Summary
Sirtuin 1 (SIRT1) is a histone deacetylase and plays diverse functions in various physiological events, from development to lifespan regulation. Here, in Parkinson’s disease (PD) model mice, we demonstrated that SIRT1 ameliorates parkinsonism, while SIRT1 knockdown further aggravates PD phenotypes. Mechanistically, SIRT1 interacts with and deacetylates pyruvate kinase M2 (PKM2) at K135 and K206, thus leading to reduced PKM2 enzyme activity and lactate production, which eventually results in decreased glial activation in the brain. Administration of lactate in the brain recapitulates PD-like phenotypes. Furthermore, increased expression of PKM2 worsens PD symptoms, and, on the contrary, inhibition of PKM2 by shikonin or PKM2-IN-1 alleviates parkinsonism in mice. Collectively, our data indicate that excessive lactate in the brain might be involved in the progression of PD. By improving lactate homeostasis, SIRT1, together with PKM2, are likely drug targets for developing agents for the treatment of neurodegeneration in PD.
Graphical abstract

Highlights
-
•
SIRT1 attenuates glial activation and rescues dopaminergic neurons in PD model mice
-
•
SIRT1 deacetylates PKM2 at K135/206 to represses its enzyme activity
-
•
Overexpression of PKM2 or lactate infusion induces PD-like phenotypes in mice
-
•
PKM2 inhibitors play a beneficial role in PD model mice
Lian et al. found that, upon various risk factors of PD, SIRT1 expression in DAs was reduced, leading to an increase in PKM2 activity. As a consequence, lactate accumulates in the brain to induce glial activation and sustained neuroinflammation. These outcomes eventually induce DA degeneration and the progression of PD.
Introduction
Parkinson’s disease (PD) is the second common neurodegenerative disease, preceded by Alzheimer’s disease. Clinical survey data suggest that approximately 6 million people worldwide are known to suffer from this disease.1 Along with rapidly aging populations, a growing number of individuals will be affected by PD. Notably, dopaminergic neuronal loss in the substantia nigra pars compacta (SNpc) is a common feature of PD.2 Consequently, loss of dopaminergic neurons causes motor symptoms, including tremor, imbalance, slowness, and stiffness, which are a hallmark and diagnostic standard of PD.3 As the disease progresses, pathological changes will occur in the cerebral cortices, resulting in cognitive impairment and hallucinations.4 To date, the precise causative factors and underlying molecular mechanisms that result in PD progression remain unclear. In general, aging, genetic mutations, and environmental toxins are all considered as risk factors for PD.5
The brain is an energy-demanding organ, and glucose is a major energy source utilized by both neurons and astrocytes. Through glycolysis, the Krebs cycle, and oxidative phosphorylation, glucose undergoes complete oxidation and produces ATP. In these processes, pyruvate is a key metabolite, linking glycolysis and subsequent oxidative phosphorylation. Pyruvate kinase (PK) is responsible for catalyzing the final step of glycolysis to produce pyruvate and ATP. In mammals, there are four isoforms of PK, namely PKL, PKR, PKM1, and PKM2. Growing evidence has shown that ectopic alterations in PKM2 activity are often associated with various pathologic progressions, including tumorigenesis,6 diabetic nephropathy,7 and autoimmunity disease.8 By utilizing PK, pyruvate is accumulated in cells, which can, in turn, be used to fuel the tricarboxylic acid cycle or be converted into lactate. Under the latter conditions, excess lactate accumulates in the brain, eventually resulting in detrimental consequences such as energy deficits and neuroinflammation.9,10 Notably, these two outcomes are closely related to the pathological progression of PD.11,12,13 Hence, we predict that PK activity, as well as subsequent lactate homeostasis, in the brain might be crucial for the onset and progression of PD.
Sirtuin 1 (SIRT1) (a member of the sirtuin family) is an NAD+-dependent protein deacetylase, highly conserved during evolution.14 Through deacetylation of histone and non-histone proteins, including p53, peroxisome proliferator-activated receptor gamma coactivator 1-alpha, nuclear factor κB (NF-κB), FoxOs (forkhead box O family), Ku70, and peroxisome proliferator-activated receptor gamma, SIRT1 plays essential roles in a broad variety of cellular processes such as DNA repair, cell apoptosis, senescence, inflammation, metabolism, and autophagy.15 By regulating these cellular events, SIRT1 prevents aging and extends lifespan in several species, including yeast, flies, worms, and mammals.16,17,18 Furthermore, SIRT1 has been shown to play a pivotal role in energy balance by affecting mitochondrial biogenesis, calcium homeostasis, and oxidative phosphorylation.19,20,21,22 Both aging and energy deficits are considered as risk factors for PD progression.2 Most recently, a cross-sectional study revealed that serum SIRT1 levels were reduced in patients with PD,23,24 implying that decreased expression of SIRT1 is likely involved in PD etiology.
Based on the involvement of SIRT1 in aging prevention, energy regulation, and PD pathogenesis, we hypothesized that SIRT1 may play a beneficial role for treating PD. To assess this hypothesis, we examined the potential neuroprotective roles of SIRT1 in cellular and animal models of PD. Our data indicated that the benefits of SIRT1 against PD are possibly attributed to deacetylation and inhibition of PKM2, which in turn improves lactate homeostasis in the brain and rescues dopaminergic neuron loss in the SNpc.
Results
SIRT1 attenuates parkinsonism in PD model mice
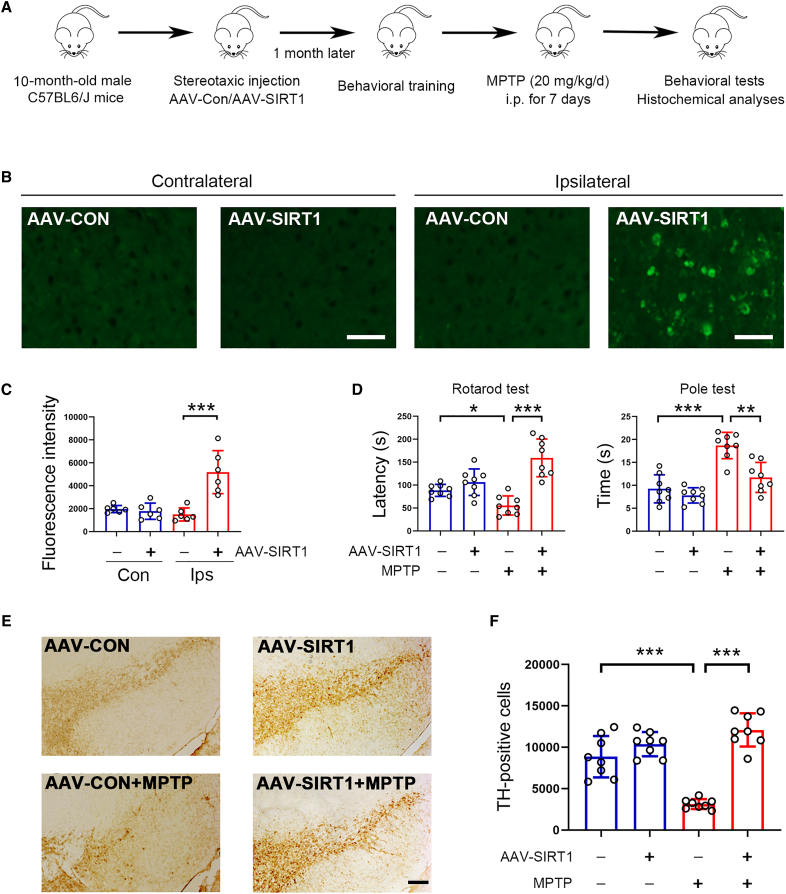
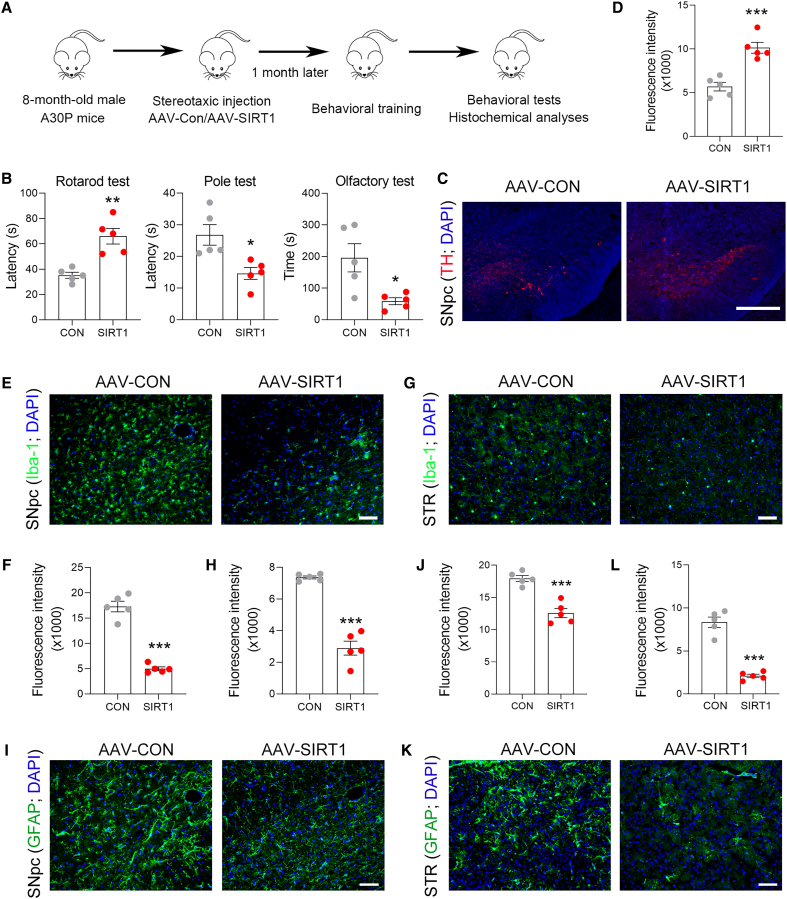
To examine the potential role of SIRT1 in PD, we first analyzed SIRT1 expression in the progression of PD. In MPTP-induced PD model mice, tyrosine hydroxylase (TH) expression was decreased in the SNpc; similar to TH, SIRT1 expression was also reduced (Figure S1A). In addition, our data showed that TH expression was markedly decreased in aged mice, and similar changes in SIRT1 expression were also observed (Figure S1B). These data indicate that SIRT1 might be involved in the progression of PD. To address this notion, we increased SIRT1 expression in the midbrain by adenovirus-associated virus (AAV)-SIRT1 and examined its potential role in PD (Figure 1A). In the ipsilateral region, SIRT1 expression was markedly increased, while its expression at contralateral sites was rather low (Figures 1B and 1C). Of note, SIRT1 was mainly expressed in dopaminergic neurons (Figure S2A). Rotarod and pole tests showed that SIRT1 improved behavioral performances in PD mice (Figure 1D). TH-positive cells in the SNpc were remarkably reduced by MPTP; however, in the presence of exogenous SIRT1, this reduction was substantially prevented (Figures 1E and 1F). To further confirm the anti-PD activity of SIRT1, a genetic PD mouse model was employed, in which alanine in 30 position of α-synuclein was substituted with proline (A30P). The expression of SIRT1 in the SNpc was markedly reduced in A30P mice (Figure S2B). Next, we delivered AAV-SIRT1 to the SNpc of A30P mice by stereotaxic injection and analyzed the potential benefits of SIRT1 in parkinsonism (Figure 2A). The movement defects in A30P mice were significantly improved by SIRT1 due to the prolonged duration in rotarod and the reduced time in pole test (Figure 2B). In addition, the sense of smell in A30P mice was also improved by SIRT1 (Figure 2B). TH expression in the SNpc was increased in AAV-SIRT1-treated A30P mice (Figures 2C and 2D). The ionized calcium-binding adaptor molecule 1 (Iba-1)-positive cells in the SNpc and striatum were reduced by SIRT1, indicating the activation of microglia was attenuated (Figures 2E–2H). Similar changes in the glial fibrillary acidic protein (GFAP)-positive cells in the SNpc and striatum were also observed (Figures 2I–2L). Together, these data strongly indicate that SIRT1 plays a beneficial role in alleviating parkinsonism in PD model mice.
