Abstract
Hepatolenticular degeneration (HLD), also known as Wilson's disease (WD), is a rare autosomal recessive disorder regarding copper metabolism. Whether gut microbiota imbalance is involved in developing HLD remains unknown. A comprehensive 16S rRNA amplicon sequencing, metagenomic sequencing, and metabonomic analysis were undertaken in patients with WD to analyze the composition and function profiles of gut microbiota in patients with WD. The data demonstrated differences in gut microbiota and metabolic pathways between WD patients and normal individuals, significantly decreasing bacterial richness and diversity. The levels of Selenomonaceae and Megamonas in WD patients are significantly higher than those in healthy individuals. The relative abundances of Roseburia inulinivorans in patients with WD are lower than in healthy individuals. Compared with healthy people, the level of metabolites in patients with WD is abnormal. Leucylproline, 5-Phenylvaleric Acid and N-Desmethylclobazam, which have nutritional and protective effects, are significantly reduced fecal metabolites in patients with WD. D-Gluconic acid, which can chelate metal ions, may be a potential treatment for WD. The positive correlation it demonstrates with Alistipes indistinctus and Prevotella stercora indicates potential bacteria able to treat WD. These metabolites are mainly related to the biosynthesis of antibiotics, alpha-linolenic acid metabolism, one carbon pool by folate, nicotinate and nicotinamide metabolism. In conclusion, the data from this study elucidate novel mechanisms describing how abnormal gut miccrobiota contribute to the pathogenesis of WD and outlines new molecules for the treatment of WD.
Keywords: Hepatolenticular degeneration, Wilson's disease, Gut microbiota, 16s rRNA sequencing, Metagenomics, Metabolomics
Subject terms: Microbiology, Gastroenterology
Introduction
Hepatolenticular degeneration (HLD) is an autosomal recessive disorder of copper metabolism caused by mutations in the ATP7B gene on chromosome 13, resulting in ATP7B functional defects. It is characterized by insufficient binding of copper to ceruloplasmin, impaired bile copper excretion, and accumulation of copper in the liver, brain, kidney, cornea, and other body organs. The clinical manifestations of this disease are diverse and can manifest varying degrees of severity. Currently, there is no radical cure for WD, and once diagnosed, a lifelong low copper diet and copper resistance treatment are required1. Early diagnosis and corrective treatments can improve the prognosis of hepatolenticular degeneration. Conversely, WD may develop into end-stage liver disease or severe motor dysfunction, seriously affecting the quality of life of patients2,3.
Gut microbiota can affect human health by providing critical benefits for developing the immune system, preventing infection, nutritional access, and neurological function4,5. Since Marshall proposed the concept of the intestinal liver axis in 1998, research on the relationship between the intestinal tract and liver diseases has attracted increasing attention6. The liver interacts directly with the intestinal tract through the hepatic hilus and bile secretion system7. During gut microbiota colonization, their metabolites are obtained through self-transformation, such as short-chain fatty acids (SCFAs), secondary bile acids, and ionic polysaccharide A (PSA). These metabolites directly activate intestinal epithelial cell receptors through the mucosal layer, such as the activation of signal pathways mediated by G protein-coupled receptors (GPRs) in intestinal epithelial cells, which stimulate the transcription process of regulatory T cells (Treg cells), promote the synthesis and release of anti-inflammatory cytokines, and improve the immune tolerance of the organism. Such metabolite receptors exist in different intestinal epithelial cells, indicating that metabolites act widely on the immune process of the body8. Therefore, gut microbiota and its metabolites are crucial in regulating the host immune system. Hepatolenticular degeneration (HLD) occurs with decreased liver function, which destroys the intestinal mucosal barrier, ultimately leading to internal environmental disturbances and a series of symptoms9. Targeting the intestinal tract to treat hepatolenticular degeneration may improve symptoms by, for example, regulating gut microbiota, avoiding the translocation of intestinal bacteria and their products to the liver, activating immune cells, producing inflammatory factors, inhibiting the liver immune response, delaying the progression of hepatitis, and cirrhosis.
Previous studies have shown that the diversity and composition of gut microbiota in WD patients are significantly lower than in healthy individuals, and there is a significant difference in gut microbiota composition between the two groups10,11. However, metagenomic sequencing and metabolomic analysis have not previously been undertaken. Whether it is possible for specific bacteria or metabolites to treat WD is still unknown. Therefore, this study conducted a comprehensive 16S rRNA amplicon sequencing, metagenomic sequencing, and metabonomic analysis of healthy individuals and WD patients to analyze the composition and function of intestinal microorganisms and characterize the metabonomic stool characteristics of WD patients. A correlation analysis was conducted between fecal metabolites and individual bacteria to find the relationship between gut microbiota functions and their metabolites and WD, explore potential bacterial biomarkers, and develop possible new therapeutic strategies.
Materials and methods
Ethics approval
All experimental protocols were approved by Shenzhen Hospital of the University of Chinese Academy of Sciences and all methods were carried out in accordance with relevant guidelines and regulations. Each patient signed a written informed consent form before being included in the study.
Study subject recruitment and fecal sample collection
Fecal samples were obtained from 11 confirmed WD patients and 20 healthy participants for subsequent 16S rRNA sequencing, metagenomic sequencing, and metabolomic analysis. The ATP7B gene confirms the diagnosis of WD and meets clinical diagnostic criteria, including family history, clinical manifestations, neurological examinations, low serum ceruloplasmin levels, high 24-h urinary copper excretion, liver function testing, liver ultrasound, and magnetic resonance imaging (MRI) of the brain. Patients with the following conditions are excluded: respiratory or renal failure, congestive heart disease, and severe liver dysfunction. The control group comprised 20 healthy participants who took no probiotics or antibiotics within the first month of inclusion. The clinical characteristics of all subjects are summarized in Table 1, and the participant demographics, such as age, gender, are comparable (P > 0.05). Fresh stool samples from all participants were collected and immediately frozen in a refrigerator at − 80 °C until extracted.
