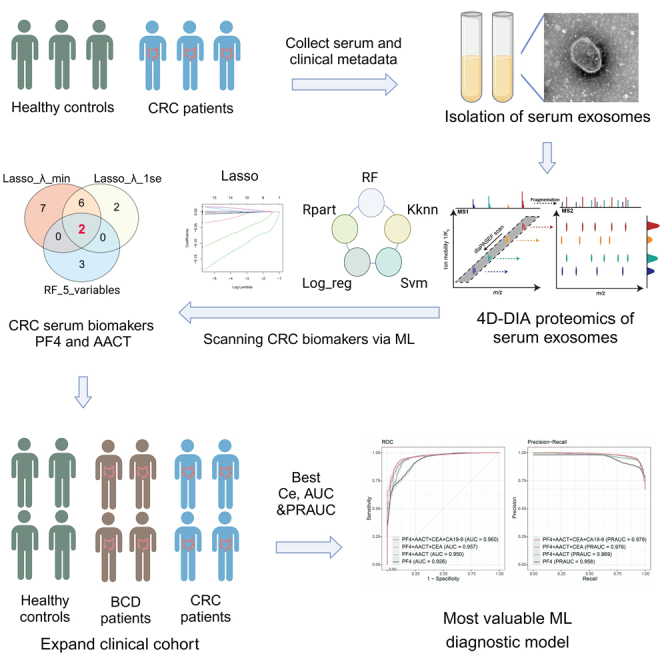
Summary
The potential of serum extracellular vesicles (EVs) as non-invasive biomarkers for diagnosing colorectal cancer (CRC) remains elusive. We employed an in-depth 4D-DIA proteomics and machine learning (ML) pipeline to identify key proteins, PF4 and AACT, for CRC diagnosis in serum EV samples from a discovery cohort of 37 cases. PF4 and AACT outperform traditional biomarkers, CEA and CA19-9, detected by ELISA in 912 individuals. Furthermore, we developed an EV-related random forest (RF) model with the highest diagnostic efficiency, achieving AUC values of 0.960 and 0.963 in the train and test sets, respectively. Notably, this model demonstrated reliable diagnostic performance for early-stage CRC and distinguishing CRC from benign colorectal diseases. Additionally, multi-omics approaches were employed to predict the functions and potential sources of serum EV-derived proteins. Collectively, our study identified the crucial proteomic signatures in serum EVs and established a promising EV-related RF model for CRC diagnosis in the clinic.
Keywords: machine learning, extracellular vesicles, colorectal cancer, biomarker, diagnosis
Graphical abstract
Highlights
-
•
4D-DIA proteomic profiles of serum EVs in CRC patients and healthy controls
-
•
Identification of proteomic signatures in serum EVs for CRC diagnosis
-
•
Development of diagnostic model distinguishing CRC from healthy controls and BCD
-
•
Prediction of functions and potential cell sources of serum EV-derived proteins
Yin et al. utilizes 4D-DIA proteomics and machine learning to identify key biomarkers PF4 and AACT in serum extracellular vesicles for colorectal cancer (CRC) diagnosis. Their random forest model demonstrates superior diagnostic performance for early-stage CRC and distinguishing CRC from benign colorectal diseases, offering a promising tool for clinical application.
Introduction
Colorectal cancer (CRC) ranks among the most common cancers worldwide, with approximately 1.8 million new cases and 900,000 deaths reported annually.1 Unfortunately, due to the insidious symptoms of early-stage CRC, more than 50% of patients are diagnosed at the progression stage with 5-year survival rate of 20%. However, early diagnosis of CRC enables patients to receive timely and optimal treatment, improving the 5-year survival rate to 90%.2 Although colonoscopy remains the gold standard for diagnosing CRC, its invasiveness and challenges of repeated examination limit its widespread use as a screening method.3 Additionally, carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), which are the two most commonly applied biomarkers for CRC diagnosis, have been reported to present insufficient sensitivity and be more suitable for dynamic monitoring of CRC patients during treatment.4,5 Consequently, the development of non-invasive methods for early diagnosis of CRC is an urgent goal that needs to be addressed.
Liquid biopsy has recently emerged as a prominent area of research in the field of cancer diagnosis and routine management, owing to its noninvasive, sensitive, and dynamic characteristics.6,7 Extracellular vesicles (EVs), important biomarkers in liquid biopsy, carry essential biological information such as proteins and nucleic acids and serve as critical mediators in intercellular communication.8 The development of digital droplet PCR technology has further advanced the study of non-coding RNAs in EVs. Researches has shown that non-coding RNAs carried by EVs derived from various body fluids hold potential to serve as candidate biomarkers for tumor diagnosis.9 In early-stage CRC, plasma EV-derived microRNAs (miRNAs), including let-7b-3p, miR-125a, and miR-320c, exhibit high diagnostic performance for CRC diagnosis.8,10 EV-derived proteins offer greater suitability for clinical examination due to their stability in comparison to RNAs.11 However, research progress in EV-derived protein profiling has been constrained by limitations of previous proteomics technology. Hence, utilizing advanced proteomics approaches such as four-dimensional independent data acquisition (4D-DIA) to uncover key biomarkers packaged in blood-derived EVs holds important implications for scanning CRC.
Machine learning (ML), as an essential branch of artificial intelligence, has garnered increasing attention in tumor diagnosis and treatment management recently.12 Compared to conventional diagnostic models, ML approaches are flexible and more suitable for capturing non-linear associations and integrating vast amounts of medical data including medical imaging and multi-omics data.13 Multiple ML methods can be employed to robustly extract crucial features from liquid biopsy analytes and build diagnostic models, thereby achieving superior specificity and sensitivity for cancer diagnosis.14 For instance, random forest (RF) algorithm-based diagnostic models exhibit remarkable performances across more than twenty types of cancer by utilizing microbes from tissues and blood.15 Thus, a ML model based on the optimal algorithm has the potential to enhance the accuracy of cancer diagnosis by profiling tissue and blood materials.
In this study, the 4D-DIA technology was employed to perform in-depth profiling of serum EV-proteomics data. Subsequently, the most valuable protein signatures were identified by utilizing ML-based pipeline and validated by ELISA detection. The object of our study was to develop a reliable EV-related RF model based on the identified protein signatures for clinical CRC diagnosis.
Results
Identification and characterization of serum EVs in HC and CRC patients
The serum EVs from the discovery set (25 cases CRC, 12 cases healthy control [HC]) were collected for candidate biomarkers screening. An expansion cohort comprising 338 cases in the train set and 328 cases in the test set was recruited for RF diagnostic model construction and validation (Table 1). Separated EVs from serum were subjected to subsequent experiments for validation (Figures 1A–1C). Nanoparticle tracking analysis (NTA) exhibited that the average diameter and distribution of EVs were compliant (Figure 1A). In western blot assay, the EV markers CD63 and TSG101 were present in isolated EVs, but not in protein lysate of CRC cell lines SW480, SW620, and HCT116. As the negative control marker expressed intracellularly, GRP94 and calnexin were not exposed in separated EVs (Figure 1B). In addition, the vesicle-like particles were confirmed by applying transmission electron microscopy (TEM) (Figure 1C).
Table 1.
