Summary
Pancreatic ductal adenocarcinoma (PDAC) still has a poor response to therapies, partly due to their cancer-associated fibroblasts (CAFs). Here, we investigate the synergistic impact of a combinatory approach between a known chemotherapy agent, such as gemcitabine (GEM), and gene-modified human mesenchymal stromal/stem cells (MSCs) secreting the pro-apoptotic soluble (s)TRAIL (sTRAIL MSCs) on both PDAC cells and CAFs. The combo significantly impacts on PDAC survival in 2D and 3D models. In orthotopic xenograft models, GEM and sTRAIL MSCs induce tumor architecture shredding with a reduction of CK7- and CK8/18-positive cancer cells and the abrogation of spleen metastases. A cytotoxic effect on primary human CAFs is also observed along with an alteration of their transcriptome and a reduction of the related desmoplasia. Collectively, we demonstrate a promising therapeutic profile of combining GEM and sTRAIL MSCs to target both tumoral and stromal compartments in PDAC.
Graphical abstract

Highlights
-
•
MSC sTRAIL and GEM combo kills TRAIL-sensitive and resistant PDAC lines
-
•
GEM sensitizes CAF to MSC sTRAIL apoptosis impacting on their transcriptome
-
•
The combo reduces desmoplastic reaction in a 3D tumor avatar model
-
•
The combo induces tumor shredding and reduction of metastases in murine models
By 2D and 3D in vitro models combined with 2 orthotopic xenografts, Grisendi et al. demonstrate that chemotherapy with gemcitabine in combo with an MSC-delivered TRAIL efficiently targets both pancreatic cancer cells and their fibroblasts (CAFs), inducing apoptosis, modifying CAF transcriptome, reducing tumor-driven desmoplastic reaction, and, ultimately, abrogating metastases.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a leading cause of cancer-related mortality.1,2 Histologically, PDAC organizes into duct-like structures embedded into stroma, mainly composed by cancer-associated fibroblasts (CAFs) and extracellular matrix (ECM) accounting for up to 75% of tumor bulk.3,4 Polychemotherapy based on gemcitabine (GEM), oxaliplatin, 5-fluorouracil (5-FU), irinotecan, and Nab-paclitaxel (Nab-PTX) marginally impacts on advanced PDAC prognosis, with a 5-year survival of less than 10%.5,6,7,8,9 This poor outcome is primarily due to intrinsic or acquired resistance.10,11 Although it is known that stroma contributes to drug resistance, acting as physical barrier and immunosuppressant, the precise reasons behind the inadequate response to treatments are not yet fully understood. Importantly, the PDAC stroma releases soluble mediators and exosomes, remodels ECM, reprograms metabolic processes, and induces epigenetic modifications.11,12,13,14 Thus, strategies targeting both malignant and stromal compartments in PDAC shall be considered for better outcomes.
TRAIL/Apo2L (tumor necrosis factor-related apoptosis-inducing ligand [TNFSF10]) production is a way in which human body reacts against tumors, killing transformed cells without damaging healthy tissues.15,16 TRAIL triggers apoptosis by functional receptors expressed by target cells, Death Receptor 4 (DR4) and 5 (DR5),16,17,18 also acting on decoy receptors: Decoy receptor 1 (DcR1), Decoy receptor 2 (DcR2), and Osteoprotegerin.19 High levels of decoy receptors and anti-apoptotic molecules, including cFLIP, Bcl-2, and XIAP, may explain why healthy tissues are protected from TRAIL-induced apoptosis.20,21,22 The recombinant human TRAIL (rhTRAIL) has been already challenged in trials,23,24 providing evidence of safety and tolerability but with negligible impact on tumors linked with poor bioavailability due to low half-life.25,26,27 To overcome these limitations, cell-based delivery systems by gene-modified mesenchymal stromal/stem cells (MSCs) have been developed to express membrane-bound/soluble TRAIL forms.28,29,30,31,32,33,34,35 We reported that MSC releasing multimeric sTRAIL (sTRAIL MSC) increases bioavailability and maximizes its effects in PDAC.36 We additionally demonstrated how, combining conventional Nab-PTX with sTRAIL MSC, it is possible to improve single-agent efficacy and overcome resistances.37 Here, we further investigated the effectiveness of a synergistic approach combining the known GEM with sTRAIL MSC not only against different PDAC cell lines, having different sensitivity to both TRAIL and GEM, but also against primary CAF from PDAC patients. Using a bioreactor,38 we additionally generated 3D PDAC avatar models to mimic cellular interactions between malignant cells and CAF to be then treated by GEM+sTRAIL MSC. Two challenging orthotopic models were further implemented to validate the hypotheses, demonstrating safety and efficacy of an intratumor delivery of gene-modified MSC combined with a systemically delivered chemotherapy.
Results
sTRAIL and GEM in combo kill both TRAIL-sensitive and resistant PDAC lines
To challenge PDAC sensitivity to conditioned medium (CM) from MSC sTRAIL or GEM as single agents, dose-response assays were carried out using wild-type (WT) BxPC-3, MIA PaCa-2 cell lines, and a previously generated sTRAIL-resistant clone of BxPC-3 (sT-resistant BxPC-337) (Figure S1). We previously demonstrated high expression of DR5 and low expression of decoys DcR1 and DcR2 in all selected lines.36,37 Here, PDAC cells were cultured for 24 h with increasing concentrations of sTRAIL (0–2,500 pg/mL; Figures S1A, S1B, and S1C). Viability decreased down to 95% ± 3% compared to controls in WT BxPC-3 and MIA PaCa-2 after incubation with sTRAIL CM (Figures S1A and S1B). Conversely, sT-resistant BxPC-3 showed a weaker response (58% ± 1%) to sTRAIL highest concentration (2,500 pg/mL; Figure S1C). CM from empty vector (EV) MSC did not induce apoptosis in any lines. As previously demonstrated for other cancer types,28 and to further confirm that PDAC cell mortality was specifically due to antitumoral effect of sTRAIL contained in CM from engineered MSC, a TRAIL-neutralizing antibody was introduced in our killing assays (Figure S1D). WT BxPC-3 cultured for 24 h with CM containing sTRAIL in presence of different concentrations of anti-human TRAIL antibody were protected from death starting from the lowest antibody concentration (0.6 μg/mL), suggesting sTRAIL specificity in inducing PDAC death.
