Abstract
TFIIIA is required to activate RNA polymerase III transcription from 5S RNA genes. Although all known TFIIIA homologs harbor nine zinc fingers that mediate DNA binding, very limited sequence homology is found among these proteins, which reflects unique properties of some TFIIIA homologs. For example, the Acanthamoeba castellanii homolog directly regulates 5S RNA transcription. We have purified and characterized A.castellanii TFIIIA (AcTFIIIA) as a step toward obtaining a clearer understanding of these differences and of the regulatory process. AcTFIIIA is 59 kDa, significantly larger than all other TFIIIA homologs isolated to date. Nevertheless, it exhibits a DNase I footprint very similar to those produced by the smaller vertebrate TFIIIA homologs, but distinct from the smaller footprint of the 51 kDa TFIIIA from Saccharomyces cerevisiae. Similar footprinting is not reflected in greater sequence similarity between the A.castellanii and vertebrate promoters.
INTRODUCTION
TFIIIA activates RNA polymerase III (Pol III)-mediated transcription of 5S RNA genes by first contacting the DNA promoter and allowing for the subsequent and sequential recruitment of TFIIIC, TFIIIB and Pol III (1,2). In Xenopus the 5S promoter consists of three elements: a 5′ proximal A-box, an intermediate element and a 3′ proximal C-box (3). TFIIIA has been found to bind all three elements, extending over the entire length of the ∼50 bp type I promoter found within the transcribed portion of the gene (4).
Because of the large amount of TFIIIA in Xenopus oocytes (up to 15% of the total soluble protein) this protein was the first eukaryotic transcription factor to be purified and cloned (4,5), which led to its extensive characterization. Xenopus laevis TFIIIA was found to have nine sequential C2H2-type zinc fingers that are essential for DNA binding, but expendable for transcription (6,7). Changes in the TFIIIA footprint effected by truncating the protein showed that the N-terminal fingers were positioned at the 3′ end of the promoter and the C-terminal fingers were positioned at the 5′ end (8). A polypeptide with only the N-terminal three fingers retained the ability to bind DNA with high affinity, illustrating the role of these fingers in promoter interactions (9,10). In contrast, disrupting the structure of any of the C-terminal three fingers was found to have small effects on DNA binding, but caused transcriptional inactivation (11).
The functional properties of other TFIIIA homologs vary from those of X.laevis TFIIIA. TFIIIA has been cloned from only four other frog species (12,13), catfish (14), human (15) and Saccharomyces cerevisiae (16). Interestingly, the amphibian homologs produce slightly different footprints on the Xenopus borealis somatic 5S RNA gene, and the ability to activate transcription in a X.laevis egg extract also differed with the substitution of TFIIIA homologs from two amphibian genera (17), showing that even among closely related species the properties of TFIIIA can differ.
Functional differences are further supported by the lack of amino acid sequence conservation among the homologs, which has hindered identification of other TFIIIA genes. Saccharomyces cerevisiae TFIIIA was identified as a nine zinc finger-containing gene flanking a cloned RNA polymerase subunit (16). Although S.cerevisiae TFIIIA, like the other homologs, has nine zinc fingers, it is only ∼23% homologous to the Xenopus factor at the amino acid level. One significant difference between Xenopus and S.cerevisiae TFIIIA is their size; yeast TFIIIA is approximately one third larger than the 39 kDa Xenopus protein primarily due to an 81 amino acid insertion between zinc fingers 8 and 9 (16) that is not found in other homologs. This domain is needed for transcription in yeast (18), while in Xenopus and other higher vertebrates, transcription requires a 15 amino acid sequence near the C-terminus (19) that is not found in the yeast or catfish homolog. Despite being larger, S.cerevisiae TFIIIA protects 16 fewer base pairs at the 5′ end of the promoter from DNase I cleavage than Xenopus TFIIIA (20), suggesting that similarly positioned zinc fingers function differently in the two proteins. Indeed, the first zinc finger in Xenopus TFIIIA is primarily used for DNA binding (8), but in yeast it is also required for the recruitment of TFIIIC and transcriptional activation (21).
Because TFIIIA has only been characterized from a small number of vertebrates and S.cerevisiae, a complete understanding of the possible variations in this protein has not yet been achieved. Indeed, database searches reveal sequences of only eight TFIIIA genes. As a prelude to cloning, we report here the purification and characterization of TFIIIA from Acanthameoba castellanii (AcTFIIIA), an organism in which TFIIIA has key regulatory functions (22). AcTFIIIA is 59 kDa, significantly larger than any of its known homologs, yet it produces a DNase I protection pattern similar to those produced by higher vertebrate TFIIIA homologs.
MATERIALS AND METHODS
Purification of TFIIIA
Acanthameoba castellanii trophozoite cells were grown and harvested as described except cells were collected at 1840 g (23). Nuclear extract was prepared from batches of 50–55 l of cells with 260 l processed in total (∼5.2 × 1011 cells), as described (23,24), but with three modifications. First, cells were homogenized with three passages through a LSC Homogenizer LH-21 (Yamato Scientific, Japan) at 1000 r.p.m., resulting in breakage of >90% of cells. Second, the cell homogenate was centrifuged at 8700 g for 15 min. Third, the ammonium sulfate-precipitated proteins were collected by centrifugation at 4420 g for 30 min.
Basic chromatography buffer (HEG) consisted of 50 mM HEPES, pH 7.9, 20% glycerol, 0.2 mM phenylmethane sulfonyl fluoride (PMSF), 0.2 mM EDTA and 1 mM dithiothreitol (DTT). Additional components supplemented this buffer depending on the chromatography step. All columns were equilibrated and washed with at least 5 column volumes of starting buffer prior to loading. Whatman P11 (phosphocellulose) was prepared per manufacturer’s instructions and additionally treated overnight with two changes, 13 bed volumes each, of 1.5 M KCl, 250 mM HEPES, pH 7.9 and 0.2 mM EDTA prior to equilibration. Chromatography procedures were performed at 4°C, and KCl concentrations were verified by conductivity measurements. The first P11 column (2.5 cm diameter, 390 ml bed volume) was equilibrated using HEG/100 mM KCl. The total nuclear extract (~3.8 g of protein, 213 ml) was applied to the column at a linear flow rate of 13.4 cm/h. Eluting protein was detected by absorbance at 280 nm. The flow-through fraction (570 ml, 1041 mg of total protein) containing TFIIIA was collected and the KCl concentration was increased to 450 mM by adding HEG/1.7 M KCl and benzamidine to a final concentration of 1 mM. This fraction was then gently mixed on ice for 1 h to dissociate TFIIIA from 5S RNA.