Clinicopathologic characteristics of patients included in the study
| Characteristics | Discovery set (n = 37) |
Train set (n = 338) |
Test set (n = 328) |
External set (n = 246) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HC (n = 12) | CRC (n = 25) | HC (n = 96) | BCD (n = 47) | CRC (n = 195) | HC (n = 112) | BCD (n = 55) | CRC (n = 161) | HC (n = 60) | Enteritis (n = 42) | Hepatitis B (n = 46) | CRC (n = 98) | |
| Age, y, mean ± SD | 50.0 ± 9.3 | 56.5 ± 13.5 | 57.3 ± 8.4 | 51.4 ± 11.5 | 58.2 ± 13.5 | 56.6 ± 8.5 | 50.1 ± 11.4 | 60.1 ± 13.7 | 63.2 ± 10.5 | 54.8 ± 9.9 | 54.7 ± 9.7 | 62.2 ± 13.4 |
| Gender, n (%) | ||||||||||||
| Male | 4 (33.3) | 13 (52.0) | 52 (54.2) | 26 (55.3) | 111 (56.9) | 60 (53.6) | 38 (69.1) | 98 (60.9) | 42 (70.0) | 31 (73.8) | 31 (67.4) | 47 (48.0) |
| Female | 8 (66.7) | 12 (48.0) | 44 (45.8) | 21 (44.7) | 84 (43.1) | 52 (46.4) | 17 (30.9) | 63 (39.1) | 18 (30.0) | 11 (26.2) | 15 (32.6) | 51 (52.0) |
| Clinical stage, n (%) | ||||||||||||
| I | – | 3 (12.0) | – | – | 22 (11.3) | – | – | 19 (11.8) | 17 (17.3) | |||
| II | – | 6 (24.0) | – | – | 48 (24.6) | – | – | 31 (19.3) | 22 (22.4) | |||
| III | – | 8 (32.0) | – | – | 83 (42.6) | – | – | 47 (29.2) | 47 (48.0) | |||
| IV | – | 4 (16.0) | – | – | 42 (21.5) | – | – | 64 (39.8) | 12 (12.2) | |||
| Unknown | – | 4 (16.0) | – | – | 0 (0.0) | – | – | 0 (0.0) | 0 (0.0) | |||
| CEA, ng/mL, n (%) | ||||||||||||
| <5 | 12 (100.0) | 14 (56.0) | 94 (97.9) | 47 (100.0) | 129 (66.2) | 108 (96.4) | 53 (96.4) | 90 (55.9) | 54 (90.0) | 41 (97.6) | 45 (97.8) | 65 (66.3) |
| ≥5 | 0 (0.0) | 11 (44.0) | 2 (2.1) | 0 (0.0) | 66 (33.8) | 4 (3.6) | 2 (3.6) | 71 (44.1) | 6 (10.0) | 1 (2.4) | 1 (2.2) | 33 (33.7) |
| CA19-9, ng/mL, n (%) | ||||||||||||
| <35 | 12 (100.0) | 18 (72.0) | 94 (97.9) | 45 (95.7) | 153 (78.5) | 111 (99.1) | 54 (98.2) | 112 (69.6) | 59 (98.3) | 40 (95.2) | 46 (100.0) | 82 (83.7) |
| ≥35 | 0 (0.0) | 7 (28.0) | 2 (2.1) | 2 (4.3) | 42 (21.5) | 1 (0.9) | 1 (1.8) | 49 (30.4) | 1 (1.7) | 2 (4.8) | 0 (0.0) | 16 (16.3) |
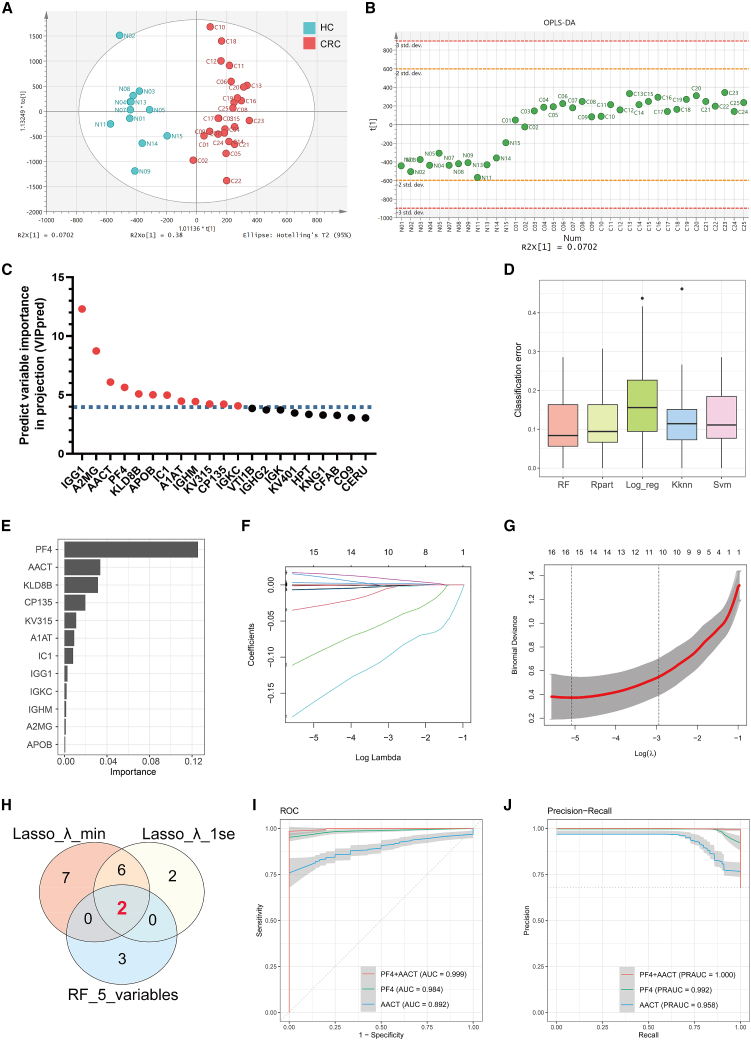
Figure 2.
Screening EV-derived biomarkers for CRC diagnosis via the ML pipeline
(A and B) Score plot (A) and scatterplot (B) exhibited significant discrimination between CRC and HC subjects via OPLS-DA analysis.
(C) Twelve candidate proteins selected based on their VIPpred scores >4.
(D) Bar plot showed the value of CE in ML diagnostic models based on different algorithms.
(E) Variable importance score plot showed the contribution of twelve candidate proteins in the RF diagnostic model.
(F and G) The Lasso regression analysis based on 4D-DIA proteomics and partial likelihood deviance on the prognostic genes. The minimum criteria and the 1-standard error (1SE) criteria were used to draw the dotted vertical lines at the optimal values of variables.
(H) Venn plot displayed the intersection of candidate proteins from the RF model and the Lasso regression models based on minimum and 1SE criteria.
(I and J) ROC curve (I) and PR curve (J) of RF diagnostic models based on PF4, AACT, and combined PF4 and AACT levels of 4D-DIA proteomics.
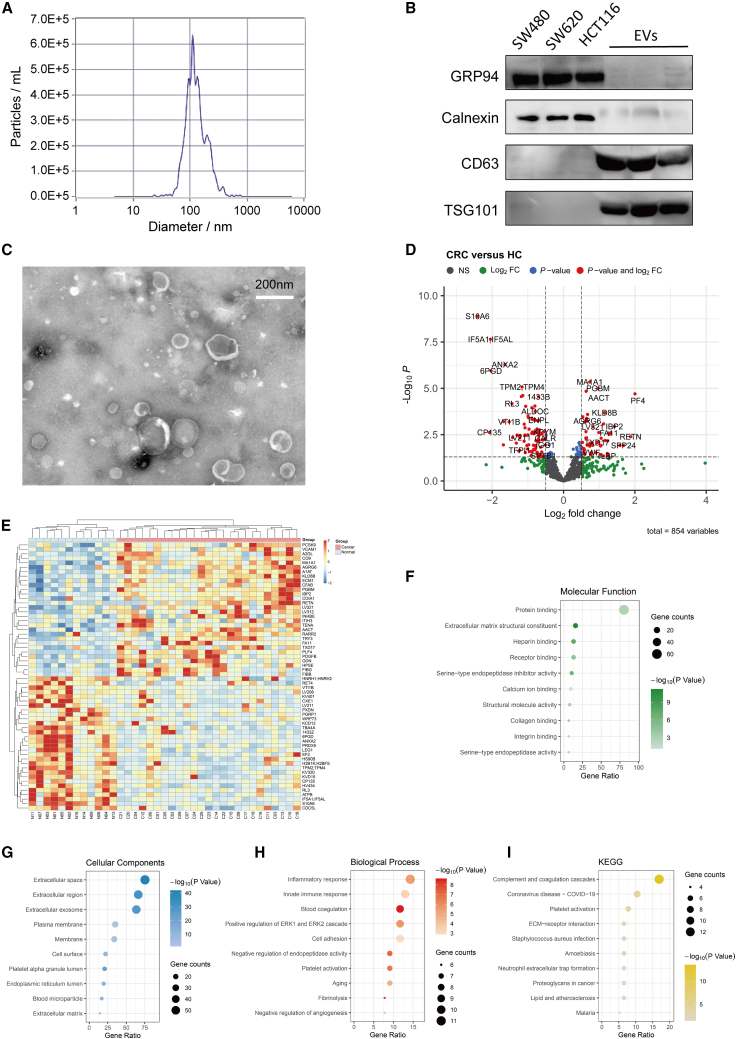
Figure 1.
Identification and characterization of serum EVs in HC and CRC patients by 4D-DIA proteomics analysis
(A) NTA showed the mode size and particle concentration of separated EVs by Flow NanoAnalyzer.
(B) Western blot detected EV markers CD63 and TSG101 in serum EVs. GRP94 and calnexin were used as negative control proteins.
(C) TEM image displayed the morphology of isolated EVs.
(D and E) DEPs from EVs between the CRC and HC groups were illustrated by volcano plot (D, p < 0.05, log2 fold change > 0.5, n = 37) and heatmap (E).
(F–I) Upregulated protein enrichment analysis revealed the potential molecular function (F), cellular component (G), biological process (H), and KEGG pathways (I) enriched in the CRC group compared to the HC group.
In further in-depth EV proteome analysis, 4D-DIA technology identified a total of 5,851 peptides and 854 proteins (Figure S1A), which included 75 upregulated and 91 downregulated proteins in the CRC group compared with the HC group (Figure S1B). Aberrant expression profiles of EVs were illustrated by volcano plot and heatmap (Figures 1D and 1E). Further, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were employed to characterize the potential function of differentially expressed proteins (DEPs). Molecular function of GO analysis revealed upregulated EV proteins related to protein binding (Figure 1F), while downregulated EV proteins associated with RNA and DNA binding (Figure S1C). Cellular components analysis showed that DEPs were localized in both the extracellular space and exosome (Figures 1G and S1D). Additionally, biological processes and KEGG pathway analysis indicated that upregulated proteins were enriched in inflammatory, immune response, blood coagulation, and platelet activation (Figures 1H and 1I). Downregulated proteins also related to certain immune response pathways including adaptive immune response (Figures S1E and S1F).
Screening proteomic biomarkers of serum EVs for CRC diagnosis via ML
To further identify the critical biomarkers of serum EVs for CRC diagnosis, we performed orthogonal partial least squares discriminant analysis (OPLS-DA) to clarify the contribution of different variables in distinguishing CRC patients from HC. The score and scatterplot displayed significant discrimination between CRC and HC subjects based on 4D-DIA proteomics (Figures 2A and 2B). 12 candidate EV proteins, including IGG1, A2MG, AACT, PF4, KLD8B, APOB, IC1, A1AT, IGHM, KV315, CP135, and IGKC, were identified as the core contributors to distinguishing CRC patients from HC by using predictive variable importance in projection (VIPpred) analysis (Figure 2C). Subsequently, ML diagnostic models based on 12 candidate EV proteins were constructed to scan the most valuable variables. Among 5 different ML algorithms, the RF model yielded the best results in terms of classification error (CE), area under the ROC curve (AUC), and area under the precision-recall curve (PRAUC) (Figures 2D, S1G, and S1H; Table S1). Hence, we opted to use the RF algorithm for subsequent ML model construction. In the RF variable importance analysis, 5 variables, including PF4, AACT, KLDB, CP135, and KV315, exhibited the highest rankings (Figure 2E). Further analysis using least absolute shrinkage and selection operator (Lasso) logistic regression identified PF4 and AACT as the top two ranking variables, which were determined to be the most valuable EV proteins for CRC diagnosis (Figures 2F–2H ; Table S2). The combined evaluation of PF4 and AACT exhibited superior diagnostic performance in the RF diagnostic model (Figures 2I and 2J).