Dealing with GEM as a single agent, after 48 h of incubation, we could significantly reduce WT BxPC-3 and sT-resistant BxPC-3 viability even at the lowest tested concentration (i.e., 0.5 μM). This increased at 72 h, with a drop of cell viability down to 42% ± 3% for WT BxPC-3 and 56% ± 3% for sT-resistant BxPC-3 at 0.5 μM GEM (Figures S1E and S1G). Conversely, MIA PaCa-2 demonstrated the highest resistance to GEM without significant mortality in 48 h, while a slightly decrease in cell viability was observed after 72 h, alongside the increase in GEM concentration. However, at the highest GEM concentration (2,000 μM), 52% of MIA PaCa-2 cells remained alive (Figure S1F). These data outlined how intrinsic and induced sTRAIL resistance did not affect GEM sensitivity in all PDAC lines, with both WT and sT-resistant BxPC-3 more sensitive than MIA PaCa-2.
These different sensitivities by PDAC lines reflect patient heterogeneity,9 thus representing a reliable platform to investigate the impact of a combo treatment by GEM+sTRAIL. Thus, we selected appropriate concentrations of GEM and MSC-derived sTRAIL to assess the synergy of their combination (Figures 1 and S2; Table S1). The combined treatment of GEM (10 μM) and sTRAIL (250 pg/mL) yielded a significant increase in WT BxPC-3 death versus either sTRAIL or GEM alone (Figure 1A). Similarly, co-treatment of MIA PaCa-2 with GEM (100 μM) and sTRAIL (500 pg/mL) doubled mortality compared to single agents (Figure 1B). This enhanced pro-apoptotic effect was also observed in sT-resistant BxPC-3 line after the combo GEM (10 μM) and sTRAIL (300 pg/mL), compared to single treatments (Figure 1C). To further confirm this, we introduced a proliferation assay on both WT BxPC-3 and MIA PaCa-2 demonstrating a decrease of the proliferation marker Ki67 (Figure 1D) and a reduction of actively proliferating tumor cells (Figures S3A and S3B) in the combo conditions. In addition, BxPC-3 line was tested in a clonogenicity assay (Figures S3C and S3D) with evidence of different levels of sensitivity to single agents. Collectively, these data value the combo treatment in proliferation rate reduction next to apoptosis induction.
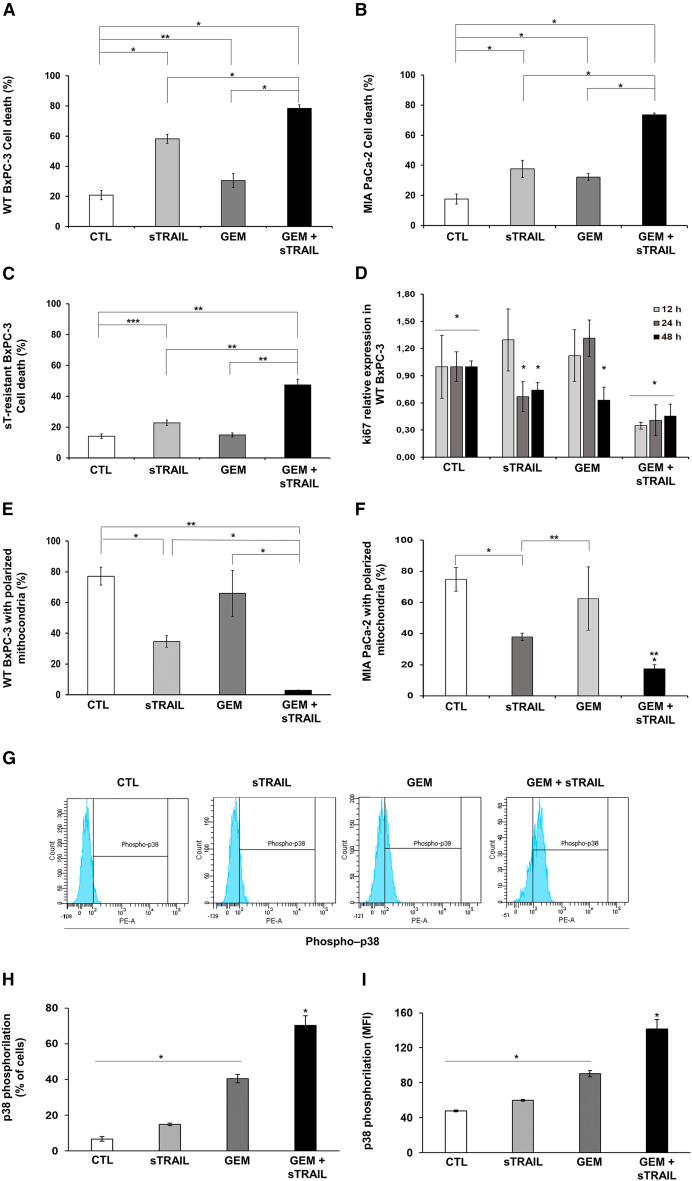
Figure 1.
sTRAIL and GEM combo impacts on PDAC survival provoking a loss of mitochondrial membrane potential and p38 phosphorylation
(A–C) Cytotoxicity assay performed in 2D by propidium Iodide PI (FACS analysis) on WT BxPC-3 (A), MIA PaCa-2 (B), and sT-resistant BxPC-3 (C) pre-treated with 10 μM (WT BxPC-3 and sT-resistant BxPC-3) or 100 μM (MIA PaCa-2) GEM for 24 h and subsequently treated for 24 h with CM collected from sTRAIL MSC containing 250 (WT BxPC-3), 500 (MIA PaCa-2), or 300 (sT-resistant BxPC-3) pg/mL sTRAIL, respectively. Culture media alone as a control (CTL). p values by t test: ∗p < 0.00001, ∗∗p < 0.005, ∗∗∗p < 0.05.
(D) Relative expression of Ki67 in WT BxPC-3 after 12, 24, and 48 h with different treatments. p values by t test for GEM, sTRAIL, and GEM+sTRAIL compared to the control. ∗p values ≤ 0.001.
