Abstract
SARS-CoV-2 caused the pandemic situation experienced since the beginning of 2020, and many countries faced the rapid spread and severe form of the disease. Mechanisms of interaction between the virus and the host were observed during acute phase, but few data are available when related to immunity dynamics in convalescents. We conducted a longitudinal study, with 51 healthy donors and 62 COVID-19 convalescent patients, which these had a 2-month follow-up after symptoms recovery. Venous blood sample was obtained from all participants to measure blood count, subpopulations of monocytes, lymphocytes, natural killer cells and dendritic cells. Serum was used to measure cytokines, chemokines, growth factors, anti-N IgG and anti-S IgG/IgM antibodies. Statistic was performed by Kruskal–Wallis test, and linear regression with days post symptoms and antibody titers. All analysis had confidence interval of 95%. Less than 35% of convalescents were anti-S IgM+, while more than 80% were IgG+ in D30. Anti-N IgG decreased along time, with loss of seroreactivity of 13%. Eosinophil count played a distinct role on both antibodies during all study, and the convalescence was orchestrated by higher neutrophil-to-lymphocyte ratio and IL-15, but initial stages were marked by increase in myeloid DCs, B1 lymphocytes, inflammatory and patrolling monocytes, G-CSF and IL-2. Later convalescence seemed to change to cytotoxicity mediated by T lymphocytes, plasmacytoid DCs, VEGF, IL-9 and CXCL10. Anti-S IgG antibodies showed the longest perseverance and may be a better option for diagnosis. The inflammatory pattern is yet present on initial stage of convalescence, but quickly shifts to a reparative dynamic. Meanwhile eosinophils seem to play a role on anti-N levels in convalescence, although may not be the major causative agent. We must highlight the importance of immunological markers on acute clinical outcomes, but their comprehension to potentialize adaptive system must be explored to improve immunizations and further preventive policies.
Keywords: Severe acute respiratory syndrome (SARS), Immune hallmarks, Antibody, Brazil
Subject terms: Immunology, Infectious diseases, Inflammation
Introduction
COVID-19 is a global viral disease caused by the Betacoronavirus known as Severe Acute Respiratory Syndrome Coronavirus- 2 (SARS-CoV-2). It primarily affects the lungs, leading to both local and systemic complications. Among individuals with no prior immunity, or those with comorbidities associated to worse outcomes, an increased risk of death has been observed1–3. The incubation period I typically around 5 days, with transmission possible within a 14-day period through the respiratory tract. This facilitates its rapid spread from an infected individual, even if asymptomatic, to an uninfected person4–6.
Main symptoms include dry cough, dyspnea, pneumonia, and in severe cases, respiratory syndrome4,6,7. Among infected, 3–20% require hospitalization, with most going to the ICU, and 1% die from complications in the acute phase8,9. Studies described human body's viral response to understand the dynamics of cells and proteins, to prevent recurrence and reduce risks during acute phases10–14.
Innate immune cells have been proposed as markers of severity during acute disease11,15,16. However, by 2022, many people had already been exposed to the virus, and the convalescence stage became a central topic of discussion. This stage refers to the clinical improvement after COVID-19, and there is still a lack of discussion regarding the immune factors that coordinate antibody and memory cell production17,18. Numerous studies have evaluated the interaction between cells, cytokines, chemokines, and growth factors, but their correlation with antibody production has not been fully elucidated yet19–22.
The World Health Organization (WHO) has proposed that individuals who have recovered from SARS-CoV-2 may donate plasma for a convalescent plasma therapy, which consists of transfusion of plasma enriched with antibodies from cured individuals into patients infected with SARS-CoV-2 and with severe form of the disease23. This approach is based on passive immunization, where antibodies specific to viral epitopes are transferred from convalescent individuals to acute patients. However, this procedure is still under study, and certain criteria must be met. Males are the primary population to plasma donation, once woman may present specific antibodies that can enhance transfusion reaction acute lung injury. Despite gender, the donor also must have sufficient concentration of antibodies in the serum24,25. This treatment is experimental, but there is not enough studies to standardize a protocol and with higher. Understand the major mechanisms related to antibody production in convalescent patients from COVID-19 can promote new therapeutic strategies to prompt higher antibody titers and so, reduce the severity of population and improve treatment to those with immunocompromising diseases.
Our aim was to conduct a longitudinal study with COVID-19 convalescent patients who had recovered from the acute phase at 30, 60, and 90 days. Understanding the involvement of cells and soluble proteins in antibody surveillance will contribute to developing further strategies for patient follow-up after the acute disease, provide important insights into convalescent progression, improve vaccination approaches, and ultimately enhance the quality of life for patients.
Results
Sociodemographic data
Fifty-one healthy donors were enrolled in this study. The mean age was 32.39 years (SD ± 11.63), with 36 (70.6%) males and 15 (29.4%) females. The majority were of admixed ethnicity (n = 45 [88.3%]), followed by 4 (7.8%) Caucasians and 2 (3.9%) African Americans. Five (9.8%) had blood type A positive, 27 (52.9%) had blood type O positive, and 19 (37.3%) had blood type O negative.
The mean age of the convalescent group was 39.94 years (SD ± 11.56), with 51 (82.3%) males and 11 (17.7%) females. In terms of ethnicity, 42 (67.8%) were admixed, 18 (29%) were Caucasians, and only 2 (3.2%) were African Americans. Regarding blood type, 17 were type A (14 [22.6%] positive and 3 [4.9%] negative), 2 were type B (1 [1.6%] positive and 1 [1.6%] negative), 2 were type AB (both positive [3.2%]), and 41 were type O (40 [64.5%] positive and 1 [1.6%] negative). The majority were overweight or obese (n = 23 [37.1%] each), followed by 15 (24.2%) with a normal BMI, and only one [1.6%] had a low BMI. Statistical analysis showed significant difference only in age (p = 0.0009) and blood type (p < 0.0001) between both groups, however we believe the blood type difference is related to higher prevalence of O and A types in the population studied (Table 1).
Table 1.
Sociodemographic data of healthy donors and convalescent patients.
| Sociodemographic data | Healthy donors (n = 51) | Convalescents (n = 62) | p value |
|---|---|---|---|
| Age, mean ± SD | 32.39 ± 11.63 | 39.94 ± 11.56 | 0.0009 |
| Gender | |||
| Male, n (%) | 36 (70.6) | 51 (82.3) | 0.1794 |
| Female, n (%) | 15 (29.4) | 11 (17.7) | |
| Ethnicity | |||
| Caucasians, n (%) | 4 (7.8) | 18 (29) | 0.0182 |
| Admixed, n (%) | 45 (88.3) | 42 (67.8) | |
| African Americans, n (%) | 2 (3.9) | 2 (3.2) | |
| Blood type | |||
| A+/A−, n (%) | 5 (9.8)/0 (0) | 14 (22.6)/3 (4.9) | < 0.0001 |
| B+/B−, n (%) | 0 (0)/0 (0) | 1 (1.6)/1 (1.6) | |
| AB+/AB−, n (%) | 0 (0)/0 (0) | 2 (3.2)/0 (0) | |
| O+/O−, n (%) | 27 (52.9)/19 (37.3) | 40 (64.5)/1 (1.6) | |
| Body mass index (kg/m2) | |||
| Low (< 18.5), n (%) | 1 (2.0) | 1 (1.6) | 0.6504 |
| Normal (18.5–24.9), n (%) | 17 (33.3) | 15 (24.2) | |
| Overweigh (25–29.9), n (%) | 19 (37.3) | 23 (37.1) | |
| Obesity (> 30), n (%) | 14 (27.4) | 23 (37.1) | |
SD standard deviation, Pos positive, Neg negative. Chi-square test was performed isolating blood groups (A, B, AB and O), and subgroups (positive or negative), and including only those with observation > 1 for statistical purposes. Significant values are in bold.