The second P11 column (2.5 cm diameter, 104 ml bed volume) was equilibrated using HEG/450 mM KCl/1 mM benzamidine. The first P11 flow-through fraction was applied to the column at a linear flow rate of 13.4 cm/h. The column was then washed and developed with a 500 ml linear salt gradient from 0.45 to 1.2 M KCl. TFIIIA eluted predominantly between 500 and 600 mM KCl. The peak TFIIIA-containing fractions were pooled (~81 ml and 4.3 mg total protein) and dialyzed for 6 h against 2 l HEG/110 mM KCl/1 mM benzamidine/5 mM DTT, reducing the KCl concentration to 140–150 mM. The higher DTT concentration stimulated DNA binding of TFIIIA in the next step.
Calf thymus DNA (21.6 mg; Sigma, St Louis) in 16 ml of 50 mM HEPES, pH 7.9, was added to the dialyzed pool from the second P11 column, reducing the KCl concentration to ∼120 mM. MgCl2 and ZnCl2 were then added to final concentrations of 7.5 mM and 2.5 µM, respectively, and the fraction was incubated at room temperature for 20 min with gentle agitation. Any precipitates that had formed were removed by centrifugation for 10 min at 7000 r.p.m. in a JA10 rotor (Beckman, Fullerton) at 4°C. The supernatant is the affinity load.
A promoter–DNA affinity column (1.5 cm diameter, 7 ml bed volume) was equilibrated using HEG/5 mM DTT/110 mM KCl/7.5 mM MgCl2/2.5 µM ZnCl2/1 mM benzamidine (without EDTA). The affinity load was applied to the column at a linear flow rate of 5.66 cm/h. The column was washed and developed with a 35 ml linear salt gradient from 110 mM KCl to 1 M KCl. TFIIIA eluted between 340 and 470 mM KCl.
Preparation of the DNA–promoter affinity matrix
The DNA coupled to the matrix consisted of five head-to-tail repeats of the A.castellanii 5S RNA gene from +15 to +96, which was originally cloned into the BamHI site of pT7T3α18 (Life Technologies, Rockville, MD) and was a generous gift from Mike Zwick. A fragment containing the multimer was excised using EcoRI and HindIII, purified as described (25) and subsequently coupled to cyanogen bromide-activated Sepharose CL-4B (Pharmacia, Piscataway) as described (26).
Determination of protein concentrations
The protein concentration of nuclear extract was determined using the enhanced protocol of the BCA protein assay (Pierce, Rockford) using the supplied bovine serum albumin (BSA) protein standard. Protein concentrations of the flow-through fraction and the pooled fractions from the second P11 column were determined by a modified Bradford microassay procedure (Bio-Rad, Hercules) using BSA as the standard. The total protein purified TFIIIA pool was determined from a Coomassie Brilliant Blue G-250-stained 11% SDS–polyacrylamide gel used to resolve a substantial portion of this protein. Prior to being loaded on the gel, proteins were precipitated using a chloroform–methanol procedure (27) with 100 ng of cofilin added as a carrier. The relative intensities of Coomassie-stained protein bands were determined using the Photometrix 1D Gel Analysis Program (version 2.51, Non Linear Dynamics, UK). β-galactosidase, carbonic anhydrase and myoglobin served as protein standards on the gel.
Fractionation of TFIIIB, TFIIIC and Pol III for DNA–protein photo-cross-linking
Nuclear extract applied to a P11 column was fractionated as described previously (22). TFIIIB, Pol III and TFIIIC were found in the 0.3, 0.45 and 0.65 M KCl step elutions, respectively.
Fractionation of TFIIIB, TFIIIC and Pol III for in vitro transcription assays
Protein retained on the first P11 column (described under ‘Purification of TFIIIA’) was eluted using HEG with 700 mM KCl and detected by absorbance at 280 nm. This fraction was diluted to 450 mM KCl with HEG and batch-loaded for 1 h onto 90 ml of P11 resin equilibrated with HEG/450 mM KCl. The slurry was then filtered through a 417 qualitative filter disc (VWR, San Francisco, CA), and all three components in the filtrate were precipitated by adding ammonium sulfate to 70% saturation, stirring overnight and centrifuging at 12 100 g for 10 min in a Beckman JA17 rotor (22). Although TFIIIC normally elutes at 0.65 M KCl from phosphocellulose (PC), it does not bind efficiently to the matrix at elevated KCl concentrations, and the majority of TFIIIC is found in the filtrate. The precipitate was resuspended in dialysis buffer (50 mM HEPES, pH 7.9, 120 mM KCl, 10% glycerol, 0.2 mM PMSF, 0.2 mM EDTA, 1 mM benzamidine and 1 mM DTT) at a volume of 0.25× load volume. The solution was then dialyzed against 2 l of dialysis buffer at 4°C for 6 h. This fraction was relatively free of contaminating TFIIIA and had a low background of 5S RNA transcription.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were done essentially as described (22). A 260 bp fragment of pAc5S.3dl5′-34 (24) was obtained by EcoRI–HindIII digestion and labeled by a fill-in reaction. EMSA reactions contained 100 mM KCl, 50 mM HEPES (pH 7.9), 7.5 mM MgCl2, 2.5 µM ZnCl2, 5 mM DTT, 10% glycerol, 4 µg of acetylated BSA and 20 000 c.p.m. of probe in a total volume of 20 µl. Reactions were incubated at 25°C for 20 min and then loaded directly on a 5% non-denaturing polyacrylamide gel as described (22) that had been prerun for 20 min at 200 V. Reactions were electrophoresed for 1.3 h at 200 V at room temperature. To quantify DNA-binding activity, a smaller fragment (–2 to +136) was used in order to reduce non-specific binding by other proteins.