(E and F) Mitochondrial depolarization (MD) by MitoStatus TMRE staining on cell lines. MD was evaluated after 12 h using FACSAria III. Untreated cells were used as a control. p value by t test for GEM, sTRAIL, and GEM+sTRAIL versus control: ∗p ≤ 0.05 for BxPC-3, ∗p ≤ 0.001 for MIA PaCa-2. p values by t test for GEM+sTRAIL compared to the single treatments: ∗∗p ≤ 0.005 for BxPC-3, ∗∗p ≤ 0.002 for MIA PaCa-2.
(G) Representative FACS analysis of p38 phosphorylated in WT BxPC-3 after treatment with GEM, sTRAIL, or GEM+sTRAIL.
(H) Percentage of cells showing p38 phosphorylated. p values by t test: ∗p < 0.04.
(I) Mean fluorescence intensity (MFI) of phosphor-p38 in WT BxPC-3 after indicated treatments. p value by t test: ∗p < 0.02. All experiments were performed in triplicate and repeated at least two times.
sTRAIL and GEM induce PDAC apoptosis promoting mitochondrial depolarization involving p38 MAPK and Bcl-2
To explore the mechanisms behind the observed synergy, we first evaluated the loss of mitochondrial membrane potential of treated WT BxPC-3 and MIA PaCa-2. The induction of mitochondrial membrane permeabilization with cytochrome c release and subsequent activation of caspase cascade is a mechanism by which GEM and TRAIL mediate apoptosis.39,40,41 The co-treatment with GEM (24 h) + sTRAIL CM (12 h) yielded a greater mitochondrial depolarization (MD) compared to single agents, supporting the observed cooperation (Figures 1E and 1F). To further elucidate the pathways involved in the synergy, we investigated the phosphorylation state of p38 mitogen-activated protein kinase (MAPK) which is known to be shared in both TRAIL and GEM antitumor activity, as one of the key players in MD acting on Bcl-2.42,43 Co-treatment by GEM+sTRAIL (4 h) led to a significant elevation in p38 phosphorylation, compared to single agents (Figures 1G–1I).
In addition, to determine whether MD is a necessary trigger for cell death following the combo treatments or simply represents a consequence, we examined the potential protective effect of Bcl-2 overexpression, known to counteract TRAIL-induced apoptosis.44 For this purpose, by lentiviral transduction a stable Bcl-2-overexpressing BxPC-3 line was created, as a model to test whether an increased Bcl-2 can interfere in cell death. Transduction efficiency was monitored by GFP expression (Figure S4A), with more than 80% of both Bcl-2 BxPC-3 and control (LC) BxPC-3 cells infected. Fluorescence-activated cell sorting (FACS) analysis was then introduced to detect Bcl-2 protein expression in WT and gene-modified BxPC-3 (Figure S4B). While WT BxPC-3 and LC BxPC-3 exhibited comparable levels of endogenous Bcl-2, as reported,44 in Bcl-2 BxPC-3 we detected a marked increase of Bcl-2.
Having generated the Bcl-2 BxPC-3 line and to assess whether MD might be affected by Bcl-2 overexpression, we evaluated the loss of mitochondrial membrane potential after GEM and sTRAIL, as single agents or in combo. Bcl-2 BxPC-3 did not exhibit relevant mitochondrial damage, maintaining viability (>80%) of PDAC cells across all treatment conditions (Figures S4C and S4D). In single agent-treated Bcl-2 BxPC-3 cells, the levels of mitochondrial polarization remained high and consistent with those observed in untreated controls (Figure S4D). A slightly although significant decrease in loss of polarization was observed in GEM+sTRAIL samples (10% ± 3% of depolarized cells compared to CTL, Figure S4D); however, the cytotoxic effect of combo treatment was largely reduced by Bcl-2 overexpression in BxPC-3 compared to WT BxPC-3, as described in Figure 1E. These data suggest that GEM+sTRAIL acts synergistically for PDAC death by activation of both intrinsic and extrinsic apoptosis pathways, identifying how Bcl-2 overexpression may protect PDAC from GEM+sTRAIL.
