Abstract
There is a collective push to diversify human genetic studies by including underrepresented populations. However, analyzing DNA sequence reads involves the initial step of aligning the reads to the GRCh38/hg38 reference genome which is inadequate for non-European ancestries. In this study, using long-read sequencing technology, we constructed de novo genome assemblies from two indigenous Americans from Arizona (IAZ). Each assembly included ∼17 Mb of DNA sequence not present [nonreference sequence (NRS)] in hg38, which consists mostly of repeat elements. Forty NRSs totaling 240 kb were uniquely anchored to the hg38 primary assembly generating a modified hg38-NRS reference genome. DNA sequence alignment and variant calling were then conducted with whole-genome sequencing (WGS) sequencing data from 387 IAZ using both the hg38 and modified hg38-NRS reference maps. Variant calling with the hg38-NRS map identified ∼50,000 single-nucleotide variants present in at least 5% of the WGS samples which were not detected with the hg38 reference map. We also directly assessed the NRSs positioned within genes. Seventeen NRSs anchored to regions including an identical 187 bp NRS found in both de novo assemblies. The NRS is located in HCN2 79 bp downstream of Exon 3 and contains several putative transcriptional regulatory elements. Genotyping of the HCN2-NRS revealed that the insertion is enriched in IAZ (minor allele frequency = 0.45) compared to other reference populations tested. This study shows that inclusion of population-specific NRSs can dramatically change the variant profile in an underrepresented ethnic groups and thereby lead to the discovery of previously missed common variations.
Keywords: de novo assembly, HiFi sequencing, genomic diversity, indigenous American, genome alignment, HCN2
Significance.
In recent years, there has been a growing effort to sequence and analyze DNA samples from underrepresented populations to combat the lack of diversity in human genetic studies. However, the process of analyzing DNA sequence data begins with aligning the reads to the GRCh38/hg38 reference genome, which does not adequately represent non-European ancestries. To bridge this gap, we created de novo genome assemblies from two indigenous Americans from Arizona to inspect nonreference genomic segments and to use these segments to enhance the reference for variant-calling pipeline. Our study is the first genomics report to identify previously unknown regions in indigenous American genomes, contributing to the broader effort to address the diversity gap in human genetics research.
Introduction
The lack of diversity in human genetic studies is of growing concern, particularly as personalized medicine is becoming a reality. The majority of genomics data, approximately 80%, predominantly represents individuals of European ancestry, highlighting a significant underrepresentation of other populations (Gurdasani et al. 2019). This lack of diversity has had adverse consequences in genetic testing and variant interpretation, particularly for patients of African and Asian ancestry (Manrai et al. 2016; Popejoy and Fullerton 2016). Recent large-scale genome studies, such as the All of Us initiative (All of Us Research Program Investigators et al. 2019), the Human Heredity and Health in Africa project (GenomeAsia100K Consortium 2019), and the GenomeAsia100K project (Mulder et al. 2018), have made significant efforts to address this disparity by actively increasing the representation of diverse populations. However, aside from the challenges associated with sample collection for sequencing, these endeavors face two main limitations when generating genomic data for underrepresented populations. One limitation is due to short-read sequencing being the main method in large population studies. While short-reads offers valuable insights through comparisons of allele frequencies and functional annotations, they struggle to capture rare variants and accurately characterize structural variants (SVs) in isolated populations (Chheda et al. 2017; Beyter et al. 2021; Wu et al. 2021). This restricts the comprehensive understanding of genetic diversity and hampers the identification of variants associated with diseases. The second limitation stems from the reliance on a human genome reference during DNA sequence read analysis. The current reference maps widely used in genetic studies, the GRCh38/hg38 and the telomere-to-telomere CHM13 (T2T-CHM13) reference genomes, primarily comprise sequences of European origin (Schneider et al. 2017; Nurk et al. 2022). Consequently, the current reference maps still exhibit limitations in adequately representing the genetic diversity found within diverse populations. It is important to recognize that individuals may carry unique sequences that are not represented in the reference map, thereby harboring genomic regions with potential disease associations and other phenotypic relevance. Therefore, it has been imperative for the genomic research in isolated populations to move beyond the traditional short-read sequencing and linear reference genome paradigm (Reis et al. 2023; Groza et al. 2024).
Long-read sequencing technologies have boosted our power in resolving complex regions of the genome, detecting SVs, and assembling complete chromosomes (Chaisson et al. 2015; Logsdon et al. 2020; Marx 2023). In the recent years, these technologies have advanced in accuracy and yield, enabling variant detection at a large scale, with recent studies showcasing their potential in population-scale analyses (De Coster et al. 2021; Marx 2023).
Significant steps have also been taken to address the issue of the low ancestral diversity in the human genome reference. The Human Pangenome Reference Consortium (HPRC) has been working on constructing a reference that encompasses diverse genomes, and they have published the first draft of the pangenome, consisting of 47 genome assemblies from a diverse cohort (Liao et al. 2023). However, there are still challenges to overcome in growing and refining this reference, both in sampling and in computational analysis. The current sampling efforts do not include indigenous American or Aboriginal peoples (https://humanpangenome.org/samples/). Indigenous American people are genetically distinct compared to other populations in the United States, with a significant variation within the group itself (Redd et al. 2006). Principal component analysis from a previous study including three ancestral populations from the 1,000 genomes database (European, East Asian, and African ancestries) and our indigenous American population from Arizona (IAZ) showed a distinct cluster for IAZ on the principal component space (Kim et al. 2020). Therefore, exploratory genomics studies using novel methods are warranted in indigenous populations to uncover previously unrecognized genetic variations and capture novel disease associations.
In this study, we aimed to improve identification of genetic variations that have not been previously studied among IAZ population. We utilized de novo genome assemblies to identify nonreference sequences (NRSs) in this cohort. These NRSs represent novel genomic segments not present in the existing reference map. By analyzing these segments, both directly and by incorporating them into our whole-genome sequencing (WGS) variant-calling pipeline, we sought to uncover previously unrecognized polymorphisms which can be assessed for a potential role in metabolic disease enriched in this underrepresented ethnic group.
