contributing to the reduction in HBV replication are not well understood (7,8). In cell culture systems IFN generally reduces HBV transcription and replication to only a limited extent (27,53–55). This suggests that the mechanism of inhibition of HBV replication observed in vivo may be more complex than those demonstrated in cell culture. This is supported by studies using HBV transgenic mice, where treatment with poly(I)·poly(C), which stimulates IFN synthesis, or direct administration of IFN results in significant inhibition of viral replication (56). In HBV transgenic mice inhibition of HBV DNA synthesis occurs in the absence of any major effect on viral transcription, suggesting that post-transcriptional mechanisms may be responsible for the reduction in viral replication (56). The mechanism of the putative post-transcriptional inhibition of viral replication is unknown.
In contrast to the proposed post-transcriptional inhibition of viral replication by IFN in HBV transgenic mice, it has been suggested that IFN might modulate HBV transcription directly by activating ISGF3 or members of the IRF family of transcription factors (25,27). The presence of an ISRE/IRE sequence in the enhancer 1/X gene promoter region of the HBV genome supports this contention (25,27). However, the effect of IFN treatment on the level of transcription from the HBV promoters using reporter gene constructs and greater-than-genome length HBV DNA constructs capable of supporting viral replication indicated that IFN has a relatively modest effect on viral transcription and replication (25,27). Our analysis supports this suggestion (Fig. 4). These observations do not appear to support the suggestion that the enhancer 1/X gene promoter ISRE/IRE sequence is a functionally important element capable of responding to IFN signaling in cell culture.
In this study the enhancer 1/X gene promoter ISRE/IRE sequence was also analyzed in the context of a minimal promoter–reporter gene construct (Figs 2 and 3). This analysis demostrated that the enhancer 1/X gene promoter ISRE/IRE sequence was capable of responding to IFN treatment in HepG2 cells (Fig. 2). In addition, ectopic expression of ISGF3 also supported transcription from a minimal promoter construct containing the enhancer 1/X gene promoter ISRE/IRE sequence, indicating that IFN treatment probably mediated its effects by directly activating the ISGF3 transcription factor. The IRF1 and IRF7 transcription factors also increase transcription from the enhancer 1/X gene promoter ISRE/IRE sequence in the context of a minimal promoter element (Fig. 3). These findings support the previous observations that members of the IRF family of transcription factors can directly bind to the enhancer 1/X gene promoter ISRE/IRE sequence (25,27). However, it is noteworthy that IFN gene expression is activated by IRFs and interferon can activate IRF gene expression (31,32,57). The complex interplay between IFN expression, ISGF3 activation and IRF gene expression suggests that several of these transcription factors may modulate HBV gene expression in response to IFN treatment by interacting with the enhancer 1/X gene promoter ISRE/IRE sequence. It is possible that these interactions may play a role in modulating HBV replication in vivo during IFN therapy despite the absence of an effect of IFN on viral replication in cell culture.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Stephen Goodbourn (University of London, London, UK) for the plasmid p[(AAGTGA)4]5tkΔ(–39)lucter, Dr David E. Levy (New York University School of Medicine, New York, NY) for the plasmids ISG54LUC, pCMVSTAT1α, pCMVSTAT2, pCMVISGF3γ p48, pBPSRT1IRF3 and pCMVIRF7, and Dr John Hiscott (McGill University, Montreal, Canada) for the plasmids pCMVIRF1 and pCMVIRF2. This work was supported by a post-doctoral fellowship from the West China University of Medical Sciences of the People’s Republic of China to H.T. and Public Health Service Grant AI30070 from the National Institutes of Health.