Table 1.
Descriptive data of included subjects in the study.
| Control (n = 20) | LC (n = 11) | P values | |
|---|---|---|---|
| Age (year) | 24.5 ± 5.0 | 27.2 ± 4.1 | 0.1777 |
| Gender | |||
| Female | 10 | 8 | 0.2755 |
| Male | 10 | 3 | |
| Urinary copper excretion (μg/24 h) | 605.78 ± 540.67 | 23.82 ± 7.96 | 0.0051 |
Fecal sample collection
As instructed, fecal samples were collected into sterile Eppendorf tubes and immediately transported at low temperatures. The frozen feces are transported to the Shenzhen Hospital of the University of Chinese Academy of Sciences using dry ice. After receiving a stool sample, they were all stored at − 80 °C until extraction.
DNA extraction and sequencing
The genomic DNA was extracted using the CTAB method. Then the purity and concentration of DNA are detected using agarose gel electrophoresis, and an appropriate amount of sample DNA is taken into the centrifuge tube. The 16S rRNA gene sequence was amplified by PCR using primers corresponding to the 16S V3V4 region (341F: CCTAYGGGRBGCASCAG and 806R: GGACTACNNGGGTATCTAAT). Using TruSeq ®, the DNA PCR-Free Sample Preparation Kit was used to construct the library. After the library was qualified, NovaSeq6000 was used for machine sequencing.
16S rRNA sequencing data analysis
Sequences analysis was performed using the Uparse algorithm12. The sequence is clustered by default into OTUs (Operational Taxonomic Units) with 97% identity. Species annotations were analyzed using the Mothur method and SILVA138 SSUrRNA database. QIIME software was used to obtain species classification information corresponding to each feature, the composition of each sample community, and estimate α and β diversity index. Dominant bacteria were assessed using linear discriminant analysis (LDA) to detect community difference between groups.
Metagenomic Analysis
Microbial DNA was fragmented, metagenomic sequencing was performed, the clean raw reads were then assembled. Trimmomatic software13 was used to filter raw tags and get high-quality sequencing data. MMseqs2 software14 was used to remove redundancy, and the similarity threshold and coverage threshold were set to 95 and 90%, respectively. The functional profiles were analyzed using KEGG (Kyoto Encyclopedia of Genes and Genomes) database, eggNOG database, and CAZyme (carbohydrate-active enzyme) database.
Fecal metabolic analysis
Non-targeted metabonomic analysis was performed using LC/MS methods. The experimental process mainly includes sample metabolite extraction, LC–MS/MS detection, and data analysis. In short, chromatographic separation is performed on the Vanquish UHPLC system, and mass spectrometry detection uses the Q Exactive ™ HF-X system(Thermo Scientific). The offline data (. raw) file is imported into the CD search software for further data analysis, finally obtaining data identification and quantitative results.
Statistical analysis
Statistical analysis was conducted using R software (Version 2.15.3), using T-tests and Wilcox tests. In all analyses, values with P < 0.05 were considered to have statistically significant differences between the groups.
Results
Comparison of gut microbiota composition between C group and WD group
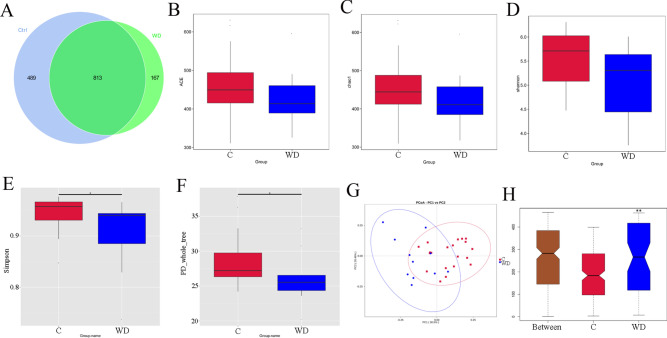
To determine whether intestinal microbial changes are related to WD, 16S rRNA was sequenced in 20 healthy individuals and 11 patients with Wilson's disease to determine the diversity and composition of fecal bacterial communities in the C and WD groups (Fig. 1). The total number of OTUs between the C group and the WD group is 813, the number of unique OTUs for the C group is 489, and the number of unique OTUs for the WD group is 167 (Fig. 1A). Alpha diversity statistics showed that there is no significant difference in the ACE index (P = 0.258), chao1 index (P = 0.261), and Shannon index (P = 0.07). However, there is a significant difference in the Simpson index (P = 0.0429) and PD_whole_tree-two index (P = 0.0487) between the two groups (P < 0.05). The species richness and diversity of the C group are significantly higher than the WD group (Fig. 1B–F). Principal Coordinate Analysis (PCoA) results show differences in gut microbiota composition between groups (Fig. 1G). Analysis of similarities (ANOSIM) showed significant differences between the groups (P = 0.007) (Fig. 1H).
Fig. 1.
Comparison of fecal microbial community structure between C and WD groups. (A) Venn diagram based on out, (B) ACE, (C) Chao1, (D) Shannon, (E) Simpon, (F) PD_whole _tree, (G) PCoA, and (H) Analysis of differences between anosim groups, *P < 0.05,**P < 0.01.