Materials and Methods
Long-read Sequencing and De Novo Assembly
Two DNA samples isolated from unrelated individuals (one female and one male) were sequenced using the PacBio HiFi SMRT sequencing platform at the DNA Sequencing Center at Brigham Young University (Provo, UT). SMRT bell adapted DNA libraries were size selected and ∼15-kb fractions were run on three SMRT cells (8-M trays) for a duration of 30 h for each sample. Bioinformatic analysis for the construction of the IAZ de novo assemblies was done by the DNA Sequencing Center at Brigham Young University. Selected quality reads were assembled using Hifiasm version 0.11 (r302). Contig continuity and statistics of read-to-contig alignments were calculated using Inspector (https://github.com/Maggi-Chen/Inspector) Area under the N curve was calculated using the caln50 package (https://github.com/lh3/calN50).
Detection of NRSs and SVs in the De Novo Assemblies
All computational analyses were run on the high-performance computing environment NIH HPC Biowulf cluster (http://hpc.nih.gov). Visualization was performed on RStudio using ggplot2 package of R (version 4.3.1).
The de novo assemblies were compared to the Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13/hg38) using the alignment script of NUCmer from the MUMmer package (version 4.0.0beta2). Each assembly was aligned to GRCh38.p13/hg38 with the parameters maxmatch -l 150 -c 400. The resulting alignments were filtered to retain only the single best alignment for each contig using the delta-filter -q command. The show-coords tool was then used to extract the coordinates and percent identity of the aligned contigs. To identify the contigs that failed to align to hg38 (<80% sequence identity), a custom script (available at https://github.com/cigdemkoroglu/NRS/blob/main/unaligned_contigs.R) was used to parse the output of the alignment and extract the unaligned contigs and contigs with low identity. Bedtools version 2.29.2 was employed to generate fasta files containing only the unaligned portions of the assemblies (NRSs). To capture shorter contigs matching reference segments and filter them out from the NRSs, the alignment workflow was repeated on the initial output of NRSs with relaxed NUCmer settings (−maxmatch -l 100 -c 200). This allowed for a more permissive alignment approach to increase the number of contigs that could be aligned to the reference. Final NRSs were aligned both to GRCh38.p13/hg38 and T2T-CHM13 v2.0/hs1 using MUMmer NUCmer (−maxmatch -l 100 -c 200) and filtered to select unique alignments between reference and query (NRS) with >95% sequence identity (delta-filter -q -i 95 -l 100 -u 100). NRSs were analyzed for repetitive sequences using RepeatMasker and Assemblytics was used to detect SVs >50 bp (http://assemblytics.com/).
Anchoring the NRSs to the GRCh38.p13/hg38 Primary Assembly
For each NRS, 100 bp of flanking sequences either side of the NRSs were aligned to the nucleotide collection database using the blastn. Contigs from the de novo assemblies with both flanking sites anchoring to the primary assembly chromosomes of hg38 were selected and appended to hg38 to generate an extended reference genome (hg38-NRS) using perEditor (https://systemsbio.ucsd.edu/perEditor/). The appended 40 pieces of segments were checked for alignment to T2T-CHM13 using Human BLAT Search (https://genome.ucsc.edu/cgi-bin/hgBlat). NRSs spanning a reference region longer than the NRS itself and those aligning to the same region, indicating repeat expansions, were not incorporated into the hg38-NRS reference genome. In cases where NRSs overlapped between both assemblies, only the region from Assembly 1 was included.
Variant Calling Using hg38 and hg38-NRS Reference Genomes
Whole genomes for 387 IAZ individuals were previously sequenced (Koroglu et al. 2020; Day et al. 2022) using Illumina short-read chemistry (30× average coverage). Characteristics of the participants are summarized in supplementary table S3, Supplementary Material online. Variant discovery was performed in parallel on WGS data from 387 IAZ samples using either hg38 or hg38-NRS as the reference. GATK 4.2.6 was used following the best practices workflow, which includes base quality score recalibration, variant calling and joint genotyping by HaplotypeCaller and GenotypeGVCFs, and variant quality score recalibration. Databases for the known variation in the GATK resource bundle were position adjusted based on hg38-NRS using a custom lift-over script (available at https://github.com/cigdemkoroglu/NRS/blob/main/lift_database.R). Monomorphic variants, variants with a call rate <95%, variants with quality scores below the truth sensitivity level 99% threshold, and variants with discordant genotypes between the duplicate pairs were removed from the final vcf files using VCFtools. Integrated Genomics Viewer was used to visualize read alignments (https://igv.org/).
Identification of Gained SNVs in the hg38-NRS Reference Genome
To compare variant calling between hg38 and hg38-NRS reference genomes, ANNOVAR (https://annovar.openbioinformatics.org/en/latest) was first used to annotate the variants identified with hg38. Next, to annotate the variants identified with hg38-NRS and to ensure compatibility between the two reference maps, the hg38-NRS variant dataset was lifted-over using custom scripts (https://github.com/cigdemkoroglu/NRS). Lastly, to identify lost and gained SNVs, the hg38 and hg38-NRS annotated datasets were compared using the LostGained tool (https://github.com/cigdemkoroglu/NRS/blob/main/LostGained.R). Among the gained variants, 15 missense variants that are reported in dbSNP but were not detected by our in-house genotyping studies using the Affymetrix SNP Array 6.0 or our WGS and whole-exome sequencing studies along with 10 randomly selected novel SNVs were verified by sequencing.
Genotyping of the HCN2-NRS
Genotyping of the HCN2-NRS in the Indigenous Americans from Arizona (IAZ) was performed using DNA isolated from 3,410 individuals who participated in a community-based longitudinal study examining type 2 diabetes-related traits conducted by the National Institute of Diabetes and Digestive and Kidney Diseases in Phoenix, Arizona between the years 1965 and 2007 (Knowler et al. 1978). All participants in the study were self-reported full-heritage Indigenous American (all eight great grandparents identified as IAZ). The HCN2-NRS was also genotyped in the ASW and GBR DNA panels from the 1,000 genomes project. The ASW and GBR DNA samples were obtained from the NHGRI Sample Repository for Human Genetic Research at the Coriell Institute for Medical Research: Repository IDs MGP00015 and MGP00003, respectively. Genotyping of the HCN2-NRS was done using custom designed TaqMan probes that detected the presence of either the RS or NRS (Thermo Fisher Scientific, Waltham, MA). Statistical analyses were performed using the software of the SAS Institute (Cary, NC, USA) as previously described (Day et al. 2022).