Once there was seen difference among age, ethnicity and blood type between HD and Convalescent (D30) groups, we conducted an analysis of immunological parameters segregating our groups based on blood type and age. Any difference was seen in the comparison of blood groups inside HD or Convalescent group. The pattern seen in the general observation seemed not to change when related to any of these three characteristics, and so, we believe that none of them had any or few interferences in the immune system in our participants. Heatmap and PCA analysis segregated by these parameters are shown in Fig. S1 for age (S1A), ethnicity (S1B) and blood type (S1C).
Analyzing the clinical data from the convalescent group, 5 out of 62 patients (8.1%) required hospitalization and mechanical ventilation. The mean hospital length of stay (LOS) was 15.82 days (SD ± 10.49), ranging from 1 to 51 days.
Antibody dynamic in convalescence
The antibody concentration was evaluated in the convalescent groups and compared to the number of days after the end of symptoms. Anti-nucleocapsid (anti-N) IgG showed a significant decrease (p = 0.0017) from 30 to 90 days after clinical recovery, which was further confirmed by correlation analysis. The analysis revealed a negative and significant reduction in anti-N concentration with an increase in the number of days after the end of symptoms (p = 0.0056), as shown in Fig. 1A. Although anti-Spike (anti-S) IgG also decreased, the concentration levels showed no significant difference, even in the correlation analysis (Fig. 1B). This suggests that the presence of anti-S antibodies in the serum persists, while anti-N antibodies decrease slowly after recovery (Fig. 1B). It should be noted that immunity remains active for a few days after viral clearance, and some patients seroconverted from a positive state (at D30) to a negative state (at both D60 and D90) during the study period for both anti-N and anti-S antibodies, using the manufacturer's cut-off.
Fig. 1.
Serum antibody analysis during convalescence. (A) Comparison of IgG anti-nucleocapsid protein (OD) and correlation under anti-N concentration and days post symptoms; (B) Comparison of IgG anti-Spike protein (AU/mL) and correlation between anti-S concentration and days post symptoms. The cutt-off 1.280 AU/mL was highlighted to demonstrate the participants that were eligible to convalescent plasma donation in D30 (n = 30/62 [48.4%]), D60 (n = 20/48 [41.7%]) and D90 (n = 14/47 [29.8%]); (C) Percentage of participants with a qualitative (pos/neg) antibody production among study period, based on anti-S immunochromatographic test, and CMIA anti-S and anti-N, with absolute and relative values on table below; (D) Fold change comparison between D60/D30, D90/D30 and D90/60 IgG anti-S antibody concentration, using quantitative result from patients with all follow-up (n = 45). Statistical analysis was performed with One-Way ANOVA followed by Turkey’s Multiple comparison test, considering significative when p < 0.05. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Serum samples from 20 out of 62 participants (32.3%) with a previous diagnosis of COVID-19 and a symptomatic period tested positive for IgM using an immunochromatographic test at D30. This number decreased to 11 out of 48 participants (22.9%) at D60, and 8 out of 47 participants (17%) at D90. IgG detection tests showed higher sensitivity. All three tests (both anti-S immunochromatographic and CMIA, and anti-N CMIA) detected more than 80% seroreactivity in convalescent individuals at D30. The D60 and D90 groups still had a seroconversion rate higher than 80% for anti-S antibody detection, while anti-N antibody detection decreased to 70.2% at D90 (Fig. 1C).
The fold change comparison shows that the antibody concentration changes gradually from 1 month to the next, but there is a more significant difference at the third month. A similar pattern was observed when comparing D30 to D60 and D60 to D90, indicating that the antibody concentration from D30 to D60 did not vary significantly compared to the change from D60 to D90. However, there was an overall decrease in antibody concentration from D30 to D90 (Fig. 1D).
WHO released a list of approved tests to detect anti-S IgG antibodies, which can help identify potential convalescent blood donors. The qualitative and quantitative CMIA test has been approved for monitoring potential blood donors who meet the eligibility criteria for antibody concentration (> 1.280 AU/mL). Among all the convalescent individuals who agreed to participate, only 30 out of 62 (48.4%) had a sufficient concentration at D30. By D60, only 20 out of 48 participants (41.7%) remained eligible, and this number further decreased to 14 out of 47 participants (29.8%) at D90 (Fig. 1B).
Inflammatory profile is mediated by memory cells and patrolling monocytes
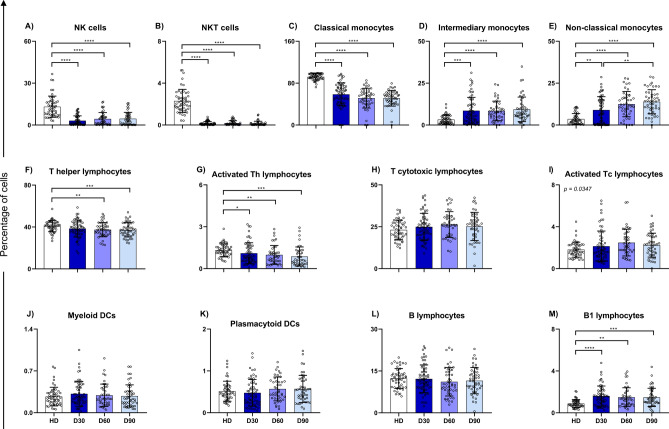
Cell counts and subpopulations were evaluated over time using blood count and flow cytometry, respectively. NK and NKT cells were significantly reduced after COVID-19 and showed no significant signs of recovery even after 90 days (Fig. 2A,B). Although the monocyte count increased in D30 patients compared to healthy controls, this increase appeared to be driven by inflammatory and patrolling monocytes. The absolute monocyte count (AMC) decreased in the second month after symptom resolution (Table 2), but patrolling monocytes continued to increase over time, reaching even higher levels at D90 (Fig. 2C–E). On the other hand, both total T helper cells and activated T helper cells decreased as convalescence progressed (Fig. 2F,G). Total T cytotoxic lymphocytes did not vary significantly between the analyzed groups, but the median of activated T cytotoxic lymphocytes showed a significant difference, although statistical analysis could not determine which groups differed (Fig. 2H,I). The subpopulation of circulating dendritic cells showed no difference between healthy donors and convalescent individuals, nor between convalescent subgroups (Fig. 2J,K). Although B lymphocytes showed no difference, an increase in B1 lymphocytes was observed during convalescence, which appeared to persist throughout the 3-month period analyzed (Fig. 2L,M).
Fig. 2.
Phenotypic analysis of immune cells, comparing HD, D30, D60 and D90 groups. Data is expressed as median and interquartile range in percentage of cells. Bar graphs represent analysis of: (A) NK cells; (B) NKT cells; (C) Classical monocytes; (D) Inflammatory monocytes; (E) Patrolling monocytes; (F) T helper lymphocytes; (G) Activated T helper lymphocytes; (H) T cytotoxic lymphocytes; (I) Activated T cytotoxic lymphocytes; (J) Plasmacytoid dendritic cells; (K) Myeloid dendritic cells; (L) B lymphocytes; and (M) B1 lymphocytes. Statistical analysis was conducted with Kruskal–Wallis and Dunn’s Multiple Comparisons tests, considering significative when p < 0.05. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Table 2.