sTRAIL MSC and GEM combo has a massive antitumor impact in a 3D culture
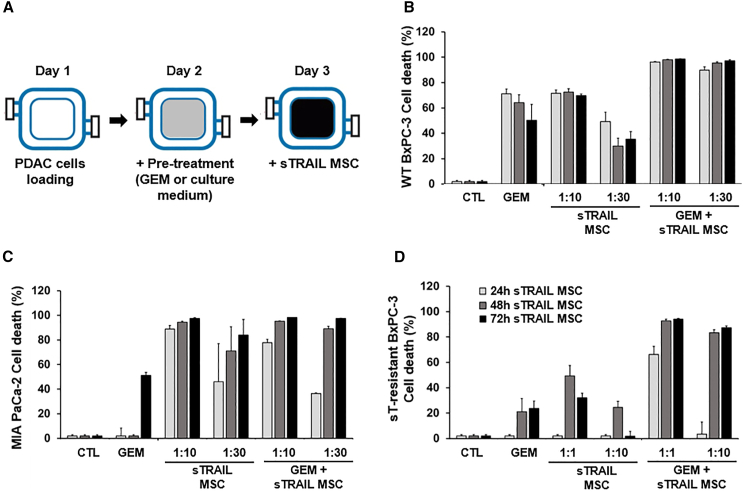
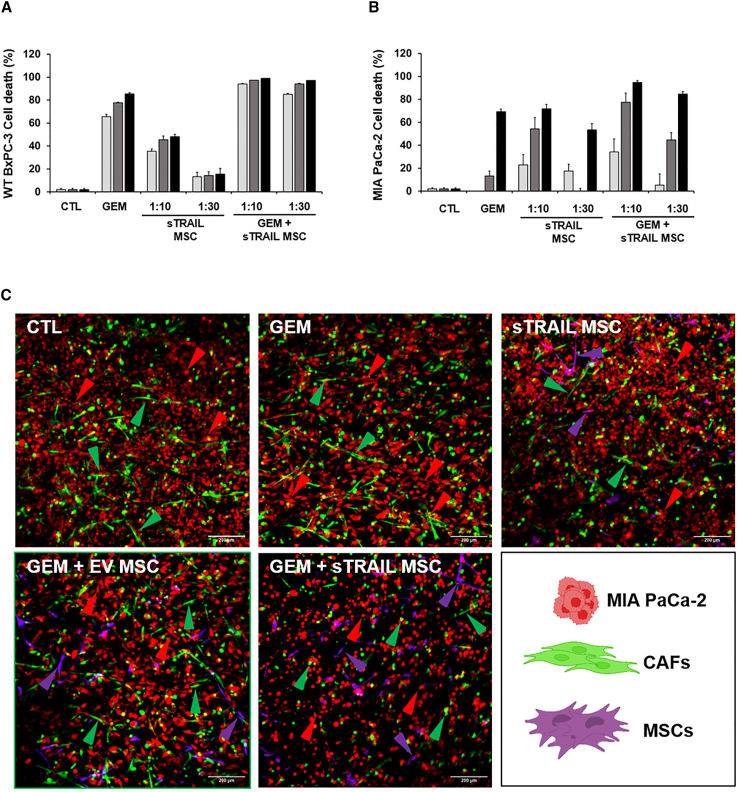
We then challenged the observed therapeutic potential by a PDAC 3D model introducing sTRAIL MSC. VITVO bioreactor recreated a 3D culture system (Figure 2).38 Luc+ PDAC cell lines were loaded and treated by GEM (10 μM for WT- and sT-resistant BxPC-3, 100 μM for MIA PaCa-2) for 24 h. sTRAIL MSCs were afterward injected at different effector:target ratios (E:T 1:1 and 1:10 for sT-resistant BxPC-3, 1:10 and 1:30 for other lines) (Figure 2A). Notably, for sT-resistant BxPC-3, we selected higher E:T ratios due to the outlined resistance to sTRAIL. For WT BxPC-3, the combination was more efficient versus single drugs after only 24 h of treatment. GEM increased sTRAIL sensitivity, and GEM and sTRAIL MSC combined at the lowest dose (1:30) yielded a cell death rate of 90% ± 2%, while only 49 (±7%) of WT BxPC-3 cells were killed by sTRAIL MSC 1:30 alone (Figure 2B). Similarly, GEM+sTRAIL MSC 1:10 and 1:30 provoked apoptosis in 98% ± 1% (1:10) and 97% ± 1% (1:30) in MIA PaCa-2 after 72 h of treatment, improving GEM-mediated cytotoxic impact (51% ± 2%, Figure 2C). GEM alone did not determine a significant cell death in sT-resistant BxPC-3 (48 h, 21% ± 10%; 72 h, 24% ± 6%) but was able to revert sTRAIL resistance, and, after 48 h of treatment with GEM+sTRAIL MSC 1:10, a cell death rate of 83% ± 2% was observed (Figure 2D).
Figure 2.
sTRAIL MSC and GEM generate a robust anti-PDAC effect in 3D cultures
(A) Cartoon of the experimental layout: luciferase+ PDAC cell lines (B, WT BxPC-3; C, MIA PaCa-2; D, sT-resistant BxPC-3) were cultured in VITVO (day 1), pre-treated (day 2) with GEM (10 μM for WT BxPC-3 and sT-resistant BxPC-3, 100 μM for MIA PaCa-2), or with plain medium for 24 h, and subsequently treated (day 3) up to 72 h with different E:T ratios of sTRAIL MSC (WT BxPC-3 and MIA PaCa-2 E:T = 1:10 and 1:30; sT-resistant BxPC-3 E:T = 1:1 and 1:10). Data are represented as mean ± SD (n = 3 experimental replicates). p values by t test: (B), CTL/GEM/sTRAIL MSC (1:30 and 1:10) vs. GEM+sTRAIL MSC (1:30 and 1:10) 24 h p ≤ 0.01, 48 h p < 0.005, 72 h p < 0.01; (C), CTL/GEM vs. GEM+sTRAIL MSC (1:30 and 1:10) 24 h p < 0.005, 48 h p < 0.0005, 72 h p < 0.005; (D), CTL/GEM/sTRAIL MSC 1:1 vs. GEM+sTRAIL MSC 1:1 24 h p 0.0005; CTL/GEM/sTRAIL MSC (1:10 and 1:1) vs. GEM+sTRAIL MSC (1:10 and 1:1) 48 h p 0.0005, 72 h p 0.00001.
We then investigated whether GEM affects sTRAIL MSC viability themselves to anticipate possible detrimental effects of chemotherapy given on sTRAIL MSC in combo. Interestingly, sTRAIL MSCs were less sensitive to GEM than WT MSC (up to 200 μM GEM), with over 60% viability after 72 h of exposure (Figure S5A). More importantly, sTRAIL release by MSC was not compromised by GEM treatment (Figure S5B). Indeed, sTRAIL levels were comparable in CM harvested after 48 h from cells exposed and unexposed to 10 μM GEM, while 100 μM GEM doubled sTRAIL secretion (Figure S5B). These data together in a more complex 3D model confirmed the synergistic effect of GEM+sTRAIL MSC combo for all PDAC lines, independently of their specific sensitivities to sTRAIL or GEM as single agents.