Results
De Novo IAZ Genome Assemblies
DNA samples from one female (Sample 1) and one male (Sample 2) IAZ were sequenced using the PacBio HiFi SMRT sequencing platform. Summary reports for the raw PacBio sequencing data are shown in supplementary table S1, Supplementary Material online. For both samples, the SMRT sequencing reactions produced approximately 64 Gb of high-fidelity subreads (>Q20) with a mean length of ∼12 kb, which were used to generate the de novo genome assemblies. Primary contigs with high contiguity were generated using HiFiasm assembler (Table 1). Assembly 1 (female IAZ) and Assembly 2 (male IAZ) yielded a total genome length of 3.05 and 3.07 Gb, respectively, indicating high assembly quality regarding genome completeness when compared to the 2.96-Gb GRCh38/hg38 reference genome excluding gaps. Read mapping rate calculated with the Inspector tool (https://github.com/Maggi-Chen/Inspector) showed that >97% of HiFi reads could be aligned to assembled contigs (Table 1). The low fraction of reads with split alignments (∼5%) calculated by Inspector indicates good consistency between reads and de novo assemblies and suggests low rate of assembly errors (Chen et al. 2021). Subsequent rounds of MUMmer alignment identified 19 and 16 Mb of DNA sequence for Assembly 1 and Assembly 2, respectively, that could not be aligned to the hg38 reference genome. These NRSs have a minimum length of 100 bps and a hg38 sequence identity <80%. A total of 9.2-Mb NRSs are present in both Assemblies 1 and 2, with >85% sequence identity.
Table 1.
Summary statistics for the IAZ de novo assemblies
| Statistics of contigs | Assembly 1 | Assembly 2 |
|---|---|---|
| Number of contigs | 1,000 | 1,115 |
| Number of contigs >10 kb | 1,000 | 1,115 |
| Number of contigs >1 Mb | 161 | 233 |
| Total length (Gb) | 3.05 | 3.07 |
| Total length of contigs >10 kb (Gb) | 3.05 | 3.07 |
| Total length of contigs >1 Mb (Gb) | 2.97 | 2.94 |
| Longest contig (Mb) | 136.6 | 102 |
| Second longest contig (Mb) | 92.3 | 100.8 |
| N50 (Mb) | 48.04 | 36.34 |
| NG50 (Mb) | 41.32 | 32.19 |
| N50 of contigs >1 Mb | 48.04 | 36.34 |
| Area under the N curve (Mb) | 46.30 | 39.89 |
| Read-to-contig alignment | Assembly 1 | Assembly 2 |
|---|---|---|
| Mapping rate (%) | 97.43 | 98.13 |
| Split-read rate (%) | 5.34 | 4.99 |
| Depth | 702 | 377 |
| Mapping rate in large contigs (%) | 95.1 | 94.66 |
| Split-read rate in large contigs (%) | 5.29 | 4.95 |
| Depth in large contigs | 703 | 380 |
Assembly 1, female IAZ individual; Assembly 2, male IAZ individual. N50 is the length of the shortest assembled contig at 50% of the total assembly length. NG50 is the length of the shortest assembled contig at 50% of the total genome length. For NG50 and area under the N curve, 3.3 Gb of genome size is assumed. Both assemblies are haploid.
NRS Size Distribution for the IAZ De Novo Assemblies
The two IAZ de novo assemblies had similar numbers of NRSs and size distribution patterns with a total length of 19.3 Mb for Assembly 1 (hg38 alignment, n = 2,177 NRSs) and 16.34 Mb for Assembly 2 (hg38 alignment, n = 1,908 NRSs) (supplementary fig. S1a–f, Supplementary Material online). For both assemblies, most of the NRSs are 10 kb and shorter but only accounted for 14.4% and 13.4% of the total lengths for Assembly 1 and Assembly 2 respectively (supplementary fig. S1a and d, Supplementary Material online). There were 130 NRSs for both assemblies with sizes greater than 10 to 100 kb with medium sizes of 20.5 kb for Assembly 1 and 22.7 kb for Assembly 2 (supplementary fig. S1b and e, Supplementary Material online). Ninety-three NRSs are larger than 100 kb (Assembly 1, n = 50; Assembly 2 n = 43), including 3 outlier NRSs larger than 500 kb (supplementary fig. S1c and f, Supplementary Material online).
Identification of Repeat Elements and Alignments to T2T-CHM13 Reference Genome
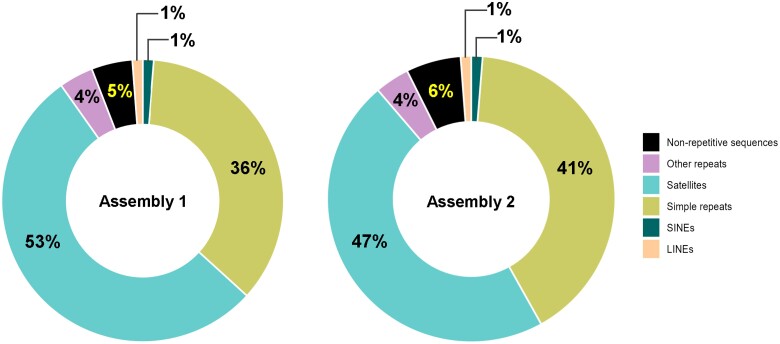
Screening the NRSs for repeat and transposable elements using RepeatMasker revealed that approximately 95% of the NRSs in both assemblies primarily consisted of satellites, simple repeats, and a small fraction of LINE and SINE transposable elements (Fig. 1). The three largest NRSs with lengths of 637 kb, 1.88 Mb (Assembly 1), and 1.47 Mb (Assembly 2) were individually analyzed for their repeat content. They consisted almost entirely of satellites and simple repeats (ranging from 99.7% to 99.9%) rendering their alignment to chromosomes challenging.