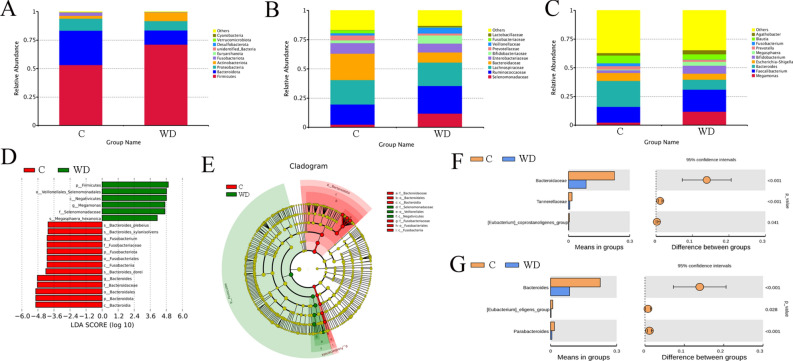
Differences between the C and WD groups in the primary classification of fecal microbiota (Fig. 2) were also reported in this study. Overall, the results of species annotation demonstrate gut microbiota in 20 phyla, 136 families, and 274 genera have been inferred. At the phyla level, the abundances of Firmicutes, Actinobacteriota and Campylobacterota in the WD group are higher than in the C group. The abundance of Bacteroidota, Proteobasteria, and Fusobasteriota in the WD group is lower than in the C group (Fig. 2A). At the family level, Bacteroidaceae and Lachnospiraceae are most abundant in the C group, followed by Ruminoccaceae. In contrast, Lachnospiraceae are the most abundant in the WD group (Fig. 2B). At the genus level, Bacteroides and Faecalibacterium are the dominant bacteria in the C group, while Faecalibacterium and Megamonas dominate in the WD group. The abundance of Bacteroides decreases in the WD group (Fig. 2C). Linear discriminant analysis (LDA) distribution maps show significant differences in the abundance of 19 species between the C and WD groups. The abundance of p_Firmicutes, f_Selenomonadaceae, and g_Megamonas are increased in the WD group. However, the levels of p_Bacteroidota, p_Fusobacteriota, f_Bacteroidaceae, g_Bacteroides and others decreased significantly in the WD group (Fig. 2D). The evolutionary branch diagram shows that Selenomonaceae plays a crucial role in the WD group at the family level and Bacteroridaceae plays a key role in the C group (Fig. 2E). At the family level, the abundance of Bacteroidaceae (P < 0.001), Tannellaceae (P < 0.001), Eubacterium coprostanoligenes_group (P = 0.041) in the WD group is significantly lower than the C group (Fig. 2F). At the genus level, the abundance of the Bacteroides (P < 0.001), Eubacterium eligens_group (P = 0.028) and Paraacteroides (P < 0.001) in the WD group is significantly lower than in the C group (Fig. 2G).
Fig. 2.
Taxonomic characteristics of fecal bacteria from the C and WD groups derived from 16S rRNA gene sequencing. (A) The relative abundance of the first ten phyla, (B) The relative abundance of the top 10 families, (C) The relative abundance of the first ten genera, (D) Histogram of LDA value distribution (LDA Score > 4), (E) Evolution branch diagram, (F) T_ Analysis chart of family-level species differences among test groups, and (G) T_ Analysis chart of species difference at genus level among test groups.
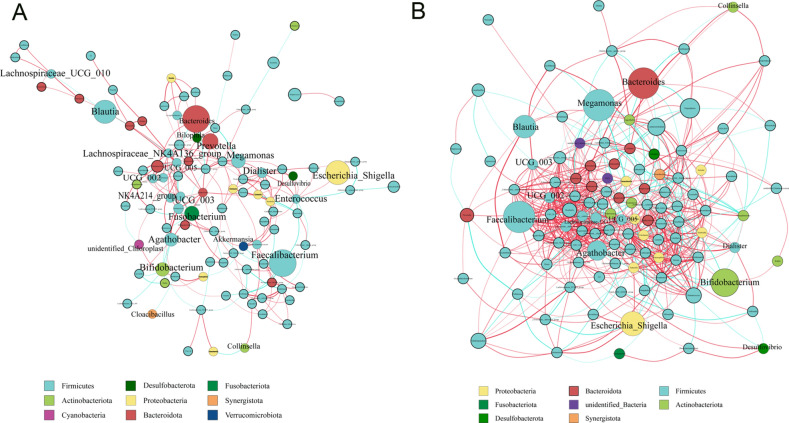
A co-occurrence network diagram based on significant Spearman correlations was constructed to describe the potential relationship between bacteria in the gut microbial community. The C and WD groups primarily have two co-occurrence networks distributed across ten main genera. The C group exhibits a substantial positive correlation symbiotic network among genera (Fig. 3A). The microbial community of the WD group has a more complex network (Fig. 3B). The correlation between the microbiota in the WD group was significantly increased compared to the C group. Additionally, UCG-003, Lachnospiraceae NK4A136 group, Dialister, UCG-005, NK4A214 group, UCG-002, Lachnospiraceae UCG-010, Agathobacter, for example, are at key positions in the correlation analysis and highly correlated with other bacterial groups. Correlation analysis shows that they play an important role in maintaining the gut microbiota structure in the subjects.
Fig. 3.
Network diagram. (A) C Group Network Diagram, and (B) WD Group Network Diagram.
Species differences at the metagenomic level
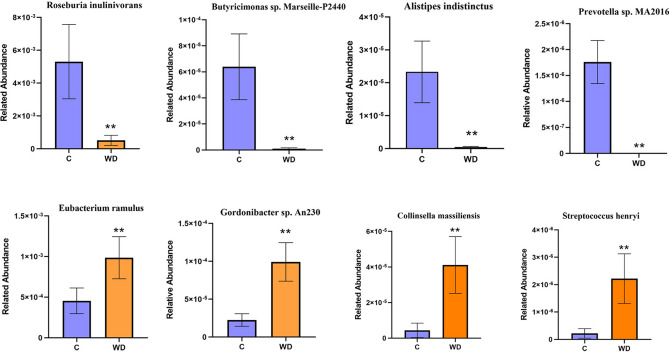
Differentially abundant bacterial species between C and WD group were also analyzed (Fig. 4 and Table S1), the abundance of Alistipes indistinctus, Roseburia inulinivorans, Butyrivibrio sp. INlla16, Butyrivibrio sp. WCD3002, Sporosarcina sp. HYO08, Burkholderiales bacterium 1_1_47, Gabonia massiliensis, Butyrivibrio sp. NC2007, Burkholderiales bacterium 21-58-4, Firmicutes bacterium GWE2_51_13, Candidatus Gastranaerophilales bacterium HUM_17, Butyricimonas sp. Marseille-P2440, Paenibacillus mucilaginosus, Alistipes sp. ZOR0009, Butyricimonas sp. An62 are significantly lower in the WD group than the C group (P < 0.01). The abundance of Gordonibacter sp. An230, Collinsella sp. An271, Collinsella massiliensis, Streptococcus henryi, Streptococcus ferus are significantly higher in the WD group than the C group (P < 0.01).