Figure 1.
Increased expression of SIRT1 plays beneficial roles against parkinsonism in mice
(A) Schematic diagram of experiments. i.p. means intraperitoneal injection.
(B) Immunostaining of SIRT1 in the brain for showing efficacy of AAV-SIRT1 transduction. Scale bar, 50 μm. n = 6 for each group.
(C) Quantitative analysis of immunostaining data shown in (B). n = 6 for each group.
(D) SIRT1 attenuates behavioral defects in PD model mice. n = 8 for each group.
(E) Histochemical analysis of tyrosine hydroxylase (TH) expression in the SNpc. Scale bar, 200 μm. n = 8 for each group.
(F) Quantitative analysis of TH-positive neurons as shown in (E). n = 8 for each group. Error bar represents ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Two-tailed Student’s t test was used to calculate statistical difference in (C). Two-way ANOVA with Tukey’s test was used to calculate statistical difference in (D) and (F).
Figure 2.
SIRT1 rectifies parkinsonism-related dysfunctions in A30P mice
(A) Schematic diagram of experiments.
(B) Defected behavioral performances in A30P mice were improved by SIRT1. n = 5 for each group.
(C and D) TH expression was increased by SIRT1 in the SNpc. TH expression (red) was analyzed by immunostaining. DAPI (blue) was used to identify the nucleus. Scale bar, 500 μm. n = 5 for each group.
(E–H) Immunostaining for Iba-1 (green) in the SNpc (E and F) and striatum (G and H). DAPI (blue) was used to identify the nucleus. Scale bar, 50 μm. n = 5 for each group.
(I–L) Immunostaining for GFAP (green) in the SNpc (I and J) and striatum (STR; K and L). DAPI (blue) was used to identify the nucleus. Scale bar, 50 μm. n = 5 for each group.
Error bar represents ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-tailed Student’s t test.
SIRT1 knockdown aggravates behavioral defects and dopaminergic neuron loss in mice
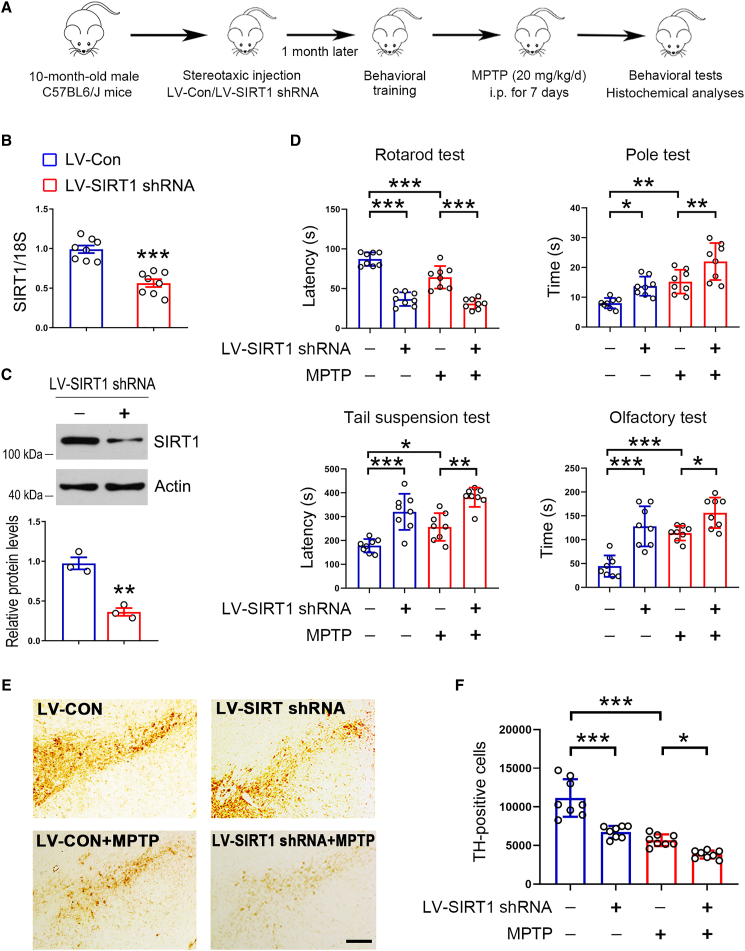
To further examine the potential roles of SIRT1 against PD, we knocked down SIRT1 by using lentivirus (LV)-SIRT1 short hairpin RNA (shRNA). The experimental design is shown in Figure 3A. Transduction of LV-SIRT1 shRNA decreased Sirt1 expression in dopaminergic neurons (Figures 3B, 3C, and S2C). Behavioral tests revealed that SIRT1 knockdown impaired behavioral performances in mice (Figure 3D). In MPTP-treated mice, decreased SIRT1 expression further aggravated behavioral defects (Figure 3D). TH-positive cells in the SNpc were reduced by SIRT1 knockdown in the presence or absence of MPTP (Figures 3E and 3F). These results indicate that decreased SIRT1 expression aggravates parkinsonism in mice, further suggesting SIRT1 plays a key role in the pathogenesis of PD.
Figure 3.
Reduced SIRT1 expression aggravates behavioral defects in mice
(A) Schematic diagram of experiments.
(B) Real-time qPCR analysis of Sirt1 expression in the midbrain. 18S was used as a house-keeping gene. n = 8 for each group.
(C) Western blot analysis of SIRT1 in the midbrain. Actin was used as a loading control. Blots are representative of 3 independent experiments.
(D) SIRT1 knockdown further impairs behavioral performances in PD model mice. n = 8 for each group.
(E) Histochemical analysis of TH expression in the SNpc. Representative images were shown. Scale bar, 200 μm. n = 8 for each group.
(F) Quantitative analysis of TH-positive neurons in the SNpc. n = 8 for each group. Error bar represents ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Two-tailed Student’s t test was used to calculate statistical difference in (B) and (C). Two-way ANOVA with Tukey’s test was used to calculate statistical difference in (D) and (F).
SIRT1 deacetylates and inhibits PKM2 activity
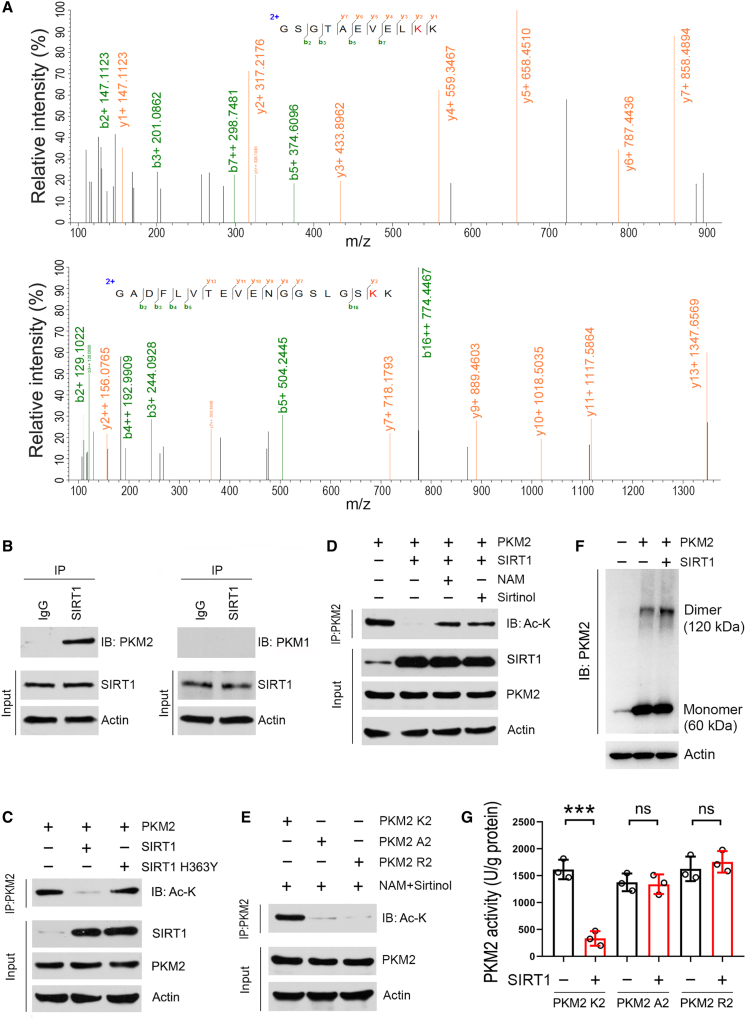
We next attempted to explore the underlying molecular mechanism. Therefore, to determine the substrates of SIRT1, we performed co-immunoprecipitation by using an antibody against SIRT1, and the resulting immunocomplex was subjected to mass spectrometric analysis. In total, 421 proteins were identified in the immunocomplex. Of these proteins, one protein, namely pyruvate kinase M2 (PKM2), drew our attention as it is a key enzyme in glycolysis. Recently, a growing number of reports have shown that glycolysis intervention is a useful strategy for treating PD.25,26 Mass spectrometry data revealed that two lysine residues, K135 and K206, in PKM2 were acetylated when SIRT1 activity was inhibited (Figure 4A). The detection of PKM2 in the co-immunocomplex further confirmed the mass spectrometry data (Figure 4B). Unlike PKM2, PKM1 does not physiologically interact with SIRT1 (Figure 4B). To analyze whether SIRT1 deacetylates PKM2, SH-SY5Y cells were co-transfected with PKM2 and SIRT1 or catalytically inactive SIRT1 mutant (H363Y),27 further subjected to the immunoprecipitation assay. Expectedly, SIRT1 resulted in a decrease in acetylated PKM2, while SIRT1 H363Y had no such an effect (Figure 4C). Moreover, inhibition of SIRT1 by nicotinamide or sirtinol increased acetylated PKM2 (Figure 4D). Notably, substitution mutants of these two acetylated lysine residues in PKM2, with alanine (A2) or arginine (R2), were not acetylated by SIRT1 inhibition (Figure 4E). Additionally, PKM2 dimer was increased in the presence of SIRT1, while its monomer was not affected (Figure 4F). As for PKM2 cellular distribution, it was not altered by SIRT1 (Figure S2D). Functional analysis showed that PKM2 enzyme activity was suppressed by SIRT1; however, PKM2 A2 and PKM2 R2 were not affected (Figure 4G). These data indicate that PKM2 is a substrate of SIRT1 in dopaminergic neurons, and SIRT1 deacetylates PKM2 at K135 and K206 to block its enzyme activity.