Fig. 1.
Repeat content for the NRSs detected in both de novo assemblies.
To verify the uniqueness of NRSs, MUMmer alignment was repeated for Assembly 1 and Assembly 2 NRSs only. Whereas no portion of Assembly 2 NRSs matched any sequence on hg38, a 585-bp piece of an Assembly 1 NRS aligned to chr 4 with 98% identity. NRS alignment using T2T-CHM13 genome as reference showed that 1.2 Mb of the total 19.3-Mb Assembly 1 NRS and 0.9 Mb of the 16.3-Mb Assembly 2 NRS have unique matches on T2T-CHM13.
Mapping the NRSs to the GRCh38/hg38 Reference Genome
NRSs totaling 241 kb had precise anchor positions at 40 distinct locations within the primary scaffold of GRCh38/hg38. Of this 241 kb, 5 kb also aligned to eight regions on the T2T-CHM13 map (Table 2 and supplementary table S2, Supplementary Material online). Namely, 236 kb of nonrepetitive, precisely locatable sequences were not present in both reference maps. Since we use hg38 genome reference in variant-calling pipeline for short-read sequencing data, we inserted 40 NRSs (241 kb) into the primary chromosomes of hg38.
Table 2.
Nonreference sequences anchoring within genes in hg38
| Gene | Insertion regiona | NRS size (bps) | Both assemblies | Alignment to T2T-CMH13 |
|---|---|---|---|---|
| LINC02270 | Chr4:12237666–12237666 | 139 | No | No |
| F11-AS1 | Chr4:186435038–186435150 | 4,714 | No | No |
| BTNL9 | Chr5:181046016–181046016 | 386 | No | No |
| DYNC2I1 | Chr7:158915724–158915724 | 679 | No | Yes (590 bp) |
| MROH5 | Chr8:141492602–141492602 | 1,722 | No | No |
| GTPBP4 | Chr10:1011004–1011004 | 341 | No | No |
| ART1 | Chr11:3653879–3653937 | 877 | Yesb | No |
| PDGFD | Chr11:104078565–104078565 | 579 | No | No |
| LOC728715 | Chr12:9404585–9404585 | 836 | Yesb | No |
| GNPTAB | Chr12:101747637–101747644 | 658 | No | No |
| MPG | Chr16:81854–81857 | 344 | No | No |
| CACNA1H | Chr16:1186044–1186087 | 466 | No | No |
| HCN2 | Chr19:605301–605528 | 187 | Yes | No |
| ADGRG2 | ChrX:19074564–19074564 | 429 | No | No |
| RGN | ChrX:47089179–47089179 | 1,127 | No | Yes |
| CRLF2 | ChrY:1198256–1198271c | 287 | No | No |
| P2RY8 | ChrY:1496850–1496850c | 136 | No | No |
NRS, nonreference sequence.
aPosition based on GRCh38/hg38. Indicated intervals are replaced by NRSs.
bNRS sizes slightly different between the two assemblies.
cPseudoautosomal region.
We inserted 33 NRSs into 16 different autosomes and 12 of the NRSs totaling 11 kb were detected in both assemblies with >99% sequence identity. Two NRSs aligned to chr X and both were located within intronic regions and the remaining 5 NRSs aligned to chrY, including a large 210-kb insertion (Table 2). The 210-kb NRS was detected in Assembly 2 (male individual) but not detected in hg38 and CHM13 after MUMmer and BLAT analyses (supplementary table S2, Supplementary Material online). Two other NRSs that anchored to chrY were detected in Assembly 2, and one of them is inserted to P2RY8, a gene located in pseudoautosomal region 1 (PAR1). The remaining two NRSs anchoring to chrY were detected in the female-derived Assembly 1. One of these is placed in another PAR1 gene, CRLF2; however, the other one sized 420 bp aligned outside of PARs, suggesting a possible misalignment. In fact, T2T-CHM13 alignment for this NRS showed 99% identity match for a 418-bp region on chrX at 85.6 Mb (supplementary table S2, Supplementary Material online).
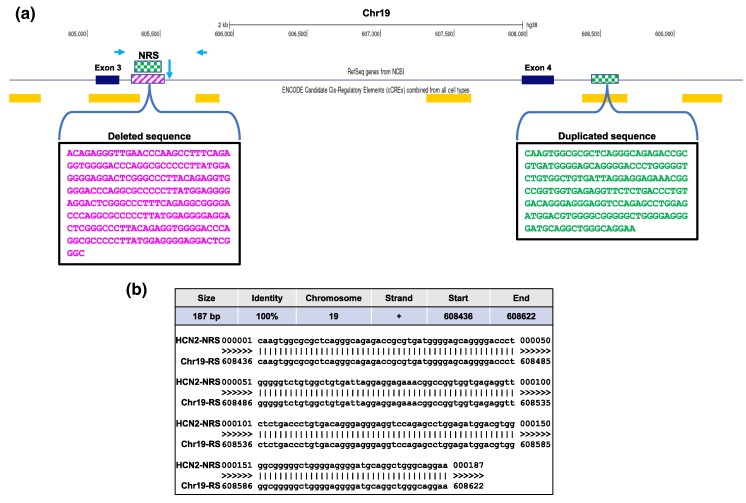
Whereas none of the 40 NRSs mapped to protein-coding regions, 17 NRSs are placed within intronic regions of 13 protein-coding and 4 nonprotein-coding genes (Table 2) and the remaining 23 aligned to intergenic regions (supplementary table S2, Supplementary Material online). Two intronic segments detected only in Assembly 2 that inserted into DYNC2I1 and RGN genes aligned to T2T-CMH13 sequences with >95% identity (Table 2). The rest of the intronic NRSs were unique. Three intronic NRSs were detected in both assemblies and only one of them, HCN2-NRS, was identical in size and therefore was selected for further characterization (Table 2). The HCN2-NRS replaces a 228-bp long sequence in hg38, between the 100-bp anchor sites completely matching the reference. This 228-bp sequence and the flanking sequences are identical in the T2T-CHM13 map except for one base (supplementary fig. S2, Supplementary Material online), suggesting that the HCN2-NRS may not exist in the DNA samples used for reference genome construction. On the other hand, multiple alignment of the replaced HCN2 region to HPRC T2T assemblies (https://genome.ucsc.edu) showed that the region is highly polymorphic among different populations (supplementary fig. S2, Supplementary Material online). HCN2-NRS is located 79 bp downstream of Exon 3. The replaced 228-bp reference sequence (RS) includes a small portion of a predicted cis-regulatory element (CRE). Individuals who carry the 187-bp HCN2-NRS also have an adjacent 100-bp deletion (Fig. 2a). The HCN2-NRS matches (100% identity) with 187 bp downstream of Exon 4 spanning a putative CRE containing various transcription factor binding sites suggesting that HCN2-NRS may have functional importance (Fig. 2a and b).