Fig. 4.
Metagenomic sequencing species shows the species differences between C and WD.
Functional differences in the metagenomic
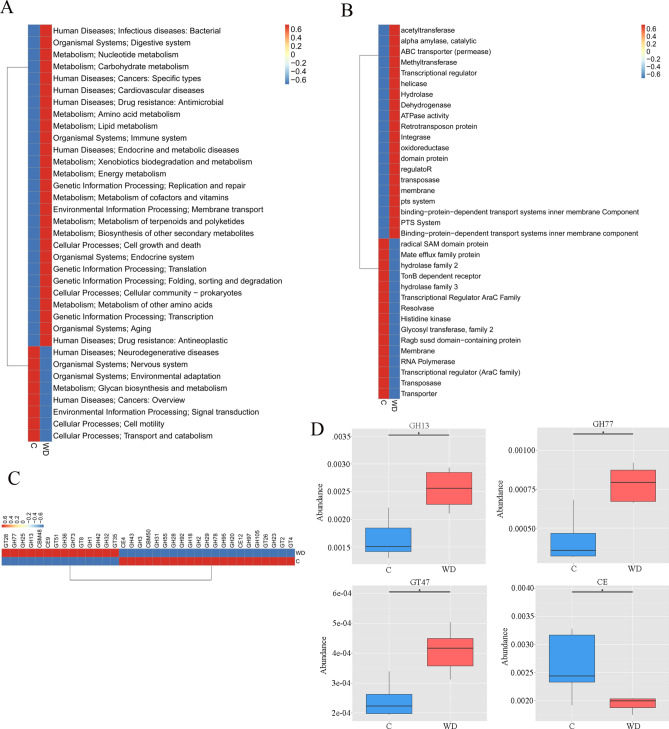
Through metagenomic sequencing analysis, WD patients showed dysfunction in several pathways of the KEGG, eggNOG, and CAZy databases.
After KEGG pathway annotation, the metabolic pathway activities of Energy, Nucleotide, Lipid, Carbohydrate, cofactors, and vitamins in the WD group increased. Human Diseases, Neurodegenerative diseases, Organismal Systems, Nervous Systems and Organizational Systems all increase in participants with WD (Fig. 5A and Table S2). The eggNOG orthologous group (og) in the WD group was also significantly separated from the C group (Fig. 5B, P = 0.045). The level of eggNOG shows a decrease in functional abundance in the WD group, such as radial SAM domain protein, Mate efflux family protein, Hydrogenase family 2, and TonB-dependent receptor.
Fig. 5.
Microbial gene function annotations for the C and WD groups KEGG. (A) The average abundance difference of KEGG functions at Level 2 is enriched in the heat map, (B) Comparison of eggNOG homologous groups (og) between the C and WD groups, (C) CAZy annotates microbial gene functions in C and WD groups, and (D) Distribution of LDA values for differential functions. Only the taxa with an LDA > 3.0 are shown.
The CAZy database shows the highest glycoside hydrolases (GH) coding gene sets, followed by glycosyltransferase (GT) coding gene sets. The level 2 cluster heat map for the relative abundance of functions between groups shows the top 35 functions in abundance and their relative abundance information between groups. Most of them come from GTs and GHs. Compared with the C group, the abundance of 14 enzymes in the WD group increased, while the abundance of 21 enzymes decreased (Fig. 5C). As shown in Fig. 5D, the abundance of GH13, GH77, and GT47 in the WD group is significantly higher than those in the C group (P < 0.05), the abundance of CE (carbohydrate esterase) in the WD group is significantly lower than that in the C group.
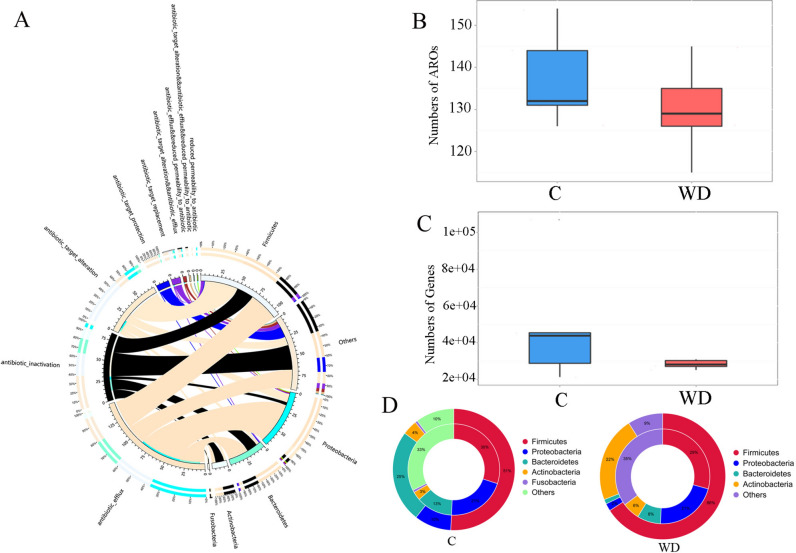
A distribution map of resistance mechanisms is plotted (Fig. 6A) to study antibiotic resistance genes (ARG) distribution in the C and WD groups. The total number of genes and resistance genes in the C group is higher than in the WD group (Fig. 6B). The abundance of tetW/N/W, tetW, efrB, and LEN-4 in the WD group increased compared to the C group (Fig. 6C). No matter how the specifications attribute is translated, the ARO distribution for each specification remains stable in both C and WD (Fig. 6D). The mechanism still needs to be further elucidated.
Fig. 6.