Figure 4.
SIRT1 deacetylates PKM2 and inhibits the activity of PKM2
(A) Identification of SIRT1-mediated deacetylated lysine residues in PKM2. SH-SY5Y cells were transfected with the plasmid expressing S-tag fused PKM2. 12 h post-transfection, cells were treated with NAM and sirtinol for an additional 12 h. PKM2 protein was purified by S-protein agarose and the purified protein was separated by SDS-PAGE. The gel was stained with colloidal Coomassie blue dye and the band for PKM2 was collected and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Spectrometry data showing lysine residues of 135 and 206 in PKM2 were acetylated by inhibition of SIRT1.
(B) Co-immunoprecipitation analysis. SH-SY5Y cells were transfected with the plasmid expressing Sirt1. 24 h post-transfection, cells were harvested for co-immunoprecipitation analysis by using anti-SIRT1 antibody. IgG was used as a negative control. The immunocomplex was subsequently analyzed by western blot.
(C) SIRT1 decreases PKM2 acetylation. SH-SY5Y cells were co-transfected with PKM2 with SIRT1 or SIRT1 H363Y. 24 h post-transfection, cells were harvested for immunoprecipitation assay by using anti-PKM2 antibody. The immunocomplex was subsequently analyzed by western blot.
(D) Inhibition of SIRT1 increases PKM2 acetylation. SH-SY5Y cells were co-transfected with PKM2 and SIRT1. 12 h post-transfection, cells were incubated with NAM or sirtinol as indicated. After additional 12 h incubation, cells were harvested for immunoprecipitation by using anti-PKM2 antibody and the immunocomplex was analyzed by western blot. NAM, nicotinamide.
(E) Mutations of K135 and K206 in PKM2 with alanine (R2) or arginine (A2) abolish acetylation modification induced by inhibition of SIRT1. SH-SY5Y cells were transfected with wild-type PKM2 (PKM2 K2) or PKM2 R2 or PKM2 R2. 12 h post-transfection, cells were treated with NAM and sirtinol for additional 12 h, and then cells were harvested for immunoprecipitation using anti-PKM2 antibody and the immunocomplex was analyzed by western blot. NAM, nicotinamide.
(F) SIRT1 induces PKM2 dimerization. SH-SY5Y cells were transfected with SIRT1 and PKM2 as indicated. 24 h post-transfection, cells were collected and incubated with DSS solution for 30 min. After cross-linking, cells were subjected to cell lysate preparation and resulting protein samples were analyzed by western blot. Actin was used a loading control.
(G) SIRT1 inhibits PKM2 activity. SH-SY5Y cells were transfected with SIRT1, SIRT1 H363Y, and PKM2 as indicated. 24 h post-transfection, cells were harvested for PKM2 purification and PKM2 activity assay. n = 3 for each group. Blots are representative of 3 independent experiments. Error bar represents ± SD. ns means no significance. ∗∗∗p < 0.001, two-tailed Student’s t test.
Lactate production was increased in PD model cells and mice
Since PKM2 is a key enzyme in glycolysis, we therefore examined glucose metabolites and related enzyme activities in PD diseased cells. To this aim, rat primary dopaminergic neurons were prepared (Figure S3A) and treated with 1-methyl-4-phenylpyridinium (MPP+). In PD modeled dopaminergic neurons, we observed that the lactate concentration was gradually increased upon the application of MPP+ (Figure S3B). Meanwhile, the lactate production-related metabolites and enzymes, including pyruvate, PK, and lactate dehydrogenase (LDH) were also increased by MPP+ (Figures S3C–S3E). In addition to MPP+, 6-hydroxydopamine (6-OHDA) and rotenone are widely used drugs in PD models.28 Our data showed that these two drugs also induced lactate, pyruvate, PK, and LDH activities in primary dopaminergic neurons (Figures S3F–S3I and S4A–S4D). Furthermore, in MPTP-induced PD model mice, lactate and LDH activity in the cerebral spinal fluid (CSF) were increased (Figures S4E and S4F). Similar findings were also observed in A30P mice (Figures S4G and S4H). These data strongly suggest that lactate accumulation in the brain may account for the pathogenesis of PD.
Lactate infusion induces PD-like phenotypes in mice
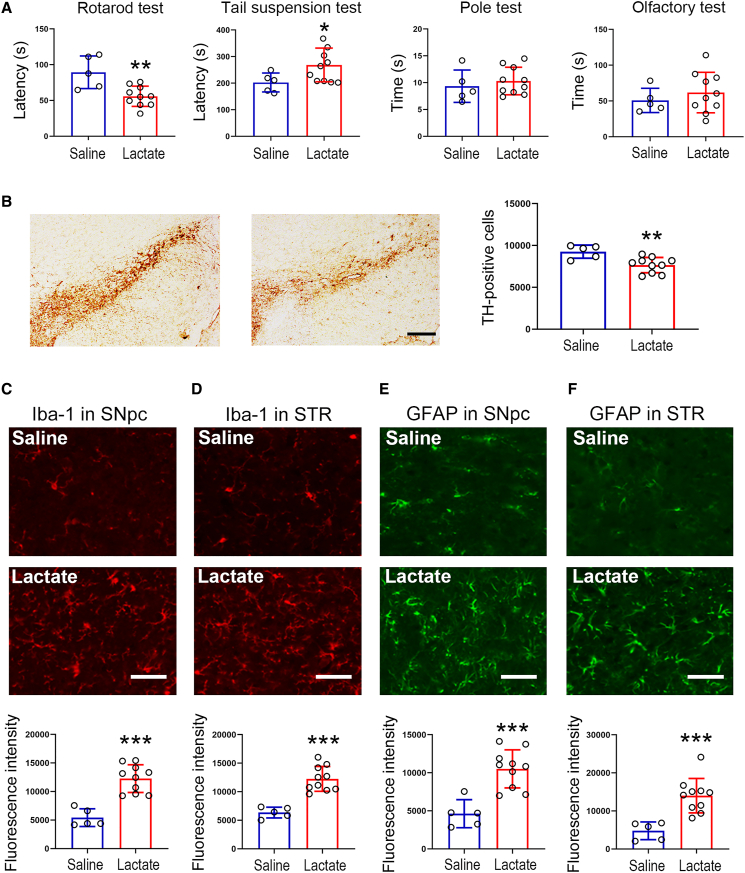
In the aforementioned data, we observed that lactate was increased in cellular and animal models of PD. Hence, we next examined whether exogenous lactate could induce parkinsonism in vivo. To this end, we administered lactate to mice in the third ventricle by stereotaxic injection. Our findings revealed that the lactate infusion caused behavioral defects in the rotarod and tail suspension tests, while the time for pole climbing down and for finding hidden food were not altered (Figure 5A). These changes induced by the lactate infusion were gradually reduced (Figures S5A–S5C). In the SNpc, TH-positive cells were reduced by lactate (Figure 5B). In addition, the lactate infusion induced microglia and astrocyte activation in both the SNpc and striatum (Figures 5C–5F). Collectively, these findings further indicate that lactate accumulation in the brain is a possible risk factor for inducing PD.
Figure 5.
Lactate infusion induces parkinsonism-like phenotypes in mice
Mice were administered with sodium lactate (300 mM, 10 μL for each mouse) into the third ventricle. 7 days post-injection, mice were subjected to behavioral tests.
(A) Lactate infusion aggravates behavioral defects in mice. n = 5 for saline group; n = 10 for lactate group.
(B) Lactate infusion induces dopaminergic neuron loss in the SNpc. Scale bar, 200 μm. n = 5 for saline group; n = 10 for lactate group.
(C and D) Lactate infusion induces microglia activation in the SNpc (C) and striatum (D). Scale bar, 50 μm. n = 5 for saline group; n = 10 for lactate group.
(E and F) Lactate infusion induces astrocyte activation in the SNpc (E) and striatum (F). Scale bar, 50 μm. n = 5 for saline group; n = 10 for lactate group. Error bar represents ± SD. STR: striatum. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-tailed Student’s t test.
To further confirm lactate-induced glial cell activation, we treated primary microglia with lactate and found that this treatment induced the nuclear translocation of NF-κB p65, and thereby increased Il-1a and Tnfa expression in microglia (Figures S6A and S6B). However, these effects did not occur in primary astrocytes (Figures S6C and S6D). Moreover, the conditioned medium from lactate-treated microglia induced cell insults in dopaminergic neurons (Figures S7A–S7C). The conditioned medium from lactate-treated astrocytes had no such effects (Figures S7D–S7F). Moreover, we co-cultured microglia and dopaminergic neurons, in which microglia or neurons were subjected to genetic manipulation. The results showed that SIRT1 overexpression in dopaminergic neurons increased the viability of neurons, while SIRT1 knockdown decreased the neuron viability (Figures S8A and S8B). Increased expression of PKM2 in dopaminergic neurons repressed the viability of neurons in the co-culture (Figure S8C). Meanwhile, instead of dopaminergic neurons, microglia were genetically manipulated as mentioned earlier; we found that these genetic manipulations had no effects on the viability of neurons in the co-culture (Figures S9A–S9C).
To reinforce that the observed benefits in PD conferred by SIRT1 are due to reduced lactate production, we treated A30P mice with AAV-SIRT1 and lactate and examined PD-related phenotypes (Figure S10A). As expected, increased SIRT1 expression improved the behavioral performances in A30P mice including motor and olfactory assays (Figures S10B–S10D). The application of lactate largely counteracted the improved performances in rotarod and pole tests, while olfactory function was not altered (Figures S10B–S10D). The histochemical analyses showed that the increased expression of TH in AAV-SIRT1-treated A30P mice was largely prevented by lactate (Figure S10E). Moreover, lactate application abolished the repressed Iba-1 expression induced by SIRT1 (Figure S10F). These data clearly indicate that the anti-PD effects of SIRT1 rely on restricted lactate production in the brain.