Fig. 2.
a) Schematic representation showing the location of the 187-bp HCN2-NRS adjacent to Exon 3. The NRS is identical to a 187-bp region in a predicted cis-transcriptional regulatory element (first rectangle after Exon 4) downstream of Exon 4. Horizontal arrows facing each other indicate the locations of the primers used to screen for the presence of the 187-bp NRS. The vertical arrow in between indicates the approximate start site of an additional 104-bp deletion present in individuals who carry the 187-bp NRS. b) HCN2-NRS and chr19-RS side by side alignment. Source: UCSC Genome Browser (GRCh38/hg38).
Regarding cytogenetic locations, only six of the hg38 inserted NRSs totaling 4.7 kb were placed into centromeric and telomeric regions (supplementary table S2, Supplementary Material online) suggesting that there was no enrichment of centromeric/telomeric DNA which is in contrast to long-read sequencing projects involving other populations (Ameur et al. 2018; Gao et al. 2023). However, when the alignments for all of the NRSs with at least one anchored flanking site were analyzed, the percentage of NRSs aligning to centromeric or telomeric regions was ∼30%.
Population Frequency and Association Analyses for HCN2-NRS
To confirm the presence of the HCN2-NRS, the region encompassing the putative NRS was PCR amplified and sequenced in the two IAZ individuals used for the de novo genome assembly along with some of their family members. Of the 10 individuals sequenced, 1 was heterozygous and 9 were homozygous for HCN2-NRS suggesting that the NRS segment was common in the IAZ population. To determine the frequency of HCN2-NRS in IAZ, we designed a custom TaqMan probe that detects the presence of the NRS and genotyped ∼3,400 IAZ individuals. We found that the HCN2-NRS was the minor allele with a frequency of 0.45. Genotyping was repeated for several individuals by PCR to verify that the custom probe was working properly (supplementary fig. S3, Supplementary Material online). For comparison, the HCN2-NRS was also genotyped in the African Ancestry in Southwest USA (ASW) and British from England and Scotland (GBR) DNA panels from the 1,000 genomes project. HCN2-NRS was not as common in these two ethnic groups with frequencies of 0.03 and 0.15 in the ASW and GBR populations, respectively. Despite being enriched in IAZ, our statistical analyses showed that the HCN2-NRS allele was not significantly associated with any obesity or type 2 diabetes-related traits which are highly prevalent in this population (data not shown).
Structural Variation in the De Novo Assemblies
SVs were analyzed directly from the de novo assemblies using Assemblytics and hg38 map as reference. Assemblytics software distinguishes between insertions/deletions and contractions/expansions of repeat elements (Nattestad and Schatz 2016). A total of 12,265 and 12,133 SVs were detected for Sample 1 and Sample 2, respectively, which is similar to an Assemblytics report for a human reference assembly (11,206 SVs) [20] but lower than the number of SVs reported in other studies that used assembly-based or long-read mapping approaches (Ameur et al. 2018; Ebert et al. 2021). The lower number of SVs in our study can be attributed to the stringency of our SV detection method. For both samples, the larger SVs (500 to 10,000 bp) are predominately composed of tandem expansions (supplementary fig. S4, Supplementary Material online). The distribution graphs also show peaks of SV counts at around 300 and 6,000 bp (supplementary fig. S4, Supplementary Material online), possibly indicating enrichment of transposable elements. This size distribution pattern is similar to a previous report (Ameur et al. 2018).
Variant Calling Using the Amended hg38-NRS Map
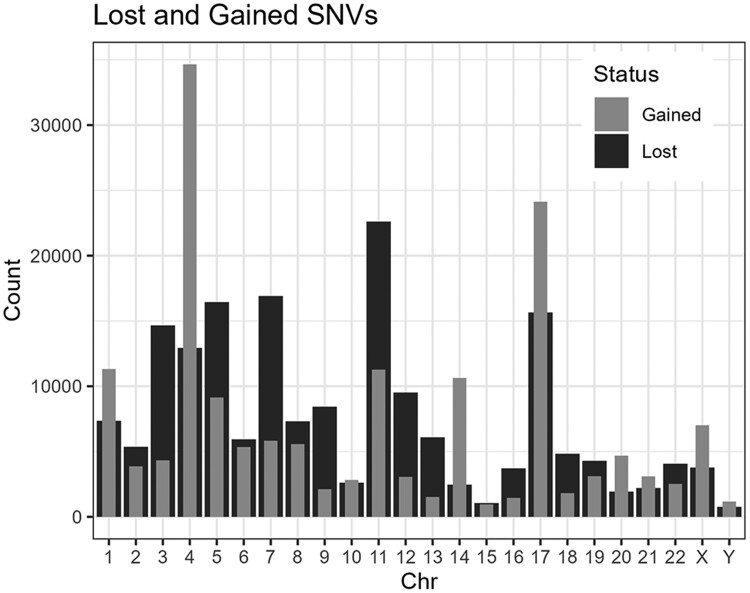
Variant calling using our short-read WGS data generated for 387 IAZ individuals (supplementary table S3, Supplementary Material online) and the amended hg38-NRS map identified 11,130,171 variants. Using the new hg38-NRS map identified 161,502 gained single-nucleotide variants (SNVs) including 695 missense substitutions that were not captured when variant calling was performed using the hg38 reference map (Fig. 3). Additionally, 180,940 SNVs that were captured using the hg38 map were not detected (lost) with the hg38-NRS map (Fig. 3). Visualization of read mapping around these NRS regions, using the Integrative Genomics Viewer, showed that for some regions the insertion of an NRS improves mapping of good quality reads as suggested by several lost or gained SNPs in the NRS site (supplementary fig. S5a, Supplementary Material online), while in other regions, the inserted NRS falls within regions of poor quality mapping and likely has a minimal effect on variant calling in the surrounding sites (supplementary fig. S5b and c, Supplementary Material online). Most of the gained SNVs (n = 111,976) were low-frequency variants; while the remaining gained SNVs (n = 49,526) were detected in ≥5% of the WGS samples. The accuracy of our variant calling with the hg38-NRS map was assessed by sequencing 25 gained SNVs (15 reported and 10 not reported in dbSNP). For the 15 dbSNPs, 13 of the gained SNVs were validated and for the 10 non-dbSNPs, 6 were validated. Most of the non-dbSNPs SNVs reside in repetitive regions of the genome which may account for the reduced accuracy.