In vitro transcription assays
Transcription reactions contained 50 mM HEPES (pH 7.9), 100 mM KCl, 7.5 mM MgCl2, 10% glycerol, 2 mM DTT, 2 µg/ml of α-amanitin, 600 µM each of ATP, CTP and GTP, 50 µM UTP, 5 µCi [α-32P]UTP (3000 Ci/mmol), 42 fmol pBS+5S.3 (22), 7.2 µl of the TFIIIB/TFIIIC/Pol III fraction and 2 µl of an affinity-purified TFIIIA fraction in a final volume of 30 µl. Reactions were started by adding the NTP mixture (3.5 µl) and were incubated at 25°C for 1.5 h. They were stopped by adding 11 µl of a freshly prepared mix of 5 µl proteinase K (10 mg/ml) plus 6 µl stop mix [250 mM NaCl, 1% SDS, 20 mM Tris–HCl (pH 7.6), 5 mM EDTA] and incubated at 50°C for 30 min. The RNA was precipitated by adding 5 µl of linear polyacrylamide (5 mg/ml), 154 µl 10 mM Tris–HCl (pH 7.5), 1 mM EDTA (pH 8.0), 100 µl of 7.5 M ammonium acetate, 1000 c.p.m. of the standard EMSA probe (as a recovery standard) and 900 µl of 95% ethanol. Samples were chilled at –20°C for 10 min, and then the RNA was collected by centrifugation for 20 min at maximum speed in an Eppendorf centrifuge. The samples were then processed as described (22).
SDS–PAGE and silver-staining of proteins
SDS–PAGE was performed as described (28). To each sample, 6× SDS loading buffer (29) was added to a final concentration of 1× in the sample. Samples were resolved on an 11% separating gel with a 4% stacking gel. The gel was stained with silver as described (30).
DNase I footprinting
The EcoRI–HindIII pAc5S.3dl5′-34 fragment was 5′ end-labeled at one end and bound to TFIIIA under conditions essentially the same as those used for the EMSA, except the reaction volume was 50 µl and 10 000 c.p.m. of probe was added to each reaction. Following protein–DNA association, 0.1 U of DNase I (Roche, Indianapolis) was added to each sample and reactions were incubated at 25°C for 2 min. Reactions were then transferred to a dry ice/ethanol bath to inactivate the enzyme and were treated with proteinase K and precipitated as described under ‘In vitro transcription assays’. Digestion products were suspended in 7 µl of loading solution (90% formamide, 10 mM EDTA, 0.1% bromophenol blue, 0.1% xylene cyanol) and resolved on an 8% polyacrylamide, 7 M urea, 1× TBE gel as described (31). Labeled digestion fragments were visualized as described previously (22) and compared to a G+A chemical sequencing ladder.
Site-specific DNA photo-cross-linking
Two oligodeoxyribonucleotides were synthesized with a phosphorothioate replacing the phosphodiester linkage between the second and third nucleotides at the 5′ end of the molecules. The oligodeoxyribonucleotides corresponded to the RNA-like strand of the 5S RNA gene from +65 to +80 and +85 to +100. Oligodeoxyribonucleotides were suspended to a concentration of 100 pmol/µl in 100 mM triethylammonium bicarbonate, pH 8.0, and 75 µl of this solution was combined with 75 µl of 100 mM azidophenacyl bromide in acetonitrile to link the azidophenacyl group to the phosphorothioate. This and all subsequent steps were performed under reduced light. The reaction was incubated at 25°C for 4 h. The solvent was then evaporated in a SpeedVac, and the oligodeoxyribonucleotide pellets were suspended in 100 µl of sterile, deionized water. The solutions were subsequently extracted three times with 100 µl of water-saturated butanol and four times with 200 µl of water-saturated diethyl ether and dried at 37°C for 15 min. The oligodeoxyribonucleotides were then diluted with 10 mM Tris, 1 mM EDTA, pH 8.0 to a final volume of 600 µl. Aliquots of 2 µl (∼20 pmol) were dried under vacuum and stored at –20°C.
The preparation of the probes has been described (32). The derivatized primers were annealed to pBS+5S.3 (22) single-stranded plasmid DNA of the template strand. The finished probe was the EcoRI–HindIII fragment used for EMSAs that contained –126 to +219 of the 5S RNA gene.
The conditions used for DNA–protein binding were the same as those used for EMSAs. Following binding, reactions were UV irradiated with 11 mJ/mm2 at 365 nm for 4 min. The cross-linked protein–DNA in the samples was treated and analyzed as described previously (33).
RESULTS
Purification of TFIIIA
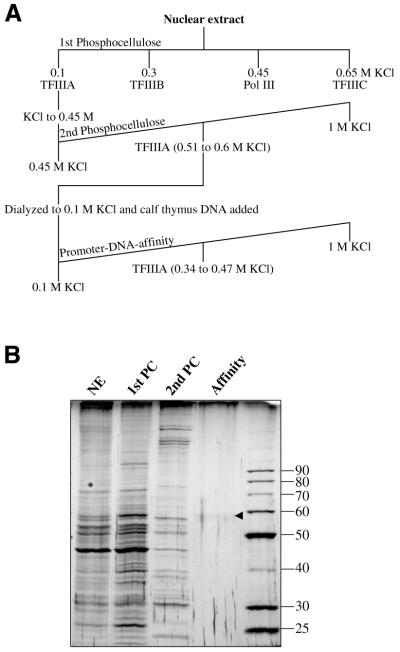
AcTFIIIA was purified to near homogeneity from a nuclear extract by three chromatography steps (Fig. 1A). Enrichment of the 59 kDa TFIIIA protein was observed on a silver-stained SDS–polyacrylamide gel (Fig. 1B) of each fraction. Overall, A.castellanii TFIIIA was purified 5037-fold from the first PC flow-through fraction to a specific activity of 196 000 U/mg protein (Table 1).
Figure 1.