Resistance mechanism and species overview diagram. (A) The circle diagram is divided into two parts, with phylum-level species information on the right and resistance mechanism information on the left, (B) Comparison of the number of antibiotic genes between C and WD, (C) Comparison of antibiotic gene types between C and WD, (D) The relationship between species attributes at the C group level and the distribution of resistance genes in the gut microbiota, and the relationship between species attributes at the WD phylum level and the distribution of resistance genes in the gut microbiota.
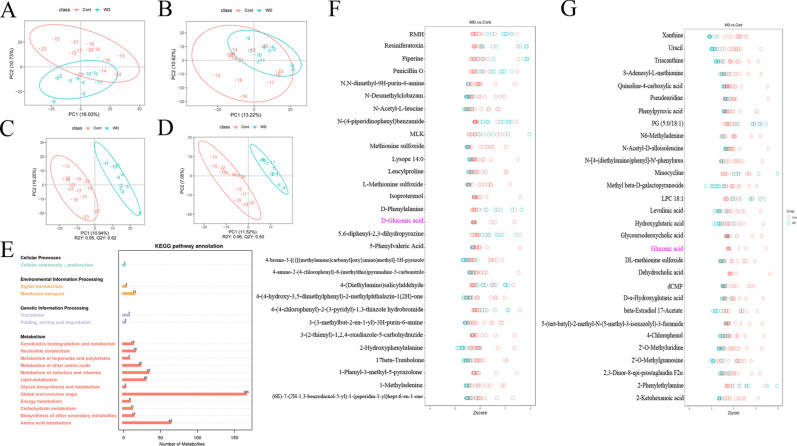
Functional indicators of the faecal metabolome
The principal component analysis results show an inherent metabolic difference between the C and WD groups (Fig. 7A,B). To further identify metabolites that can distinguish between the C and WD groups, indicating a predictive and reliable model and highlighting differences in metabolite abundance between the C and WD groups is significant (Fig. 7C,D). Seventeen KEGG pathways significantly differ between the two groups in each secondary classification under the primary classification (Fig. 7E). The first three enrichment pathways are Global and overview maps, Amino acid metabolism, and Metabolism of cofactors and vitamins. Four hundred thirty-eight metabolic products changed. Levels of metabolites such as 5,6-diphenyl-2,3-dihydropyrazine, N-(4-piperidinophenyl) benzamide, 4-(4-chlorophenyl)-2-(3-pyridyl)-1,3-thiazole hydrobromide in WD samples have increased. Levels of metabolites such as all-trans-retinal, 5-Phenylvaleric Acid, N-Desmethylcyclobazam, Leucylproline, N-acetyl-L-leucine, Uraci, Xanthine, Hypoxanthine, D-Gluconic acid are decreased (Fig. 7F,G and Table S3-4).
Fig. 7.
Multivariate statistical analysis of fecal metabolites in C and WD groups. (A) PCA analysis (positive ion mode), (B) PCA analysis (negative ion mode), (C) Scatter plot of PLS-DA scores (positive ion mode), (D) PLS-DA score scatter plot (negative ion mode), (E) KEGG pathway map, (F) Differential metabolite cluster heat map (positive ion mode), and (G) Differential metabolite cluster heat map (negative ion mode ion mode) *P < 0.05, ** < 0.01.
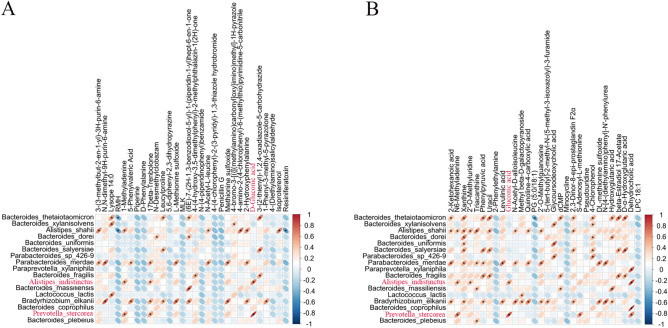
A Pearson Correlation Analysis was performed between metagenome and metabolites with significant differences (Fig. 8A,B) to explore the potential relationship between gut microbiota and metabolites. Among the metabolites significantly reduced in WD patients' feces, 3- (3-methylbut-2-en-1-yl) -3H-purin-6-amine is significantly positively associated with Alistipes shahii, and Parabacteroides Merdae (P < 0.05). N, N-dimethyl-9H-purin-6-amine is significantly positively associated with Paraacteroides merdae, and Bradyrhizobium Elkanii (P < 0.05). 5-Phenoylvaleric Acid is significantly positively correlated with Paraacteroides merdae, and Bradyrhizobium Elkanii (P < 0.05). N-acetyl-L-leucine (NALL) is the N-acetyl derivative of L-leucine and is significantly positively correlated with Alistipes shahii and Parabacteroides merdae (P < 0.05). Leucylproline is significantly positively correlated with Bacteroides thetaiotaomicron, Bacteroides salyersiae, Bradyrhizobium elkanii, and Gabonia Massiliensis, respectively (P < 0.05). D-Gluconic acid is significantly positively correlated with Alistipes indistinctus and Prevotella stercora (P < 0.05). Alistipes indistinctus is not detected in WD patients, while Prevotella stercora had an extremely low abundance in WD patients. N-Desmethylchlorozam is significantly positively correlated with Bacteroides Dorei (P < 0.05). 2-Hydroxyphenylalanine, a phenylalanine derivative, is significantly reduced in fecal metabolites of WD patients, is significantly positively correlated with Alistipes shahii, Parabacteroides merdae, and Bradyrhizobium Elkanii (P < 0.05). Lysope 14:0 is significantly positively correlated with Bacteroides thetaiotaomicron, Bacteroides xylanisolvens and Lactococcus Lactis (P < 0.05).
Fig. 8.