Validation of aberrant PF4 and AACT levels in expansion cohorts
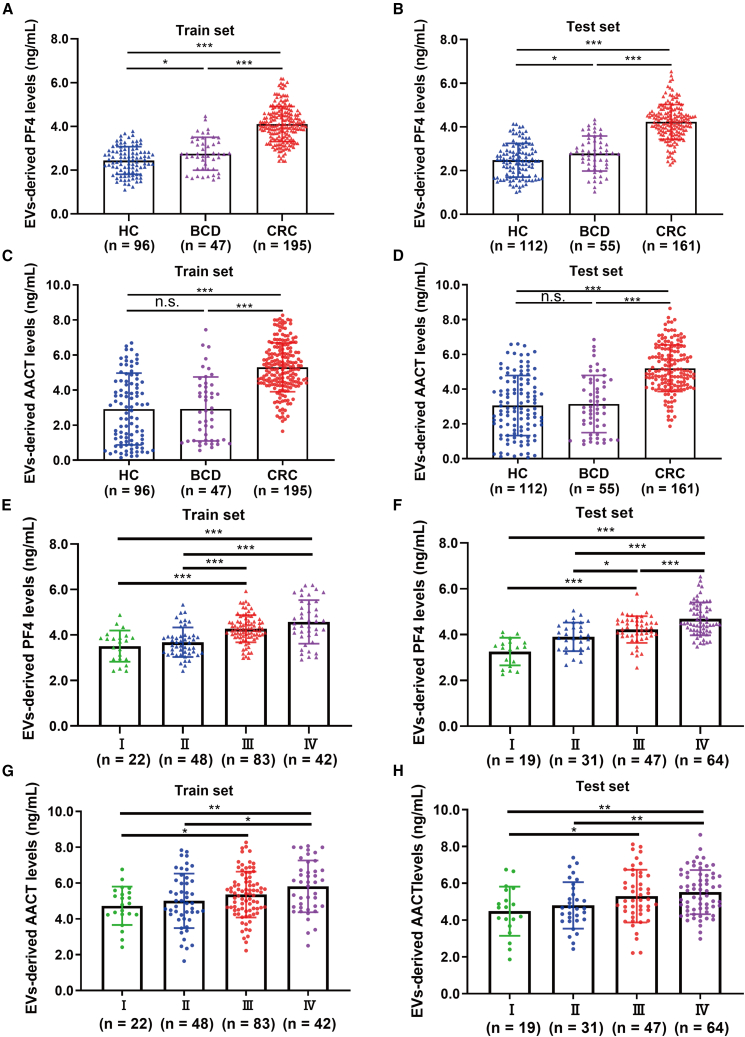
To validate the aberrant elevation of PF4 and AACT identified in the discovery set, EVs from an expansion of 912 individuals, comprising 338 cases in the train set, 328 cases in the test set, and 246 cases in the external set were collected for ELISA detection (Table 1). Distinctly, compared with HC or patients with benign colorectal disease (BCD) or inflammatory disease, the levels of EV-derived PF4 were significantly increased in the train, test, and external sets (Figures 3A, 3B, and S2A). Consistently, elevated AACT levels were also observed in CRC patients compared to HC or patients with BCD or inflammatory disease (Figures 3C, 3D, and S2B). Additionally, statistical analysis of PF4 and clinicopathological characteristics indicated that the levels of PF4 were significantly associated with clinical stage, tumor-node-metastasis (TNM) classification, and differentiation (Table S3). AACT levels were also related to clinical stage and TNM classification (Table S4). Impressively, the levels of serum EV-derived PF4 gradually elevated with clinical staging (Figures 3E, 3F, and S2C). In line with PF4, AACT levels also incrementally increased with the progression of clinical stages (Figures 3G, 3H, and S2D). Intriguingly, EV-derived PF4 and AACT levels were notably reduced in CRC patients after treatment (Figures S2E–S2H). Taken together, these results confirmed the potential of EV-derived PF4 and AACT as important biomarkers for CRC diagnosis and post-treatment monitoring.
Figure 3.
The aberrant levels of PF4 and AACT in expansion cohorts
(A and B) EV-derived PF4 levels detected by ELISA in HC (train set: n = 96, test set: n = 112), BCD (train set: n = 47, test set: n = 55), and CRC (train set: n = 195, test set: n = 161) groups from the train set (A) and the test set (B).
(C and D) EV-derived AACT levels detected by ELISA in HC, BCD, and CRC groups from the train set (C) and test set (D).
(E and F) The levels of EV-derived PF4 at different clinical stages of CRC patients in the train set (E, I: n = 22, II: n = 48, III: n = 83, IV: n = 42) and the test set (F, I: n = 19, II: n = 31, III: n = 47, IV: n = 64).
(G and H) The levels of EV-derived AACT at different clinical stages of CRC patients in the train set (G) and the test set (H). Data are shown as mean ± SD; n.s., not significant, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Development and validation of the EV-related RF diagnostic model for CRC diagnosis
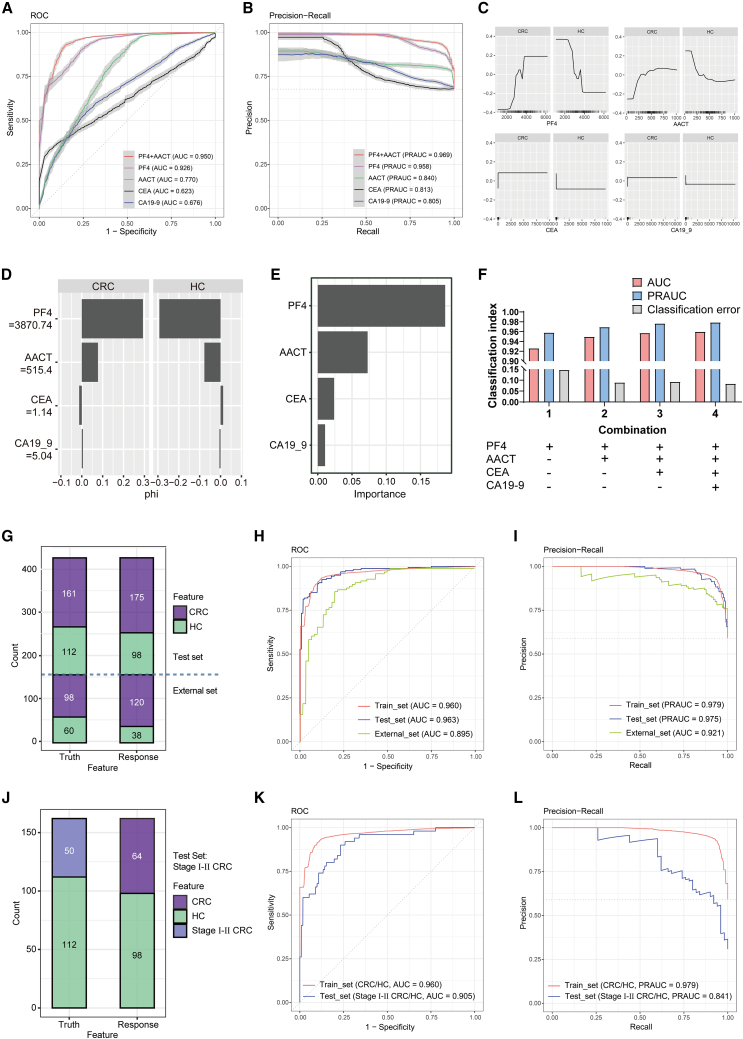
Subsequently, RF diagnostic models were constructed to evaluate the diagnostic efficiency of EV-derived PF4 and AACT compared with traditional CRC biomarkers CEA and CA19-9. In the train set, receiver operating characteristics (ROC) curves displayed significantly higher AUC values for both PF4 (AUC = 0.926) and AACT (AUC = 0.770) compared to CEA (AUC = 0.623) and CA19-9 (AUC = 0.676). Moreover, the combination of PF4 and AACT yielded an impressive AUC of 0.950 (Figure 4A). Consistently, in precision-recall (PR) curve analysis, PF4 and AACT demonstrated superior PRAUC compared with CEA and CA19-9, and the combined PF4 and AACT model even achieved a higher PRAUC of 0.969 (Figure 4B). Accumulated local effects (ALEs) analysis confirmed that both PF4 and AACT had a more pronounced effect on predicting CRC compared to CEA and CA19-9 (Figure 4C). The Shapley value also showed that higher PF4 (≥3870.74 pg/mL) and AACT (≥515.4 ng/mL) levels made the largest contribution in discriminating CRC from HC (Figure 4D), which aligns with the results from the importance analysis (Figure 4E).
Figure 4.
Construction and validation of the EV-related RF diagnostic model for CRC detection
(A and B) ROC curve (A) and PR curve (B) of RF diagnostic models based on indicated variables in the train set.
(C) The ALE curve depicts the accumulated local effects of PF4, AACT, CEA, and CA19-9. The x axis represents the feature values, and the y axis represents the accumulated local effects.
(D) Shapley values bar plot illustrates the Shapley values for each feature in the RF diagnostic model. Each bar represents the average contribution on discriminating CRC patients from HC.
(E) Variable importance score plot showed the contribution of 4 variables in the RF diagnostic model.
(F) CE, AUC, and PRAUC values of the RF diagnostic models with different variable combinations.
(G) Confusion matrix displayed the prediction results for 273 test set sample (161 CRC and 112 HC) and 158 external set sample (98 CRC and 60 HC) through the EV-related diagnostic model.