REFERENCES
- 1.Ganem D. (1982) Persistent infection of humans with hepatitis B virus: mechanisms and consequences. Rev. Infect. Dis., 4, 1026–1047. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D. and Varmus,H.E. (1987) The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem., 56, 651–693. [DOI] [PubMed] [Google Scholar]
- 3.Beasley R.P., Hwang,L.-Y., Lin,C.-C. and Chien,C.-S. (1981) Hepatocellular carcinoma and hepatitis B virus—a prospective study of 22707 men in Taiwan. Lancet, 2, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 4.Beasley R.P. (1988) Hepatitis B virus: the major etiology of hepatocellular carcinoma. Cancer, 61, 1942–1956. [DOI] [PubMed] [Google Scholar]
- 5.Malik A.H. and Lee,W.M. (2000) Chronic hepatitis B virus infection: treatment strategies for the next millennium. Ann. Intern. Med., 132, 723–731. [DOI] [PubMed] [Google Scholar]
- 6.Lin O.S. and Keeffe,E.B. (2001) Current treatment strategies for chronic hepatitis B and C. Annu. Rev. Med., 52, 29–49. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle J.H. and Di Bisceglie,A.M. (1997) The treatment of chronic viral hepatitis. N. Engl. J. Med., 336, 347–356. [DOI] [PubMed] [Google Scholar]
- 8.Niederau C., Heintges,T., Lange,S., Goldmann,G., Niederau,C.M., Mohr,L. and Haussinger,D. (1996) Long-term follow-up of HBeAg-positive patients treated with interferon alfa for chronic hepatitis B. N. Engl. J. Med., 334, 1422–1427. [DOI] [PubMed] [Google Scholar]
- 9.Krogsgaard K., Bindslev,N., Christensen,E., Craxi,A., Schlichting,P., Schalm,S., Carreno,V., Trepo,C., Gerken,G., Thomas,H.C. et al. (1994) The treatment effect of alpha interferon in chronic hepatitis B is independent of pre-treatment variables. Results based on individual patient data from 10 clinical controlled trials. J. Hepatol., 21, 646–655. [DOI] [PubMed] [Google Scholar]
- 10.Narkewicz M.R., Smith,D., Silverman,A., Vierling,J. and Sokol,R.J. (1995) Clearance of chronic hepatitis B virus infection in young children after alpha interferon treatment. J. Pediatr., 127, 815–818. [DOI] [PubMed] [Google Scholar]
- 11.Naoumov N.V., Thomas,M.G., Mason,A.L., Chokshi,S., Bodicky,C.J., Farzaneh,F., Williams,R. and Perrillo,R.P. (1995) Genomic variations in the hepatitis B core gene: a possible factor influencing response to interferon alfa treatment. Gastroenterology, 108, 505–514. [DOI] [PubMed] [Google Scholar]
- 12.Raney A.K. and McLachlan,A. (1991) In McLachlan,A. (ed.), Molecular Biology of the Hepatitis B Virus. CRC Press, Boca Raton, FL, pp. 1–37.
- 13.Schaller H. and Fischer,M. (1991) Transcriptional control of hepadnavirus gene expression. Curr. Top. Microbiol. Immunol., 168, 21–39. [DOI] [PubMed] [Google Scholar]
- 14.Yen T.S.B. (1993) Regulation of hepatitis B virus gene expression. Semin. Virol., 4, 33–42. [Google Scholar]
- 15.Kosovsky M.J., Qadri,I. and Siddiqui,A. (1998) In Koshy,R. and Caselmann,W.H. (eds), Hepatitis B Virus: Molecular Mechanisms in Disease and Novel Strategies for Therapy. Imperial College Press, London, UK, pp. 21–50.