PKM2 induces PD-like phenotypes in mice
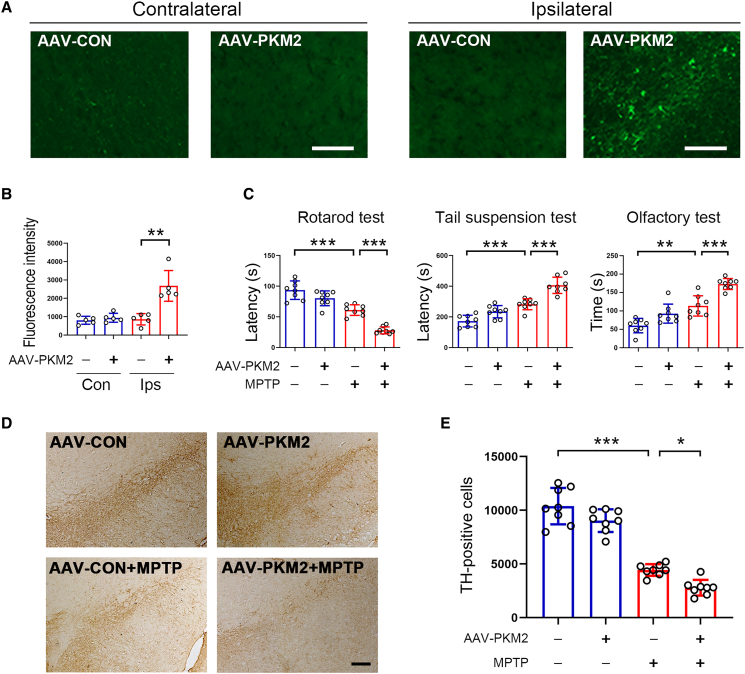
To further confirm the notion that excessive lactate is a causative factor for inducing PD, we increased PKM2 expression in the SNpc by stereotaxic injection with AAV-PKM2 to induce lactate production. Immunofluorescence analysis showed that PKM2 expression was markedly increased by AAV-PKM2 (Figures 6A and 6B). Dopaminergic neurons are the main target cells of AAV-PKM2 (Figure S11A). Increased expression of PKM2 further worsened behavioral defects induced by MPTP (Figure 6C). Loss of dopaminergic neurons in the SNpc of PD model mice was also worsened by PKM2 (Figures 6D and 6E). These results indicate that ectopically increased PKM2 expression might be involved in the progression of PD.
Figure 6.
PKM2 worsened PD symptoms in MPTP-treated mice
10-month-old male C57BL6/J mice received AAV-PKM2 or AAV-Con virus in the SNpc by stereotaxic injection. One month later, mice were subjected to PD model generation by intraperitoneal injection with MPTP at the dosage of 20 mg/kg/day for consecutive 7 days
(A) Immunofluorescence analysis for PKM2 expression in the contralateral and ipsilateral regions. Scale bar, 100 μm. n = 5 for each group.
(B) Quantitative analysis of PKM2 expression based upon immunofluorescence data as shown in (A). n = 5 for each group.
(C) Behavioral performance tests. n = 8 for each group.
(D) Immunohistochemical staining for TH-positive cells in the SNpc. Scale bar, 200 μm.
(E) Quantitative analysis of TH-positive cells in the SNpc based upon immunohistochemical staining as shown in (D). n = 8 for each group. Error bar represents ± SD. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Two-tailed Student’s t test was used to calculate statistical difference in (B). Two-way ANOVA with Tukey’s test was used to calculate statistical difference in (C) and (E).
Inhibition of PKM2 alleviates parkinsonism in mice
Next, we administered shikonin, a potent inhibitor of PKM2, to PD model mice (Figure 7A). Behavioral tests showed that shikonin treatment improved the performance on rotarod, tail suspension, and olfaction (Figure 7B). The loss of TH-positive neurons in the SNpc in PD model mice was attenuated by shikonin (Figure S11B). Furthermore, PKM2-IN-1, a recently synthesized specific inhibitor of PKM2,29 was also employed to confirm the roles of PKM2 in the progression of PD. The results showed that the behavioral defects in tail suspension, pole, and olfactory tests in PD model mice were improved by PKM2-IN-1 (Figure 7C). The loss of dopaminergic neurons in the SNpc was largely prevented by PKM2-IN-1 (Figure S11C). The chemical structures of shikonin and PKM2-IN-1 were shown in Figures 7D and 7E. To examine whether these two chemicals can across the blood-brain barrier to play a role in the brain, we measured shikonin and PKM2-IN-1 in brain tissue using high-performance liquid chromatography-mass spectrometry. 1-h post-gavage, both shikonin and PKM2-IN-1 can be detected in brain tissues with values of 1.269 and 2.738 ng/g, respectively (Figures 7F and S11D–S11G).
Figure 7.
PKM2 inhibition attenuates PD-related phenotypes
(A) Experimental outlines showing PD model mouse generation and pharmacological interventions.
(B) Behavioral tests showing the defects in rotarod, pole, and olfactory tests were improved by shikonin. n = 5 for each group.
(C) The behavioral defects in MPTP-treated mice were alleviated by PKM2-IN-1. n = 5 for each group.
(D and E) The chemical structures of shikonin (D) and PKM2-IN-1 (E).
(F) The levels of shikonin and PKM2-IN-1 in the brain. n = 5 for each group. Error bar represents ± SD. ns means no significance. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, two-way ANOVA with Tukey’s test.
Furthermore, we also analyzed whether PKM2 inhibition alleviates parkinsonism in A30P mice (Figure S12A). The movement defects and olfactory dysfunction in A30P mice were improved by shikonin (Figure S12B). The Iba-1 expression in the SNpc was reduced by shikonin (Figure S12C). Lactate in the CSF was reduced in shikonin-treated A30P mice (Figure S12D). Like shikonin, PKM2-IN-1 also exhibited anti-PD activities in A30P mice (Figures S12E and S12F). A reduction in lactate in the CSF was observed in PKM2-IN-1-treated A30P mice (Figure S12G). Together, these data further indicate that lactate accumulation is a detrimental factor for the progression of PD, and PKM2 is likely a drug target with a great therapeutic potential for developing agents against PD.
Discussion
SIRT1 is an NAD-dependent deacetylase, and its overexpression increased the survival of dopaminergic neurons in PD model cells; and in contrast, SIRT1 knockdown further aggravated MPP+-induced cell injury.30 In the present study, we observed that the behavioral defects in PD model mice were improved by SIRT1. In line with our data, increasing evidence has revealed the beneficial roles of SIRT1 in PD model animals.31,32,33,34 However, the underlying molecular mechanisms remain elusive. Using co-immunoprecipitation and mass spectrometry, PKM2 was identified as a downstream target of SIRT1. Moreover, two lysine residues in PKM2, K135 and K206, were deacetylated by SIRT1. These deacetylation modifications markedly blocked PKM2 enzyme activity. In line with our findings, post-translational modification has been considered as an efficient strategy for manipulating PKM2 enzyme or transcription activity.35,36,37,38,39,40,41
PK is an enzyme responsible to produce pyruvate, with two known isoforms, namely PKM1 and PKM2. Along with LDH, PK catalyzes glucose into pyruvate and lactate. All three tested PD-inducing drugs including MPP+, 6-OHDA, and rotenone are capable of stimulating PK and LDH activities in dopaminergic neurons. The reason for the enhanced PK and LDH is likely due to neurotoxin-induced repression of SIRT1. As a result, PKM2 activity was increased, leading to pyruvate production, which may activate LDH activity by a positive feedback loop. Interestingly, a large-scale proteomic analysis has shown that PKM2 and LDHB are two key factors for early changes in energy metabolism associated with neuroinflammation.42 This clinical survey data further substantiated our findings that PK and LDH activation are likely involved in the pathogenesis of PD, since neuroinflammation has been shown to be involved in the progression of PD.43,44
The ectopic fluctuation of pyruvate disturbs energy balance by inducing mitochondrial dysfunction. Inhibition of the mitochondrial pyruvate carrier by MSDC-160 alleviates PD symptoms.45 Lactate is derived from pyruvate through LDH, which could be used as energy source in many tissues except in the brain.46,47 Reportedly, excessive lactate accumulation results in mitochondrial dysfunction and eventually leads to energy defects.48 In this regard, excessive lactate may have pathogenetic relevance for PD. Indeed, our data demonstrated that lactate administration in the third ventricle induced PD-like phenotypes in mice. In dopaminergic neurons, treatment with MPP+, 6-OHDA, or rotenone substantially increased pyruvate and lactate production, implying that excessive pyruvate and/or lactate might be mediators of PD induced by these neurotoxins. Consistent with our findings, lactate is reportedly increased in the CSF of patients with AD, another neurodegenerative disease.48 Furthermore, CSF lactate is gradually enhanced in aging populations.49 Serum lactate was found to be higher in patients with PD than in healthy controls.50 All these reports, together with our findings, strongly suggest that excessive lactate in the brain might be a risk factor for the development of PD.
PKM2 triggers the synthesis of pyruvate, which was further converted into lactate with the aid of LDH. Hence, in this pathway, PKM2 is responsible for the first step in manipulating lactate production. SIRT1 curbs lactate accumulation via the deacetylation-induced PKM2 inhibition, thus alleviating neuroinflammation. It has been suggested that PKM2 dimerization represses its enzyme activity.7,51 In line with this notion, we observed that PKM2 dimer was increased in the presence of SIRT1. Lactate has been shown to activate Toll-like receptor 4 signaling and the NF-κB pathway, resulting in the upregulation of inflammatory gene expression in macrophages.52 In the present study, we also observed that lactate induces NF-κB p65 nuclear translocation in primary microglia. Macrophages with high Ldha expression have shown potent capacities for migration and pro-inflammatory functions.53
Neuroinflammation is a contributing factor for the development of PD, and inhibition of neuroinflammation is a useful approach to slow or prevent this disease.43 Our data revealed that excessive lactate is capable of inducing neuroinflammation. Therefore, strategies associated with inhibition of lactate production might hold therapeutic potential in PD. For instance, PKM2 inhibition appears to be a useful strategy against PD. Indeed, we treated PD model mice with PKM2 inhibitors shikonin/PKM2-IN-1 and found that these therapies could partially alleviate PD phenotypes. In addition to neuroinflammation induction, lactate may affect dopaminergic neuron viability in other ways. For instance, the increased uptake of lactate has been shown to promote reactive oxygen species production to induce oxidative stress in neurons, resulting in axon degeneration in the peripheral nervous system.54 In addition, the continued lactate efflux could induce intracellular acidification that led to a greater loss in NAD+, ATP depletion, and cell death.55
By producing ATP, increased glycolysis has been shown to attenuate the progression of PD.25 Several reports showed that astrocytic lactate could be used as a fuel for neuronal survival.56,57,58,59 Therefore, glycolysis inhibition such as PKM2 inhibitor application seems to be a detrimental intervention for PD, which is different from our conclusion that PKM2 inhibition plays a beneficial role in PD model mice. It should be mentioned that in PD diseased cells induced by commonly used drugs such as MPP+, rotenone, and 6-OHDA, mitochondrial function was markedly destroyed. In familial forms of PD, Parkin, α-synuclein, DJ-1, UCHL-1, LRRK2, PINK1, NURR1, VPS35, and HtrA2 have pathogenic mutations that directly or indirectly associate with mitochondrial dysfunction.60 Under these pathophysiological conditions, therefore, excessive pyruvate produced in astrocyte glycolysis cannot be consumed by neurons, most of which is metabolized into lactate with the aid of LDH. On the other hand, based on our data in this study, SIRT1 expression was decreased by aging, MPTP, and A30P in dopaminergic neurons, which weakens its inhibitory effect on PKM2, leading to increased PKM2 activity and pyruvate production. By a positive feedback loop, LDH activity was induced to metabolize pyruvate into lactate. Together, cumulative accumulation of lactate can induce multiple deleterious outcomes such as inflammation.61 In this regard, therefore, PKM2 inhibition is likely an effective way against PD. In agreement with this notion, it has been shown that activation of hexokinase 2 (HK2), the first enzyme in glycolysis, induces dopaminergic neuron apoptosis by promoting lactate production.62 Most recently, induction of HK2 expression has been shown to induce hepatic stellate cell activation, in which lactate plays a pivotal role.63 Therefore, we propose that there is a threshold of lactate in the brain, which governs its role as a friend or foe. This notion can be verified by using newly developed lactate sensors for tracking lactate fluctuation in the brain.64
Collectively, our data demonstrated that SIRT1 alleviates PD symptoms by improving lactate homeostasis in the brain. PKM2 is a downstream target of SIRT1, and K135 and K206 in PKM2 were deacetylated by SIRT1, thus resulting in the inhibition of PKM2 enzyme activity. Consequently, lactate production in the brain was reduced. In the brain, excessive lactate can induce microglia activation to produce pro-inflammatory cytokines, which may trigger astrocyte activation to produce more pro-inflammatory cytokines. The resulting neuroinflammation induces various detrimental outcomes, such as loss of dopaminergic neurons and the onset of PD. Therefore, CSF lactate might be a useful index for the early diagnosis of PD. Moreover, inhibition of PKM2 is an attractive strategy for treating PD. These findings strongly suggest that PKM2 may be a drug target for developing agents to treat PD or other neurodegenerative diseases presenting an energy imbalance.