Fig. 3.
Number of SNVs gained (light gray) and lost (dark gray) per chromosome for the cohort of 387 short-read WGS samples when analyzed with the hg38-NRS map compared to hg38 map.
Discussion
In this study, our goal was to enhance the identification of genetic variations that remain largely unexplored within an indigenous American cohort from Arizona. We focused on identifying NRSs in de novo genome assemblies constructed for the two participants from the IAZ cohort. The newly identified NRSs signify novel genomic regions not found in the GRCh38/hg38 reference map. The two de novo assemblies we studied had 19.3 and 16.3 Mb (9.2 Mb common) of NRSs that are not present in hg38. However, approximately 5% of NRSs from each assembly aligned to unique regions on the T2T-CHM13 genome reference. Relative to the hg38 genome, the recent T2T-CHM13 assembly is a more complete and accurate reference (Nurk et al. 2022). Therefore, it is expected that de novo assemblies will have a smaller number of NRS when aligned to T2T-CHM12. However, our repeated alignments show that >15 Mb of sequences of indigenous American genomes are not represented in either reference map, underscoring the need for population-specific genome references in genetic studies.
The de novo assemblies were constructed using long-read PacBio HiFi sequencing which produces highly accurate long-read (>10 kb) sequencing data (Hon et al. 2020). The sizes for the de novo genome assemblies were similar to hg38 and exhibited high contiguity with a mean N50 value of 42.2 Mb. We identified >10,000 SVs per de novo genome assembly which is in line with the expectations for long-read sequencing (Ho et al. 2020). This assembly-based approach for SV detection proved particularly advantageous for capturing variants of substantial size (Mahmoud et al. 2019), and the categorization facilitated by Assemblytics allowed us to differentiate between SVs occurring in repetitive regions, such as tandem expansions and repeat contractions, and those in nonrepetitive regions (insertions/deletions). It is worth noting that the largest insertions and deletions (>500 kb) often resided in regions characterized by high repeat content. We discovered 220 shared deletions (>500 kb) in both de novo assemblies with 90 SVs located within genes (data not shown). The application of a high-throughput genotyping approach with custom CNV probes could potentially reveal commonly occurring deletions and uncover potential associations with diseases. However, probe designs for these regions, predominantly repetitive in content, would pose a challenge.
The detected NRSs in two assemblies were found to be highly enriched in repeat sequences, especially satellites and simple repeats. This high repeat content was consistent with the previous literature on nonreference genomic parts (Li et al. 2019; Ebert et al. 2021), and rendered the precise placement of NRSs into chromosomes challenging. We identified 1.5 Mb of nonrepetitive NRS per assembly and 241 kb was anchored to specific chromosomal regions in the primary assembly of hg38. The largest NRS (215 kb) was anchored to chrY. We anticipated that this large NRS was a result of chrY being incomplete in the current hg38 map due to its complex repeat structure (Skaletsky et al. 2003). However, neither MUMmer alignment nor BLAT search for this NRS found a highly identical match in T2T-CHM13, suggesting that this largest segment anchoring to chr Y is unique to the studied genome.
Several of the NRSs anchored to intronic regions and we focused on a particular NRS in HCN2. HCN2-NRS was detected in both de novo assemblies and genotyping showed that the HCN2-NRS allele is common, with a minor allele frequency (MAF) of 0.45 in the IAZ population. We also determined that HCN2-NRS is present in Caucasians and African Americans but at lower frequencies (MAF = 0.15 and 0.03, respectively).
The HCN2-NRS matches (100% identity) with a region located downstream of Exon 4 that encompasses a putative CRE suggesting that individuals carrying the HCN2-NRS may have additional transcription factor binding sites which may potentially influence gene expression. HCN2 encodes for a voltage-gated cation channel predominantly expressed in the heart and the brain and plays a crucial role in migraine, inflammatory, and neuropathic pain including diabetic neuropathy (Young et al. 2014; Tsantoulas et al. 2017, 2022). HCN2 ion channels initiate neuropathic pain by modulating action potential firing in nociceptor neurons involved in sensing pain intensity (Emery et al. 2011).
Compared to Caucasians, chronic widespread and localized musculoskeletal pain disorders are uncommon among IAZ (Jacobsson et al. 1996), however, IAZ have high rates of diabetic neuropathy and pain from rheumatoid arthritis (Del Puente et al. 1989; Jaiswal et al. 2016). It is possible that the common HCN2-NRS allele might modestly contribute to diabetic neuropathy since there are multiple rare intronic variants with nominal associations with diabetic neuropathy reported in the T2D Knowledge Portal (https://t2d.hugeamp.org/).