(H and I) ROC curve (H) and PR curve (I) were plotted for the EV-related diagnostic model using the train and test sets.
(J) Confusion matrix displayed the prediction results for 162 untrained test samples comprising 50 individuals of stages I and II CRC patients and 112 individuals of HC through the EV-related diagnostic model.
(K and L) ROC curve (K) and PR curve (L) were plotted for the EV-related diagnostic model using the train and test sets.
To achieve the best combination of the EV-derived and traditional CRC biomarkers, RF diagnostic models were developed using different combinations of variables. As shown in Figure 4F, the combination of PF4, AACT, CEA, and CA19-9 achieved the best diagnostic performance with an AUC of 0.960, PRAUC of 0.979, and CE of 0.08 (Figures S3A and S3B; Table S5). Thus, the optimal diagnostic model based on the 4 variables was defined as the EV-related model and subsequently validated in the test set (Table S6). The confusion matrix illustrated that the EV-related model exhibited superior accuracy of 0.883 in the test set and 0.810 in the external set (Figures 4G and S3C; Table S6). Furthermore, the excellent diagnostic performance was also confirmed by other metrics including ROC and PRAUC curves with an AUC of 0.963 and 0.895 and a PRAUC of 0.975 and 0.921 in the test set and external set, respectively (Figures 4H and 4I; Table S6).
The diagnosis of early-stage tumors poses greater challenges in comparison to advanced-stage tumors due to the scarcity of suitable tumor markers. To evaluate the diagnostic efficacy of the EV-related model for early-stage CRC, we extracted data from patients with stage I and stage II CRC in the test set for further validation. In confusion matrix analysis, the EV-related model demonstrated reliable accuracy in discriminating patients with stage I and stage II tumors from HC subjects in the test set (Figures 4J and S3D). Consistently, ROC and PRAUC curves further validated the excellent diagnostic efficacy of the EV-related model (Figure 4K and 4L; Table S6). Moreover, the ability of the EV-related model was also tested in distinguishing patients with CRC from patients with BCD or inflammatory disease. Notably, the model presented outstanding diagnostic performance in discriminating between CRC and other patients (Figures S3E–S3J; Table S6). Collectively, EV-derived PF4 and AACT outperformed CEA and CA19-9 as biomarkers for CRC diagnosis. The EV-related model exhibited superior diagnostic performance for CRC, including early-stage diagnosis and differential diagnosis from patients with BCD or inflammatory disease.
Functional enrichment analysis of EV-derived PF4 and AACT
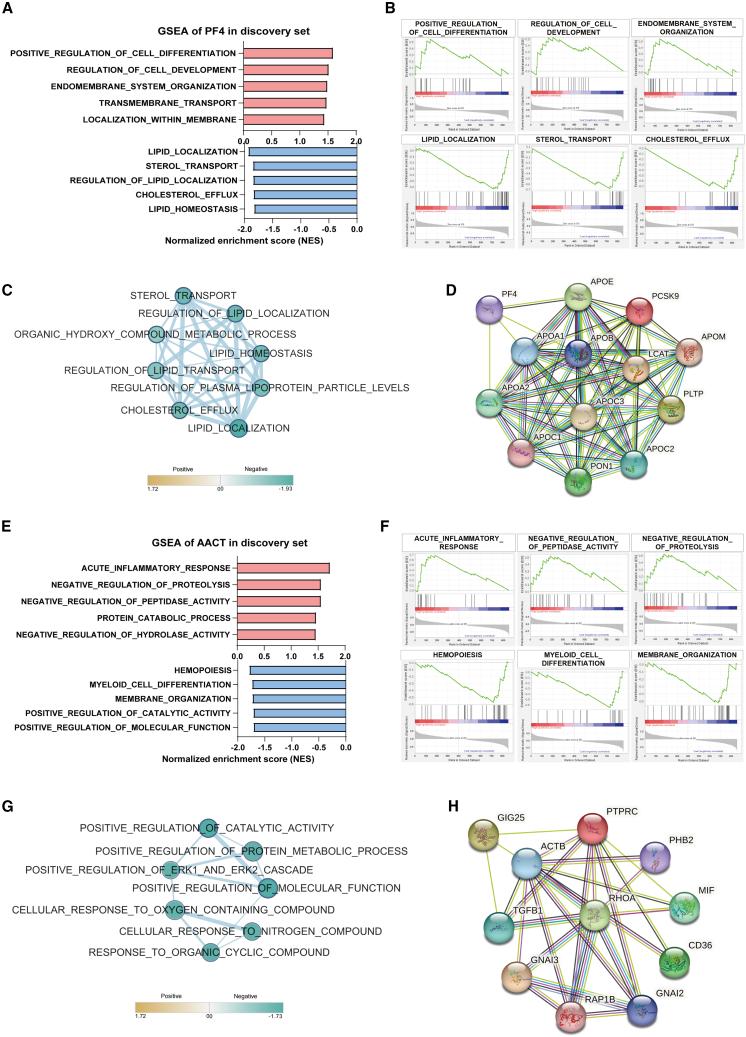
To gain insights into the potential functions of EV-derived PF4 and AACT in CRC, we performed gene set enrichment analysis (GSEA). The results showed that EV-derived PF4 in the discovery set was enriched in pathways related to cell differentiation, cell development, and transmembrane transport. Particularly, lipid localization and cholesterol efflux pathways were negatively correlated with PF4, and similar pathway enrichment results were also obtained in The Cancer Genome Atlas (TCGA) database (Figures 5A, 5B, and S4A). EnrichmentMap analysis was utilized to research the associations between these enriched terms. Likewise, the EV-derived PF4 low-expressed phenotype exhibited strong associations between the localization and homeostasis of lipid, cholesterol efflux, and sterol transport pathways in corresponding association network (Figure 5C). Furthermore, core genes in the EnrichmentMap network were extracted using leading edge analysis and integrated with PF4 to construct the protein-protein interaction (PPI) networks in the STRING database (Figures S4C and 5D). The PPI network indicated that PF4 might interact with the core genes including APOA1, APOA2, and APOE (Figure 5D).
Figure 5.
Functional prediction of EV-derived PF4 and AACT
(A and B) GSEA displayed the top ranking pathways based on EV-derived PF4-high (red, n = 13) and PF4-low (blue, n = 12) phenotypes.
(C) EnrichmentMap network analysis of associated pathways enriched in EV-derived PF4-low phenotype.
(D) STRING database analysis revealed the potential interaction between PF4 and the key proteins involved in enriched pathways.
(E and F) GSEA displayed the top ranking pathways based on EV-derived AACT-high (red, n = 13) and AACT-low (blue, n = 12) phenotypes.
(G) EnrichmentMap network analysis of associated pathways enriched in EV-derived AACT-low phenotype.
(H) STRING database analysis revealed the potential interaction between AACT and the key proteins involved in enriched pathways.
The same analysis pipeline was also processed in EV-derived AACT. GSEA results displayed that the AACT-high phenotype was enriched in several pathways, including “Acute inflammatory response,” “Negative regulation of proteolysis,” and “Negative regulation of peptidase activity” (Figures 5E and 5F). The same enriched pathways were also validated in the TCGA database (Figure S4B). Consistently, EV-derived AACT was negatively associated with the metabolic process and metabolites pathways involved in proteolysis (Figures 5E and 5F), whose association was also reflected in the EnrichmentMap network (Figure 5G). Moreover, leading edge analysis was performed to obtain hub genes in the EnrichmentMap network (Figure S4D). The PPI network comprising AACT and hub genes revealed that AACT might interact with transforming growth factor (TGF)-β1, ACTB, and PTPRC, suggesting its potential role in inflammatory, cytoskeleton, and protein metabolic pathways (Figure 5H).
Deciphering specific cell types releasing EV-derived PF4 and AACT
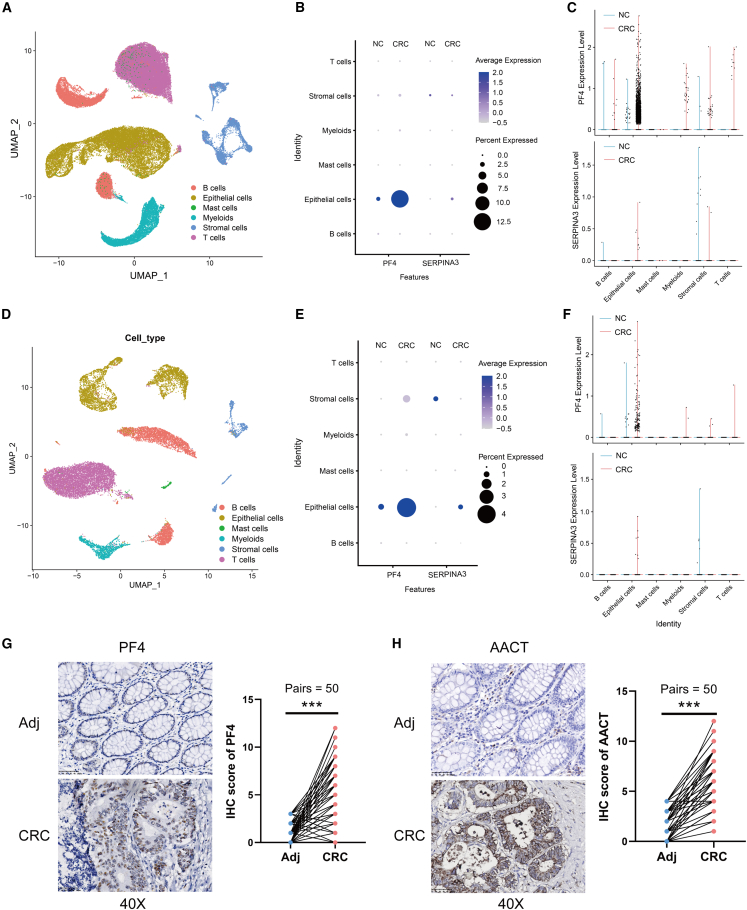
Next, single-cell transcriptome analysis of the GEO: GSE132465 and GEO: GSE132257 datasets was employed to identify the specific cell types responsible for releasing PF4 and AACT packaged in EVs. When analyzing the GEO: GSE132465 dataset comprising normal and CRC tissues (Figures 6A and S4E), PF4 exhibited a dramatic elevation in CRC epithelial cells compared to normal epithelial cells. Additionally, PF4 was also slightly upregulated in myeloid cells, stromal cells, and T cells (Figures 6B and 6C). Consistently, in the GEO: GSE132257 dataset (Figures 6D and S4F), PF4 expression was markedly elevated in CRC epithelial cells and slightly elevated in stromal cells, myeloid cells, and T cells compared to normal tissues (Figures 6E and 6F). As for AACT, its expression was significantly higher in CRC epithelial cells compared to normal epithelial cells in both the GEO: GSE132465 (Figures 6B and 6C) and GEO: GSE132257 datasets (Figures 6E and 6F). Furthermore, immunohistochemistry (IHC) was performed to detect PF4 and AACT expression in 50 paired CRC and adjacent tissues. Consistent with single-cell transcriptome analysis, the expression levels of PF4 and AACT were abnormally elevated in CRC epithelial cells compared to adjacent normal epithelial cells (Figures 6G and 6H). Taken together, aberrant elevation of EV-derived PF4 and AACT might release from CRC epithelial cells, and PF4 might also originate from myeloid cells, stromal cells, and T cells.