- 16.Ou J.-H., Bao,H., Shih,C. and Tahara,S.M. (1990) Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J. Virol., 64, 4578–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsch R.C., Lavine,J.E., Chang,L.-J., Varmus,H.E. and Ganem,D. (1990) Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature, 344, 552–555. [DOI] [PubMed] [Google Scholar]
- 18.Junker-Niepmann M., Bartenschlager,R. and Schaller,H. (1990) A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J., 9, 3389–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartenschlager R., Junker-Niepmann,M. and Schaller,H. (1990) The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol., 64, 5324–5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Will H., Reiser,W., Weimer,T., Pfaff,E., Buscher,M., Sprengle,R., Cattaneo,R. and Schaller,H. (1987) Replication strategy of human hepatitis B virus. J. Virol., 61, 904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada G., Sakamoto,Y., Mizuno,M., Nishihara,T., Kobayashi,T., Takahashi,T. and Nagashima,H. (1982) Electron and immunoelectron microscopic study of Dane particle formation in chronic hepatitis B virus infection. Gastroenterology, 83, 348–356. [PubMed] [Google Scholar]
- 22.Gerber M.A., Sells,M.A., Chen,M.-L., Thung,S.N., Tabibzadeh,S.S., Hood,A. and Acs,G. (1988) Morphologic, immunohistochemical and ultrastructural studies of the production of hepatitis B virus in vitro. Lab. Invest., 59, 173–180. [PubMed] [Google Scholar]
- 23.Kamimura T., Yoshikawa,A., Ichida,F. and Sasaki,H. (1981) Electron microscopic studies of Dane particles in hepatocytes with special reference to intracellular development of Dane particles and their relation with HBeAg in serum. Hepatology, 1, 392–397. [DOI] [PubMed] [Google Scholar]
- 24.Tang H. and McLachlan,A. (2001) Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl Acad. Sci. USA, 98, 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakao K., Nakata,K., Yamashita,M., Tamada,Y., Hamasaki,K., Ishikawa,H., Kato,Y., Eguchi,K. and Ishii,N. (1999) p48 (ISGF-3γ) is involved in interferon-α-induced suppression of hepatitis B virus enhancer-1 activity. J. Biol. Chem., 274, 28075–28078. [DOI] [PubMed] [Google Scholar]
- 26.Fujii Y., Shimizu,T., Kusumoto,M., Kyogoku,Y., Taniguchi,T. and Hakoshima,T. (1999) Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J., 18, 5028–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rang A., Heise,T. and Will,H. (2001) Lack of a role of the interferon-stimulated response element-like region in interferon α-induced suppression of hepatitis B virus in vitro. J. Biol. Chem., 276, 3531–3535. [DOI] [PubMed] [Google Scholar]
- 28.Levy D.E., Kessler,D.S., Pine,R., Reich,N. and Darnell,J.E.,Jr (1988) Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev., 2, 383–393. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka N., Kawakami,T. and Taniguchi,T. (1993) Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol., 13, 4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler D.S., Veals,S.A., Fu,X.Y. and Levy,D.E. (1990) Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev., 4, 1753–1765. [DOI] [PubMed] [Google Scholar]
- 31.Marie I., Durbin,J.E. and Levy,D.E. (1998) Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J., 17, 6660–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harada H., Fujita,T., Miyamoto,M., Kimura,Y., Maruyama,M., Furia,A., Miyata,T. and Taniguchi,T. (1989) Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell, 58, 729–739. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Dubois M.F., Pourcel,C., Rousset,S., Chany,C. and Tiollais,P. (1980) Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl Acad. Sci. USA, 77, 4549–4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raney A.K., Milich,D.R., Easton,A.J. and McLachlan,A. (1990) Differentiation specific transcriptional regulation of the hepatitis B virus large surface antigen gene in human hepatoma cell lines. J. Virol., 64, 2360–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raney A.K., Easton,A.J. and McLachlan,A. (1994) Characterization of the minimal elements of the hepatitis B virus large surface antigen promoter. J. Gen. Virol., 75, 2671–2679. [DOI] [PubMed] [Google Scholar]
- 37.Wathelet M.G., Lin,C.H., Parekh,B.S., Ronco,L.V., Howley,P.M. and Maniatis,T. (1998) Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell, 1, 507–518. [DOI] [PubMed] [Google Scholar]
- 38.Reich N., Evans,B., Levy,D., Fahey,D., Knight,E.,Jr and Darnell,J.E.,Jr (1987) Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl Acad. Sci. USA, 84, 6394–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bluyssen H.A. and Levy,D.E. (1997) Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem., 272, 4600–4605. [DOI] [PubMed] [Google Scholar]
- 40.Whiteside S.T., King,P. and Goodbourn,S. (1994) A truncated form of the IRF-2 transcription factor has the properties of a postinduction repressor of interferon-beta gene expression. J. Biol. Chem., 269, 27059–27065. [PubMed] [Google Scholar]