Purification of AcTFIIIA. (A) Purification strategy of the Pol III factors. (B) A silver-stained 11% SDS–polyacrylamide gel shows the relative purity of TFIIIA at each stage of the purification. NE, 0.5 µg nuclear extract; 1st PC, 0.5 µg of the 0.1 M KCl flow-through fraction from the first PC column; 2nd PC, 0.5 µg of TFIIIA pooled from the second PC column; affinity, 19 µl of fraction 21 from the promoter–DNA affinity column (Fig. 2). The arrowhead shows the protein band correlated with TFIIIA activity.
Table 1. Purification of TFIIIA from A.castellanii nuclear extract.
| Fraction | Volume (ml) | Total protein (mg) | Units | Specific activity (units/mg) | Fold purification | % |
|---|---|---|---|---|---|---|
| Nuclear extract | 213 | 3797 | – | – | – | – |
| 1st PC flow-through | 470 | 1041 | 147 828 | 142 | 3.65 | 100 |
| 2nd PC pool | 81.3 | 4.3 | 28 268 | 6574 | 169 | 19 |
| Affinity pool | 6.13 | 0.003 | 588 | 196 000 | 5037 | 0.4 |
Protein concentrations were estimated as described in Materials and Methods. TFIIIA activity was determined by EMSA, with 1 unit of activity defined as the amount of protein required to shift 1 fmol of a radiolabeled 5S RNA gene fragment from –2 to +136.
Nuclear extract was prepared from cells harvested in early log phase to ensure the full activity of AcTFIIIA, which becomes inactivated later in the growth cycle (22). As with Xenopus and human TFIIIA, AcTFIIIA in a crude extract will not bind PC at 0.1 M KCl and is found in the flow-through fraction (22), which is attributed to its binding to 5S RNA (34). However, at 0.45 M KCl AcTFIIIA–nucleic acid interactions were disrupted, and TFIIIA was able to bind a second PC column, while 83% of contaminating proteins did not bind. The peak of TFIIIA eluted between 0.5 and 0.6 M KCl. Although the pool of TFIIIA exhibited a 46-fold increase in activity over the PC flow-through fraction (Table 1), fractions were not sufficiently pure to correlate a specific protein with activity (data not shown). When free of nucleic acid, AcTFIIIA does not bind as tightly to cation exchangers as human TFIIIA, so PC and BioRex 70 chromatography is much less efficient in enriching for AcTFIIIA.
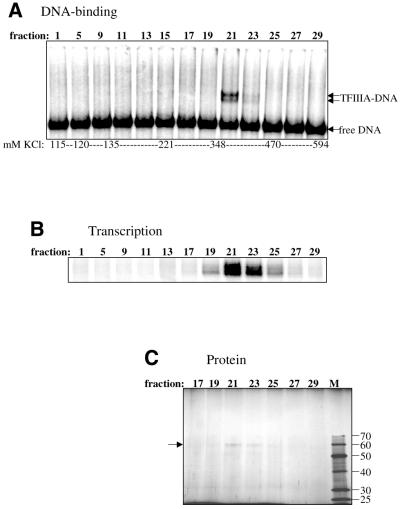
A pentamer of head-to-tail repeats of the A.castellanii 5S RNA gene (35) from +17 to +96 (+1 denotes the transcription start site) was used for the promoter–DNA affinity column, as repeats of the minimal promoter (+47 to +100) exhibited much weaker interactions with AcTFIIIA. Calf-thymus DNA was added to the TFIIIA-containing fraction as a non-specific competitor prior to promoter–DNA affinity chromatography. The quantity of competitor DNA used was the amount required to produce a distinct TFIIIA–DNA complex in an EMSA (data not shown). Other competitor DNAs tested [poly(dI–dC), salmon sperm, poly(dA–dT), pBS plasmid] did not result in the same high purification, but sometimes led to higher yields (data not shown). TFIIIA eluted between 340 and 470 mM KCl, and its DNA-binding activity was localized to fractions 19–23, with peak activity in fraction 21 (Fig. 2A). Two complexes were observed from the EMSA; the lower complex is presumably similar to the Xenopus proteolytic fragment missing its C-terminus (J.Gottesfeld, personal communication). The same fractions with DNA-binding activity also activated 5S RNA transcription in vitro when combined with a fraction containing TFIIIB, TFIIIC and Pol III (Fig. 2B). Transcription activity was observed in additional fractions outside those with DNA-binding activity because of the greater sensitivity of the transcription assay. The DNA-binding and transcription activities correlated exactly with a protein of 59 kDa, which was the only major protein observed in the column fractions (Fig. 2C).
Figure 2.
AcTFIIIA activity correlates with a 59 kDa protein in promoter–DNA affinity fractions. (A) DNA-binding activity was tested by EMSA using 3 µl of each column fraction. (B) The fractions were tested for the ability to activate 5S RNA transcription in the presence of TFIIIB, TFIIIC and Pol III. (C) Fractions encompassing the peak of TFIIIA activity were resolved on an 11% SDS–polyacrylamide gel (19 µl of the indicated fraction/lane) that was then stained with silver. ‘M’ denotes 0.25 µl of the Benchmark Protein Ladder (Life Technologies) with protein molecular weights shown at the right. The arrow identifies TFIIIA.
DNase I footprinting
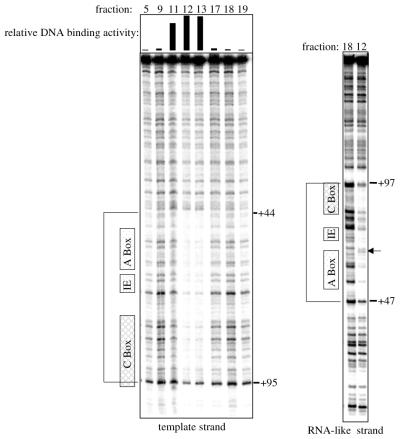
DNase I footprinting was performed to identify the region of the 5S RNA gene to which AcTFIIIA bound. Using fractions eluted from a promoter–DNA affinity column, the footprint was found to extend from approximately +44 to +97 of the 5S RNA gene (Fig. 3). A hypersensitive cleavage site at +64 on the RNA-like strand was also observed. This protection pattern is nearly identical to those observed using TFIIIA from higher eukaryotes: TFIIIA from X.laevis protects +47 to +96 of the 5S RNA gene from DNase I cleavage and produces hypersensitive cleavage at +62 on the RNA-like strand (4). The similarities in the AcTFIIIA and X.laevis TFIIIA footprints indicate that these proteins form similar complexes with the DNA. The known DNA-binding properties of X.laevis TFIIIA were, therefore, used to further characterize AcTFIIIA.