Correlation between gut microbiota abundance and differential metabolite levels. (A) Heatmap of correlation analysis considering gut microbiota with significant differences at the species level, and metabolites with significant differences in C and WD groups (Positive ion mode), and (B) Heatmap of correlation analysis considering gut microbiota with significant differences at the species level, and metabolites with significant differences in C and WD groups (Negative ion mode).
In negative ion mode, 2-Ketohexanoic acid is functionally related to caproic acid, which is significantly positively associated with Bacteroides thetaiotaomicron, Alistipes shahii, Parabacteroides merdae, and Bradyrhizobium Elkanii (P < 0.05). Uracil correlates significantly positively with Bacteroides thetaiotaomicron, Alistipes shahii, Bacteroides salyersiae and Bacteroides fragilis (P < 0.05). Xanthine is significantly positively correlated with Bacteroides thetaiotaomicron, Alistipes shahii, Bacteroides dorei, Bacteroides uniformis, Bacteroides salyersiae, Parabacteroides merdae and Bradyrhizobium Elkanii (P < 0.05). Gluconic acid correlates significantly positively with Prevotella stercorea and Alistipes indistinctus (P < 0.05). Dehydrocholic acid is significantly positively correlated with Bacteroides coprophilus, Prevotella stercorea, Alistipes indistinctus and Paraprevotella xylaniphila (P < 0.05). Glycoursodeoxycholic acid significantly decreased and is significantly positively correlated with Bacteroides xylanisolvens, Bacteroides uniformis and Parabacteroides sp_426-9 (P < 0.05).
Discussion
Hepatolenticular degeneration (HLD) is an autosomal recessive disorder of copper metabolism. There have been some reports detailing the gut microbiota of WD. However, multiple omics approaches have not elucidated the relationship between WD and gut microbiota. This study aims to investigate, for the first time, the gut microbiota of WD patients using multi-omics, including 16S rRNA amplicon sequencing, metagenomic sequencing, and metabonomic analysis.
The human gut microbiota participates in nutritional metabolism, inhibits pathogen growth, and matures and maintains the immune system to ensure host balance4,15,16. Microbiological research found that the abundance and diversity of gut microbiota in WD patients are significantly lower than those in the C group. The abundance of Bacteroides fragilis in the intestine of patients with WD is significantly reduced. Bacteroides fragilis produces unique inhibitory sphingolipids to supplement the endogenous antigen environment of the body and maintain the stable balance of the host's constant natural killer T cells17. The WD group also showed reduced levels of Verrucomicrobia, a mucin-degrading bacterium present in the intestinal mucosa, which contributes to intestinal health and glucose homeostasis, and plays an interface role between the human intestinal microbiome and host tissue18. Therefore, decreased Verrucomicrosbia may lead to physiological dysfunction in patients with WD. The levels of Selenomonaceae and Megamonas in WD patients are significantly higher than in healthy individuals. Studies have shown that the abundance of Selenomonaceae and Megamonas increases in patients with cholestatic liver disease19. Such increases infer that Selenomonaceae and Megamonas are associated with biliary copper excretion disorders in patients with WD, resulting in excessive copper ion deposition on hepatocytes and cytotoxic effects, leading to decreased liver function.
Changes in bacterial composition may lead to a decrease in the concentration of SCFAs in the intestine, leading to deviations in the physiological functions of patients with WD. Studies have shown that SCFAs affect the development of Treg cells by inhibiting HDAC activity20. In this study, the relative abundance of Roseburia inulinivorans in the WD group is lower than in the C group at the species level. Roseburia inulinivorans produces short-chain fatty acids. Important short-chain fatty acid butyrates activate various physiological signaling pathways, such as anti-inflammatory activity, regulatory T cell differentiation, and proliferation21,22.
Compared with the C group, WD patients have abnormal metabolite levels. The content of all-trans retinal in WD patients decreased. Studies have shown that all trans retinoic acid can regulate pigmentation, neuroretinal maturation, and corneal transparency in human ocular organs23. Retinol RA has been shown to promote the differentiation of immature T and B cells and to perform multiple functions in the presence of different cytokines24. All trans-retinal may be related to the formation of k-f rings in the eyes of WD patients. These metabolites are mainly related to pathways such as the biosynthesis of antibiotics, alpha-linolenic acid metabolism, one carbon pool by foam, nicotinate and nicotinamide metabolism.
In this study, Leucylproline is significantly reduced in fecal metabolites in patients with WD. Metabolic and hormonal disorders caused by chronic liver disease ultimately lead to increased protein hydrolysis and decreased protein synthesis, which leads to the development and progression of malnutrition25. Leucylproline is a dipeptide formed from L-leucine and L-proline residues, a secondary metabolite that is functionally related to L-leucine and L-proline. Leucylproline is significantly positively correlated with Bacteroides taiotaomicron, Bacteroides saliersiae, Bradyrhizobium elkanii, Gabonia massiliansis, and Clostridium polynesiense.
The abundance of medium-chain fatty acids, such as 5-Phenylvaleric acid, is significantly downregulated in the WD group. 5-Phenylvaleric acid protects cells against endoplasmic reticulum stress-induced neuronal cell death and is a potential candidate for treating neurodegenerative diseases26. 5-Phenylvaleric acid is significantly positively correlated with Parabacteroides merdae and Bradyrhizobium elkanii. The abundance of these species in the WD group is also lower than that in the C group and is not even detectable. The abundance of Parabacteroides merdae and Bradyrhizobium elkanii in the WD group is lower than that in the C group.
N-Desmethylcyclobazam has anticonvulsant pharmacological effects27. 1-Phenyl-3-methyl-5-pyrazolone is a free radical scavenger and neuroprotective agent used to treat amyotrophic lateral sclerosis and stroke28. Studies have shown that N-acetyl-L-leucine treatment can improve neural function after injury by limiting cortical cell death and neuroinflammation29. Therefore, N-acetyl-L-leucine is a promising candidate drug for neuroprotection. Studies have shown that elevated phenylalanine impacts nervous system performance30. D-phenylalanine, a metabolite of phenylalanine metabolism, is significantly elevated in the feces of patients with WD. Bacteria significantly associated with the above metabolites may have protective outcomes on neural function. Clostridium IV and XV a can stimulate the production of TGF- β by colon epithelial cells, inducing T cells to differentiate into Treg cells31.