Laboratorial parameters of healthy donors and convalescent patients at D30, D60 and D90.
| Variables | HD n = 51 |
D30 n = 62 |
D60 n = 48 |
D90 n = 47 |
p value |
|---|---|---|---|---|---|
| RBC (× 106/µL, median [IQR]) | 4.99 [4.59–5.40] | 4.90 [4.56–5.20] | 4.89 [4.47–5.15] | 4.89 [4.71–5.29] | 0.4676 |
| Hemoglobin (g/dL, median [IQR]) | 14.90 [13.60–16.00] | 14.40 [13.38–15.03] | 14.35 [13.23–14.90] | 14.40 [13.50–15.10] | 0.0602 |
| Hematocrit (%, median [IQR]) | 44.70 [40.6–47.4] | 42.60 [40.45–45.45] | 42.75 [39.13–44.50] | 43.90 [41.50–46.20] | 0.0660 |
| MCV (fL, median [IQR]) | 87.80 [84.70–90.40] | 88.15 [84.85–90.80] | 87.90 [84.35–91.08] | 89.10 [85.30–91.40] | 0.7292 |
| MCH (pg, median [IQR]) | 29.70 [28.8–30.70] | 29.60 [28.38–30.60] | 29.85 [28.10–30.70] | 29.80 [27.90–30.80] | 0.8607 |
| MCHC (g/dL, median [IQR]) | 34.05 [33.10–34.60] | 33.70 [32.78–34.450] | 33.59 [32.75–34.48] | 33.40 [32.60–34.10] | 0.1667 |
| RDW (%, median [IQR]) | 13.70 [13.10–14.00] | 14.40 [13.75–14.95] | 14.40 [13.90–15.08] | 14.35 [13.68–15.03] | < 0.0001a,b,c |
| WBC (× 106/µL, median [IQR]) | 6.33 [5.17–6.95] | 6.78 [5.87–7.51] | 6.34 [5.75–7.57] | 6.65 [5.61–7.97] | 0.2906 |
| Neutrophil (× 103/µL, median [IQR]) | 3.34 [2.8–4.17] | 4.00 [3.21–4.65] | 3.80 [3.14–4.61] | 3.79 [3.04–4.84] | 0.0757 |
| Lymphocyte (× 103/µL, median [IQR]) | 1.85 [1.59–2.18] | 1.98 [1.61–2.22] | 1.93 [1.6–2.3] | 1.98 [1.61–2.23] | 0.8081 |
| Monocyte (× 103/µL, median [IQR]) | 0.38 [0.28–0.42] | 0.43 [0.37–0.48] | 0.37 [0.33–0.42] | 0.38 [0.28–0.43] | 0.0028a,d,e |
| Basophil (× 103/µL, median [IQR]) | 0.03 [0.02–0.05] | 0.04 [0.02–0.06] | 0.03 [0.02–0.04] | 0.03 [0.02–0.05] | 0.2906 |
| Eosinophil (× 103/µL, median [IQR]) | 0.19 [0.12–0.38] | 0.19 [0.13–0.26] | 0.18 [0.13–0.26] | 0.16 [0.12–0.26] | 0.6799 |
| Platelet count (× 103/µL, median [IQR]) | 243.0 [210–282] | 259.5 [212–290] | 245.0 [215–266] | 245.00 [209–272] | 0.7717 |
aSignificant difference for HD vs D30; bSignificant difference for HD vs D60; cSignificant difference for HD vs D90; dSignificant difference for D30 vs D60; eSignificant difference for D30 vs D90; fSignificant difference for D60 vs D90. Statistical analysis was performed with Kruskal–Wallis test, and Dunn’s Multiple Comparison Test to compare hematological values from all groups. A p < 0.05 was considered significant and highlighted by bold type font.
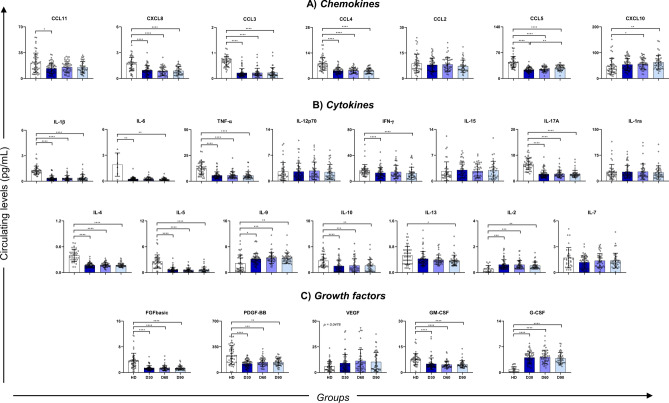
To evaluate the involvement of cytokines, chemokines, and growth factors in convalescent individuals, we quantified these soluble proteins and compared them to the healthy donor group. The convalescent stage appeared to be characterized by higher levels of VEGF, G-CSF, IL-2, IL-9, and CXCL10, but also lower serum concentrations of FGF basic, PDGF-BB, GM-CSF, IL-1β, IL-6, TNF-α, IFN-γ, IL-4, IL-5, IL-17A, IL-10, CXCL8, CCL3, CCL4, and CCL5 (Fig. 3). CCL5 were significantly low at D30 but appeared to increase until D90, serving as early markers of return to normality.
Fig. 3.
Circulating level of soluble molecules comparing HD, D30, D60 and D90 groups. Data is expressed as median and interquartile range in pg/mL. Circulating levels of chemokines (A), cytokines (B) and growth factors (C). Statistical analysis was conducted with Kruskal–Wallis and Dunn’s Multiple Comparisons tests, considering significative when p < 0.05. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
The correlation matrix demonstrates that healthy donors have fewer interactions between cytokines and more interactions with circulating cells and subpopulations, while convalescent individuals exhibit more immune dynamics between molecules. Patients in the initial stages of convalescence, represented by D30, showed a greater number of interactions between molecules, mainly cytokines.
The number of correlations appears to decrease as convalescence progresses until the late stage, although it can be observed that anti-S antibodies have a positive correlation with AEC and CXCL10 in D30 (alongside anti-N antibodies) and a negative correlation with patrolling monocytes. In D60, the immunomodulation was related to inflammatory molecules such as IL-6 and CXCL8, and negatively correlated with B lymphocytes. In D90, the participation seems to be guided by pDC and CXCL8 (Fig. 4A,B). AEC showed a positive and significant correlation with both anti-N and anti-S antibodies in D30 and D90 (Fig. 4), which may be related to its functionality.
Fig. 4.
Biomarker correlation matrix indicating difference in patter of healthy donors (A), and convalescence in D30 (B), D60 (C) and D90 (D). Networks were based on Spearman’s correlation indices (r). Association was significant when p < 0.05 between all markers analyzed. Blue scale, ranging from − 1.0 to 1.0, shows correlation strength, as represented on image. WBC white blood count, ANC absolute neutrophil count, ALC absolute lymphocyte count, AMC absolute monocyte count, AEC absolute eosinophil count, ABC absolute basophil count, NK natural killer, Chemokines CXCL8, CXCL10, CCL3, CCL4, CCL2, CCL5 and CCL11, Cytokines IL-1β, IL-1ra, IL-6, TNF-α, IL-12p70, IFN-γ, IL-2, IL-7, IL-9, IL-15, IL-4, IL-5, IL-13, IL-17A, IL-10, Growth factors VEGF, FGF basic, PDGF-BB, GM-CSF, G-CSF, HD healthy donors.
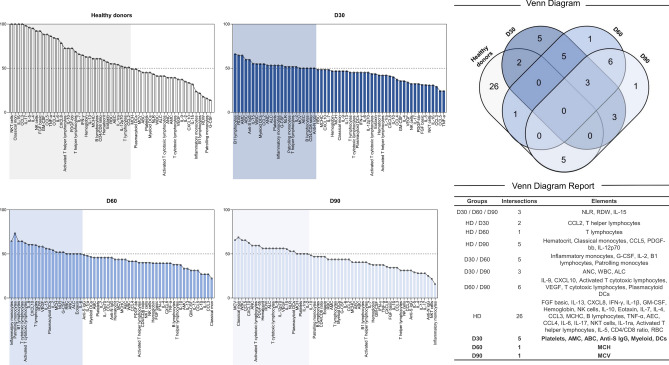
Inflammatory markers still may be used to characterize the initial stage of convalescence
Biomarker analysis and the Venn diagram revealed that convalescent individuals are typically characterized by higher levels of IL-15, NLR, and RDW. However, when we segregate the convalescent stage and inspect for biomarkers in each timepoint, we observe that in the beginning of convalescence, 30 days after clinical recovery, high levels of AMC, ABC, platelets, myeloid DCs and anti-S IgG were also observed. Furthermore, immunomodulation from the first (D30) to the second month (D60), can be observed through increased production of inflammatory and patrolling monocytes, B1 lymphocytes, G-CSF, IL-2, and the IFN-γ/IL4 ratio. Although B1 lymphocytes are elevated in circulation, only D30 was marked by a higher increase in anti-S antibodies.