Figure 3.
The DNase I footprint of AcTFIIIA is similar to that of Xenopus TFIIIA. Promoter–DNA affinity fractions indicated above each gel were used for footprinting. The relative DNA binding activity determined by EMSA is graphically represented above each lane. The gel on the left shows DNase I cleavage of the template strand, and the gel on the right shows cleavage of the RNA-like strand. The positions of the A-box, the intermediate element (IE) and the C-box are shown to the left of each gel. Footprinted regions are identified by brackets. Nucleotide positions +44 and +95 are indicated on the template strand, and nucleotide positions +47 and +97 are indicated on the RNA-like strand. The arrow identifies a hypersensitive cleavage site at +64 on the RNA-like strand.
Protein–DNA photo-cross-linking using purified TFIIIA fractions
Site-specific photo-cross-linking was used to verify that the 59 kDa protein present in the purified TFIIIA preparation interacts specifically with the promoter. A photo-active azidophenacyl group was synthetically added to a phosphorothioate in DNA radiolabeled two nucleotides upstream of this site (36). Upon UV irradiation, the azide is converted to a nitrene that covalently cross-links proteins within 11 Å of the phosphate backbone (37). Following digestion of the DNA, proteins interacting with the DNA near the modified site are identified as radiolabeled bands on an SDS–polyacrylamide gel. This technique has been used to determine the positions of transcription complexes on several different promoters (33,36,38–40).
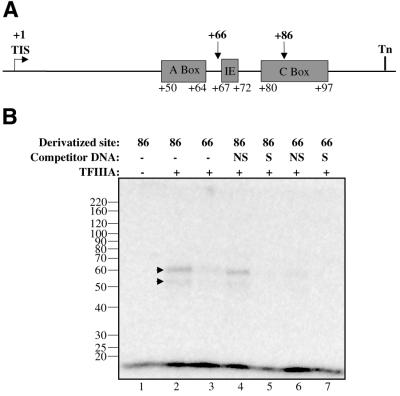
Two photoaffinity probes were used: one modified within the C-box (+86) and the other modified between the A-box and the intermediate element (+66; Fig. 4A). The +86 probe should be highly effective in capturing AcTFIIIA on the DNA since the C-box is indispensable for TFIIIA binding in Xenopus (3), and zinc fingers 1 and 2 are in close proximity to the phosphate backbone of the DNA at +86 (41). The +66 probe is expected to be less efficient in contacting TFIIIA, because it is not within any of the three promoter elements, although it is within the footprinted region of the DNA (Fig. 4A) and within 11 Å of residues in zinc finger 6 (42).
Figure 4.
The 59 kDa protein is specifically UV photo-cross-linked to the A.castellanii 5S RNA gene promoter. (A) A schematic representation of the 5S RNA gene shows the positions of each photoderivative. IE, intermediate element; TIS, transcription initiation site; Tn, termination site. (B) Affinity-purified TFIIIA (+) was cross-linked to 7500 c.p.m. of the indicated photoaffinity probe in a 30 µl reaction. Protein markers are indicated at the left of the panel. NS, reactions preincubated with 8 ng of pBS(–) non-specific competitor DNA; S, reactions preincubated with 8 ng pBS+5S.3 specific competitor DNA; –, reactions lacking TFIIIA or competitor DNA. Labeled peptides are indicated with arrows.
Each probe was found to cross-link the 59 kDa protein from the purified AcTFIIIA fraction (Fig. 4B, lanes 2–4 and 6). Another protein migrating at 54 kDa was also observed, which was probably the degradation product related to the lower complex seen in the EMSA (Fig. 2A). Cross-linking of the 59 kDa protein to the +86 photoaffinity probe was more efficient than cross-linking to the +66 probe (Fig. 4B, cf. lanes 2 and 3 and lanes 4–6), but this difference could also be caused by variability in cross-linking efficiency based on different amino acids in proximity to each derivative (43). The specificity of cross-linking was shown by the ability of plasmid DNA containing the 5S RNA gene to abolish cross-linking (lanes 5 and 7), while plasmid DNA lacking the insert only slightly reduced the signal (lanes 4 and 6). Furthermore, photo-cross-linking was not observed using a probe with a truncated C-box (data not shown). These results show that the 59 kDa protein in purified TFIIIA fractions is able to interact specifically with the 5S RNA gene in regions footprinted similarly by Xenopus and AcTFIIIA. Furthermore, similar photo-cross-linking and resistance to competition of the proteolytic product suggests that the DNA-binding region was not cleaved. In similarly degraded Xenopus TFIIIA, the C-terminus outside the zinc fingers is cleaved.
Protein–DNA photo-cross-linking using crude preparations of TFIIIA
Cross-linking in fractions from earlier stages in the purification was done to confirm the specificity of the 59 kDa protein–promoter interaction. In nuclear extract, the first PC flow-through fraction, and the second PC pool, a pair of proteins near 60 kDa specifically cross-linked to the 5S RNA gene (Fig. 5). In these fractions the size of the labeled protein was sometimes larger as a result of variations in the size of the DNA tag covalently linked to the protein. Differential digestion of the DNA cross-linked to the polypeptides has been shown to alter the electrophoretic mobility of labeled polypeptides (33). Indeed, digestion of the DNA following UV irradiation may have been partially quenched by greater amounts of non-specific competitor DNA used with crude fractions. Alternatively, a component in the cruder fractions may have interfered with digestion. Regardless of the small differences in the electrophoretic mobilities of the radiolabeled bands, the specificity of cross-linking of the characteristic ‘doublet’ detected using purified TFIIIA was consistently observed.