Figure 6.
scRNA-seq analysis reveals CRC epithelial cells as the major source of EV-derived PF4 and AACT production
(A) Uniform manifold approximation and projection (UMAP) plot showed different cell types in CRC (n = 23) and normal (n = 10) tissues via single-cell RNA sequencing (scRNA-seq) analysis from the GEO: GSE132465 dataset.
(B) Dot plot showed the expression of PF4 and AACT in normal and CRC tissues from the GEO: GSE132465 dataset.
(C) Violin plot exhibited the expression of PF4 and AACT in normal and CRC tissues from the GEO: GSE132465 dataset.
(D) UMAP plot showed different cell types in CRC (n = 5) and normal (n = 5) tissues via scRNA-seq analysis from the GEO: GSE132257 dataset.
(E) Dot plot showed the expression of PF4 and AACT from the GEO: GSE132257 dataset.
(F) Violin plot exhibited the expression of PF4 and AACT from the GEO: GSE132257 dataset.
(G and H) Representative images and statistical analysis of PF4 (G) and AACT (H) IHC staining in 50 paired adjacent and CRC specimens (400× magnification). Scale bar: 50 μm.
Discussion
CRC is a common malignant tumor with high incidence and mortality rates, ranking third among different types of cancers.16 Due to the lack of effective approaches for early diagnosis, the 5-year survival rate of CRC patients is approximately 50%–60%, which even plunges to 14% for patients with metastasis.17 Non-invasive approaches with accurate and repeatable characteristic are in high demand for early detection to improve patient outcome. In this context, our study identified two biomarkers, PF4 and AACT, by deeply profiling 4D-DIA proteomics data of EVs with a ML pipeline. Subsequently, the optimal RF model based on PF4 and AACT was constructed and yielded the superior diagnostic performance. The identified EV-proteomic signatures and developed RF model provide valuable tools for enhancing early detection and management of CRC in clinical settings.
As an emerging liquid biopsy technology, EVs constitute significant potential for clinical applications in drug delivery therapy and cancer diagnosis.18 Analyzing EVs from serum offers several advantages compared to direct serum testing.9 Primarily, the lipid bilayer structure of EVs shields their cargo from degradation, offering a more accurate representation of the body’s state. In addition, the proteins in the serum of CRC patients are enriched by EVs, which substantially augment detection efficacy. Consequently, EVs have garnered increasing attention in the realm of liquid biopsy. The application of ultracentrifugation for EV extraction we employed is widely recognized as a robust extraction method.19 To verify the reproducibility of our experiments, we utilized a commercial extraction kit based on size exclusion chromatography (SEC) principles for EV isolation. Correlation analysis demonstrated a strong correlation between biomarkers isolated by ultracentrifugation and SEC methods (Figures S5A and S5B). Additionally, aberrant levels of PF4 and AACT isolated by SEC were also observed in the CRC group compared to the HC group (Figures S5C and S5D). RF models based on both EV extraction methods exhibited robust diagnostic performance (Figures S5E and S5F). These results demonstrate the reproducibility of our experiments and the reliability of the proteomic signatures we identified.
A previous study on using serum EVs for CRC diagnosis showed significant limitations, including testing mixed samples and employing unstable TMT-tagged mass spectrometry with instability and limited proteome coverage.20 In contrast, our study conducted separate testing for multiple samples and utilized the most recent mass spectrometry methods, resulting in significantly different protein markers compared to theirs. Moreover, we also identified two biomarkers proposed in this study, namely FN1 and HSP90AA1. The results indicated that FN1 levels were slightly elevated in the CRC group compared to the HC group (Figure S6A), while HSP90AA1 showed no difference between the two groups (Figure S6B). ROC and PR curves demonstrated that the diagnostic efficacy of RF models based on FN1, HSP90AA1, and their combination was markedly inferior compared to that of the models based on PF4, AACT, and their combination (Figures S6C and S6D). Consistent results were also obtained from ALE (Figure S6E) and variable importance analysis (Figure S6F). These data further validated the proteomic signatures we identified as robust biomarkers for CRC diagnosis.
Recently, the application of ML in the field of oncology has been increasingly highlighted. By performing robust feature selection from analytes of liquid and tissue biopsy, we can use ML approaches to improve the accuracy and efficiency of cancer diagnosis, treatment decision, and prognostic prediction.13,21 In our research, various ML algorithms, including support vector machine, k-nearest neighbor, decision tree (Rpart), RF, and logistic regression, were employed to construct diagnostic models. Compared to conventional linear analysis of logistic regression, models based on other ML algorithms such as RF demonstrated a significant improvement in terms of AUC from 0.887 to 0.993 (Figure S2A). Consequently, ML is capable of profiling crucial proteomic features from EVs and developing more robust and reliable models compared to traditional linear regression models.
PF4, also known as CXCL4, was a chemokine mainly produced by activated platelets participating in numerous biological processes, including host inflammatory response promotion, hematopoiesis, and angiogenesis inhibition.22 In addition to platelets, PF4 is also produced and secreted by other cells, such as somatic cells and cancer cells.23,24 Our IHC results indicate that PF4 is highly expressed in CRC epithelial cells compared to adjacent tissues (Figures 6E and 6J). Moreover, single-cell transcriptome analysis revealed that the elevated EV-derived PF4 in the serum of CRC patients may originate from CRC epithelial cells, with a slight increase also observed in myeloid, stromal, and T cells (Figure 6). Our findings suggest that the abnormally elevated PF4 in serum EVs may originate from both tumor cells and the tumor microenvironment. A growing number of studies focus on the role of PF4 in reshaping the immune microenvironment.25 PF4 has been shown to not only drive macrophage migration during tumor progression but also induce differentiation of recruited monocytes into myeloid-derived suppressor cells, thereby suppressing CD8+ T cell function.26,27 Furthermore, PF4 promoted regulatory T cell (Treg) production in a mouse model of sepsis through activation of the STAT5/FOXP3 pathway.28 In addition, PF4 reduced the proliferation of cytotoxic T lymphocytes and promoted the proliferation of Tregs, thereby suppressing the immune response to CRC in transplanted tumor-bearing mice.23 Besides, PF4 deletion abrogated SPP1+ macrophage differentiation and improved fibrosis after cardiac and renal injury.29 PF4 impaired the phagocytic capacity of macrophages by reducing CD36 levels, leading to the development of cardiovascular disease.30 CD36 was a key transporter protein in maintaining lipid homeostasis, and several studies suggested that CD36 was involved in reprogramming lipid metabolism of the tumor microenvironment in CRC.31,32,33 Our bioinformatics results also suggested that EV-derived PF4 was responsible for regulating lipid homeostasis and lipid localization (Figures 5A–5D). It would be intriguing to further explore whether EV-derived PF4 regulated lipid metabolism homeostasis through CD36 to reshape the tumor microenvironment. Additionally, the PPI network indicated that PF4 might interact with several apolipoproteins, including APOA1, APOA2, and APOE, suggesting the potential mechanism of PF4 participating in lipid homeostasis and cholesterol efflux (Figure 5D). Moreover, in our single-cell transcriptome analysis, the elevated EV-derived PF4 in the serum of CRC patients may release from CRC epithelial cells, with a slight increase also observed in myeloid, stromal, and T cells (Figure 6). However, PF4 has not yet been a therapeutic target due to the absence of a defined receptor to explain its regulatory function on immune cells. A recent study revealed that PF4 bound to glycosaminoglycan sugars on proteoglycans in the endothelial extracellular matrix, leading to increased adhesion of leukocytes to blood vessels and causing a series of non-specific recruitment of leukocytes.34 Further studies are needed to fully understand the mechanism of PF4’s regulatory effects on immune cells.