Limitations of the study
Due to time and resource limitations, we did not examine our key findings in human samples, such as the lactate levels, SIRT1 expression, and deacetylated PKM2 in the brains of patients with PD. In addition, we did not construct genetic mice with dopaminergic neuron-specific overexpression or deficiency of Sirt1 and Pkm2 to validate our conclusions. Moreover, the detailed molecular mechanisms for SIRT1-induced repression in PKM2 enzyme activity are still unclear, which might be resolved by analyzing the conformational changes in deacetylated PKM2 at K135/206.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SIRT1 | Cell Signaling Technology | Cat# 8469; RRID: AB_10999470 |
| Anti-SIRT1 | Abcam | Cat# ab110304; RRID: AB_10864359 |
| Anti-TH | Abcam | Cat# ab137869; RRID: AB_2801410 |
| Anti-TH | Invitrogen | Cat# MA1-24654; RRID: AB_795666 |
| Anti-PKM1 | Cell Signaling Technology | Cat# 7067; RRID: AB_2715534 |
| Anti-PKM2 | Cell Signaling Technology | Cat# 4053; RRID: AB_1904096 |
| Anti-GFAP | Abcam | Cat# ab68428; RRID: AB_1209224 |
| Anti-Iba-1 | Wako Chemicals | Cat# 019–19741; RRID: AB_839504 |
| Anti-p65 | Santa Cruz | Cat# sc-8008; RRID: AB_628017 |
| Anti-Acetylated lysine | Cell Signaling Technology | Cat# 9441; RRID: AB_331805 |
| Anti-Actin | Cell Signaling Technology | Cat# 3700; RRID: AB_2242334 |
| Bacterial and virus strains | ||
| AAV-SIRT1 | HanBio | https://hanbio.net/ |
| LV-SIRT1 shRNA | Huang et al.65 | N/A |
| AAV-PKM2 | HanBio | https://hanbio.net/ |
| Chemicals, peptides, and recombinant proteins | ||
| MPTP | Sigma-Aldrich | Cat# M0896 |
| Lactate | Sigma-Aldrich | Cat# 71720 |
| Leupeptin | Sigma-Aldrich | Cat# L5793 |
| Aprotinin | Sigma-Aldrich | Cat# A6106 |
| NP-40 | Sigma-Aldrich | Cat# NP40 |
| PMSF | Sigma-Aldrich | Cat# 52332 |
| Okadaic acid | Sigma-Aldrich | Cat# O9381 |
| TSA | Sigma-Aldrich | Cat# T1952 |
| Sodium butyrate | Sigma-Aldrich | Cat# B5887 |
| Sirtinol | Sigma-Aldrich | Cat# S7942 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| HBSS | Sigma-Aldrich | Cat# H9269 |
| Poly-L-lysine | Sigma-Aldrich | Cat# P4832 |
| Coomassie Blue dye | Beyotime | Cat# P0017 |
| Shikonin | MedChemExpress | Cat# HY-N0822 |
| PKM2-IN-1 | MedChemExpress | Cat# HY-103617 |
| Lipofectamine | Invitrogen | Cat# 11668019 |
| Trypsin | Promega | Cat# V5111 |
| Penicillin/streptomycin | Gibco | Cat# 15140122 |
| B27 supplement | Gibco | Cat# 17504044 |
| FBS | Gibco | Cat# 10099158 |
| Disuccinimidyl suberate | Thermo Scientific | Cat# 21655 |
| Critical commercial assays | ||
| Cell counting kit-8 | MedChemExpress | Cat# HY-K0301 |
| Protein assay kit | Pierce | Cat# 23225 |
| cDNA Synthesis Kit | Vazeme | Cat# R211-01 |
| RNA isolator | Vazeme | Cat# RC202-01 |
| Pyruvate assay kit | Sigma-Aldrich | Cat# MAK071 |
| Lactate assay kit | Sigma-Aldrich | Cat# MAK064 |
| Pyruvate kinase activity assay kit | Sigma-Aldrich | Cat# MAK072 |
| Co-immunoprecipitation assay kit | Pierce | Cat# 26149 |
| FastStart Essential DNA Green Master | Roche | Cat# 06402712001 |
| Experimental models: Cell lines | ||
| SH-SY5Y | ATCC | Cat# CRL-2266 |
| HEK293 | ATCC | Cat# CRL-1573 |
| Primary dopaminergic neurons | This paper | N/A |
| Primary microglia | This paper | N/A |
| Primary astrocytes | This paper | N/A |
| Experimental models: Organisms/strains | ||
| SNCA-A30P (B6.Cg-Tg(THY1-SNCA∗A30P)TS2Sud/J) transgenic mice | Jackson Laboratory | ATCC 012265 |
| C57BL/6 mice | Animal Laboratory of Nantong University (Nantong, China) | N/A |
| SD rat pups | Animal Laboratory of Nantong University (Nantong, China) | N/A |
| Recombinant DNA | ||
| pECE-SIRT1 | Addgene | Cat# 1791 |
| pECE-SIRT1 H363Y | Addgene | Cat# 1792 |
| pcDNA3.1(+)-PKM2 | This paper | N/A |
| pcDNA3.1(+)-PKM2 A2 | This pager | N/A |
| pcDNA3.1(+)-PKM2 R2 | This paper | N/A |
| Software and algorithms | ||
| ImageJ | NIH Image | https://imagej.nih.gov/ij/index.html |
| MASCOT engine (version 2.4) | Matrix Science | https://www.matrixscience.com/ |
| Other | ||
| PVDF membrane | Millipore | Cat# IPVH00010 |
| S-protein agarose | Novagen | Cat# 3226253 |
| Neurobasal medium | Gibco | Cat# 21103049 |
| 0.22 μm syringe filters | Millipore | Cat# SLGVR33RS |
| Transwell insert | Corning | Cat# 3422 |
Resource availability
Lead contact
Further information and request of resources can be reached by the lead contact, Dr. Cheng Sun (suncheng1975@ntu.edu.cn).
Materials availability
All materials generated in this study are available upon request to the lead contact with a completed material transfer agreement.
Data and code availability
-
•
All data reported in this paper will be available by contacting the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Cell culture
SH-SY5Y cells and HEK293 cells were obtained from ATCC (Manassas, VA, USA). Cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and incubated at 37°C in a humidified atmosphere with 5% CO2. Primary rat dopaminergic neurons were prepared as described previously.66 In brief, pregnant Sprague Dawley (SD) rats with E14.5 pups were anesthetized by isoflurane and then euthanized by cervical dislocation. An abdominal incision was made to obtain the uterus, which was then placed in ice-cold HBSS in a 100 mm Petri dish. Embryos were removed from the uterus using forceps and placed in fresh ice-cold HBSS. The mesencephalic arch was dissected under a dissection microscope (10× magnification), and the entire midbrain was taken out. The midbrain tissue was flattened on the Petri dish and the ventral midbrain was collected and placed in ice-cold HBSS in a 15 mL conical tube. After removing the HBSS, the ventral midbrain was incubated with pre-warmed (37°C) 0.05% trypsin-EDTA at 37°C for 5–10 min. Neurobasal medium containing 50 U/ml penicillin and streptomycin, 1x B27 supplement, 10% FBS was added to de-activate the digestion. The deactivation medium was removed and the tissue was washed twice in neurobasal medium with the supplements as mentioned above. Complete dissociation was achieved by 8–10 passes through a fire-polished glass pipette in the complete medium. The resulting dissociated neurons were seeded on a poly-L-lysine coated cell culture dish. The maturation of mesencephalic neurons required 7–8 days with medium changes every 2 days. Primary microglia and astrocytes were prepared from postnatal day 1 SD rat pups. Briefly, meninges-free cortices were collected and trypsinized. The resulting cells were cultured in DMEM with 10% FBS and 1% penicillin-streptomycin at 37°C under humidified 5% CO2. Cell culture medium was changed every 2 days. After 10–12 days, the cultured cells were subjected to shaking at 200 rpm for 2 h. The cells floated in culture medium were collected after centrifugation and used as microglia. For astrocytes, the cultured cells were subjected to shaking at 250 rpm for 12–16 h, the remaining attached cells were used as astrocytes.
Animals
C57BL/6 mice were obtained and bred at Animal Laboratory of Nantong University. SNCA-A30P (B6.Cg-Tg(THY1-SNCA∗A30P)TS2Sud/J; 012265) transgenic mice were purchased from the Jackson Laboratory (USA). All of the mice used in this study are male, which were maintained under specific pathogen-free conditions. To generate PD model mice, MPTP was intraperitoneally (i.p.) injected at the dosage of 20 mg/kg/day for consecutive 7 days67 MPTP was dissolved in saline at 2 mg/mL, which was given to each mouse at 10 μL/g body weight. Control group mice were received saline at the same volume. To inhibit PKM2, mice were treated with shikonin or PKM2-IN-1 at the dosage of 50 mg/kg by gavage. To increase lactate in the brain, 10 μL of 300 mM lactate sodium was injected into the third ventricle by stereotaxic injection. Alternatively, each mouse was given 10 μL of 60 mM lactate sodium by intranasal administration for consecutive 5 days. All experiments were performed in accordance with the approved guidelines of the Institutional Ethical Committee of the Nantong University (Approval ID: SYXK [SU] 2017-0046).
Virus production and injection
Sirt1 knockdown in vivo by the stereotaxic injection with lentivirus expressing Sirt1 shRNA (LV-SIRT1 shRNA). Each mouse was received 4 μL of LV-SIRT1 shRNA (1 x 1012 VG/ml). Control mice were treated with the same dosage of lentivirus expressing shRNA targeting LacZ (LV-Con). LV-SIRT1 shRNA and LV-Con were provided by Dr. Xinzhong Huang.68 A Hamilton syringe was used to inject lentivirus at a rate of 1 μL/min to the right SNpc at the following stereotaxic coordinates: 1.3 mm rostral to the bregma, 3.2 mm lateral to the midline, and 4.3 mm ventral to the dura, with the tooth bar set at zero. Adeno-associated virus expressing Sirt1 (AAV-SIRT1) was produced at HanBio (Shanghai, China) by using AAV9 serotype. To increase Sirt1 expression, mice were injected with the AAV-SIRT1, AAV-PKM2 or the control virus AAV-Con. AAV-SIRT1, AAV-PKM2 and AAV-Con were constructed at HanBio (Shanghai, China). Each mouse was administered with 4 μL virus with the titration of 1 x 1012 VG/ml. The method of injection was similar to the procedures as mentioned above. One month later, mice were subjected to PD model generation.