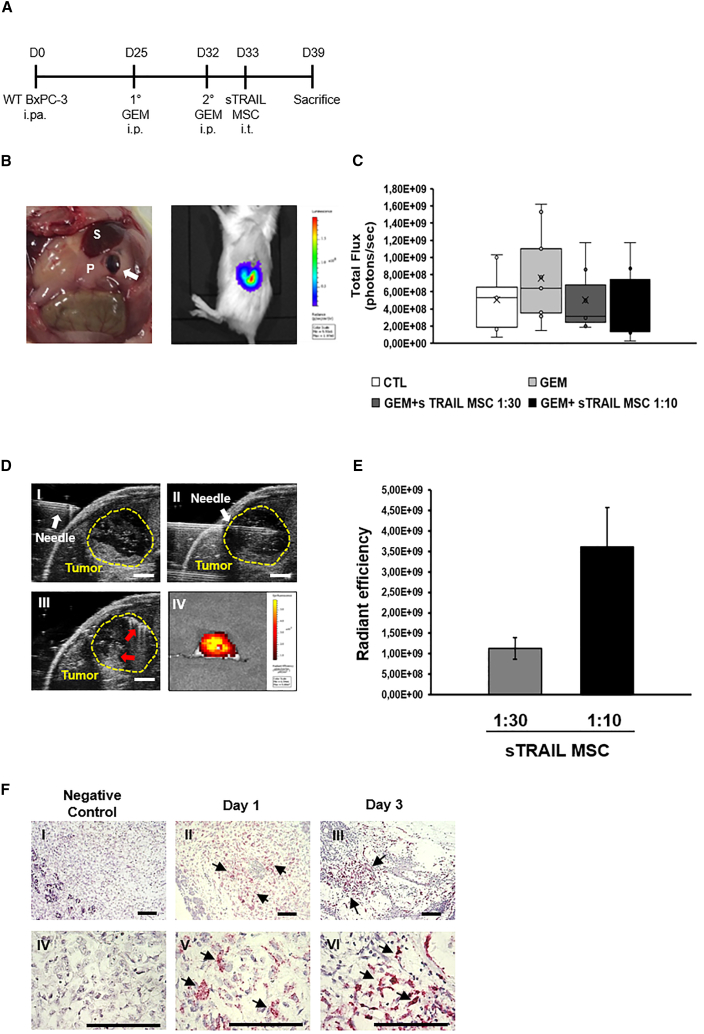
PDAC engrafts into murine pancreas to be targeted by US-guided sTRAIL MSC intratumoral delivery
A first PDAC orthotopic model was implemented to mimic a clinical situation with intratumorally delivered cells and assess sTRAIL MSC persistence, to ultimately confirm the efficacy of GEM+sTRAIL MSC. At day 0, 1 × 106 WT BxPC-3-Luc+ cells were intra-PDAC (in the tail) injected into non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) mice, with a pocket that was visible inside the pancreas after the procedure (Figures 3A and 3B, left panel). Intratumoral localization was verified by bioluminescence (BLI) starting at day 7 (Figure 3B, right panel and 3C). GEM (50 mg/kg) was intraperitoneally (i.p.) administered at days 25 and 32, while DiR-labeled sTRAIL MSCs were delivered at two different doses (1:10 and 1:30) on day 33 by ultrasound (US)-guided intratumor (i.t.) injection (Figures 3A and 3D), to enable precise syringe needle location inside the tumor and monitor cell distribution during injection (Figure 3D I–III). The presence of fluorescent areas in tumor burden, indicating the engraftment and stable persistence of DiR-labeled sTRAIL MSC in the PDAC xenotransplant, was detected up to 7 days after cell injection in all treated tumors (Figure 3D IV). After 7 days, the fluorescence intensity registered from PDAC samples detected in mice who received sTRAIL MSC at the 1:10 dose was three times higher compared to the 1:30 dose, thus proportionally reflecting the higher number of injected cells (Figure 3E). Notably, no other tissues showed fluorescence, suggesting that sTRAIL MSC did not migrate toward other organs after intratumor administration (data not shown). The BaseScope assay after necropsy confirmed that the MSC detected by fluorescence in vivo could engraft into PDAC. sTRAIL MSCs appeared either as spindle-shaped viable cells organized in small groups or as single cells (Figure 3F, arrows). The staining intensity and the abundant red dots inside the cytoplasm suggested that engineered MSC expressed the sTRAIL gene in vivo even after intratumor delivery (Figure 3F, II, III, V, and VI). sTRAIL RNA was not visible in samples collected from untreated mice (Figure 3F, I and IV). This orthotopic model confirmed the possibility to recreate human PDAC in mice, indicating the safety and feasibility of the sTRAIL MSC in vivo intra-PDAC injection.
Figure 3.
Establishment of the orthotopic model targeted by sTRAIL MSC intratumoral delivery
(A) Animal treatment schedule. WT BxPC-3-luciferase+ cells (1 × 106) were injected in NOD-SCID mice pancreas at day 0. GEM was intraperitoneally administered at days 25 and 32, followed by ultrasound (US)-guided intratumor injection of sTRAIL MSC in two doses (E:T = 1:30 and 1:10) at day 33. Number of mice/group: CTL, n = 9; GEM, n = 9; GEM+sTRAIL MSC 1:30, n = 7; GEM+sTRAIL MSC 1:10, n = 8.
(B) Left picture: after WT BxPC-3 injection by surgical procedure, a pocket within the mice pancreas was visible (white arrow). P, pancreas; S, spleen; right picture: representative image of tumor bioluminescence signal 7 days after WT BxPC-3 injection.
(C) Boxplot referring the quantitative analysis of bioluminescence signal, measured in total flux (photon/sec), in mice 7 days after WT BxPC-3 injection. p values by t test: p ≥ 0.05.
(D) Representative images of sTRAIL MSC US-guided injection in orthotopic PDAC. I, the syringe needle is entering mouse skin; II, the needle is inside the tumor; III, sTRAIL MSCs (red arrows) are located inside the tumor after injection. Yellow dashed line surrounds the tumor area. Scale bar 2 mm. IV, fluorescence image of murine pancreas with intrapancreatic tumor explanted 7 days after injection of sTRAIL MSC labeled by DiR.
(E) Fluorescence radiant efficiency signal in the GEM+sTRAIL MSC groups (1:10 and 1:30) generated by DiR-labeled sTRAIL MSC engrafted into pancreatic tumor 7 days post-injection. p value by t test: ∗p < 0.05.
(F) Representative images of BaseScope Assay on WT BxPC-3 tumors one (II and V) and three (III and VI) days after sTRAIL MSC (1:10) injection. Untreated tumor samples were also introduced as negative control (I and II). Individual sTRAIL RNA transcripts appear as distinct dots (arrows) of red chromogen precipitate. I–III, magnification 100×; IV–VI magnification 400×. Scale bar 100 μm.