Glycoprotein AACT was a serine protease inhibitor synthesized primarily in the liver and secreted into the blood.35 However, increasing evidence suggested that AACT could also serve as a tumor biomarker and played a crucial role in tumor progression. Sequential window acquisition of all theoretical (SWATH) mass spectrometry identified plasma AACT as a candidate biomarker for the early diagnosis of glioblastoma, while substantially elevated levels of AACT in the urine of patients with metastatic lung cancer have also been reported.36,37 Diagnostic panels of AACT, peptides containing single amino acid variants, and thrombospondin-1 displayed excellent diagnostic performance in identifying pancreatic cancer from HCs, and the combination of AACT and prostate-specific antigen (PSA) remarkably improved the diagnosis of prostate cancer.38,39 Although a marked rise in AACT in CRC tissue compared to paracancerous tissue was reported in previous studies, no significant differences were observed in plasma from CRC patients compared to healthy subjects.40,41
In our study, using 4D-DIA proteomics and ELISA, we demonstrated that AACT was dramatically elevated in EVs of CRC patients and was of clinical diagnostic value (Figures 1 and 3). We revealed the relationship between AACT in EVs and tumors, although the exact function of EV-derived AACT in the carcinogenesis and development of CRC requires further investigation. Previous studies presented that AACT regulated cytokine secretion by activating the nuclear factor κB (NF-κB) signaling pathway, which also promoted the growth and migration of CRC cells.41,42 Interestingly, our bioinformatics results also indicated that EV-derived AACT was most associated with the acute inflammatory response pathway. which could activate NF-κB signaling (Figures 5E and 5F). Moreover, the STRING database analysis suggested that AACT might be involved in inflammatory and NF-κB signaling pathways through the important inflammatory regulator TGF-β (Figure 5H). Nevertheless, AACT was also able to enter the nucleus and establish a strong link with chromatin, leading to the inhibition of liver cancer cell proliferation.43,44 It remained unclear whether AACT in EVs also exhibited these contradictory effects. Moreover, AACT exhibited aberrant elevation in CRC epithelial cells and negative correlations with proteolysis pathway. The potential role of AACT in cytoskeleton and protein metabolic pathways remains to be further investigated (Figures 5E–5H and 6). Taken together, AACT may hold great promise as a diagnostic and therapeutic target for CRC, although further studies are needed to fully understand its role in tumor progression.
Limitations of the study
A limitation of our research is that the sample size of our cohorts was not large enough. Nevertheless, we were able to replicate our findings in the proteomics cohort using ELISA in two independent cohorts. In addition, we needed to expand the enrolled population range in order to verify the effect of cardiovascular disease, inflammation, and other confounding factors on the identified markers. Meanwhile, the specificity and sensitivity of the combination of PF4 and AACT for other gastrointestinal tumors also required more samples to be evaluated.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Serum samples from patients | The Seventh Affiliated Hospital of Sun Yat-Sen University | This paper |
| Serum samples from patients | Shenzhen People’s Hospital | This paper |
| Serum samples from patients | Sun Yat-sen University Cancer Center | This paper |
| Critical commercial assays | ||
| Human PF4 ELISA Kit | Neobioscience | EHC135.96 |
| EVs extraction Kit | Echobiotech | ES9P11e |
| Human AACT ELISA Kit | FineTest | EH0570 |
| Deposited data | ||
| Mass spectrometry data | iProX database | IPX0008187000 |
| Single-cell RNA sequencing data | GEO database | GEO: GSE178341, GSE132465 |
| TCGA CRC dataset | TCGA database | https://gdc.cancer.gov/about-data/publications/pancanatlas |
| Software and algorithms | ||
| R (version 4.2.3) | R Project | https://www.r-project.org |
| SPSS 26.0 | IBM | RRID:SCR_002865 |
Resource availability
Lead contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Zhijian Huang (huangzhj29@mail2.sysu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier IPX0008187000.
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
The discovery cohort comprised of 12 healthy controls (HC) and 25 colorectal cancer (CRC) patients before treatment. Serum samples from HC and CRC patients were collected between May 2022 and June 2022 at Sun Yat-sen University Cancer Center.
Expansion cohorts for ELISA detection, model development, and validation include three listed cohorts as follows.
-
•
The train set composed of 96 HC, 47 patients with benign colorectal diseases (BCD), and 195 CRC patients before treatment. Serum samples from the train set were collected between August 2020 and October 2022 at Sun Yat-sen University Cancer Center.
-
•
The test set consisted of 112 HC, 55 BCD patients, and 161 CRC patients. Serum samples from the test set were collected between September 2020 and December 2022 at The Seventh Affiliated Hospital of Sun Yat-Sen University. The CRC group included 161 cases of CRC patients before treatment and 39 cases of CRC patients after treatment.
-
•
The external set consisted of 60 HC, 42 Enteritis, 46 Hepatitis B, and 98 CRC patients. Serum samples from the external set were collected between October 2023 and March 2024 at Shenzhen People’s Hospital.
The enrolled HC individuals had no history of intestinal diseases, inflammatory diseases, or other diseases. The diagnosis of CRC was confirmed through histopathological examination, and serum samples were collected at the time of diagnosis prior to tumor resection or chemoradiotherapy, except for the 39 post-treatment CRC patients in the test set. The diagnosis of BCD was based on standard endoscopic, histologic, and radiographic criteria. Informed consent was obtained from all participants, and the study was approved by the Ethics Committee of The Seventh Affiliated Hospital of Sun Yat-Sen University (KY-2020-039-01), Sun Yat-sen University Cancer Center (B2022-475-01), and Shenzhen People’s Hospital (LL-KY-2022478). The clinical and biological characteristics of the individuals from the four cohorts were described in Table 1.
Method details
Serum EVs isolation
2 mL Fresh blood collected from the individuals were centrifuged at 4000 g for 20 min in the vacuum blood tube. The supernatant serum was then stored at −80°C. The collected frozen serum was uniformly thawed at 4°C and then centrifuged at 10,000 g for 20 min. The resulting suspensions were diluted with 4 mL of cold PBS and transferred to the 0.22 μm filter. After centrifugation of the filtered supernatants at 120,000 g for 90 min, the supernatants were aspirated to collect the pellets at the bottom of the tube. The pellets were subsequently resuspended in 4 mL PBS and subjected to another centrifugation at 120,000 g for 90 min. The isolated EV samples were suspended in 100 μL of PBS and added to the mini-EVS purification column to remove free proteins and nucleic acids by adsorption. The remaining suspension represented high-purity purified EVs and was stored at −80°C for further use.
Commercially EV extraction kit was also applied for the isolation of serum EVs in the external set based on the size exclusion chromatography (SEC) principle. One milliliter of blood plasma, filtered through a 0.8 μm filter, was diluted 1.5 times with PBS and further purified using Exosupur columns (ES9P11e, Echobiotech, China). The samples were then eluted with PBS, and 2.5 mL eluate fractions were collected. Subsequently, these fractions were concentrated down to 200 μL using 100 kDa molecular weight cut-off Amicon Ultra spin filters (Millipore, Germany).
Serum EV identification
For western blotting, 90 μL of RIPA lysis buffer was mixed with 10 μL of EV samples and incubated on ice for 30 min. Afterward, the mixtures were centrifuged at 12,000 g for 5 min at 4°C, and the supernatant was collected. Protein quantification was performed using the BCA assay kit. After conducting SDS-PAGE electrophoresis, EV-derived protein samples were transferred onto a PVDF membrane and blocked with 7% skim milk at room temperature for 1–2 h. Overnight incubation at 4°C was carried out with primary antibodies. Subsequent incubation with secondary antibodies was performed at room temperature. The chemiluminescence signals were captured using the ChemiDoc Touch imaging system (Bio-rad, USA).
For transmission electron microscopy (TEM), EV suspensions were fixed using 0.1% (v/v) paraformaldehyde at a 1:1 volume ratio for 30 min. A drop of 10 μL of fixed EVs was placed on a carbon-coated copper grid for 3 min. Excess liquid was absorbed using filter paper. Subsequently, 2% phosphotungstic acid was added to the grid for staining, and excess liquid was again absorbed using filter paper. The copper grids were finally examined and photographed using TEM HT7800 (Hitachi, Tokyo, Japan).
For nanoparticle tracking analysis (NTA), the EV samples were diluted 1:1000 with PBS. The diluted samples were directly analyzed using a nanoparticle tracking analyzer ZetaVIEW S/N 21–734 (Particle Metrix, Munich, Germany).
ELISA detection
ELISA kits were applied to detect the levels of PF4 (Neobioscience, EHC135.96) and AACT (FineTest, EH0570) derived from serum EVs. A total of 10 μL of EV samples were mixed with 90 μL of RIPA lysate on ice for 60 min, and were then diluted with 200 μL of PBS. Next, 100 μL of the diluted samples was added to a 96-well plate for ELISA detection, following the manufacturer’s protocol. Finally, the absorbance at 450 nm was measured using synergyH1 multi-model readers (BioTek, Vermont, USA).
4D-DIA quantitative proteomics
4D-DIA quantitative proteomics analysis was conducted by Shanghai Genechem Co., Ltd. In this analysis, a total of 37 serum EV samples from the discovery set were subjected to protein extraction using ultrasonic lysis, with the addition of 1% protease inhibitor. 200 μg protein samples were subsequently digested into peptides by filter-aided sample preparation (FASP).
DDA mass spectrometry library construction: Each sample was loaded with 200 ng of peptides and desalted using Evotips. Separation was performed using the nanoflow Evosep One system (Evosep, Denmark), coupled to a timsTOF Pro mass spectrometer (Bruker, Bremen, Germany) equipped with a CaptiveSpray ion source. Buffer A consisted of 0.1% aqueous formic acid, while buffer B comprised 0.1% formic acid in acetonitrile. Chromatographic separation was conducted using the 30SPD method provided by Evosep One. Following chromatographic separation on Evosep One, samples underwent mass spectrometric analysis using the PASEF mode of the timsTOF Pro Mass Spectrometer (Bruker, Bremen, Germany). Ionization was conducted in positive ion mode with a mass range of 100–1700 m/z. The 1/K0 ion mobility range was set to 0.6–1.6 V⋅s/cm2, with an ion accumulation/release time of 100 ms and a 100% ion utilization rate. The capillary voltage was set to 1500 V, and the drying gas flow rate was 3 L/min with a drying temperature of 180°C. PASEF settings included 10 MS/MS scans (total cycle time: 1.16 s), a charge range of 0–5, dynamic exclusion time of 0.4 min, ion target intensity of 10,000, ion intensity threshold of 2500, and collision-induced dissociation energy of 20–59 eV.