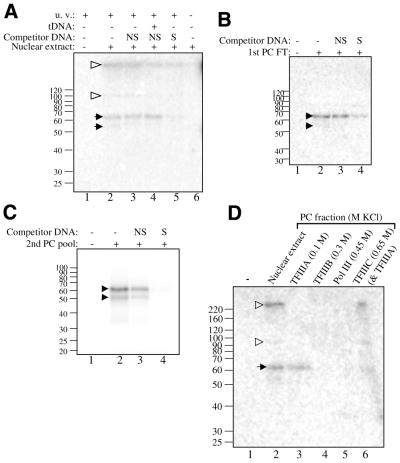
Figure 5.
The 59 KDa protein is detected in all TFIIIA-containing fractions. (A) Nuclear extract (73 µg of protein) with 12 µg of calf thymus DNA was cross-linked with 4600 c.p.m. of the +66 photoaffinity probe in a 30 µl reaction. The indicated reactions were treated with 150 ng of specific or non-specific competitor DNA. Lane 4 shows a reaction preincubated with 15 ng of a tRNAmet gene [EcoRI/HindIII digested pArabMet, 4 (22)]. Lane 6 shows a reaction that was not UV-irradiated. Black arrows identify AcTFIIIA, and open arrowheads show additional labeled polypeptides. (B) The first PC flow-through fraction (15 µg of protein) with 4 µg of calf thymus DNA was cross-linked with 4600 c.p.m. of the +66 photoaffinity probe in a 20 µl reaction. The indicated reactions were treated with 160 ng of specific or non-specific competitor DNA. (C) TFIIIA activity from the second PC column was cross-linked with the +86 photoaffinity probe essentially as described for the PC flow-through fraction. (D) PC column fractions (15 µg of protein per fraction) containing the separated Pol III factors with 4 µg of calf-thymus DNA were cross-linked with 2900 c.p.m. of the +66 photoaffinity probe. Above each lane, the KCl concentration applied to the column and the Pol III factor eluted are indicated.
The 59 kDa protein was specifically cross-linked in the crude nuclear extract using the +66 probe (Fig. 5A, lanes 3 and 5). The efficiency with which the 59 kDa subunit cross-linked to the probe in nuclear extract was unexpected, as nuclear extract has been found to induce the formation of higher-order complexes on the DNA that obscure detection of TFIIIA–DNA complexes (data not shown). However, some higher-order complexes should contain TFIIIA as well as some or all of the other Pol III transcription factors. Subunits of these factors, particularly those of TFIIIC, should cross-link to the +66 and +86 probes. Two other polypeptides were indeed detected: one in the range of 100 kDa and another above the 220 kDa marker (lanes 2–5). Detection of these polypeptides was reduced by the addition of either the Arabidopsis thalianii tRNAmet gene, which should bind TFIIIC but not TFIIIA, or plasmid DNA containing the 5S RNA gene (lanes 4 and 5). These two polypeptides were not detected in fractions free of TFIIIC (Fig. 5B and C).
Also tested by cross-linking were the Pol III factors, which are separated using PC chromatography: TFIIIA elutes with 0.1 M KCl, TFIIIB with 0.3 M KCl, Pol III with 0.45 M KCl and TFIIIC with 0.65 M KCl (22). In addition to the nuclear extract and the first PC flow-through fraction (Fig. 5D, lanes 2 and 3), the 59 kDa protein was detected in the 0.65 M KCl fraction, which contains TFIIIA that had been retained on the column in addition to TFIIIC (lane 6). The 59 kDa protein was not observed in either the TFIIIB or Pol III-containing fraction (lanes 4 and 5). Identical cross-linking of the 59 kDa was obtained using the +86 probe (data not shown). These results show that the 59 kDa protein is only found in fractions possessing TFIIIA activity. The ∼100 kDa and >220 kDa polypeptides were again detected in nuclear extract and were also detected in the 0.65 M KCl, TFIIIC-containing fraction (lanes 2 and 6), supporting the hypothesis that these are TFIIIC subunits alluded to above.
Peptide sequences
From purified AcTFIIIA (~1 pmol), three sequences were obtained from tryptic peptides by the HHMI Biopolymer/Keck Foundation Biotechnology Resource Laboratory at the Yale University School of Medicine: N,G,(T,S)-F-L-H,(E,N)-P,(E,N)-S-Q-L-(R); E,L-F,L-A,Q-R-N-T; Q,T,A,D,S,V-V-H-D,(Q)-(G),(L)-L-(R). Amino acid positions are separated by hyphens; commas separate multiple amino acids detected at a single position; weakly detected amino acids are shown in parentheses. Although a variety of conditions were tested, PCR amplification using a degenerate primer derived from the first sequence and a 3′-RACE PCR primer (Integrated DNA Technologies, Coralville, IA) did not yield any products from cDNA that had been synthesized using total RNA from A.castellanii, a cDNA cloning primer (Integrated DNA Technologies), and the M-MLV Reverse Transcriptase H(–) Point Mutant (Promega, Madison, WI). The primers derived from the second and third amino acids sequences were too degenerate to be used for PCR.
DISCUSSION
TFIIIA has been characterized from only a small number of organisms and, except for possessing nine zinc fingers, has been found to diverge significantly. This factor is the regulatory target of 5S RNA transcription in A.castellanii (22), so as part of a study of the regulatory mechanism, it was purified, characterized and compared with TFIIIA homologs from other species. We found that AcTFIIIA is a 59 kDa protein, making it the largest of the known homologs. Although AcTFIIIA is closest in size to S.cerevisiae TFIIIA (51 kDa), the promoter interaction and apparent susceptibility to proteolysis of AcTFIIIA is more similar to the vertebrate homolog from Xenopus. As in most cell types other than oocytes, A.castellanii cells contain very low quantities of TFIIIA. Based on its DNA-binding activity in the first PC flow-through fraction, only ∼170 molecules of TFIIIA were estimated to be present in each A.castellanii cell. This estimate is likely to be slightly low considering that a small portion of TFIIIA is retained on the first PC column. Nevertheless, such low abundance complicated effective purification of large amounts of the protein.