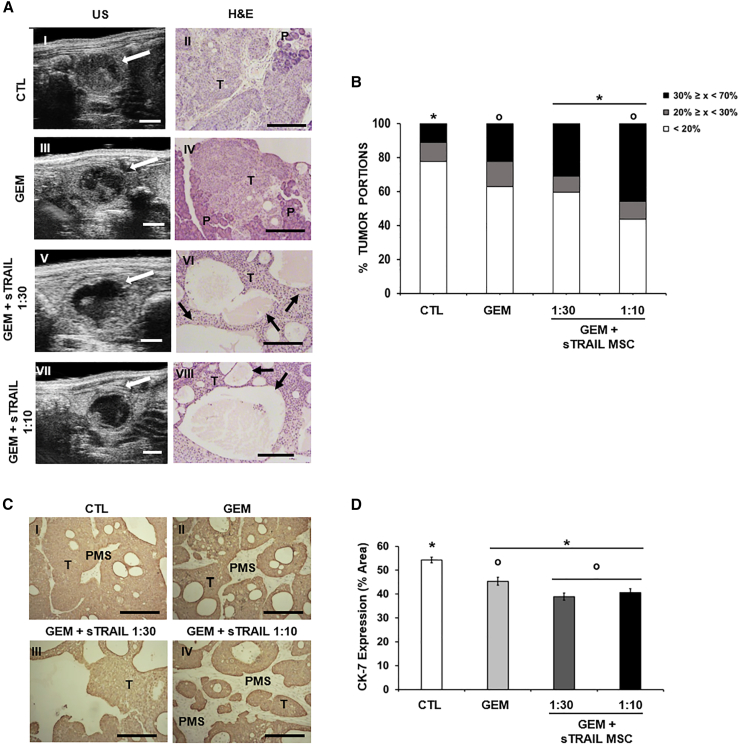
sTRAIL MSC and GEM in combo impact PDAC orthotopic models
Once created the model, we proceeded with US imaging allowing non-invasive and real-time analysis of implanted PDAC volume and neoplastic tissue composition over treatment (Figure 4A). Areas of necrosis and serum were visible by US as intense hypoechoic (dark) regions in comparison to trophic tumor parenchyma, as reported.45 A small amount of necrosis was observed in the control group, while treated mice displayed a higher degree of hypoechoic areas due to tumor tissue shredding (Figure 4A). Three categories of hypoechoic/empty areas in tumors were defined: tumor portions with 0%–20% empty area (low necrotic), tumor portions with 20%–30% empty area (intermediate necrotic), and tumor portions with over 30% empty area and up to a maximum of 70% (high necrotic) (Figure 4B). Most tumor portions in control, GEM, and GEM+sTRAIL 1:30 groups displayed a level of black/hypoechoic/empty tissue between 0% and 20% (78%, 63%, and 60%, respectively). However, the latter demonstrated a slight increase in the number of tumor portions with a range of black/hypoechoic/empty ≥30%, suggesting an antitumor effect of GEM+sTRAIL MSC starting from at the lowest dose. Importantly, in the group treated with GEM+sTRAIL MSC 1:10, the majority (56%) of tumor portions presented more than 20% necrotic area, and there we observed a 4- and 2-folds increase of tumor portions with ≥30% of necrosis versus control and GEM groups, respectively.
Figure 4.
sTRAIL MSC and GEM generate a robust anti-PDAC effect in vivo
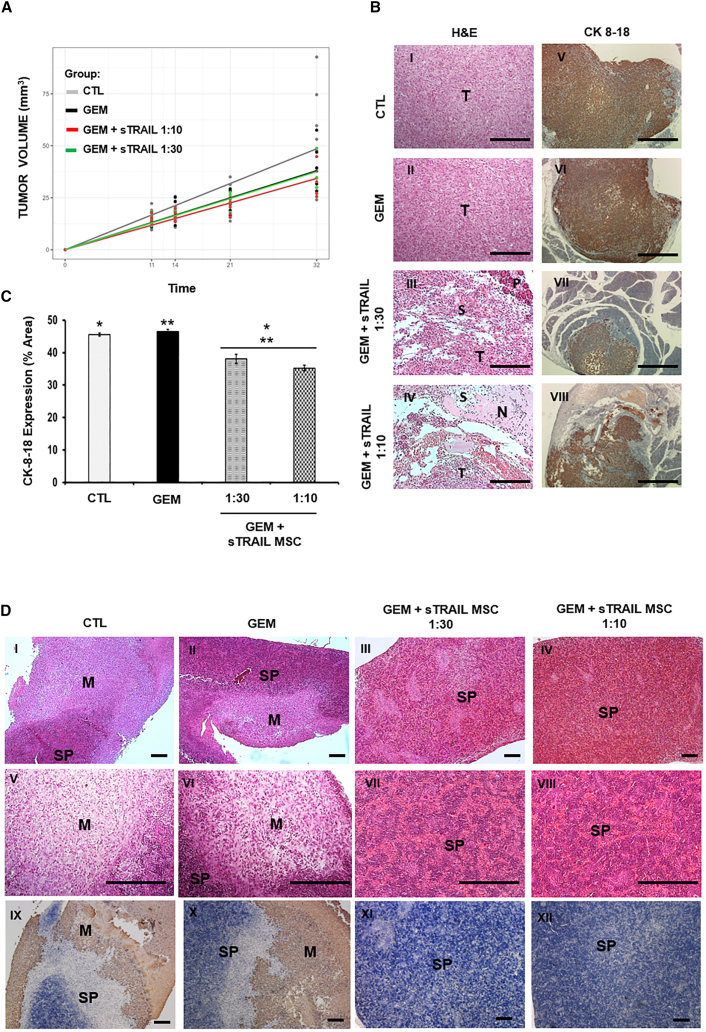
(A) Representative US and histological H&E images of orthotopic-implanted PDAC. In the left column US pictures of control (I) GEM alone treated (III) or with GEM+sTRAIL MSC 1:30 (V) or 1:10 (VII). White arrows indicate the intrapancreatic tumor. Compact and trophic neoplastic tissue was visible as bright hyperechoic regions in the control (I), while degenerated tumor appeared as hypoechoic (dark) regions in the treated mice (III, V, VII). Scale bar 2 mm. In the right column, microphotographs of H&E staining of tumors. T, tumor parenchyma; P, pancreas; black arrow, necrotic areas. Magnification 100×, scale bar 200 μm.
(B) Percentage of tumor portions per group with black/hypoechoic/empty areas measured via US. p values by chi-squared test: x < 20%, CTL/GEM vs. GEM+sTRAIL MSC 1:10 p ≤ 0.05; 30% ≤ x < 70%, CTL vs. GEM+sTRAIL MSC 1:30 p < 0.05, CTL/GEM vs. GEM+sTRAIL MSC 1:10 p ≤ 0.01.
(C) Histology of PDAC in mice stained by anti-CK7 IHC; pancreatic murine stroma (PMS) is visible among human tumor cells (T). Magnification 100×, scale bar 200 μm.