Method details
Plasmids
pECE-SIRT1 and pECE-SIRT1 H363Y are gifts from Michael Greenberg.27 The coding sequence of human Pkm2 was synthesized and incorporated into pcDNA3.1(+) using endonuclease sites of EcoR I and Xho I (Obio Technology, Shanghai, China). S protein (SP) tag was generated by PCR and incorporated into pcDNA3.1(+)-PKM2 at the amino terminus by EcoR I and BamH I. pcDNA3.1(+)-PKM2 A2 (K135A/K206A) and pcDNA3.1(+)-PKM2 R2 (K135R/K206R) were generated by PCR-based mutagenesis using pcDNA3.1(+)-PKM2 as a template. The primer sequences are listed in Table S1. All plasmids were further confirmed by sequencing.
Cell viability assay
Cell viability was examined with a cell counting kit-8 according to the manufacturer’s instructions. Briefly, cells were seeded in a 96-well plate, 10 μL of CCK-8 working solution was added to each well. After 2 h-incubation in a cell incubator, the absorbance values at 405 nm were recorded with a microplate reader (BioTek, Synergy H1).
LDH activity assay
LDH activity was measured by a commercial kit according to the method as described elsewhere.67 Briefly, at the end of treatments, cell culture medium was harvested and then incubated with the LDH-assay reaction mixture. The incubation was performed for 30 min at room temperature in dark. The absorbance at 490 nm was detected with a microplate reader (BioTek, USA). Cell death ratio was calculated by the formula: The cell death ratio (%) = (Asample – Ablank)/(Amax – Ablank) × 100.
Behavioral performance tests
Tail suspension test: Mice were suspended from the hook of a tail suspension test box (45 cm in height, 15 cm in width, 11.5 cm in depth), 60 cm above the surface of a platform, using adhesive tape placed 1 cm from the tip of the tail. After acclimatization, the process was recorded by a camera for at least 10 min and the time of immobility was measured. Mice were considered immobile only when they hung passively and were completely motionless. Pole test: A metal rod (75 cm long with a 9 mm diameter) wrapped with bandage gauze was used as the pole. Before the actual test, the mice were trained for two consecutive days and each training session consisted of three test trials. Mice were placed on the top of the pole. The time to turn and total time to reach the base of the pole were recorded. The end of test was defined by placement of all 4 paws on the base. The maximum cutoff time to stop the test and recording was 60 s. Olfactory test: Mice were fed with cheese pellets before olfactory test to make sure they are familiar with the pellet odor. Mice were fasted for 20 h prior to the test. One cheese pellet was buried 1.0 cm below the bedding on a corner of cage. In each test, mouse was placed at the center of cage at the beginning. The time was recorded when the cheese pellet was detected and eaten by mice. If mouse failed to find the pellets within 5 min, the test stopped and a score of 300 s was awarded. The bedding and the cage were cleaned for each trial. The test was performed for 3 consecutive days and each mouse received one trial per day. Rotarod test: The rotarod test was conducted to evaluate the motor deficits, as previously described with slight modification.67 The rotarod machine detects mouse drop and records the time and acceleration each mouse passes on its corresponding channel. Before the injection of MPTP, the model mice were pre-trained within 5 min (2–20 rpm) acceleration mode for 3 consecutive days. One week after the injection of MPTP, the time required for each mouse to fall was recorded and analyzed. For each behavioral test, mice with ectopic performances in behavioral training were excluded. The remaining animals with similar behavioral values were included and randomly grouped, and no animals were excluded in the formal analyses.
Immunohistochemistry and immunofluorescence
Brain tissues were fixed in phosphate-buffered 4% paraformaldehyde, pH7.4, at 4°C, and then tissues were cut on a vibratome set at 12 μm. The sections were incubated in Quick/Block buffer (Beyotime, Shanghai, China; Cat. No. P0260) for 1 h at room temperature. After 3 time-washing in PBST, the sections were incubated with an anti-Tyrosine hydroxylase (anti-TH) antibody, followed by incubation with biotin-conjugated antibody (Zsbio, Beijing, China; Cat. No. sp-9000) and ABC reagents (Zsbio, Beijing, China; Cat. No. pk-4001). Sections were developed using Fast DAB Peroxidase Substrate (Sigma-Aldrich; Cat. No. D0426). Immunofluorescence was performed on 12 μm thick serial brain sections. After blocking, the sections were incubated with the primary antibodies including anti-Iba-1 antibody, anti-GFAP antibody, anti-SIRT1, anti-PKM2, and anti-TH for overnight at 4°C. The sections were rinsed 3 times in PBST and then incubated with the second antibodies for 2 h at room temperature. After removing the second antibodies, the sections were rinsed 3 times in PBST and the images were taken with a fluorescence microscope (Zeiss, Axio Image M2). For immunofluorescence analysis in cells, cells were seeded into a 24-well plate with glass coverslips (3 × 104 per well). Next day, cells were treated with lactate for 24 h. Cells were fixed with methanol at room temperature for 15 min and washed 3 times in PBS. Then cells were blocked with 5% BSA solution containing 0.1–0.5% Triton X-100 for 2 h at room temperature. After blocking, cells were incubated with primary antibodies including anti-Iba-1, GFAP and NF-κB p65 overnight at 4°C. After 3 time-washing, cells were incubated with an Alexa Fluor 488/594-conjugated secondary antibody for 2 h at room temperature. Finally, the nuclei were stained with DAPI. Photographs (400X) were taken with a fluorescence microscope (Olympus, BX51). The total numbers of TH-positive cells were counted by unbiased stereology method as described elsewhere.65 Iba-1- and GFAP-positive cells were analyzed by the ImageJ software. Details of all antibodies are listed in key resources table.
PKM2 dimer analysis
To examine whether SIRT1 affects PKM2 dimer, SH-SY5Y cells were overexpressed with PKM2 and SIRT1 by transfection. 24 h later, cells were collected and suspended in PBS (pH 8.0). Cells were washed 3 time with ice-cold PBS (pH 8.0) to remove amine-containing culture media and proteins from the cells. Disuccinimidyl suberate was dissolved in DMSO immediately before use. For cross-linking, cells were incubated with 1 mM DSS for 30 min at room temperature, and the reaction was quenched by adding 20 mM Tris buffer for 15 min. After cross-linking, cells were washed and subjected to protein extraction and western blot analysis.
Protein extraction and western blot analysis
Cells were lysed in pre-cold lysis buffer containing 25 mM Tris-HCl (pH 7.4), 100 mM NaF, 50 mM Na4P2O7, 10 mM Na3VO4, 10 mM EGTA, 10 mM EDTA, 1% NP-40, 10 μg/mL Leupeptin, 10 μg/mL Aprotinin, 2 mM PMSF, 20 nM Okadaic acid. The lysates were centrifuged at 12,000 rpm for 15 min at 4°C. Cell debris was discarded and supernatant was kept on ice. Protein concentration in supernatant was measured by a kit (Pierce) according the manufacturer’s instructions. Equal protein concentration in each sample was prepared and 1X Laemmli buffer was added and boiled at 100°C for 5 min. The procedures for Western blot were described elsewhere.69 Briefly, protein samples were resolved by SDS-PAGE and transferred to PVDF membrane. After blocking in 5% BSA, membranes were incubated with the primary antibodies including anti-SIRT1, anti-PKM1, anti-PKM2, anti-acetylated lysine, and anti-β-Actin, at 4°C for overnight. After three-time washing in TBST, membranes were incubated with the second antibody for 1 h at room temperature. The second antibody was thoroughly removed by four-time washing in TBST. Membrane was developed using a chemiluminescence assay system (Roche) and exposed to Kodak films. Blots were quantified by the ImageJ program. Details of all antibodies are listed in key resources table.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells or tissues by using RNA isolator (Vazyme, Nanjing, China), which was then transcribed into cDNA with a cDNA Synthesis Kit (Vazeme, Nanjing, China). Levels of mRNA were analyzed with an iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad), using FASTSTART ESSENTIAL DNA GREEN MASTER (Roche). The mRNA levels were normalized to the expression of 18S rRNA. The primer sequences used in this study are listed in Table S1.
Pyruvate and lactate assay
Pyruvate and lactate were analyzed, respectively, by a pyruvate assay kit (Sigma-Aldrich; Cat. No. MAK071) and a lactate assay kit (Sigma-Aldrich; Cat. No. MAK064) according to the manufacturer’s instructions.
Pyruvate kinase activity assay
Pyruvate kinase activity was measured by a pyruvate kinase activity assay kit (Sigma-Aldrich; Cat. No. MAK072) according to the manufacturer’s instructions.
Co-immunoprecipitation and LC-MS/MS spectrometry
SH-SY5Y cells were transfected with a plasmid expressing Sirt1 by Lipofectamine. 24 h post-transfection, cells were harvested and subjected to co-immunoprecipitation analysis with a commercial kit (Pierce) by using antibodies against SIRT1 and PKM2, according to the manual instructions. The resulting immune-complex was analyzed by liquid chromatography followed by liquid chromatography-tandem mass spectrometry (LC/MS/MS).
Acetylated PKM2 preparation
HEK293 cells were transfected with a plasmid expressing PKM2-SP by Lipofectamine (Invitrogen). 12 h post-transfection, cell culture medium was replenished, in which 0.2 μM TSA, 2 mM sodium butyrate and 5 μM sirtinol were supplemented.70 After 24 h-incubation, cells were lysed in pre-cold lysis buffer containing 25 mM Tris-HCl (pH 7.4), 100 mM NaF, 50 mM Na4P2O7, 10 mM Na3VO4, 10 mM EGTA, 10 mM EDTA, 1% NP-40, 10 μg/mL Leupeptin, 10 μg/mL Aprotinin, 2 mM PMSF, 20 nM Okadaic acid, 0.2 μM TSA, 2 mM sodium butyrate and 5 μM sirtinol. Cell lysates were centrifuged at 10,000 rpm for 15 min and the supernatants were subjected to protein purification by S-protein agarose. The purified protein samples were denatured by adding 1X Laemmli buffer and boiled at 100°C for 5 min. The purified PKM2 protein was eluted, separated by SDS-PAGE. After staining with colloidal Coomassie Blue dye, PKM2 band was isolated from the gel and subjected to LC/MS/MS.