Later stages of convalescence seen in our study was marked by both total and activated T cytotoxic lymphocytes, plasmacytoid DCs, VEGF, IL-9 and CXCL10. This profile suggests a reparative process, which can be related to the extensively injury caused by the immunity priming during acute phase. None of the convalescent groups exhibited higher production of anti-N antibodies (Fig. 5).
Fig. 5.
Biomarker signature of groups represented in a Venn Diagram. (A) Frequency of subjects with biomarker level above the Cut-off; (B) Venn Diagram representing the groups, intersections, and elements, suggesting potential hallmarks for immunomodulation under convalescence. Global median for each parameter was measured and used to characterize participants as low (< 50%) or higher (> 50%) producers. HD healthy donors.
Discussion
COVID-19 has impacted numerous individuals worldwide, with its peak occurring between 2020 and 2021. This period was characterized by immense strain on healthcare systems, as efforts were made to find better treatments, identify effective drugs, and conduct improved clinical trials. The pandemic situation compelled researchers to enhance their understanding of how the body responds to the virus, particularly focusing on the dynamics of convalescence and the immunological factors involved in disease recurrence, antibody production, and prevention strategies. In this study, we propose perspectives on immune dynamics from 30 to 90 days after clinical recovery, aiming to characterize the immune response in terms of cells and molecules.
Our investigation into antibody dynamics revealed that anti-S IgG antibodies persisted longer than IgM antibodies, as expected. In our study, only 32% of participants had detectable IgM levels at D30, which is lower than what has been reported in the literature26. Conversely, more than 80% of convalescents in our study exhibited detectable IgG anti-Spike antibodies, aligning with findings from other studies26–28. Importantly, IgG anti-Spike antibodies demonstrated greater stability in serum compared to IgG anti-nucleocapsid antibodies, which exhibited a rapid decline during convalescence. The seroreactivity of anti-S antibodies varied by less than 5.2% over the 3-month period following viral clearance, whereas anti-N antibodies varied by 13.4% during the same timeframe. The peak period of antibody production occurred 10–20 days after the onset of symptoms and appeared to be primarily influenced by IgG1 and IgG3 subclasses29,30. Some studies have reported that the persistence of memory B cells and circulating Tfh cells plays a significant role in the production of anti-S antibodies, while anti-N and anti-RBD antibodies show a notable decrease31–35.
Several concerns have been raised regarding the decrease in antibodies during convalescence. One of these concerns is the mutation that occurs in the germinal center at the onset of convalescence, which has been correlated with antibody titers36. It has been observed that patients who experience reinfection within a certain period do not produce antibodies, suggesting that antibody production helps prevent new cases of infection. However, this hypothesis has been questioned due to the specificity of antibodies to other SARS-CoV-2 lineages that have undergone mutations during the pandemic. Despite the reduction in antibody titers, an increase in neutralization ability has been observed34,36.
Some studies have shown a relationship between antibody production and the severity of the disease37. Patients who experience symptoms such as fever, cough, dyspnea, and pneumonia are 50 times more likely to produce higher antibody titers34,38. However, the severity of the disease is also related to viral load and the availability of viral antigens. It is worth noting that only anti-RBD antibodies have been found to increase over time in patients with a severe condition39,40.
The immunological dynamics during convalescence involve a higher participation of inflammatory and patrolling monocytes. Total monocytes were found to regulate as convalescence progresses, but the activated inflammatory subpopulations appeared to increase, including an increase in patrolling monocytes. In the literature, both inflammatory and patrolling monocytes have been reported to decrease during convalescence and stabilize their levels around 5 months12,18,41. This difference may be related to the activation of monocytes since it has been previously shown that HLA-DR expression increases during convalescence18,42,43. However, there is no available data on which subpopulations express this marker during convalescence. The higher control of inflammatory status and tissue repair may be a clear reason for this increased involvement, as the increase in both subpopulations persists for up to 2 months after viral clearance.
Convalescence is characterized by a progressive decrease in antibody titers, but it can still activate local macrophages and induce inflammation through FcγRIIa receptors, thereby inducing proinflammatory molecules. M1 macrophages have been shown to be more sensitive to activated antibodies, but M2 macrophages may also polarize to M1 upon antibody recognition. M1 macrophages have been described to produce high levels of IFN-γ, CXCL8, and CCL244–47. However, our patients showed a reduction in the first two markers during convalescence.
Even though our results demonstrated that convalescence can be marked by cytotoxicity, it appears to be regulated by T lymphocytes. NK and NKT cells showed a significant decrease, which has been previously described48–50 and may be due to the production of other cells involved in tissue repair. This same pattern was observed during the acute phase, as low NK cell counts were associated with disease severity and compromised cytotoxicity48. Our findings suggest that this compromise in NK cell counts persists during convalescence. Interestingly, both myeloid and plasmacytoid dendritic cells showed no difference, indicating that their role in viral clearance occurs rapidly and is stimulus-dependent.
The molecular profile showed that an inflammatory pattern remains predominant over anti-inflammatory markers. However, both pro-inflammatory and anti-inflammatory mediators were reduced compared to healthy donors. Immunosurveillance during convalescence is characterized by a lower production of inflammatory and reparative factors, along with intense regulation mediated by increased concentrations of CXCL10, IL-2, and G-CSF. Anti-N was downregulated by B lymphocytes in the beginning of convalescence, however, in D60, it seems that this cell population participates also in downregulation of anti-S antibodies as well, characteristic that is not seen in D90. Some immunological cells were still present in D60, but with a considerable reduction in antibody titers, seen in the fold change analysis, probably due to a decrease in inflammatory stimuli. Memory cells were shown to persist even with a reduction in antibody production, exhibiting reactivity to most viral antigens even after 6 months26,51. In later stages of convalescence, there was a loss of intensive interactions, indicating that the immune system strives to reach homeostasis a few months after viral clearance.
Throughout the entire convalescent period, there was an increase in the production of NLR. Previous studies have suggested this marker as a prognostic factor for acute patients10,20,52–54, and our findings suggest that it remains elevated for a prolonged period during convalescence. Other studies have also reported an increase in neutrophil count with activation markers even 28 days after clinical recovery12,55. Furthermore, the production of neutrophil extracellular traps (NETs) has been detected even after months17,56,57. Our findings of increased G-CSF levels may contribute to neutrophil recruitment. The literature demonstrated that neutrophil metabolic function lowered during the acute phase and further reduced during convalescence, suggesting the production of non-reactive cells during this period. Therefore, G-CSF becomes an important factor in inducing urgent granulopoiesis, particularly in the first 2 months of convalescence12,30.
All 3 months of convalescence were characterized by a higher production of IL-15. In the initial stage, there was also involvement of G-CSF and IL-2, while the last stage was marked by mainly producing IL-9 and CXCL10. Some studies have described the IL-15/IL-15RA axis as a crucial factor in the functional exhaustion, senescence, and apoptosis of NK and NKT γδ cells, which promotes rapid control of infection through cytotoxicity and antibody response during convalescence39,47,58–61.
G-CSF, IL-2, and IFN-γ are molecules involved in the cytokine storm during the acute phase of COVID-1962–65, and as observed in our study, this profile of proliferation persists in convalescence, mainly mediated by G-CSF and IL-2. This may be attributed to the functionality of repair and the migration of granulocytes from the bloodstream to the affected tissues12. The subsequent dynamics of IL-9 and CXCL10 at D60/D90 were also observed in chronic allergic diseases and are associated with the involvement of mastocytes, the induction of lymphocyte adhesion to endothelium, and bone marrow inhibition66. CXCL10 has previously been linked to acute outcomes and the prediction of improvement in COVID-19 patients, but its role in convalescence may be associated with its anti-inflammatory properties67.