DIA mass spectrometry analysis: The peptide samples were diluted to 10 ng/μL with 0.1% formic acid and supplemented with iRT peptide mixture. 200 ng peptide sample mixed with iRT was analyzed on a Evosep One system (Evosep, Denmark) coupled to a timsTOF Pro (Bruker, Bremen, Germany) equipped with a CaptiveSpray source. Peptides were separated on a 15 cm × 150 μm analytical column, 1.9 μm C18 beads with a packed emitter tip (Evosep, Denmark). The column temperature was maintained at 50°C using an integrated column oven (Bruker, Germany). The LC-separation method was provided by Evosep One at 30 samples per day. For diaPASEF, we adapted the instrument firmware to perform data-independent isolation of multiple precursor windows within a single TIMS separation (100 ms). We used a method with two windows in each 100 ms diaPASEF scan. 100 of these scans covered the diagonal scan line for doubly and triply charged peptides in the m/z – ion mobility plane with narrow 25 m/z precursor windows.
Raw data of DDA and DIA were processed and analyzed by Spectronaut (Biognosys AG, Switzerland) with default settings. Spectronaut was set up to search the database assuming trypsin as the digestion enzyme. Carbamidomethyl (C) was specified as the fixed modification. Oxidation (M) and acetyl (Protein N-term) were specified as the variable modifications. Retention time prediction type was set to dynamic iRT. Spectronaut will determine the ideal extraction window dynamically depending on iRT calibration and gradient stability. Q value cutoff on precursor and protein level was applied 1%.
Machine learning and development of the EV-related diagnostic model
Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) was performed using SIMCA software version 14.1. The protein of predictive variable importance in projection larger than 4 (VIPpred > 4) was considered to be candidate EV biomarkers for CRC discrimination. 5 ML algorithms were used to conduct the diagnostic models based on the candidate EV biomarkers for the selection of the optimal algorithm. Based on the optimal RF algorithm, 12 candidate biomarkers were further evaluated in RF diagnostic model in a bootstrap cross-validation manner using mlr3 R package (version 0.14.1) and Lasso logistic regression analysis.
For the development of the EV-related diagnostic model, different combinations of the variables including PF4, AACT, CEA, and CA19-9 were used to conduct the RF diagnostic model. The best EV-related diagnostic model based on the optimal combination of variables was then determined by its superior diagnostic efficiency. To ensure the reliability of the developed model, the test set was employed to validate the diagnostic performance of the EV-related model by confusion matrix, receiver operating characteristics (ROC) curve, and precision-recall (PR) curve.
Bioinformatics analysis
RNA-seq data and clinical data of TCGA CRC database were obtained from The Cancer Genome Atlas (TCGA) databases (https://genome-cancer.ucsc.edu). Gene Set Enrichment Analysis (GSEA) was manipulated to predict the GO molecular function, cellular component, biological process, and Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets of the Molecular Signature Database v7.4 (http://www.broadinstitute.org/gsea/msigdb) based on PF4 or AACT high and low expressed phenotype. EnrichmentMap plugin in Cytoscape 3.8.2 software was utilized to conduct the association of the enriched pathways. Leading edge analysis was performed by GSEA 4.1.0 to elucidate key genes involved in the EnrichmentMap pathways network. The protein-protein interaction (PPI) networks were constructed using the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/).
scRNA-seq data analysis
The raw single-cell RNA sequencing data were downloaded in GEO database (GSE132465 and GSE132257) and processed using the R package Seurat (version 3.1.1) on R platform (version 4.2.1). The GEO: GSE132465 dataset comprises 23 CRC samples and 10 normal tissue samples, with a total of 63,689 single-cell transcriptomic data. The GEO: GSE132257 dataset was composed of 5 CRC samples and 5 normal tissue samples, with a total of 18,409 cells of transcriptomic data. After passing quality control, cells were merged into one count matrix and conducted normalization, dimensional reduction, and clustering by using the NormalizeData, ScaleData, and RunPCA functions according to Seurat R package. FindClusters function was used for cell clustering and FindAllMarkers function was applied for identifying the markers of clusters. Cell types were annotated by using ScType function and marker gene expression based on CellMarker database.
Immunohistochemistry staining
The sections of 50 paired CRC and adjacent tissue were collected at The Seventh Affiliated Hospital of Sun Yat-Sen University. The tissue sections were initially deparaffinized, followed by rehydration in a graded ethanol series and pretreated with 0.01 M citrate buffer (pH 6.0) using a high-pressure method. Subsequently, the sections were immersed in 3% H2O2 for 20 min to quench endogenous peroxidas, and goat serum was applied to block nonspecific background staining. Next, primary antibodies PF4 (Servicebio, GB113482) and AACT (ZSGB-BIO, ZA0006) were applied. After an overnight incubation with the primary antibodies at 4°C, the sections were treated with HRP-conjugated secondary antibody. The antigen-antibody complex was visualized by incubation with the DAB kit. The stained sections were captured using a slide scanner (Axio Scan. Z1, ZEISS). Protein expression levels were determined using the staining index (SI), calculated by multiplying the score for stained cell proportions by the staining intensity score. Stained tumor cell proportions were graded as follows: 0, <5% positive tumor cells; 1, 5%–25% positive tumor cells; 2, 26%–50% positive tumor cells; 3, 51%–75% positive tumor cells; 4, >75% positive cells. Staining intensity was scored as follows: 0, negative staining (no staining); 1, weak staining (light yellow); 2, moderate staining intensity (brown); 3, positive staining (yellow).
Quantification and statistical analysis
Statistical analyses were performed with the IBM SPSS Statistics 26.0 and R version 4.2.3. The data variability was presented as the SD (mean ± SD) and analyzed via unpaired Student’s t test between two groups for normally distributed data. Otherwise, the data were analyzed via nonparametric Mann-Whitney test. The diagnostic performance in terms of AUC, PRAUC, classification error (CE), sensitivity, specificity, precision, recall, accuracy, and F1 score was calculated by using mlr3 R package. p < 0.05 was defined statistical significance.
Acknowledgments
This study was supported by funds from the National Natural Science Foundation of China (82103346, 82202829, 82202985, and 82203661), Guangdong Basic and Applied Basic Research Foundation (2021A1515110094, 2022A1515111199, 2022A1515111062, and 2023A1515220107), General Project of Shenzhen Science and Technology Innovation Commission (JCYJ20220530145003008 and JCYJ20220530145001002), Guangdong Natural Science Fund (2023A1515010214), and Five-Three Project for Training Clinician-Scientists of Shenzhen People’s Hospital (no. SYWGSLCYJ202201).
Author contributions
Conceptualization, Z.H., H.Y., and Z.Y.; methodology, Z.H., H.Y., and Y.R.; data collection, X.L., Y.Y., J.T., G.H., and L.Z.; statistical analysis, H.Y., J.X., Z.H., and S.X.; funding acquisition, J.X., Z.H., H.Y., Z.Y., and X.Y.; study supervision, Z.Y. and X.Y. All authors reviewed the manuscript and approved the final revision.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101689.
Contributor Information
Xiaopeng Yuan, Email: yuanxp2001@126.com.
Zheng Yang, Email: yangzheng@sysush.com.