We aimed to uncover previously undetected polymorphisms by incorporating the NRS segments with precise anchor locations on hg38 into our WGS variant-calling pipeline. We chose to work with hg38 reference to ensure compatibility across different datasets we have, since the GRCh38/hg38 genome has been the standard reference for all our genetic studies in last years. In fact, due to its comprehensive annotation, hg38 is still the preferred genome reference for SNV calling using short-read sequencing data, whereas the T2T-CHM13 reference is particularly valuable for analyzing data generated from long-read sequencing technologies and for detection and characterization of large structural variation.
Our validation assessment for “gained” variants, namely the subset of genomic variants that were only called using hg38-NRS as the reference, using Sanger sequencing underlines the reliability of our WGS data analysis with the extended reference map. However, the calls arising from repetitive regions provided lower rates of true positives. Further research is needed to understand the functional significance of the gained variants and potential contributions to diseases among IAZ population. Some NRSs are inserted into complex regions with repeats; therefore, we did not observe substantial improvement in short-read mapping for every amended region on the reference (supplementary fig. S5, Supplementary Material online). This may have resulted in missing SNV calls. Analysis with longer reads could increase the number of variant calls for those regions.
Conclusion
The overall findings we presented have expanded our understanding of genetic diversity within indigenous American populations. The presence of unique genetic elements and variants unrepresented in existing reference maps demonstrates the importance of delving into the genomics of underrepresented populations. By combining long-read sequencing and novel NRS identification methods, we were able to uncover previously elusive genetic variations. This study not only highlights the value of studying isolated populations but also emphasizes the need for diverse and comprehensive reference maps to accurately capture the complexity of human genetic diversity. Future studies on genome structure of indigenous Americans should include a telomere-to-telomere reference construction and comparison/addition to the human pangenome. Our current work contributes to the broader goal of enhancing genomic research's inclusivity and represents a step toward more precise understanding of genetic factors underlying health disparities in underrepresented populations.
Supplementary Material
Acknowledgments
We thank Chris Wiedrich for his help in genotype validation, Sayuko Kobes for running association tests, Dr. Paolo Piaggi for providing statistical consultation, and Dr. Peter Audano for database inquiry. This work was supported by the Intramural Research Program of the NIDDK, NIH. We utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Contributor Information
Çiğdem Köroğlu, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Peng Chen, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Michael Traurig, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Serdar Altok, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Clifton Bogardus, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Leslie J Baier, Diabetes Molecular Genetics Section, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Phoenix, AZ 85004, USA.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
C.K. and C.B. contributed to study conception. C.K. designed the study design, analyzed data, wrote scripts, and performed troubleshooting. P.C. provided consultation regarding data analysis and contributed to custom scripting. S.A. contributed to custom scripting and troubleshooting. M.T. and C.K. performed genotyping and visualization. L.J.B. supervised the study. C.K. wrote the original draft and all authors commented on and revised previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Conflict of Interest
The authors have no relevant financial or nonfinancial interests to disclose.
Data Availability
Custom R scripts are accessible via this repository: https://github.com/cigdemkoroglu/NRS. Other available software applications are described in the “Methods” section. Individual-level sequencing data are not publicly available due to NIH exemption for indigenous genome data.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Literature Cited
- All of Us Research Program Investigators; Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, Jenkins G, Dishman E. The “All of Us” research program. N Engl J Med. 2019:381(7):668–676. 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A, Che H, Martin M, Bunikis I, Dahlberg J, Höijer I, Häggqvist S, Vezzi F, Nordlund J, Olason P, et al. De novo assembly of two Swedish genomes reveals missing segments from the human GRCh38 reference and improves variant calling of population-scale sequencing data. Genes (Basel). 2018:9(10):486. 10.3390/genes9100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyter D, Ingimundardottir H, Oddsson A, Eggertsson HP, Bjornsson E, Jonsson H, Atlason BA, Kristmundsdottir S, Mehringer S, Hardarson MT, et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat Genet. 2021:53(6):779–786. 10.1038/s41588-021-00865-4. [DOI] [PubMed] [Google Scholar]
- Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015:517(7536):608–611. 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang AY, Gao M, Chong Z. Accurate long-read de novo assembly evaluation with Inspector. Genome Biol. 2021:22(1):312. 10.1186/s13059-021-02527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chheda H, Palta P, Pirinen M, McCarthy S, Walter K, Koskinen S, Salomaa V, Daly M, Durbin R, Palotie A, et al. Whole-genome view of the consequences of a population bottleneck using 2926 genome sequences from Finland and United Kingdom. Eur J Hum Genet. 2017:25(4):477–484. 10.1038/ejhg.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day SE, Traurig M, Kumar P, Piaggi P, Koroglu C, Kobes S, Hanson RL, Bogardus C, Baier LJ. Functional variants in cytochrome b5 type A (CYB5A) are enriched in Southwest American Indian individuals and associate with obesity. Obesity (Silver Spring). 2022:30(2):546–552. 10.1002/oby.23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, Weissensteiner MH, Sedlazeck FJ. Towards population-scale long-read sequencing. Nat Rev Genet. 2021:22(9):572–587. 10.1038/s41576-021-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Puente A, Knowler WC, Pettitt DJ, Bennett PH. High incidence and prevalence of rheumatoid arthritis in Pima Indians. Am J Epidemiol. 1989:129(6):1170–1178. 10.1093/oxfordjournals.aje.a115238. [DOI] [PubMed] [Google Scholar]