(D) Quantification of CK7-positive areas within groups. p values by t test: ∗p < 0.00001; °p < 0.05.
These US observations were then validated by histology. H&E staining was compared to US imaging, and the presence of large necrotic areas in treated samples was confirmed (Figure 4A). Hypoechoic regions detected by US corresponded to the degenerated and necrotic areas in tumor specimens. All treatments modified PDAC microenvironment integrity with empty holes. However, the combo was able to empower GEM effect provoking the formation of larger, more numerous areas of necrosis together with empty space (Figures 4A and 4C). A significant reduction of CK7 expression was also observed in all treated tumors compared to the control after the combo (at both E:T ratios), having a higher reduction of tumor cells compared to GEM alone (CK7-positive cells: CTL = 54% ± 1%; GEM = 45% ± 2%; GEM+sTRAIL 1:30 = 39% ± 2%; GEM+sTRAIL 1:10 = 41% ± 2%), further confirming the synergistic antitumor impact observed in H&E analysis (Figures 4C and 4D). Curiously, histology revealed that xenotransplants were composed by WT BxPC-3 organized into tumor “islets,” together with infiltrating pancreatic murine stroma (PMS) cells (Figure 4C). Hence, we sought to evaluate the impact of the combo on PMS too, isolating them as a primary line from an explanted WT BxPC-3 tumor to then perform a cytotoxicity assay. PMS displayed resistance to sTRAIL (Figure S6), with 95% ± 0% of cells still viable after incubation with sTRAIL CM. GEM was able to reduce PMS viability without significant contribution of the GEM+sTRAIL CM combo (51% ± 5% and 44% ± 3%, respectively).
To further challenge the combo efficacy in vivo, we generated an additional orthotopic PDAC model in a less favorable setting using the highly metastasizing MIA PaCa-2.46 This line retains a lower sensitivity to GEM and sTRAIL compared to BxPC-3 (Figures S1B and S1F) and, in previous pre-clinical models, did not typically show necrosis even after treatment with promising anti-PDAC agents.47,48,49 A significant reduction of tumor volume was observed by US in mice treated with the combo GEM+sTRAIL MSC at the dose 1:10 versus the control group, while apparently no volumetric difference was evident versus the GEM group alone (Figure 5A). After sacrifice, MIA PaCa-2 tumors were collected to evaluate histopathology (tumor architecture and cell morphology): the control and GEM groups were more compact while, on the contrary, an alteration of tumor architecture with a less dense PDAC structure, empty spaces, and a higher stromal component were observed in mice treated by GEM+sTRAIL MSC, in particular at 1:10 ratio (Figure 5B, left column). To better compare the tumor burden among groups, anti-CK8/18 staining, a known PDAC marker,50 was performed outlining a significant reduction in CK8/18+ cells in mice treated with GEM+sTRAIL MSC versus both the control and GEM groups. No significant difference was noted when comparing the control group with mice solely treated by GEM (Figure 5B, right column, and 5C).
Figure 5.
sTRAIL MSC and GEM impact on MIA PaCa-2 abrogating splenic metastasis in an orthotopic model
(A) Tumor volume was measured through US scans from the day of tumor implant until sacrifice. The dependent variables were the raw measurements of the outcomes, whereas the set of independent variables includes treatment arm, time (in days from baseline), and the interaction between arm and time. A random intercept term was also included to account for repeated measurements over the same individual. Results of this analysis were expressed as mean differences and are reported with 95% confidence intervals (CIs) and p values. All treated groups show a significant reduction in tumor growth compared to CTL; in particular GEM vs. CTL p = 0.001; GEM+ sTRAIL1:30 vs. CTL p = 0.001; GEM+sTRAIL1:10 vs. CTL p = 0.000005.
(B) H&E staining and immunohistochemistry anti-CK8/18 on intrapancreatic tumors. Positive areas (brown, 3,3′-diaminobenzidine [DAB]) represent human PDAC tumor. Tumor degeneration was observed in mice treated with GEM+sTRAIL MSC. T, tumor; S, stroma; N, necrotic area; P, pancreas. Magnification 100×, scale bar 200 μm.
(C) Quantification of CK8/18-positive areas within the different groups. For each tumor, two slides were evaluated. p value by t test: ∗,∗∗p < 0.0001; ∗∗∗p < 0.05.
(D) Representative images of hematoxylin and eosin staining (upper and middle rows) and CK8/18 immunohistochemistry (lower row) on mice metastatic spleens. Tumor metastatic areas were found in the spleen of control group (CTL) and of mice treated with GEM, but not in the groups treated with the combinatorial regimen. M, metastasis; SP, spleen. I–IV and IX–XII magnification 100×, scale bar 200 μm; IX–XII magnification 200×, scale bar 100 μm.
During necropsy MIA PaCa-2 spleen localizations were noticed within the control and GEM groups. Metastases appeared as white patches in up to 43% of the control group and in 33% of the mice treated with GEM. Interestingly, mice treated with GEM+sTRAIL MSC did not reveal metastases. Spleens were then analyzed for histological confirmation by H&E staining (upper and middle panels) and anti-CK8/18 immunohistochemistry (lower panels) (Figure 5D). These findings with a highly aggressive MIA PaCa-2 model confirmed the local effectiveness of GEM+sTRAIL MSC on primary tumor, additionally suggesting the control of a metastatic disease.