- 41.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin R., Mustafa,A., Nguyen,H., Gewert,D. and Hiscott,J. (1994) Mutational analysis of interferon (IFN) regulatory factors 1 and 2. Effects on the induction of IFN-beta gene expression. J. Biol. Chem., 269, 17542–17549. [PubMed] [Google Scholar]
- 43.Sorge J., Wright,D., Erdman,V.D. and Cutting,A.E. (1984) Amphotropic retrovirus vector system for human cell gene transfer. Mol. Cell. Biol., 4, 1730–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graham F.L. and Van der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- 45.Raney A.K., Johnson,J.L., Palmer,C.N.A. and McLachlan,A. (1997) Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol., 71, 1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Wet J.R., Wood,K.V., DeLuca,M., Helinski,D.R. and Subramani,S. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol., 7, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLachlan A., Milich,D.R., Raney,A.K., Riggs,M.G., Hughes,J.L., Sorge,J. and Chisari,F.V. (1987) Expression of hepatitis B virus surface and core antigens: influences of pre-s and precore sequences. J. Virol., 61, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Summers J., Smith,P.M., Huang,M. and Yu,M. (1991) Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol., 65, 1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 50.Patel N.U., Jameel,S., Isom,H. and Siddiqui,A. (1989) Interactions between nuclear factors and the hepatitis B virus enhancer. J. Virol., 63, 5293–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Levy R., Faktor,O., Berger,I. and Shaul,Y. (1989) Cellular factors that interact with the hepatitis B virus enhancer. Mol. Cell. Biol., 9, 1804–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ori A., Zauberman,A., Doitsh,G., Paran,N., Oren,M. and Shaul,Y. (1998) p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J., 17, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caselmann W.H., Meyer,M., Scholz,S., Hofschneider,P.H. and Koshy,R. (1992) Type I interferons inhibit hepatitis B virus replication and induce hepatocellular gene expression in cultured liver cells. J. Infect. Dis., 166, 966–971. [DOI] [PubMed] [Google Scholar]
- 54.Rang A., Günther,S. and Will,H. (1999) Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J. Hepatol., 31, 791–799. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi Y. and Koike,K. (1989) Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J. Virol., 63, 2936–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McClary H., Koch,R., Chisari,F.V. and Guidotti,L.G. (2000) Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol., 74, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen H., Hiscott,J. and Pitha,P.M. (1997) The growing family of interferon regulatory factors. Cytokine Growth Factor Rev., 8, 293–312. [DOI] [PubMed] [Google Scholar]
- 58.Siddiqui A., Jameel,S. and Mapoles,J. (1987) Expression of the hepatitis B virus X gene in mammalian cells. Proc. Natl Acad. Sci. USA, 84, 2513–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Treinin M. and Laub,O. (1987) Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol. Cell. Biol., 7, 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu K.-Q. and Siddiqui,A. (1991) Regulation of the hepatitis B virus gene expression by the enhancer element I. Virology, 181, 721–726. [DOI] [PubMed] [Google Scholar]
- 61.Landschulz W.H., Johnson,P.F., Adashi,E.Y., Graves,B.J. and McKnight,S.L. (1988) Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev., 2, 786–800. [DOI] [PubMed] [Google Scholar]
- 62.Chen M., Hieng,S., Qian,X., Costa,R. and Ou,J.H. (1994) Regulation of hepatitis B virus ENI activity by hepatocyte-enriched transcription factor HNF3. Virology, 205, 127–132. [DOI] [PubMed] [Google Scholar]
- 63.Ori A. and Shaul,Y. (1995) Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology, 207, 98–106. [DOI] [PubMed] [Google Scholar]
- 64.Garcia A.D., Ostapchuk,P. and Hearing,P. (1993) Functional interaction of nuclear factors EF-C, HNF-4 and RXRα with hepatitis B virus enhancer I. J. Virol., 67, 3940–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huan B. and Siddiqui,A. (1992) Retinoid X receptor RXRα binds to and trans-activates the hepatitis B virus enhancer. Proc. Natl Acad. Sci. USA, 89, 9059–9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huan B., Kosovsky,M.J. and Siddiqui,A. (1995) Retinoid X receptor α transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J. Virol., 69, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siegrist C.A., Durand,B., Emery,P., David,E., Hearing,P., Mach,B. and Reith,W. (1993) RFX1 is identical to enhancer factor C and functions as a transactivator of the hepatitis B virus enhancer. Mol. Cell. Biol., 13, 6375–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ostapchuk P., Scheirle,G. and Hearing,P. (1989) Binding of nuclear factor EF-C to a functional domain of the hepatitis B virus enhancer region. Mol. Cell. Biol., 9, 2787–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maguire H.F., Hoeffler,J.P. and Siddiqui,A. (1991) HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science, 252, 842–844. [DOI] [PubMed] [Google Scholar]