The three metabolites, uracil, xanthine, and hypoxantine, are relatively reduced in the feces of WD patients and can form complexes with copper ions32,33. Uracil is a common and naturally occurring pyrimidine nucleobase, a component of RNA, and an essential nutrient. Reducing Uracil metabolites may also lead to immune system dysfunction, thereby increasing the risk of infection. Xanthine is an important alkaloid, and its metabolic process in the human body is very complex and correlates with various diseases. Hypoxantine is one of the products of Xanthine oxidized by xanthine oxidase. The above three metabolites all play important roles in the human body, and their reduction in fecal metabolites in WD patients may suggest that they are related to copper ion accumulation in WD patients and are potential biomarkers of WD.
D-Gluconic acid and Gluconic acid may be potential drugs for treating WD. D-Gluconic acid is an essential metabolite, a chelating agent, and a penicillium metabolite34,35. A gluconic acid aqueous solution contains cyclic ester gluconic acid δ Lactone structure, which chelates metal ions and forms very stable complexes. The reagent exhibits strong chelating activity in alkaline solutions against anions such as calcium, iron, aluminum, copper, and other heavy metals. D-Gluconic acid is also functionally related to the Microbial metabolism in diverse environments, Biosynthesis of Antibiotics pathway. D-Gluconic acid was significantly positively correlated with Alistipes indistinctus and Prevotella stercora. Alistipes indistinctus is not detected in patients with WD, while Prevotella_stercora had a very low abundance in patients with WD. These two bacteria may have potential roles in treating WD.
The main limitation of the study is the small samples size, more useful bacteria and metabolites that may be used to treat WD have not been discovered yet.
Conclusion
In summary, these results show that the fecal microbiome and metabolome of WD patients differ from those of healthy individuals. The disorder of gut microbiota in WD patients leads to abnormal metabolic products, which may worsen the symptoms and progression of the disease. Potential bacteria and metabolites as drugs for WD treatment have been identified, providing a novel and unique proposition for WD treatment. Using specific bacteria or related metabolites of the microbiome may be expected to prevent or treat WD.
Supplementary Information
Author contributions
All authors contributed to conceptualizing, drafting, and revising the manuscript. All authors have read and agreed to the final version of the manuscript.
Funding
This research was funded by the Shenzhen Science and Technology Innovation Program (No. JCYJ20220530165400002), Shenzhen Guangming District Economic Development Special Fund (No. 2021R01132).
Data availability
The data that support the findings of the study are available in BioProject at https://dataview.ncbi.nlm.nih.gov/object/PRJNA1019925?reviewer=lqflng6odon2j8k1eptrbv6n1r with reference number “PRJNA1019925”. Any other data supporting this study’s conclusions are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Shenzhen Hospital of the Chinese Academy of Sciences. All methods were carried out in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71740-5.
References
- 1.Li, W. J., Wang, J. F. & Wang, X. P. Wilson’s disease: Update on integrated Chinese and Western medicine. Chin. J. Integr. Med.19, 233–240. 10.1007/s11655-012-1089-8 (2013). 10.1007/s11655-012-1089-8 [DOI] [PubMed] [Google Scholar]
- 2.Yi, L. P. et al. Present status of diagnosis and treatment of hepatolenticular degeneration. Zhonghua Gan Zang Bing Za Zhi27, 161–165. 10.3760/cma.j.issn.1007-3418.2019.03.001 (2019). 10.3760/cma.j.issn.1007-3418.2019.03.001 [DOI] [PubMed] [Google Scholar]
- 3.Dusek, P. et al. The neurotoxicity of iron, copper and manganese in Parkinson’s and Wilson’s diseases. J. Trace Elem. Med. Biol.31, 193–203. 10.1016/j.jtemb.2014.05.007 (2015). 10.1016/j.jtemb.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 4.McKenzie, C., Tan, J., Macia, L. & Mackay, C. R. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev.278, 277–295. 10.1111/imr.12556 (2017). 10.1111/imr.12556 [DOI] [PubMed] [Google Scholar]
- 5.Zhong, H. et al. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome7, 2. 10.1186/s40168-018-0608-z (2019). 10.1186/s40168-018-0608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura, K. & Ohnishi, H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J. Gastroenterol.20, 7381–7391. 10.3748/wjg.v20.i23.7381 (2014). 10.3748/wjg.v20.i23.7381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesaro, C. et al. Gut microbiota and probiotics in chronic liver diseases. Dig. Liver Dis.43, 431–438. 10.1016/j.dld.2010.10.015 (2011). 10.1016/j.dld.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 8.Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med.22, 586–597. 10.1038/nm.4106 (2016). 10.1038/nm.4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsuoka, H. & Schmid-Schonbein, G. W. Mechanisms for blockade of in vivo activator production in the ischemic intestine and multi-organ failure. Shock14, 522–527. 10.1097/00024382-200014050-00005 (2000). 10.1097/00024382-200014050-00005 [DOI] [PubMed] [Google Scholar]