The specific higher producers at D30 were AMC, ABC, myeloid DCs, platelets, and anti-S IgG antibodies. Myeloid DCs have been reported to be reduced or have limited involvement during the acute phase45,57,68,69. Antibodies were prominently produced only at D30, and we propose that this specific period of acute disease is the main source of antibody concentration.
We observed that within our convalescent groups, D30 was the optimal time for collecting convalescent plasma. Less than 50% of our patients had a sufficient antibody concentration for plasma donation, which decreased to less than 30% after 90 days. While many studies focus on evaluating the efficacy of convalescent plasma use in acute COVID-19 patients, there is limited data available to assess the factors involved in immune dynamics and antibody production, particularly in relation to obtaining convalescent plasma32,40. However, there is an urgent need to improve the collection of plasma samples for storage, considering future pandemics and severe cases.
The convalescent period, which is still lacking in comprehensive discussions, holds valuable data, especially regarding immunity and the factors contributing to antibody production. Understanding these mechanisms can help prevent new cases, reduce the recurrence of severe cases, inform vaccination strategies, and promote a better quality of life for the exposed population. We emphasize the dynamics of immune cells, soluble molecules, and particularly the maintenance of antibodies in COVID-19 convalescent patients.
Furthermore, we identified that inflammatory and patrolling monocytes, together with B1 lymphocytes, persist in the immune system after infection. Based on literature data, the cytokine storm triggered during SARS-CoV-2 infection lead to stimulation on immune system, mainly participation of neutrophils and cytotoxicity70,71. This profile was shown in our study still present in the beginning of convalescence, which immunomodulation is primarily driven by G-CSF, IL-2 and IL-15. While inflammatory markers are still present in the initial stages of convalescence, around 60 days, the profile shifts toward a proliferative pattern characterized mainly by IL-9 and CXCL10, that tend to last till 90 days of convalescence. The mechanisms and profile seem to shift towards a proliferative and reparative process, related to lymphocyte stimulation and angiogenesis (Fig. 6). Although these statements must be confirmed by future studies, we acknowledge that there is a downregulation of inflammatory process during convalescence, but still a lack on the dynamics related to improve memory on these patients.
Fig. 6.
Concluding remarks on immunological shifts related to acute (based on literature) and convalescence COVID-19 patients. DC dendritic cells, NLR neutrophil-to-lymphocyte ratio.
As well, we do comprehend that understanding the dynamic of immune system is still a lack and require more approaches to better understand which mechanisms are related to antibody dynamics, but also immunoregulation. Characterize the participants based on clinical and laboratorial scores, to evaluate the acute phase and perform a follow-up till late stages of convalescence can validate the results presented here and determine more robust biomarkers for disease progression. Due to limitation of data regarding patient’s medical history during acute phase of COVID-19, and due to our patients be mainly mild cases during acute, we were unable to determine clinical scores from our patients and therapies that were used, which we believe it could determine better conclusions and propose novel studies. Other factors during acute phase were shown to impact on immunity in convalescent conditions, and although we considered this in our analysis, we were limited regarding the therapy used72. Only two had hospital admission, and mostly couldn’t address the therapy used during symptomatic period. None of them received neutralizing antibodies against SARS-CoV-2 neither previously to infection, nor during acute phase. Some, however, were vaccinated during the study period, after inclusion, but the data were not included in the analysis.
We highlight the dynamics of immune cells, soluble molecules and specially, the maintenance of antibodies which are produced along COVID-19 convalescent patients. We identified that anti-N IgG reduced quickly after viral clearance, while anti-S IgG changes slowly during the first 3 months, being a good proposal for diagnostics and therapeutical strategies. We could also identify that after infection, inflammatory and patrolling monocytes are still present on immunity, together with B1 lymphocytes, and immunomodulation driven mainly by G-CSF, IL-2 and IL-15 in convalescence. Inflammation is still present on initial stages of convalescence, but around 60 days, the profile seems to start to change to a proliferative pattern, characterized by IL-9 and CXCL10. Our results contribute to hallmarks involved on antibody production and evaluate higher proportion of cells and soluble immune molecules under these patients. Our findings will support a better comprehension over the major hallmarks for immune sustainment and dynamics after SARS-CoV-2 infection.
Methods
Ethical statement
The participants enrolled in this study provided written consent by formal signature on the consent form. The study was submitted and approved by the Ethical Committee of Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas (CEP-HEMOAM) under processes of #4.126.784 and #1.982.466. The protocols followed the guidelines of the Declaration of Helsinki and Resolution 466/2012 of the Brazilian National Health Council for research involving human subjects.
Participants and samples
Our study involved 51 healthy donors (HD) who were eligible blood donors recruited before the COVID-19 pandemic, on June 13th 2017 and June 30th 2017. These donors tested negative for HIV, HBV, HCV, HTLV, Chagas disease, and syphilis. They also had no clinical symptoms prior to donation. Additionally, we included 62 COVID-19 convalescent patients who had previously tested positive for RT-PCR and experienced a symptomatic period. The age range of the convalescents was 18–60 years, and they were recruited 30 days after clinical recovery. These convalescents were followed up monthly for up to 3 months, during which blood samples were collected. The convalescent group was recruited at Fundação Hospitalar de Hematologia e Hemoterapia do Amazonas (HEMOAM) in Manaus, Amazonas, Brazil, between July 20th 2020 and June 2nd 2021. These participants were identified based on a positive RT-PCR test and were invited to participate 30 days after clinical recovery, following the resolution of acute symptoms developed during COVID-19 phase (i.e. fever, cough fatigue or muscle aches), and when eligible for blood donation. Male gender was preferred for participation due to convalescent plasma collection, although women were also included but showed no significant differences in laboratory parameters. No other distinctions were made.
The convalescent group consisted of 62 patients at enrollment (D30), which completed all three follow-ups. As some individuals received the COVID-19 vaccine during the follow-up period, their results were not included in the analysis. Therefore, data from only 48 patients are reported at D60, and 47 at D90.
Patients who were asymptomatic for COVID-19, symptomatic at the time of recruitment, taking drugs that could inhibit ACE, had anti-erythrocyte antibodies, received any COVID-19 vaccine prior to inclusion, were pregnant, or were indigenous were not included in the study. Patients who tested positive for RT-PCR during recruitment or showed seroreactivity to other infectious diseases (HIV, HBV, HCV, syphilis, Chagas disease, or HTLV) were excluded. Samples obtained from patients who received the vaccine during the follow-up period were not included in the analysis.
Sociodemographic and clinical data collected from all participants included gender, age, ethnicity, blood type, body mass index (BMI), duration of symptoms during acute COVID-19, and the need for hospitalization and mechanical ventilation. Blood samples were collected through venipuncture, with 4 mL of whole blood collected in EDTA (BD Vacutainer® EDTA K2) tubes and 5 mL collected in separator gel (Gel BD SST® II Advance) tubes. Serum samples were stored at − 80 °C for further procedures.
Blood count and immune cell analysis
Fresh whole blood was used to measure the parameters of red and white blood cells (RBC and WBC, respectively) and platelets. This was done using an automatic hematological blood counter (ADVIA 2120i, Siemens, USA) located at HEMOAM. Immunophenotyping of immune cells was also performed at HEMOAM using the following antibodies: anti-CD3 (PERCP), anti-CD4 (FITC), anti-CD8 (PE), and anti-CD69 (APC) to identify total T lymphocytes (CD3+), T helper lymphocytes (CD3+CD4+), activated T helper lymphocytes (CD3+CD4+CD69+), T cytotoxic lymphocytes (CD3+CD8+), and activated T cytotoxic lymphocytes (CD3+CD8+CD69+); anti-CD5 (FITC) and anti-CD19 (PE) to identify B (CD19+) and B1 (CD19+CD5+) lymphocytes; anti-CD16 (FITC), anti-CD14 (APC), and anti-HLA-DR (PE) to classical (CD14+CD16-), inflammatory (CD14+CD16+), and patrolling monocytes (CD14lowCD16+); anti-CD123 (FITC), anti-CD11c (PE), and anti-CD14 (APC) to myeloid (CD14−CD123+) and plasmacytoid (CD14−CD11c+) dendritic cells; and anti-CD3 (PERCP), anti-CD16 (FITC), and anti-CD56 (PE) to NK (CD3−CD56+CD16+) and NKT (CD3+CD56+CD16+) cells (Fig. S2). The antibodies were purchased from BD Biosciences (San Diego, CA, USA), Beckman Coulter (Brea, California, USA), and BioLegend (San Diego, CA, USA).