Mass spectrometry analyses
For co-immunoprecipitation protein samples, 200 μg of proteins were incorporated into 30 μL SDT buffer (4% SDS, 100 mM DTT, 150 mM Tris-HCl, pH 8.0). The detergent, DTT and other low-molecular-weight components were removed using UA buffer (8 M Urea, 150 mM Tris-HCl, pH 8.0) by repeated ultrafiltration (Microcon units, 10 kD). Then 100 μL iodoacetamide (100 mM IAA in UA buffer) was added to block reduced cysteine residues and the samples were incubated for 30 min in darkness. The filters were washed with 100 μL UA buffer three times and then 100 μL 25 mM NH4HCO3 buffer twice. Finally, the protein suspensions were digested with 4 μg trypsin in 40 μL 25 mM NH4HCO3 buffer overnight at 37°C, and the resulting peptides were collected as a filtrate. For purified PKM2 protein, the gel slice was first treated with iodoacetamide and DTT, and then digested with trypsin. The tryptic digests of protein samples were then desalted on C18 Cartridges (Empore SPE Cartridges C18 (standard density), bed I.D. 7 mm, volume 3 mL, Sigma), concentrated by vacuum centrifugation and reconstituted in 40 μL of 0.1% (v/v) formic acid. The peptide content was estimated by UV light spectral density at 280 nm using an extinctions coefficient of 1.1 of 0.1% (g/L) solution that was calculated on the basis of the frequency of tryptophan and tyrosine in vertebrate proteins. The resulting samples were subjected to LC-MS/MS spectrometry analysis (Shanghai Applied Protein Technology Co., Ltd.). MS/MS spectra were searched using MASCOT engine (Matrix Science, London, UK; version 2.4) against the UniProtKB human (161584 total entries, downloaded 20180105).
Cerebral spinal fluid (CSF) collection
The procedures for CSF collection were described elsewhere.71 Briefly, mice were anesthetized by isoflurane. An incision from the top of the skull to the dorsal thorax is made. The meninges overlying the cisterna magna is exposed after removing the musculature. The tissue above the cisterna magna is carefully excised and the surrounding area is gently cleaned under a dissecting microscope. The arachnoid membrane covering the cistern was punctured using a microneedle. A polypropylene narrow bore pipette is used to collect the CSF at the punctured site.
Shikonin and PKM2-IN-1 assays
To examine shikonin and PKM2-IN-1 in the brain, 8-week-old male C57BL/6J mice were treated with shikonin and PKM2-IN-1 by gavage at the dosage of 50 mg/kg. One hour later, mice were sacrificed after anaesthetization and brain tissues were collected. To exact chemicals, brain tissues were homogenized in methanol and then subjected to centrifugation at 3000 rpm for 10 min. The resulting supernatants were filtered using 0.22 μm syringe filters. An HPLC-MS (LCMS-IT-TOF, Shimadzu, Japan) coupled with a software LC-MS solution was used to detect shikonin and PKM2-IN-1. Separation was performed on an SB-C18 column (75 × 2.1 mm id, 2.7 μm) from Agilent (CA, USA). The mobile phase was composed of 0.1% formic acid containing 10 mM ammonium formate and acetonitrile (20:80), at a flow rate of 1 mL/min, the temperature was maintained at 35°C and the acquisition wavelength was set to 518 nm for both shikonin and PKM-IN-1. For shikonin, the mass spectrometer was operated in positive ion mode with the following parameters: ionspray voltage: 4.0 kV; curtain gas pressure: 30 psi; nebulizer gas pressure: 50 psi; heating gas pressure: 50 psi; source temperature: 350°C; collision gas pressure: 8 psi. For PKM2-IN-1, the mass spectrometer was operated in positive ion mode with the following parameters: ionspray voltage: 5.5 kV; curtain gas pressure: 30 psi; nebulizer gas pressure: 50 psi; heating gas pressure: 50 psi; source temperature: 500°C; collision gas pressure: 8 psi. Multiple reaction monitoring (MRM) mode was used throughout analyses.
Quantification and statistical analysis
Data are presented as means ± SD. Statistical significance for two groups was analyzed by two-tailed Student’s t test. Statistical significance for multiple groups was calculated with one-way ANOVA or two-way ANOVA. All the statistical analyses were done in the GraphPad Prism version 9.0.0. Significance was accepted at the level of p < 0.05.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (nos. 32271193 and 82272167), the Social and Livelihood Science and Technology Projects of Nantong (MSZ2022048), the Basic Research Program of Jiangsu Education Department (22KJB31006), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX23-3383), and the project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author contributions
B.L., J.Z., X.Y., J.W., L.L., Q.J., Y.W., Y.J., and X.L. performed the experiments and collected and analyzed the data. Y.C., X.T., and C.S. conceived the idea, designed the project, supervised the study, and wrote the manuscript.
Declaration of interests
The effect of PKM2-IN-1 in treating Parkinson’s disease has been patented (WO2022022748), and C.S. and Y.W. are listed as the inventors.
Published: August 10, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101684.
Contributor Information
Yu Chen, Email: cyu1229@126.com.
Xin Tang, Email: tangxin@ntu.edu.cn.
Cheng Sun, Email: suncheng1975@ntu.edu.cn.
Supplemental information
References
- 1.GBD 2016 Neurology Collaborators Global, regional, and national burden of neurological disorders, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:459–480. doi: 10.1016/S1474-4422(18)30499-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalia L.V., Lang A.E. Parkinson's disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong M.J., Okun M.S. Diagnosis and Treatment of Parkinson Disease: A Review. JAMA. 2020;323:548–560. doi: 10.1001/jama.2019.22360. [DOI] [PubMed] [Google Scholar]
- 4.Braak H., Del Tredici K., Rüb U., de Vos R.A.I., Jansen Steur E.N.H., Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 5.Bloem B.R., Okun M.S., Klein C. Parkinson's disease. Lancet. 2021;397:2284–2303. doi: 10.1016/S0140-6736(21)00218-X. [DOI] [PubMed] [Google Scholar]
- 6.Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W.K., Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi W., Keenan H.A., Li Q., Ishikado A., Kannt A., Sadowski T., Yorek M.A., Wu I.H., Lockhart S., Coppey L.J., et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat. Med. 2017;23:753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angiari S., Runtsch M.C., Sutton C.E., Palsson-McDermott E.M., Kelly B., Rana N., Kane H., Papadopoulou G., Pearce E.L., Mills K.H.G., O'Neill L.A.J. Pharmacological Activation of Pyruvate Kinase M2 Inhibits CD4(+) T Cell Pathogenicity and Suppresses Autoimmunity. Cell Metab. 2020;31:391–405.e8. doi: 10.1016/j.cmet.2019.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proia P., Di Liegro C.M., Schiera G., Fricano A., Di Liegro I. Lactate as a Metabolite and a Regulator in the Central Nervous System. Int. J. Mol. Sci. 2016;17:1450. doi: 10.3390/ijms17091450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Passarella S., de Bari L., Valenti D., Pizzuto R., Paventi G., Atlante A. Mitochondria and L-lactate metabolism. FEBS Lett. 2008;582:3569–3576. doi: 10.1016/j.febslet.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 11.Hoyer S. Brain glucose and energy metabolism during normal aging. Aging. 1990;2:245–258. doi: 10.1007/BF03323925. [DOI] [PubMed] [Google Scholar]
- 12.Saxena U. Bioenergetics failure in neurodegenerative diseases: back to the future. Expert Opin. Ther. Targets. 2012;16:351–354. doi: 10.1517/14728222.2012.664135. [DOI] [PubMed] [Google Scholar]
- 13.Abeliovich A., Gitler A.D. Defects in trafficking bridge Parkinson's disease pathology and genetics. Nature. 2016;539:207–216. doi: 10.1038/nature20414. [DOI] [PubMed] [Google Scholar]
- 14.Sauve A.A., Wolberger C., Schramm V.L., Boeke J.D. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa T., Guarente L. SnapShot: sirtuins, NAD, and aging. Cell Metab. 2014;20:192–192.e1. doi: 10.1016/j.cmet.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Burnett C., Valentini S., Cabreiro F., Goss M., Somogyvári M., Piper M.D., Hoddinott M., Sutphin G.L., Leko V., McElwee J.J., et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouchiroud L., Houtkooper R.H., Moullan N., Katsyuba E., Ryu D., Cantó C., Mottis A., Jo Y.S., Viswanathan M., Schoonjans K., et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson R.M., Latorre-Esteves M., Neves A.R., Lavu S., Medvedik O., Taylor C., Howitz K.T., Santos H., Sinclair D.A. Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science. 2003;302:2124–2126. doi: 10.1126/science.1088697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwabu M., Yamauchi T., Okada-Iwabu M., Sato K., Nakagawa T., Funata M., Yamaguchi M., Namiki S., Nakayama R., Tabata M., et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- 20.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y., Zhu X., Zhou Y., Zhang D. Reduced serum SIRT1 levels in patients with Parkinson's disease: a cross-sectional study in China. Neurol. Sci. 2021;42:1835–1841. doi: 10.1007/s10072-020-04711-z. [DOI] [PubMed] [Google Scholar]
- 24.Maszlag-Torok R., Boros F.A., Vecsei L., Klivenyi P. Gene variants and expression changes of SIRT1 and SIRT6 in peripheral blood are associated with Parkinson's disease. Sci. Rep. 2021;11:10677. doi: 10.1038/s41598-021-90059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai R., Zhang Y., Simmering J.E., Schultz J.L., Li Y., Fernandez-Carasa I., Consiglio A., Raya A., Polgreen P.M., Narayanan N.S., et al. Enhancing glycolysis attenuates Parkinson's disease progression in models and clinical databases. J. Clin. Invest. 2019;129:4539–4549. doi: 10.1172/JCI129987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foltynie T. Glycolysis as a therapeutic target for Parkinson's disease. Lancet Neurol. 2019;18:1072–1074. doi: 10.1016/S1474-4422(19)30404-1. [DOI] [PubMed] [Google Scholar]
- 27.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 28.Dovonou A., Bolduc C., Soto Linan V., Gora C., Peralta Iii M.R., Lévesque M. Animal models of Parkinson's disease: bridging the gap between disease hallmarks and research questions. Transl. Neurodegener. 2023;12:36. doi: 10.1186/s40035-023-00368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ning X., Qi H., Li R., Li Y., Jin Y., McNutt M.A., Liu J., Yin Y. Discovery of novel naphthoquinone derivatives as inhibitors of the tumor cell specific M2 isoform of pyruvate kinase. Eur. J. Med. Chem. 2017;138:343–352. doi: 10.1016/j.ejmech.2017.06.064. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y., Jiang Y., Yang Y., Huang X., Sun C. SIRT1 Protects Dopaminergic Neurons in Parkinson's Disease Models via PGC-1alpha-Mediated Mitochondrial Biogenesis. Neurotox. Res. 2021;39:1393–1404. doi: 10.1007/s12640-021-00392-4. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y., Zhang S., Guan J., Jiang Y., Zhang J., Luo L., Sun C. SIRT1 attenuates neuroinflammation by deacetylating HSPA4 in a mouse model of Parkinson's disease. Biochim. Biophys. Acta, Mol. Basis Dis. 2022;1868:166365. doi: 10.1016/j.bbadis.2022.166365. [DOI] [PubMed] [Google Scholar]