AcTFIIIA was purified using chromatography steps similar to those used to enrich for human TFIIIA activity (34). Paralleling human TFIIIA, AcTFIIIA from a crude extract does not bind PC, presumably because of the formation of the 5S RNA–TFIIIA complex (44). Supporting this hypothesis, we found that AcTFIIIA binds specifically to 5S RNA, and this complex can be dissociated with high salt (data not shown). At 450 mM KCl, AcTFIIIA in the first PC flow-through fraction dissociates from nucleic acids and binds to a second PC column, which greatly enhances the purification but leads to a loss in yield.
Three rounds of affinity chromatography had been used successfully to purify human TFIIIA (45), but resulted in almost complete loss of AcTFIIIA activity (data not shown). Therefore, a single affinity chromatography step was optimized by using calf thymus DNA to minimize non-specific binding of proteins to the column. Affinity-purified TFIIIA was >5000-fold more pure than TFIIIA in the first PC flow-through fraction. Purification from nuclear extract could not be quantified, as DNA-binding activity is affected by nucleases and other proteins that bind the DNA probe in this fraction. Although TFIIIA was definitively identified using this purification scheme, the yield was extremely low. A significant amount of human TFIIIA activity is reportedly lost using chromatography buffers at pH 7.9, but not with buffers at pH 7.0. However, no difference in yield was detected when AcTFIIIA was purified using chromatography buffers at either pH. The greatest reduction in activity occurred during affinity chromatography and was caused by some non-specific binding of TFIIIA to the calf thymus DNA. However, though the use of other competitor DNAs led to higher yields, they also led to significantly lower fold purification.
Site-specific DNA–protein photo-cross-linking confirmed that AcTFIIIA was the 59 kDa protein. This protein was specifically cross-linked in all fractions possessing TFIIIA activity, but not detected in fractions lacking activity. Furthermore, AcTFIIIA was correlated to the 59 kDa protein by southwestern analysis and by elution/renaturation of TFIIIA from a protein gel slice (data not shown).
The results show that AcTFIIIA is the largest TFIIIA homolog that has been identified to date. The second largest is S.cerevisiae TFIIIA (51 kDa), and its large size is primarily due to an 81 amino acid insert between its C-terminal two zinc fingers. AcTFIIIA is not likely to harbor this same domain, as this expansion segment is specifically required for 5S RNA transcription in S.cerevisiae (46). We speculate that an inter-finger insertion would cause the DNase I footprint of AcTFIIIA to vary from that of the Xenopus protein. However, it remains possible that an altered mode of binding would produce the same footprint. Two additional polypeptides of ∼100 and >220 kDa were cross-linked to the 5S RNA gene in nuclear extract and the 0.65 M KCl, TFIIIC-containing PC fraction. Cross-linking of these polypeptides was found to be reduced by the addition of the 5S RNA gene or a tRNAmet gene competitor DNA. These results suggest that the two polypeptides correspond to subunits of TFIIIC. Further purification of TFIIIC in conjunction with a comprehensive set of photoaffinity probes along the 5S RNA gene might confirm these initial findings.
Acknowledgments
ACKNOWLEDGEMENTS
We thank A. Bric for technical assistance and I. Lemasson for critical review of the manuscript. This work was supported by US Public Health Service grants GM26059 and GM22580 to M.R.P.
REFERENCES
- 1.Bieker J.J., Martin,P.L. and Roeder,R.G. (1985) Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell, 40, 119–127. [DOI] [PubMed] [Google Scholar]
- 2.Setzer D.R. and Brown,D.D. (1985) Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem., 260, 2483–2492. [PubMed] [Google Scholar]
- 3.Pieler T., Hamm,J. and Roeder,R.G. (1987) The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell, 48, 91–100. [DOI] [PubMed] [Google Scholar]
- 4.Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 5.Ginsberg A.M., King,B.O. and Roeder,R.G. (1984) Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell, 39, 479–489. [DOI] [PubMed] [Google Scholar]
- 6.Miller J., McLachlan,A.D. and Klug,A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith D.R., Jackson,I.J. and Brown,D.D. (1984) Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell, 37, 645–652. [DOI] [PubMed] [Google Scholar]
- 8.Vrana K.E., Churchill,M.E., Tullius,T.D. and Brown,D.D. (1988) Mapping functional regions of transcription factor TFIIIA. Mol. Cell. Biol., 8, 1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen J.H., Hansen,P.K., Lillelund,O. and Thogersen,H.C. (1991) Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett., 281, 181–184. [DOI] [PubMed] [Google Scholar]
- 10.Liao X.B., Clemens,K.R., Tennant,L., Wright,P.E. and Gottesfeld,J.M. (1992) Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5 S RNA gene. J. Mol. Biol., 223, 857–871. [DOI] [PubMed] [Google Scholar]
- 11.Del Rio S., Menezes,S.R. and Setzer,D.R. (1993) The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol., 233, 567–579. [DOI] [PubMed] [Google Scholar]
- 12.Gaskins C.J. and Hanas,J.S. (1990) Sequence variation in transcription factor IIIA. Nucleic Acids Res., 18, 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaskins C.J., Smith,J.F., Ogilvie,M.K. and Hanas,J.S. (1992) Comparison of the sequence and structure of transcription factor IIIA from Bufo americanus and Rana pipiens. Gene, 120, 197–206. [DOI] [PubMed] [Google Scholar]
- 14.Ogilvie M.K. and Hanas,J.S. (1997) Molecular biology of vertebrate transcription factor IIIA: cloning and characterization of TFIIIA from channel catfish oocytes. Gene, 203, 103–112. [DOI] [PubMed] [Google Scholar]
- 15.Drew P.D., Nagle,J.W., Canning,R.D., Ozato,K., Biddison,W.E. and Becker,K.G. (1995) Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene, 159, 215–218. [DOI] [PubMed] [Google Scholar]
- 16.Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 17.Gaskins C.J., Fiser-Littell,R.M., Duke,A.L. and Hanas,J.S. (1989) Species variation in transcription factor IIIA. Nucleic Acids Res., 17, 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne C.A. and Segall,J. (1993) Mapping regions of yeast transcription factor IIIA required for DNA binding, interaction with transcription factor IIIC and transcription activity. J. Biol. Chem., 268, 11364–11371. [PubMed] [Google Scholar]
- 19.Mao X. and Darby,M.K. (1993) A position-dependent transcription-activating domain in TFIIIA. Mol. Cell. Biol., 13, 7496–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun B.R., Riggs,D.L., Kassavetis,G.A. and Geiduschek,E.P. (1989) Multiple states of protein-DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc. Natl Acad. Sci. USA, 86, 2530–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothfels K., Rowland,O. and Segall,J. (1998) Essential role for the amino terminal zinc finger of TFIIIA from Saccharomyces cerevisiae in formation of a functional transcription complex on the 5S RNA gene. RNA Polymerase I and III Transcription Conference, Pacific Grove, CA.