Zhijian Huang, Email: huangzhj29@mail2.sysu.edu.cn.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Biller L.H., Schrag D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA. 2021;325:669–685. doi: 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- 3.Rutter M.D., Beintaris I., Valori R., Chiu H.M., Corley D.A., Cuatrecasas M., Dekker E., Forsberg A., Gore-Booth J., Haug U., et al. World Endoscopy Organization Consensus Statements on Post-Colonoscopy and Post-Imaging Colorectal Cancer. Gastroenterology. 2018;155:909–925. doi: 10.1053/j.gastro.2018.05.038. [DOI] [PubMed] [Google Scholar]
- 4.Li C., Zhang D., Pang X., Pu H., Lei M., Fan B., Lv J., You D., Li Z., Zhang T. Trajectories of Perioperative Serum Tumor Markers and Colorectal Cancer Outcomes: A Retrospective, Multicenter Longitudinal Cohort Study. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engle D.D., Tiriac H., Rivera K.D., Pommier A., Whalen S., Oni T.E., Alagesan B., Lee E.J., Yao M.A., Lucito M.S., et al. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science. 2019;364:1156–1162. doi: 10.1126/science.aaw3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z., Hu E., Shen H., Tan J., Zeng S. The functional and clinical roles of liquid biopsy in patient-derived models. J. Hematol. Oncol. 2023;16:36. doi: 10.1186/s13045-023-01433-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clack K., Soda N., Kasetsirikul S., Mahmudunnabi R.G., Nguyen N.T., Shiddiky M.J.A. Toward Personalized Nanomedicine: The Critical Evaluation of Micro and Nanodevices for Cancer Biomarker Analysis in Liquid Biopsy. Small. 2023;19 doi: 10.1002/smll.202205856. [DOI] [PubMed] [Google Scholar]
- 8.Ebrahimi N., Faghihkhorasani F., Fakhr S.S., Moghaddam P.R., Yazdani E., Kheradmand Z., Rezaei-Tazangi F., Adelian S., Mobarak H., Hamblin M.R., Aref A.R. Tumor-derived exosomal non-coding RNAs as diagnostic biomarkers in cancer. Cell. Mol. Life Sci. 2022;79:572. doi: 10.1007/s00018-022-04552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu D., Li Y., Wang M., Gu J., Xu W., Cai H., Fang X., Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer. 2022;21:56. doi: 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Min L., Zhu S., Chen L., Liu X., Wei R., Zhao L., Yang Y., Zhang Z., Kong G., Li P., Zhang S. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J. Extracell. Vesicles. 2019;8 doi: 10.1080/20013078.2019.1643670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen I.H., Xue L., Hsu C.C., Paez J.S.P., Pan L., Andaluz H., Wendt M.K., Iliuk A.B., Zhu J.K., Tao W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clift A.K., Dodwell D., Lord S., Petrou S., Brady M., Collins G.S., Hippisley-Cox J. Development and internal-external validation of statistical and machine learning models for breast cancer prognostication: cohort study. BMJ. 2023;381 doi: 10.1136/bmj-2022-073800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 14.Halner A., Hankey L., Liang Z., Pozzetti F., Szulc D., Mi E., Liu G., Kessler B.M., Syed J., Liu P.J. DEcancer: Machine learning framework tailored to liquid biopsy based cancer detection and biomarker signature selection. iScience. 2023;26 doi: 10.1016/j.isci.2023.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W., Wang T., Wang N., Zhang H., Zha Y., Ji L., Chu Y., Ning K. Artificial intelligence-enabled microbiome-based diagnosis models for a broad spectrum of cancer types. Brief Bioinform. 2023;24 doi: 10.1093/bib/bbad178. [DOI] [PubMed] [Google Scholar]
- 16.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 17.Xie Y.H., Chen Y.X., Fang J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Targeted Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piffoux M., Silva A.K.A., Gazeau F., Salmon H. Potential of on-chip analysis and engineering techniques for extracellular vesicle bioproduction for therapeutics. View. 2022;3 [Google Scholar]
- 19.Li Z., Moniruzzaman M., Dastgheyb R.M., Yoo S.W., Wang M., Hao H., Liu J., Casaccia P., Nogueras-Ortiz C., Kapogiannis D., et al. Astrocytes deliver CK1 to neurons via extracellular vesicles in response to inflammation promoting the translation and amyloidogenic processing of APP. J. Extracell. Vesicles. 2020;10 doi: 10.1002/jev2.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y., Xie Y., Xu L., Zhan S., Xiao Y., Gao Y., Wu B., Ge W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int. J. Cancer. 2017;140:900–913. doi: 10.1002/ijc.30496. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Liu Y., Niu X. Application of artificial intelligence for improving early detection and prediction of therapeutic outcomes for gastric cancer in the era of precision oncology. Semin. Cancer Biol. 2023;93:83–96. doi: 10.1016/j.semcancer.2023.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Huang H. Platelet factor-4 (CXCL4/PF-4): an angiostatic chemokine for cancer therapy. Cancer Lett. 2013;331:147–153. doi: 10.1016/j.canlet.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Deng S., Deng Q., Zhang Y., Ye H., Yu X., Zhang Y., Han G.Y., Luo P., Wu M., Yu Y., Han W. Non-platelet-derived CXCL4 differentially regulates cytotoxic and regulatory T cells through CXCR3 to suppress the immune response to colon cancer. Cancer Lett. 2019;443:1–12. doi: 10.1016/j.canlet.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Gao J., Wang X., Deng S., Ye H., Guan W., Wu M., Zhu S., Yu Y., Han W. CXCL4 mediates tumor regrowth after chemotherapy by suppression of antitumor immunity. Cancer Biol. Ther. 2015;16:1775–1783. doi: 10.1080/15384047.2015.1095404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bikfalvi A., Billottet C. The CC and CXC chemokines: major regulators of tumor progression and the tumor microenvironment. Am. J. Physiol. Cell Physiol. 2020;318:C542–C554. doi: 10.1152/ajpcell.00378.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox J.M., Kausar F., Day A., Osborne M., Hussain K., Mueller A., Lin J., Tsuchiya T., Kanegasaki S., Pease J.E. CXCL4/Platelet Factor 4 is an agonist of CCR1 and drives human monocyte migration. Sci. Rep. 2018;8:9466. doi: 10.1038/s41598-018-27710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph R., Soundararajan R., Vasaikar S., Yang F., Allton K.L., Tian L., den Hollander P., Isgandarova S., Haemmerle M., Mino B., et al. CD8(+) T cells inhibit metastasis and CXCL4 regulates its function. Br. J. Cancer. 2021;125:176–189. doi: 10.1038/s41416-021-01338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu T., Zhao J., Wang X., Meng Y., Zhao Z., Bao R., Deng X., Bian J., Yang T. CXCL4 promoted the production of CD4(+)CD25(+)FOXP3(+)treg cells in mouse sepsis model through regulating STAT5/FOXP3 pathway. Autoimmunity. 2020;53:289–296. doi: 10.1080/08916934.2020.1777283. [DOI] [PubMed] [Google Scholar]
- 29.Hoeft K., Schaefer G.J.L., Kim H., Schumacher D., Bleckwehl T., Long Q., Klinkhammer B.M., Peisker F., Koch L., Nagai J., et al. Platelet-instructed SPP1(+) macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsey M.L., Jung M., Yabluchanskiy A., Cannon P.L., Iyer R.P., Flynn E.R., DeLeon-Pennell K.Y., Valerio F.M., Harrison C.L., Ripplinger C.M., et al. Exogenous CXCL4 infusion inhibits macrophage phagocytosis by limiting CD36 signalling to enhance post-myocardial infarction cardiac dilation and mortality. Cardiovasc. Res. 2019;115:395–408. doi: 10.1093/cvr/cvy211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gong J., Lin Y., Zhang H., Liu C., Cheng Z., Yang X., Zhang J., Xiao Y., Sang N., Qian X., et al. Reprogramming of lipid metabolism in cancer-associated fibroblasts potentiates migration of colorectal cancer cells. Cell Death Dis. 2020;11:267. doi: 10.1038/s41419-020-2434-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P., Qin H., Li Y., Xiao A., Zheng E., Zeng H., Su C., Luo X., Lu Q., Liao M., et al. CD36-mediated metabolic crosstalk between tumor cells and macrophages affects liver metastasis. Nat. Commun. 2022;13:5782. doi: 10.1038/s41467-022-33349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu S., Chaudhary O., Rodríguez-Morales P., Sun X., Chen D., Zappasodi R., Xu Z., Pinto A.F., Williams A., Schulze I., Farsakoglu Y. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8(+) T cells in tumors. Immunity. 2021;54:1561–15677. doi: 10.1016/j.immuni.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray A.L., Karlsson R., Roberts A.R.E., Ridley A.J.L., Pun N., Khan B., Lawless C., Luís R., Szpakowska M., Chevigné A., et al. Chemokine CXCL4 interactions with extracellular matrix proteoglycans mediate widespread immune cell recruitment independent of chemokine receptors. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2022.111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Y., Wang W., Wang Q., Zhang Y., Zahid K.R., Raza U., Gong Y. Alpha-1-antichymotrypsin as a novel biomarker for diagnosis, prognosis, and therapy prediction in human diseases. Cancer Cell Int. 2022;22:156. doi: 10.1186/s12935-022-02572-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyauchi E., Furuta T., Ohtsuki S., Tachikawa M., Uchida Y., Sabit H., Obuchi W., Baba T., Watanabe M., Terasaki T., Nakada M. Identification of blood biomarkers in glioblastoma by SWATH mass spectrometry and quantitative targeted absolute proteomics. PLoS One. 2018;13 doi: 10.1371/journal.pone.0193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Li Y., Qiu F., Qiu Z. Comparative analysis of the human urinary proteome by 1D SDS-PAGE and chip-HPLC-MS/MS identification of the AACT putative urinary biomarker. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010;878:3395–3401. doi: 10.1016/j.jchromb.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 38.Nie S., Yin H., Tan Z., Anderson M.A., Ruffin M.T., Simeone D.M., Lubman D.M. Quantitative analysis of single amino acid variant peptides associated with pancreatic cancer in serum by an isobaric labeling quantitative method. J. Proteome Res. 2014;13:6058–6066. doi: 10.1021/pr500934u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu L., Jäämaa S., Af Hällström T.M., Laiho M., Sankila A., Nordling S., Stenman U.H., Koistinen H. PSA forms complexes with alpha1-antichymotrypsin in prostate. Prostate. 2013;73:219–226. doi: 10.1002/pros.22560. [DOI] [PubMed] [Google Scholar]
- 40.Dimberg J., Ström K., Löfgren S., Zar N., Hugander A., Matussek A. Expression of the serine protease inhibitor serpinA3 in human colorectal adenocarcinomas. Oncol. Lett. 2011;2:413–418. doi: 10.3892/ol.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao L.L., Pei X.F., Qiao X., Yu J., Ye H., Xi C.L., Wang P.Y., Gong Z.L. SERPINA3 Silencing Inhibits the Migration, Invasion, and Liver Metastasis of Colon Cancer Cells. Dig. Dis. Sci. 2018;63:2309–2319. doi: 10.1007/s10620-018-5137-x. [DOI] [PubMed] [Google Scholar]
- 42.Alfadda A.A., Benabdelkamel H., Masood A., Jammah A.A., Ekhzaimy A.A. Differences in the Plasma Proteome of Patients with Hypothyroidism before and after Thyroid Hormone Replacement: A Proteomic Analysis. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santamaria M., Pardo-Saganta A., Alvarez-Asiain L., Di Scala M., Qian C., Prieto J., Avila M.A. Nuclear alpha1-antichymotrypsin promotes chromatin condensation and inhibits proliferation of human hepatocellular carcinoma cells. Gastroenterology. 2013;144:818–828. doi: 10.1053/j.gastro.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 44.Ko E., Kim J.S., Bae J.W., Kim J., Park S.G., Jung G. SERPINA3 is a key modulator of HNRNP-K transcriptional activity against oxidative stress in HCC. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier IPX0008187000.
-
•
This study did not generate custom computer code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.