A sample of 100 µL of whole blood was incubated with the respective antibodies, followed by a lysis solution at room temperature. The cells were washed, resuspended in PBS, and then stored at 4 °C until they were acquired by flow cytometry within the next 24 h. A total of 30,000 events were acquired using the FACSCanto II flow cytometer at Fundação HEMOAM. The analysis was conducted using FlowJo Software v. 10.8 to define subpopulations based on morphometric characteristics and fluorescence from monoclonal antibodies. The gates and strategies used are indicated in Fig. S1. The percentage of cells was used for statistical analysis.
Chemokine, cytokine, and growth factor assay
Molecules were measured using the Luminex technique. Cytokines IL-1β, IL-1ra, IL-6, TNF-α, IL-12p70, IFN-γ, IL-2, IL-7, IL-9, IL-15, IL-4, IL-5, IL-13, IL-17, and IL-10; chemokines CXCL8, CXCL10, CCL3, CCL4, CCL2, CCL5, and CCL11; and growth factors VEGF, FGF-basic, PDGF, GM-CSF, and G-CSF were measured in serum samples at Instituto René Rachou (FIOCRUZ-MG). The procedure was conducted using the Bioplex-Pro Human Cytokine 27-Plex Kit (Bio-Rad, California, USA) following the manufacturer's instructions and protocol. Acquisition and concentration measurements were performed on a Luminex 200 System and analyzed using Bioplex Manager Software with Five Parameters Logistic Regression. The concentration is expressed in pg/mL. The detection limits for the molecules were as follows: CXCL8 = 42,150 pg/mL; CXCL10 = 31,236 pg/mL; CCL2 = 24,282 pg/mL; CCL3 = 960 pg/mL; CCL4 = 11,233 pg/mL; CCL5 = 16,533 pg/mL; CCL11 = 26,842 pg/mL; IL-1β = 8608 pg/mL; IL-1ra = 91,661 pg/mL; IL-2 = 18,297 pg/mL; IL-9 = 25,642 pg/mL; IL-15 = 22,328 pg/mL; IL-4 = 4789 pg/mL; IL-5 = 23,105 pg/mL; IL-6 = 37,680 pg/mL; IL-7 = 16,593 pg/mL; IL-10 = 35,170 pg/mL; IL-12p70 = 37,684 pg/mL; IL-13 = 8090 pg/mL; IL-17A = 28,850 pg/mL; IFN-γ = 25,411 pg/mL; TNF-α = 64,803 pg/mL; PDGF-BB = 24,721 pg/mL; FGFb = 16,046 pg/mL; G-CSF = 40,049 pg/mL; GM-CSF = 12,844 pg/mL; and VEGF = 29,464 pg/mL.
Antibody measurement
Antibodies IgG anti-Spike (anti-S) and anti-nucleocapsid (anti-N) were measured using chemiluminescence with the Chemiluminescence Microparticle Immunoassay (CMIA) by Abbott test on Architect. The procedures were conducted based on the manufacturer's instructions. A qualitative test was conducted to measure anti-N IgG, and the result was obtained in Index (S/C). It was considered positive when the Index was greater than 1.4. Quantitative data of Optical Density (OD) were also recorded and used for statistical analysis. The qualitative-quantitative test of IgG anti-S was measured, and the concentration is expressed in AU/mL. The concentration was used for statistical analysis, and according to the manufacturer's instructions, samples were considered positive when the concentration was greater than 50 AU/mL. All procedures were performed on the Architect equipment, located in the serology department of Fundação HEMOAM.
Statistical analysis
All data were stored in Microsoft Excel for further analysis in GraphPad Prism v. 8.0 (San Diego, CA, USA). The Shapiro-Wilk normality test was performed to evaluate the normality of the parameters, and the median and interquartile range (IQR) (25th and 75th percentiles) were acquired. Sociodemographic data are expressed as absolute values and percentages. Analysis of these parameters were done by Mann–Whitney test, while categorical parameters were compared with Chi-square test. Heatmap and Principal Component Analysis (PCA) were performed in R Studio v.2023.12.1 software (Project for Statistical Computing Version 3.0.1). The median of cells and molecules from all groups was compared using the Kruskal–Wallis and Dunn's Multiple Comparison tests. A confidence interval of 95% was considered, and statistical values were used when p < 0.05.
The correlation analysis was performed using Spearman correlation test in GraphPad Prism v. 8.0 software (San Diego, CA, USA) for all cells, cell subtypes, and molecules within each group. Significant results were used to construct biomarker correlation matrices based on the Spearman correlation coefficient (r). Significance was identified when the p-value was < 0.05.
The median value of each parameter was calculated across all groups and used as a cutoff point to categorize each participant as a "high" or "low" producer, as described previously. Median values for each parameter used are: RBC = 4.91 106/µL; hemoglobin = 14.50 g/dL; hematocrit = 43%; MCV = 88.1 fL; MCH = 29.70 pg; RDW = 14.10%; WBC = 6.53 103/µL; ANC = 3.77 103/µL; ALC = 1.95 103/µL; AMC = 0.39 103/µL; AEC = 0.18 103/µL; ABC = 0.03 103/µL; platelet = 248 103/µL; classical monocytes = 63.90%; inflammatory monocytes 5.60%; patrolling monocytes = 8.20%; T lymphocytes = 67.85%; T helper lymphocytes = 38.70%; activated T helper lymphocytes = 1.07%; cytotoxic T lymphocytes = 25.15%; activated cytotoxic T lymphocytes = 1.82%; NK cells = 5.24%; NKT cells = 0.15%; mDCs = 0.25%; pDCs = 0.47%; B lymphocytes = 11.60%; B1 lymphocytes = 1.16%; VEGF = 6.61 pg/mL; FGF basic = 1.38 pg/mL; PDGF-BB = 141.40 pg/mL; GM-CSF = 4.52 pg/mL; G-CSF = 3.94 pg/mL; IL-1β = 0.37 pg/mL; IL-1ra = 21.59 pg/mL; IL-6 = 0.20 pg/mL; TNF-α = 5.59 pg/mL; IL-12p70 = 1.97 pg/mL; IFN-γ = 13.04 pg/mL; IL-2 = 0.48 pg/mL; IL-7 = 1.33 pg/mL; IL-9 = 4.07 pg/mL; IL-15 2.36 pg/mL; IL-4 = 0.18 pg/mL; IL-5 = 0.69 pg/mL; IL-13 = 0.38 pg/mL; IL-17A = 2.87 pg/mL; IL-10 = 1.41 pg/mL; CXCL8 = 0.98 pg/mL; CXCL10 = 52.00 pg/mL; CCL3 = 0.22 pg/mL; CCL4 = 4.70 pg/mL; CCL2 = 7.56 pg/mL; CCL5 = 28.05 pg/mL; CCL11 = 15.22 pg/mL; anti-N IgG (OD) = 4.40 Index; anti-S IgG = 872.70 AU/mL. Percentage values were then obtained from these groups, and parameters that reached more than the 50th percentile were characterized as higher producers, represented in a graphic biomarker signature. Each parameter was isolated for each group and presented in a Venn Diagram, showing the respective groups, intersections, and elements. This analysis was conducted using a public website (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Linear regression was calculated to identify the relationship between the number of days from the end of symptoms and the concentration of both anti-S IgG (AU/mL) and anti-N IgG (OD), obtained by the CMIA method. A significant value was considered when p < 0.05. To evaluate antibody concentration, qualitative values (positive/negative) were also calculated from immunochromatographic tests (IgM and IgG anti-S) and CMIA (IgG anti-S and anti-N). Percentage values were used to calculate the seroconversion rate for all convalescent groups. A fold change analysis was also performed using the mean value of the concentration of anti-S IgG from participants at D30/D60, D30/D90, and D60/D90, compared using the One-Way ANOVA and Tukey’s Multiple Comparison test. All methodological procedures are described in Fig. S3.