- 22.Matthews J.L., Zwick,M.G. and Paule,M.R. (1995) Coordinate regulation of ribosomal component synthesis in Acanthamoeba castellanii: 5S RNA transcription is down regulated during encystment by alteration of TFIIIA activity. Mol. Cell. Biol., 15, 3327–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radebaugh C.A., Matthews,J.L., Geiss,G.K., Liu,F., Wong,J.M., Bateman,E., Camier,S., Sentenac,A. and Paule,M.R. (1994) TATA box-binding protein (TBP) is a constituent of the polymerase I-specific transcription initiation factor TIF-IB (SL1) bound to the rRNA promoter and shows differential sensitivity to TBP-directed reagents in polymerase I, II and III transcription factors. Mol. Cell. Biol., 14, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zwick M.G., Imboden,M.A. and Paule,M.R. (1991) Specific transcription of an Acanthamoeba castellanii 5S RNA gene in homologous nuclear extracts. Nucleic Acids Res., 19, 1681–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radebaugh C.A., Kubaska,W.M., Hoffman,L.H., Stiffler,K. and Paule,M.R. (1998) A novel transcription initiation factor (TIF), TIF-IE, is required for homogeneous Acanthamoeba castellanii TIF-IB (SL1) to form a committed complex. J. Biol. Chem., 273, 27708–27715. [DOI] [PubMed] [Google Scholar]
- 26.Kadonaga J.T. and Tjian,R. (1986) Affinity purification of sequence-specific DNA binding proteins. Proc. Natl Acad. Sci. USA, 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wessel D. and Flugge,U.I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem., 138, 141–143. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 29.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.E., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) In Chanda,V.B. (ed.), Analysis of Proteins. Current Protocols. Vol. 2. John Wiley & Sons, Inc., New York, NY.
- 30.Blum H., Beier,H. and Gross,H.J. (1987) Improved Silver Staining of Plant Proteins, RNA and DNA in Polyacrylamide Gels. Electrophoresis, 8, 93–99. [Google Scholar]
- 31.Geiss G.K., Radebaugh,C.A. and Paule,M.R. (1997) The fundamental ribosomal RNA transcription initiation factor-IB (TIF-IB, SL1, factor D) binds to the rRNA core promoter primarily by minor groove contacts. J. Biol. Chem., 272, 29243–29254. [DOI] [PubMed] [Google Scholar]
- 32.Kim T., Lagrange,T., Naryshkin,N., Reinberg,D. and Ebright,R.H. (1999) In Taverns,A. and Buckle,M. (eds), Protein–DNA Interactions: A Practical Approach. IRL Press, Oxford, UK.
- 33.Gong X., Radebaugh,C.A., Geiss,G.K., Simon,M.N. and Paule,M.R. (1995) Site-directed photo-cross-linking of rRNA transcription initiation complexes. Mol. Cell. Biol., 15, 4956–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifart K.H., Wang,L., Waldschmidt,R., Jahn,D. and Wingender,E. (1989) Purification of human transcription factor IIIA and its interaction with a chemically synthesized gene encoding human 5 S rRNA. J. Biol. Chem., 264, 1702–1709. [PubMed] [Google Scholar]
- 35.Zwick M.G., Wiggs,M. and Paule,M.R. (1991) Sequence and organization of 5S RNA genes from the eukaryotic protist Acanthamoeba castellanii. Gene, 101, 153–157. [DOI] [PubMed] [Google Scholar]
- 36.Lagrange T., Kim,T.K., Orphanides,G., Ebright,Y.W., Ebright,R.H. and Reinberg,D. (1996) High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl Acad. Sci. USA, 93, 10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer A.N. and Barany,F. (1995) Photoaffinity cross-linking of TaqI restriction endonuclease using an aryl azide linked to the phosphate backbone. Gene, 153, 1–8. [DOI] [PubMed] [Google Scholar]
- 38.Bartholomew B., Kassavetis,G.A. and Geiduschek,E.P. (1991) Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol., 11, 5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartholomew B., Kassavetis,G.A., Braun,B.R. and Geiduschek,E.P. (1990) The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J., 9, 2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- 41.Wuttke D.S., Foster,M.P., Case,D.A., Gottesfeld,J.M. and Wright,P.E. (1997) Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. J. Mol. Biol., 273, 183–206. [DOI] [PubMed] [Google Scholar]
- 42.Nolte R.T., Conlin,R.M., Harrison,S.C. and Brown,R.S. (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tate J.J., Persinger,J. and Bartholomew,B. (1998) Survey of four different photoreactive moieties for DNA photoaffinity labeling of yeast RNA polymerase III transcription complexes. Nucleic Acids Res., 26, 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shastry B.S. (1996) Transcription factor IIIA (TFIIIA) in the second decade. J. Cell Sci., 109, 535–539. [DOI] [PubMed] [Google Scholar]
- 45.Moorefield B. and Roeder,R.G. (1994) Purification and characterization of human transcription factor IIIA. J. Biol. Chem., 269, 20857–20865. [PubMed] [Google Scholar]
- 46.Rowland O. and Segall,J. (1998) A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]