- Ebert P, Audano PA, Zhu Q, Rodriguez-Martin B, Porubsky D, Bonder MJ, Sulovari A, Ebler J, Zhou W, Serra Mari R, et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021:372(6537):eabf7117. 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery EC, Young GT, Berrocoso EM, Chen L, McNaughton PA. HCN2 ion channels play a central role in inflammatory and neuropathic pain. Science. 2011:333(6048):1462–1466. 10.1126/science.1206243. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yang X, Chen H, Tan X, Yang Z, Deng L, Wang B, Kong S, Li S, Cui Y, et al. A pangenome reference of 36 Chinese populations. Nature. 2023:619(7968):112–121. 10.1038/s41586-023-06173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenomeAsia100 K Consortium . The GenomeAsia 100 K project enables genetic discoveries across Asia. Nature. 2019:576(7785):106–111. 10.1038/s41586-019-1793-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groza C, Schwendinger-Schreck C, Cheung WA, Farrow EG, Thiffault I, Lake J, Rizzo WB, Evrony G, Curran T, Bourque G, et al. Pangenome graphs improve the analysis of structural variants in rare genetic diseases. Nat Commun. 2024:15(1):657. 10.1038/s41467-024-44980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdasani D, Barroso I, Zeggini E, Sandhu MS. Genomics of disease risk in globally diverse populations. Nat Rev Genet. 2019:20(9):520–535. 10.1038/s41576-019-0144-0. [DOI] [PubMed] [Google Scholar]
- Ho SS, Urban AE, Mills RE. Structural variation in the sequencing era. Nat Rev Genet. 2020:21(3):171–189. 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon T, Mars K, Young G, Tsai YC, Karalius JW, Landolin JM, Maurer N, Kudrna D, Hardigan MA, Steiner CC, et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data. 2020:7(1):399. 10.1038/s41597-020-00743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson LT, Nagi DK, Pillemer SR, Knowler WC, Hanson RL, Pettitt DJ, Bennett PH. Low prevalences of chronic widespread pain and shoulder disorders among the Pima Indians. J Rheumatol. 1996:23(5):907–909. [PubMed] [Google Scholar]
- Jaiswal M, Fufaa GD, Martin CL, Pop-Busui R, Nelson RG, Feldman EL. Burden of diabetic peripheral neuropathy in Pima Indians with type 2 diabetes. Diabetes Care. 2016:39(4):e63–e64. 10.2337/dc16-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HI, Ye B, Gosalia N; Regeneron Genetics Center; Köroğlu Ç, Hanson RL, Hsueh WC, Knowler WC, Baier LJ, Bogardus C, et al. Characterization of exome variants and their metabolic impact in 6,716 American Indians from the Southwest US. Am J Hum Genet. 2020:107(2):251–264. 10.1016/j.ajhg.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978:108(6):497–505. 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- Koroglu C, Gluck ME, Traurig M, Votruba SB, Krakoff J, Stinson EJ, Chen P, Bogardus C, Piaggi P, Baier LJ. Assessing established BMI variants for a role in nighttime eating behavior in robustly phenotyped Southwestern American Indians. Eur J Clin Nutr. 2020:74(12):1718–1724. 10.1038/s41430-020-0654-z. [DOI] [PubMed] [Google Scholar]
- Li R, Tian X, Yang P, Fan Y, Li M, Zheng H, Wang X, Jiang Y. Recovery of non-reference sequences missing from the human reference genome. BMC Genomics. 2019:20(1):746. 10.1186/s12864-019-6107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WW, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, Lu S, Lucas JK, Monlong J, Abel HJ, et al. A draft human pangenome reference. Nature. 2023:617(7960):312–324. 10.1038/s41586-023-05896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020:21(10):597–614. 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. Structural variant calling: the long and the short of it. Genome Biol. 2019:20(1):246. 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016:375(7):655–665. 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. Method of the year: long-read sequencing. Nat Methods. 2023:20(1):6–11. 10.1038/s41592-022-01730-w. [DOI] [PubMed] [Google Scholar]
- Mulder N, Abimiku A, Adebamowo SN, de Vries J, Matimba A, Olowoyo P, Ramsay M, Skelton M, Stein DJ. H3Africa: current perspectives. Pharmgenomics Pers Med. 2018:11:59–66. 10.2147/PGPM.S141546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M, Schatz MC. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics. 2016:32(19):3021–3023. 10.1093/bioinformatics/btw369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. The complete sequence of a human genome. Science. 2022:376(6588):44–53. 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016:538(7624):161–164. 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AJ, Chamberlain VF, Kearney VF, Stover D, Karafet T, Calderon K, Walsh B, Hammer MF. Genetic structure among 38 populations from the United States based on 11 U.S. Core Y chromosome STRs. J Forensic Sci. 2006:51(3):580–585. 10.1111/j.1556-4029.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- Reis ALM, Rapadas M, Hammond JM, Gamaarachchi H, Stevanovski I, Ayuputeri Kumaheri M, Chintalaphani SR, Dissanayake DSB, Siggs OM, Hewitt AW, et al. The landscape of genomic structural variation in Indigenous Australians. Nature. 2023:624(7992):602–610. 10.1038/s41586-023-06842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017:27(5):849–864. 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003:423(6942):825–837. 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Tsantoulas C, Laínez S, Wong S, Mehta I, Vilar B, McNaughton PA. Hyperpolarization-activated cyclic nucleotide-gated 2 (HCN2) ion channels drive pain in mouse models of diabetic neuropathy. Sci Transl Med. 2017:9(409):eaam6072. 10.1126/scitranslmed.aam6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulas C, Ng A, Pinto L, Andreou AP, McNaughton PA. HCN2 ion channels drive pain in rodent models of migraine. J Neurosci. 2022:42(40):7513–7529. 10.1523/JNEUROSCI.0721-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Jiang Z, Li T, Xie C, Zhao L, Yang J, Ouyang S, Liu Y, Li T, Xie Z. Structural variants in the Chinese population and their impact on phenotypes, diseases and population adaptation. Nat Commun. 2021:12(1):6501. 10.1038/s41467-021-26856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GT, Emery EC, Mooney ER, Tsantoulas C, McNaughton PA. Inflammatory and neuropathic pain are rapidly suppressed by peripheral block of hyperpolarisation-activated cyclic nucleotide-gated ion channels. Pain. 2014:155(9):1708–1719. 10.1016/j.pain.2014.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom R scripts are accessible via this repository: https://github.com/cigdemkoroglu/NRS. Other available software applications are described in the “Methods” section. Individual-level sequencing data are not publicly available due to NIH exemption for indigenous genome data.