Supplementary Information
Acknowledgements
Authors would like to extend their gratitude to all staff from laboratory of Genomics, from Fundação HEMOAM, for their support to this work. Financial support was provided in the form of grants from Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM) (Pró-Estado Program - #002/2008, #007/2018, #005/2019, PCTI-EMERGESAÚDE - AM Program - #005/2020 and POSGRAD Program - #002/2023 and #002/2024), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (INCT-Sangue Process - #405918/2022-4) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (PROCAD-Amazônia 2018 Program - #88881.200581/2018-01 and PDPG-CONSOLIDACAO-3-4 Program #88887.707248/2022-00). ALS-J, LSO, SD, TCCC, LAX and FAS-H have fellowships from CAPES and FAPEAM (PhD and SI students). LHF, EC and AT-C are level 1 research fellows from CNPq. OM-F is a level 1 research fellow from CNPq and a research fellow from the program supported by the Universidade do Estado do Amazonas (PROVISIT No. 005/2023-PROPESP/UEA). CAS, AGC and AM are level 2 research fellows from CNPq. The funders had no role in study design and decision to publish, or preparation of the manuscript.
Author contributions
A.L.S-J., L.S.O., S.D. and W.L.L.N. performed laboratorial procedures of flow cytometry, A.L.S.-J., T.C.C.C. and L.A.X. recruited and collected all data and blood samples, F.S.A.-H. and C.M.M.A. measured the antibodies assays, A.L.S.-J., M.A.E.C., D.M.T. P.V.S.N., M.S.S.C., A.M.T., N.A.F., A.G.C. and A.M. conceptualized the study and analyzed the data, A.L.S.-J. wrote the main manuscript, A.M.T., L.H.F., C.A.S., E.C.S., A.T.C., O.A.M.-F., A.G.C. and A.M. supervised and revised the final manuscript, M.A.E.C., N.A.F.,L.H.C., C.A.S., E.C.S., A.T.C., O.A.M.-F., A.G.C. and A.M. provided funding acquisition. All authors have read and agreed to the published version of the manuscript.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Allyson Guimarães Costa, Email: allyson.gui.costa@gmail.com.
Adriana Malheiro, Email: malheiroadriana@yahoo.com.br.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71419-x.
References
- 1.Cui, J., Li, F. & Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol.17, 181–192 (2019). 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dousari, A. S., Moghadam, M. T. & Satarzadeh, N. COVID-19 (Coronavirus Disease 2019): A new coronavirus disease. Infect. Drug Resist.13, 2819–2828 (2020). 10.2147/IDR.S259279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang, Y.-Z. & Holmes, E. C. A Genomic perspective on the origin and emergence of SARS-CoV-2. Cell181, 223–227 (2020). 10.1016/j.cell.2020.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison, A. G., Lin, T. & Wang, P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol.41, 1100–1115 (2020). 10.1016/j.it.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kevadiya, B. D. et al. Diagnostics for SARS-CoV-2 infections. Nat. Mater.20, 593–605 (2021). 10.1038/s41563-020-00906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan, A. H. et al. COVID-19 transmission, vulnerability, persistence and nanotherapy: A review. Environ. Chem. Lett.19, 2773–2787 (2021). 10.1007/s10311-021-01229-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cevik, M., Kuppalli, K., Kindrachuk, J. & Peiris, M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ371, 1–6 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Lamers, M. M. & Haagmans, B. L. SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol.20, 270–284 (2022). 10.1038/s41579-022-00713-0 [DOI] [PubMed] [Google Scholar]
- 9.Buss, L. et al. Predicting SARS-CoV-2 variant spread in a completely seropositive population using semi-quantitative antibody measurements in blood donors. Vaccines10, 1–11 (2022). 10.3390/vaccines10091437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leppkes, M. et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine58, 9 (2020). 10.1016/j.ebiom.2020.102925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai, J. et al. The neutrophil-to-lymphocyte ratio determines clinical efficacy of corticosteroid therapy in patients with COVID-19. Cell Metab.33, 258–269 (2021). 10.1016/j.cmet.2021.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwiecień, I. et al. Neutrophil maturation, reactivity and granularity research parameters to characterize and differentiate convalescent patients from active SARS-CoV-2 infection. Cells10, 1–18 (2021). 10.3390/cells10092332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lourda, M. et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. Proc. Natl. Acad. Sci.118, 1–12 (2021). 10.1073/pnas.2109123118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil-Manso, S. et al. Multidimensional analysis of immune cells from COVID-19 patients identified cell subsets associated with the severity at hospital admission. PLoS Pathog.19, 1–23 (2023). 10.1371/journal.ppat.1011432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens, R. J. H. et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the sysmex XN-10 hematology analyzer. Clin. Chem. Lab. Med.59, 783–793 (2021). 10.1515/cclm-2020-1529 [DOI] [PubMed] [Google Scholar]
- 16.Reyes, L. et al. A type I IFN, prothrombotic hyperinflammatory neutrophil signature is distinct for COVID-19 ARDS. Wellcome Open Res.6, 1–42 (2021). 10.12688/wellcomeopenres.16584.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng, H. et al. Circulating markers of Neutrophil Extracellular Traps are of prognostic value in patients with COVID-19. Arterioscler. Thromb. Vasc. Biol.41, 988–994 (2021). 10.1161/ATVBAHA.120.315267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajamanickam, A. et al. Dynamic alterations in monocyte numbers, subset frequencies and activation markers in acute and convalescent COVID-19 individuals. Sci. Rep.11, 1–9 (2021). 10.1038/s41598-021-99705-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vitte, J. et al. A granulocytic signature identifies COVID-19 and its severity. J. Infect. Dis.222, 1985–1996 (2020). 10.1093/infdis/jiaa591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez, L. et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep. Med.1, 100078 (2020). 10.1016/j.xcrm.2020.100078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winheim, E. et al. Impaired function and delayed regeneration of dendritic cells in COVID-19. PLoS Pathog.17, 1–30 (2021). 10.1371/journal.ppat.1009742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva-junior, A. L. et al. Immune dynamics involved in acute and convalescent COVID-19 patients. Immuno3, 86–111 (2023). 10.3390/immuno3010007 [DOI] [Google Scholar]
- 23.WHO. Coronavirus disease (COVID-19) outbreak. https://www.who.int (2020).