- 32.Chao C.C., Huang C.L., Cheng J.J., Chiou C.T., Lee I.J., Yang Y.C., Hsu T.H., Yei C.E., Lin P.Y., Chen J.J., Huang N.K. SRT1720 as an SIRT1 activator for alleviating paraquat-induced models of Parkinson's disease. Redox Biol. 2022;58:102534. doi: 10.1016/j.redox.2022.102534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donmez G., Outeiro T.F. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol. Med. 2013;5:344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo Y.J., Dong S.Y., Cui X.X., Feng Y., Liu T., Yin M., Kuo S.H., Tan E.K., Zhao W.J., Wu Y.C. Resveratrol alleviates MPTP-induced motor impairments and pathological changes by autophagic degradation of alpha-synuclein via SIRT1-deacetylated LC3. Mol. Nutr. Food Res. 2016;60:2161–2175. doi: 10.1002/mnfr.201600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W., Zheng Y., Xia Y., Ji H., Chen X., Guo F., Lyssiotis C.A., Aldape K., Cantley L.C., Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv L., Xu Y.P., Zhao D., Li F.L., Wang W., Sasaki N., Jiang Y., Zhou X., Li T.T., Guan K.L., et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol. Cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das Gupta K., Shakespear M.R., Curson J.E.B., Murthy A.M.V., Iyer A., Hodson M.P., Ramnath D., Tillu V.A., von Pein J.B., Reid R.C., et al. Class IIa Histone Deacetylases Drive Toll-like Receptor-Inducible Glycolysis and Macrophage Inflammatory Responses via Pyruvate Kinase M2. Cell Rep. 2020;30:2712–2728.e8. doi: 10.1016/j.celrep.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Bettaieb A., Bakke J., Nagata N., Matsuo K., Xi Y., Liu S., AbouBechara D., Melhem R., Stanhope K., Cummings B., et al. Protein tyrosine phosphatase 1B regulates pyruvate kinase M2 tyrosine phosphorylation. J. Biol. Chem. 2013;288:17360–17371. doi: 10.1074/jbc.M112.441469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu K., Li F., Han H., Chen Y., Mao Z., Luo J., Zhao Y., Zheng B., Gu W., Zhao W. Parkin Regulates the Activity of Pyruvate Kinase M2. J. Biol. Chem. 2016;291:10307–10317. doi: 10.1074/jbc.M115.703066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang J., Cao R., Zhang Y., Xia Y., Zheng Y., Li X., Wang L., Yang W., Lu Z. PKM2 dephosphorylation by Cdc25A promotes the Warburg effect and tumorigenesis. Nat. Commun. 2016;7:12431. doi: 10.1038/ncomms12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nandi S., Razzaghi M., Srivastava D., Dey M. Structural basis for allosteric regulation of pyruvate kinase M2 by phosphorylation and acetylation. J. Biol. Chem. 2020;295:17425–17440. doi: 10.1074/jbc.RA120.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson E.C.B., Dammer E.B., Duong D.M., Ping L., Zhou M., Yin L., Higginbotham L.A., Guajardo A., White B., Troncoso J.C., et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020;26:769–780. doi: 10.1038/s41591-020-0815-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 44.Glass C.K., Saijo K., Winner B., Marchetto M.C., Gage F.H. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh A., Tyson T., George S., Hildebrandt E.N., Steiner J.A., Madaj Z., Schulz E., Machiela E., McDonald W.G., Escobar Galvis M.L., et al. Mitochondrial pyruvate carrier regulates autophagy, inflammation, and neurodegeneration in experimental models of Parkinson's disease. Sci. Transl. Med. 2016;8:368ra174. doi: 10.1126/scitranslmed.aag2210. [DOI] [PubMed] [Google Scholar]
- 46.Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Le Z., Yanxiang Guo J., et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabinowitz J.D., Enerbäck S. Lactate: the ugly duckling of energy metabolism. Nat. Metab. 2020;2:566–571. doi: 10.1038/s42255-020-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liguori C., Stefani A., Sancesario G., Sancesario G.M., Marciani M.G., Pierantozzi M. CSF lactate levels, tau proteins, cognitive decline: a dynamic relationship in Alzheimer's disease. J. Neurol. Neurosurg. Psychiatry. 2015;86:655–659. doi: 10.1136/jnnp-2014-308577. [DOI] [PubMed] [Google Scholar]
- 49.Schirinzi T., Di Lazzaro G., Sancesario G.M., Summa S., Petrucci S., Colona V.L., Bernardini S., Pierantozzi M., Stefani A., Mercuri N.B., Pisani A. Young-onset and late-onset Parkinson's disease exhibit a different profile of fluid biomarkers and clinical features. Neurobiol. Aging. 2020;90:119–124. doi: 10.1016/j.neurobiolaging.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 50.Nagesh Babu G., Gupta M., Paliwal V.K., Singh S., Chatterji T., Roy R. Serum metabolomics study in a group of Parkinson's disease patients from northern India. Clin. Chim. Acta. 2018;480:214–219. doi: 10.1016/j.cca.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 51.Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., et al. Pyruvate kinase M2 regulates Hif-1alpha activity and IL-1beta induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samuvel D.J., Sundararaj K.P., Nareika A., Lopes-Virella M.F., Huang Y. Lactate boosts TLR4 signaling and NF-kappaB pathway-mediated gene transcription in macrophages via monocarboxylate transporters and MD-2 up-regulation. J. Immunol. 2009;182:2476–2484. doi: 10.4049/jimmunol.0802059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaushik D.K., Bhattacharya A., Mirzaei R., Rawji K.S., Ahn Y., Rho J.M., Yong V.W. Enhanced glycolytic metabolism supports transmigration of brain-infiltrating macrophages in multiple sclerosis. J. Clin. Invest. 2019;129:3277–3292. doi: 10.1172/JCI124012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia L., Liao M., Mou A., Zheng Q., Yang W., Yu Z., Cui Y., Xia X., Qin Y., Chen M., Xiao B. Rheb-regulated mitochondrial pyruvate metabolism of Schwann cells linked to axon stability. Dev. Cell. 2021;56:2980–2994.e6. doi: 10.1016/j.devcel.2021.09.013. [DOI] [PubMed] [Google Scholar]
- 55.Benjamin D., Robay D., Hindupur S.K., Pohlmann J., Colombi M., El-Shemerly M.Y., Maira S.M., Moroni C., Lane H.A., Hall M.N. Dual Inhibition of the Lactate Transporters MCT1 and MCT4 Is Synthetic Lethal with Metformin due to NAD+ Depletion in Cancer Cells. Cell Rep. 2018;25:3047–3058.e4. doi: 10.1016/j.celrep.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi S. Neuroprotective Function of High Glycolytic Activity in Astrocytes: Common Roles in Stroke and Neurodegenerative Diseases. Int. J. Mol. Sci. 2021;22:6568. doi: 10.3390/ijms22126568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roumes H., Dumont U., Sanchez S., Mazuel L., Blanc J., Raffard G., Chateil J.F., Pellerin L., Bouzier-Sore A.K. Neuroprotective role of lactate in rat neonatal hypoxia-ischemia. J. Cereb. Blood Flow Metab. 2021;41:342–358. doi: 10.1177/0271678X20908355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muraleedharan R., Gawali M.V., Tiwari D., Sukumaran A., Oatman N., Anderson J., Nardini D., Bhuiyan M.A.N., Tkáč I., Ward A.L., et al. AMPK-Regulated Astrocytic Lactate Shuttle Plays a Non-Cell-Autonomous Role in Neuronal Survival. Cell Rep. 2020;32:108092. doi: 10.1016/j.celrep.2020.108092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fedotova E.I., Dolgacheva L.P., Abramov A.Y., Berezhnov A.V. Lactate and Pyruvate Activate Autophagy and Mitophagy that Protect Cells in Toxic Model of Parkinson's Disease. Mol. Neurobiol. 2022;59:177–190. doi: 10.1007/s12035-021-02583-8. [DOI] [PubMed] [Google Scholar]
- 60.Bose A., Beal M.F. Mitochondrial dysfunction in Parkinson's disease. J. Neurochem. 2016;139:216–231. doi: 10.1111/jnc.13731. [DOI] [PubMed] [Google Scholar]
- 61.Ayyangar U., Karkhanis A., Tay H., Afandi A.F.B., Bhattacharjee O., Ks L., Lee S.H., Chan J., Raghavan S. Metabolic rewiring of macrophages by epidermal-derived lactate promotes sterile inflammation in the murine skin. EMBO J. 2024;43:1113–1134. doi: 10.1038/s44318-024-00039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J., Chen L., Qin Q., Wang D., Zhao J., Gao H., Yuan X., Zhang J., Zou Y., Mao Z., et al. Upregulated hexokinase 2 expression induces the apoptosis of dopaminergic neurons by promoting lactate production in Parkinson's disease. Neurobiol. Dis. 2022;163:105605. doi: 10.1016/j.nbd.2021.105605. [DOI] [PubMed] [Google Scholar]
- 63.Rho H., Terry A.R., Chronis C., Hay N. Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis. Cell Metab. 2023;35:1406–1423.e8. doi: 10.1016/j.cmet.2023.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X., Zhang Y., Xu L., Wang A., Zou Y., Li T., Huang L., Chen W., Liu S., Jiang K., et al. Ultrasensitive sensors reveal the spatiotemporal landscape of lactate metabolism in physiology and disease. Cell Metab. 2023;35:200–211.e9. doi: 10.1016/j.cmet.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nam J.H., Park E.S., Won S.Y., Lee Y.A., Kim K.I., Jeong J.Y., Baek J.Y., Cho E.J., Jin M., Chung Y.C., et al. TRPV1 on astrocytes rescues nigral dopamine neurons in Parkinson's disease via CNTF. Brain. 2015;138:3610–3622. doi: 10.1093/brain/awv297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaven F., Marin P., Claeysen S. Primary culture of mouse dopaminergic neurons. J. Vis. Exp. 2014;◼:e51751. doi: 10.3791/51751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng S., Wang C., Ma J., Jiang K., Jiang Y., Gu X., Sun C. Achyranthes bidentata polypeptide protects dopaminergic neurons from apoptosis in Parkinson's disease models both in vitro and in vivo. Br. J. Pharmacol. 2018;175:631–643. doi: 10.1111/bph.14110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang X.Z., Wen D., Zhang M., Xie Q., Ma L., Guan Y., Ren Y., Chen J., Hao C.M. Sirt1 activation ameliorates renal fibrosis by inhibiting the TGF-beta/Smad3 pathway. J. Cell. Biochem. 2014;115:996–1005. doi: 10.1002/jcb.24748. [DOI] [PubMed] [Google Scholar]
- 69.Tan R., Krueger R.K., Gramelspacher M.J., Zhou X., Xiao Y., Ke A., Hou Z., Zhang Y. Cas11 enables genome engineering in human cells with compact CRISPR-Cas3 systems. Mol. Cell. 2022;82:852–867.e5. doi: 10.1016/j.molcel.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dioum E.M., Chen R., Alexander M.S., Zhang Q., Hogg R.T., Gerard R.D., Garcia J.A. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 71.DeMattos R.B., Bales K.R., Parsadanian M., O'Dell M.A., Foss E.M., Paul S.M., Holtzman D.M. Plaque-associated disruption of CSF and plasma amyloid-beta (Abeta) equilibrium in a mouse model of Alzheimer's disease. J. Neurochem. 2002;81:229–236. doi: 10.1046/j.1471-4159.2002.00889.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be available by contacting the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.