- 10.Cai, X. et al. Altered diversity and composition of gut microbiota in Wilson’s disease. Sci. Rep.10, 21825. 10.1038/s41598-020-78988-7 (2020). 10.1038/s41598-020-78988-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng, H. et al. Association study of gut flora in Wilson’s disease through high-throughput sequencing. Medicine (Baltimore)97, e11743. 10.1097/MD.0000000000011743 (2018). 10.1097/MD.0000000000011743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas, B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res.21, 494–504. 10.1101/gr.112730.110 (2011). 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120. 10.1093/bioinformatics/btu170 (2014). 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirdita, M., Steinegger, M. & Söding, J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics35, 2856–2858. 10.1093/bioinformatics/bty1057 (2019). 10.1093/bioinformatics/bty1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirata, S. I. & Kunisawa, J. Gut microbiome, metabolome, and allergic diseases. Allergol. Int.66, 523–528. 10.1016/j.alit.2017.06.008 (2017). 10.1016/j.alit.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan, B. & Taneja, V. Microbial modulation of the gut microbiome for treating autoimmune diseases. Expert Rev. Gastroenterol. Hepatol.12, 985–996. 10.1080/17474124.2018.1517044 (2018). 10.1080/17474124.2018.1517044 [DOI] [PubMed] [Google Scholar]
- 17.Hapil, F. Z. & Wingender, G. The interaction between invariant Natural Killer T cells and the mucosal microbiota. Immunology155, 164–175. 10.1111/imm.12958 (2018). 10.1111/imm.12958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson, J. R. et al. A preliminary examination of gut microbiota, sleep, and cognitive flexibility in healthy older adults. Sleep Med.38, 104–107. 10.1016/j.sleep.2017.07.018 (2017). 10.1016/j.sleep.2017.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, T. et al. Comprehensive analysis of gut microbiota and fecal bile acid profiles in children with biliary atresia. Front Cell Infect. Microbiol.12, 914247. 10.3389/fcimb.2022.914247 (2022). 10.3389/fcimb.2022.914247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, L. et al. Histone/protein deacetylase inhibitor therapy for enhancement of Foxp3+ T-regulatory cell function posttransplantation. Am. J. Transplant.18, 1596–1603. 10.1111/ajt.14749 (2018). 10.1111/ajt.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vuik, F. et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United European Gastroenterol. J.7, 897–907. 10.1177/2050640619852255 (2019). 10.1177/2050640619852255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagpal, R., Neth, B. J., Wang, S., Craft, S. & Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine47, 529–542. 10.1016/j.ebiom.2019.08.032 (2019). 10.1016/j.ebiom.2019.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isla-Magrane, H., Zufiaurre-Seijo, M., Garcia-Arumi, J. & Duarri, A. All-trans retinoic acid modulates pigmentation, neuroretinal maturation, and corneal transparency in human multiocular organoids. Stem Cell Res. Ther.13, 376. 10.1186/s13287-022-03053-1 (2022). 10.1186/s13287-022-03053-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, M. et al. Prevalence of hyperuricemia among Chinese adults: Findings from two nationally representative cross-sectional surveys in 2015–16 and 2018–19. Front. Immunol.12, 791983. 10.3389/fimmu.2021.791983 (2021). 10.3389/fimmu.2021.791983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dos Santos, A. L. S. & Anastácio, L. R. The impact of L-branched-chain amino acids and L-leucine on malnutrition, sarcopenia, and other outcomes in patients with chronic liver disease. Expert Rev. Gastroenterol. Hepatol.15, 181–194. 10.1080/17474124.2021.1829470 (2021). 10.1080/17474124.2021.1829470 [DOI] [PubMed] [Google Scholar]
- 26.Mimori, S. et al. Protective effects of 4-phenylbutyrate derivatives on the neuronal cell death and endoplasmic reticulum stress. Biol. Pharm Bull.35, 84–90. 10.1248/bpb.35.84 (2012). 10.1248/bpb.35.84 [DOI] [PubMed] [Google Scholar]
- 27.Arfman, I. J. et al. Therapeutic drug monitoring of antiepileptic drugs in women with epilepsy before, during, and after pregnancy. Clin. Pharmacokinet.59, 427–445. 10.1007/s40262-019-00845-2 (2020). 10.1007/s40262-019-00845-2 [DOI] [PubMed] [Google Scholar]
- 28.Turnbull, J. Reappraisal of an ALS trial: Unaccounted procedural risk. Lancet Neurol.19, 717–718. 10.1016/S1474-4422(20)30265-9 (2020). 10.1016/S1474-4422(20)30265-9 [DOI] [PubMed] [Google Scholar]
- 29.Martakis, K. et al. Efficacy and safety of N-acetyl-L-leucine in children and adults with GM2 gangliosidoses. Neurology10.1212/WNL.0000000000201660 (2022). 10.1212/WNL.0000000000201660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazi, T. A. et al. Dysregulated choline, methionine, and aromatic amino acid metabolism in patients with wilson disease: Exploratory metabolomic profiling and implications for hepatic and neurologic phenotypes. Int. J. Mol. Sci.20, 5937. 10.3390/ijms20235937 (2019). 10.3390/ijms20235937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science331, 337–341. 10.1126/science.1198469 (2011). 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavelle, A. & Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol.17, 223–237. 10.1038/s41575-019-0258-z (2020). 10.1038/s41575-019-0258-z [DOI] [PubMed] [Google Scholar]
- 33.Mikulski, C. M., Tran, T. B., Mattucci, L. & Karayannis, N. M. Xanthine, hypoxanthine and guanine copper(II) complexes. Inorganica Chimica Acta78, 211–218 (1983). 10.1016/S0020-1693(00)86515-9 [DOI] [Google Scholar]
- 34.Mehtio, T. et al. Production and applications of carbohydrate-derived sugar acids as generic biobased chemicals. Crit. Rev. Biotechnol.36, 904–916. 10.3109/07388551.2015.1060189 (2016). 10.3109/07388551.2015.1060189 [DOI] [PubMed] [Google Scholar]
- 35.Kim, H. Y., Park, H. M. & Lee, C. H. Mass spectrometry-based chemotaxonomic classification of Penicillium species (P. echinulatum, P. expansum, P. solitum, and P. oxalicum) and its correlation with antioxidant activity. J. Microbiol. Methods90, 327–335. 10.1016/j.mimet.2012.06.006 (2012). 10.1016/j.mimet.2012.06.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of the study are available in BioProject at https://dataview.ncbi.nlm.nih.gov/object/PRJNA1019925?reviewer=lqflng6odon2j8k1eptrbv6n1r with reference number “PRJNA1019925”. Any other data supporting this study’s conclusions are available from the corresponding author upon request.