- 24.Juskewitch, J. E., Stubbs, J. R. & Gandhi, M. J. Elevated rate of HLA antibodies in male COVID-19 convalescent plasma donors: A risk factor for transfusion-related acute lung injury. Mayo Clin. Proc.96, 500–502 (2021). 10.1016/j.mayocp.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yiğenoğlu, T. N. et al. Convalescent plasma therapy in patients with COVID-19. J. Clin. Apher.35, 367–373 (2020). 10.1002/jca.21806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dan, J. M. et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science371, eabf4063 (2021). 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bošnjak, B. et al. Low serum neutralizing anti-SARS-CoV-2 S antibody levels in mildly affected COVID-19 convalescent patients revealed by two different detection methods. Cell. Mol. Immunol. Mol. Immunol.18, 936–944 (2020). 10.1038/s41423-020-00573-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang, T. et al. Declining levels of neutralizing antibodies against SARS-CoV-2 in convalescent COVID-19 patients one year post symptom onset. Front. Immunol.12, 1–10 (2021). 10.3389/fimmu.2021.708523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, D.-M. et al. Eosinophil-mediated lung inflammation associated with elevated natural killer T cell response in COVID-19 patients. Korean J. Intern. Med.37, 201–209 (2022). 10.3904/kjim.2021.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, D.-M. et al. Enhanced eosinophil-mediated inflammation associated with antibody and complement-dependent pneumonic insults in critical COVID-19. Cell Rep.37, 1–14 (2021). 10.1016/j.celrep.2021.109798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung, J. H. et al. SARS-CoV-2-specific T cell memory is sustained in COVID-19 convalescent patients for 10 months with successful development of stem cell-like memory T cells. Nat. Commun.12, 1–12 (2021). 10.1038/s41467-021-24377-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson, R. W. et al. SARS-CoV-2 epitope-specific CD4+ memory T cell responses across COVID-19 disease severity and antibody durability. Sci. Immunol.7, abl9464 (2022). 10.1126/sciimmunol.abl9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prete, C. A. et al. SARS-CoV-2 antibody dynamics in blood donors and COVID- 19 epidemiology in eight Brazilian state capitals: A serial crosssectional study. Elife11, 1–53 (2022). 10.7554/eLife.78233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokal, A. et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell184, 1201–1213 (2021). 10.1016/j.cell.2021.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chansaenroj, J. et al. Long-term persistence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein-specific and neutralizing antibodies in recovered COVID-19 patients. PLoS One17, 1–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaebler, C. et al. Evolution of antibody immunity to SARS-CoV-2. Nature591, 639–644 (2021). 10.1038/s41586-021-03207-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, F. et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. Lancet Infect. Dis.152, 82–87 (2020). [Google Scholar]
- 38.Wardhani, S. O. et al. The predictors of high titer of anti-SARS-CoV-2 antibody of convalescent plasma donors. Clin. Epidemiol. Glob. Health11, 1–4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreras, C. et al. SARS-CoV-2-specific memory T lymphocytes from COVID-19 convalescent donors: Identification, biobanking, and large-scale production for adoptive cell therapy. Front. Cell Dev. Biol.9, 1–12 (2021). 10.3389/fcell.2021.620730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, R. et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity53, 864-877.e5 (2020). 10.1016/j.immuni.2020.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, D. et al. COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol.1, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neeland, M. R. et al. Innate cell profiles during the acute and convalescent phase of SARS-CoV-2 infection in children. Nat. Commun.10.1038/s41467-021-21414-x (2021). 10.1038/s41467-021-21414-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin, S. et al. Dynamic changes in monocytes subsets in COVID-19 patients. Hum. Immunol.82, 170–176 (2021). 10.1016/j.humimm.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv, J. et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov.7, 1–12 (2021). 10.1038/s41421-021-00258-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niles, M. A. et al. Macrophages and dendritic cells are not the major source of pro-inflammatory cytokines upon SARS-CoV-2 infection. Front. Immunol.12, 1–12 (2021). 10.3389/fimmu.2021.647824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoepel, W. et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med.13, 1–17 (2021). 10.1126/scitranslmed.abf8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, F. et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct. Target. Ther.5, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taghiloo, S. et al. Apoptosis and immunophenotyping of peripheral blood lymphocytes in Iranian COVID-19 patients: Clinical and laboratory characteristics. J. Med. Virol.93, 1589–1598 (2021). 10.1002/jmv.26505 [DOI] [PubMed] [Google Scholar]
- 49.Antonioli, L., Fornai, M., Pellegrini, C. & Blandizzi, C. NKG2A and COVID-19: Another brick in the wall. Cell. Mol. Immunol.17, 672–674 (2020). 10.1038/s41423-020-0450-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leem, G. et al. Abnormality in the NK-cell population is prolonged in severe COVID-19 patients Galam information. J. Allergy Clin. Immunol.148, 996-1006.e18 (2020). 10.1016/j.jaci.2021.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sosa-Hernández, V. A. et al. B cell subsets as severity-associated signatures in COVID-19 patients. Front. Immunol.11, 1–12 (2020). 10.3389/fimmu.2020.611004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asghar, M. S. et al. Hematological parameters predicting severity and mortality in COVID-19 patients of Pakistan: A retrospective comparative analysis. J. Community Hosp. Intern. Med. Perspect.10, 514–520 (2020). 10.1080/20009666.2020.1816276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Man, M. A. et al. Neutrophil-to-Lymphocyte Ratio, platelets-to-lymphocyte ratio, and eosinophils correlation with high-resolution computer tomography severity score in COVID-19 patients. PLoS One16, 1–12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taj, S. et al. Role of hematological parameters in the stratification of COVID-19 disease severity. Ann. Med. Surg.62, 68–72 (2021). 10.1016/j.amsu.2020.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chao, Y. et al. Distinct phenotypes of platelet, monocyte, and neutrophil activation occur during the acute and convalescent phase of COVID-19. Platelets32, 1092–1102 (2021). 10.1080/09537104.2021.1921721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Middleton, E. A. et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood136, 1169–1179 (2020). 10.1182/blood.2020007008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parackova, Z. et al. Disharmonic inflammatory signatures in COVID-19: Augmented neutrophils’ but impaired monocytes’ and dendritic cells’ responsiveness. Cells9, 1–17 (2020). 10.3390/cells9102206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masselli, E. & Vitale, M. NK cells on the ViP stage of COVID-19. EBioMedicine69, 103458 (2021). 10.1016/j.ebiom.2021.103458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Notarbartolo, S. et al. Integrated longitudinal immunophenotypic, transcriptional, and repertoire analyses delineate immune responses in patients with COVID-19. Sci. Immunol.5021, 1–19 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Flament, H. et al. Outcome of SARS-CoV-2 infection is linked to MAIT cell activation and cytotoxicity. Nat. Immunol.22, 322–335 (2021). 10.1038/s41590-021-00870-z [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J. Y. et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol.21, 1107–1118 (2020). 10.1038/s41590-020-0762-x [DOI] [PubMed] [Google Scholar]
- 62.Varchetta, S. et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol.18, 604–612 (2021). 10.1038/s41423-020-00557-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, Y. et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J. Allergy Clin. Immunol.146, 119–127 (2020). 10.1016/j.jaci.2020.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng, M. et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol.10.1038/s41423-020-0402-2 (2020). 10.1038/s41423-020-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burgos-Blasco, B. et al. Hypercytokinemia in COVID-19: Tear cytokine profile in hospitalized COVID-19 patients. Exp. Eye Res.200, 1–6 (2020). 10.1016/j.exer.2020.108253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abbas, A. K., Lichtman, A. H. & Pillai, S. Cellular and Molecular Immunology Vol. 9 (Elsevier, 2018). [Google Scholar]
- 67.Wu, Z. et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis.3099, 1–9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arunachalam, P. S. et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science369, 1–11 (2020). 10.1126/science.abc6261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, G. et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov.6, 1–14 (2020). 10.1038/s41421-020-00225-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu, G., Zhang, S., Hu, H., Liu, T. & Huang, J. The role of neutrophil-lymphocyte ratio and lymphocyte–monocyte ratio in the prognosis of type 2 diabetics with COVID-19. Scott. Med. J.65, 154–160 (2020). 10.1177/0036933020953516 [DOI] [PubMed] [Google Scholar]
- 71.Zuo, Y. et al. Neutrophil extracellular traps and thrombosis in COVID-19. J. Thromb. Thrombolysis51, 446–453 (2021). 10.1007/s11239-020-02324-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rotundo, S. et al. Spike-specific T-cell responses in patients with COVID-19 successfully treated with neutralizing monoclonal antibodies against SARS-CoV-2. Int. J. Infect. Dis.124, 55–64 (2022). 10.1016/j.ijid.2022.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.