Myofibroblastic and inflammatory CAF subtypes are isolated from PDAC samples
Human primary CAFs were obtained from four patients (PZ1–PZ4). These fibroblast-shaped cells (Figure S7A, arrows) were characterized for CD73 (98% ± 1%), CD90 (99%), and CD105 (87% ± 12%) expression and the lack of CD45, EPCAM, and HLA-DR markers, proving their stromal cell lineage as reported,51 and without leukocyte or epithelial contaminants (Figure S7B). Moreover, to predict their sTRAIL sensitivity, we investigated key TRAIL receptors showing high levels of DR5 (60% ± 3%) and negligible expression of DR4 and decoy DcR1 and 2 receptors (<10%; Figure S7C). The isolated CAFs were further characterized by RNA sequencing (RNA-seq) for subtypes definition, which encompasses myofibroblastic CAFs (myCAFs), inflammatory CAFs (iCAFs), and antigen-presenting CAFs (apCAFs), in accordance with reported classifications.47,48,49 As illustrated in Figure S7, our CAFs exhibited distinct transcriptome markers, such as ACTA2 (α-SMA), POSTN (periostin), TAGLN (transgelin), COL1A1, COL10A1, COL12A1, THY1, and FAP, indicating a myCAFs-dominant signature (Figures S7D and S7E).52,53,54 Moreover, the IL6, CXCL1, CXCL12, and LIF expression, alongside a lack of DPT and HAS1 expression,55 pointed to the co-presence of an iCAFs subpopulation. Conversely, an absence of the HLA-DR expression implies that apCAFs constitute a negligible subpopulation within isolated CAFs (Figure S7D).52,53,54 It is also noteworthy that markers associated with other CAF subpopulations, including ENG, GLI1, HOXB6, and ISLR, were found to be expressed at considerable levels in the isolated CAFs (Figure S7D).54
sTRAIL MSC and GEM in combo eradicate PDAC in a 3D human PDAC avatars
Having isolated human CAF, considering their PDAC protective role14 and the TRAIL resistance observed in their murine counterpart (PMS), we generated a 3D fully human PDAC avatar recreating the complex PDAC architecture using CAF combined with PDAC lines. Cells were mixed at the same PDAC/CAF ratio, as reported in PDAC specimens from patients.4 Once established, 3D PDAC avatars were treated by GEM+sTRAIL MSC to verify whether CAF could have a protective effect. Despite their presence, the synergistic impact mediated by GEM+sTRAIL MSC 1:10 (T:E) and 1:30 was effective provoking at 72 h more than 95% ± 1% of cell death in PDAC lines (Figures 6A and 6B). The MIA PaCa-2 avatar co-treatment with GEM+sTRAIL MSC 1:10 significantly improved the antitumor effect displayed by single agents, reverting the observed GEM resistance and doubling the impact of sTRAIL MSC alone at 24 h (Figure 6B).
Figure 6.
Primary CAFs do not interfere on GEM and sTRAIL MSC cytotoxicity in 3D PDAC avatars
Luc+ BxPC-3 WT (A) and MIA PaCA-2 (B) pre-treated with 10 or 100 μM GEM for 24 h and subsequently treated up to 72 h with different concentrations of sTRAIL MSC (E:T = 1:10 and 1:30). Tumor viability was quantified by BLI signal intensity. Data are represented as mean ± SD (n = 2 experimental replicates). p values by t test: (A) 24, 48, and 72 h CTL/GEM/sTRAIL MSC (1.30 and 1:10) vs. GEM+sTRAIL MSC (1:30 and 1:10) p < 0.00005; (B), 24 h CTL/GEM vs. GEM+sTRAIL MSC (1:30 and 1:10) p ≤ 0.05, 24 h sTRAIL MSC 1:30 vs. GEM+sTRAIL MSC 1:10 p < 0.05; 48 h CTL/GEM/sTRAIL MSC 1:30 vs. GEM+sTRAIL MSC (1:30 and 1:10) p < 0.05; 72 h CTL/GEM/sTRAIL MSC (1.30 and 1:10) vs. GEM+sTRAIL MSC (1:30 and 1:10) p < 0.05.
(C) Confocal microscopy imaging of co-culture with MIA PaCA-2 (Orange CMRA Dye, red arrows) and CAF (CellTrace CFSE, green arrows) cells loaded into a VITVO and treated with GEM alone, sTRAIL MSC (CellTracker Deep Red dye, purple arrows) alone, or GEM+ sTRAIL MSC. Untreated MIA PaCa-2 or MIA PaCa-2 treated with GEM+EV MSC were used as controls. Images were acquired at the end of 72 h with Nikon A1 Plus confocal microscope; objective 10×; z stack step 5 μm; scale bar 200 μm. Experiments are expressed as duplicates.
Live imaging performed on 3D human PDAC avatars revealed the presence of both PDAC (MIA PaCa-2) and CAF cells homogenously distributed (Figure 6C). In the control (CTL), MIA PaCa-2 (red cells) efficiently colonized the 3D matrix appearing as a dense mesh of cellular elements at high density with a typical polygonal morphology (Figure 6C, red arrows; Figure S8). Differently, CAFs (in green) adopted an elongated shape, embedding among tumor cells and embracing scaffold fibers (Figure 6C, green arrows; Figure S8). Live images taken from VITVO revealed that treatment by single agents had only a slight impact on cells morphology: MIA PaCa-2 cells and CAF retained their elongated, viable cell shape after treatment with GEM or sTRAIL MSC alone (E:T ratio of 1:10, purple cells; Figure 6C, upper central and right panels). In these conditions, only a minor population of rounded cells with pycnotic nuclei was observed by microscopy corroborating the low level of mortality measured by BLI (Figure 6B). An apparently visible PDAC cell toxicity was not detectable after GEM+EV MSC treatment (Figure 6C, lower left panel). On the contrary, GEM+sTRAIL MSC combo approach provoked marked changes in MIA PaCa-2 and CAF morphologies with both cell types losing the classical phenotype of adherent cells to acquire a round shape largely with pycnotic nuclei, suggestive of cell death (Figure 6C, lower central panel; Figure S8, bottom panels). These data from complex in vitro models are consistent with those observed in the previous 3D cultures without CAF, indicating their negligible role in the anti-PDAC combo strategy.
GEM sensitizes CAF to sTRAIL impacting on their transcriptome and mitigating desmoplastic reaction in a 3D human PDAC avatar
Having excluded CAF interference in our approach, we investigated whether sTRAIL (0–2,500 pg/mL) may even trigger apoptosis in CAF. For all tested conditions, CAF viability after 24 h was not significantly reduced compared to untreated cells (Figures S9A–S9D), indicating a resistance to sTRAIL despite the observed DR5 expression. We then considered GEM as possible sensitizer for TRAIL resistance in CAF, as for cancer lines. Cells were pre-treated by GEM (10 μM) for 24 h, and CM containing sTRAIL (1,000 pg/mL) was added for 24 additional hours. Despite a cytotoxic effect of GEM as a single treatment (>40% of CAF death), the combo GEM+sTRAIL significantly reinforced GEM effect in all CAF samples (Figures S9E–S9H). Moreover, GEM+sTRAIL had an impact on CAF proliferation with a significant reduction of Ki67, associated with MD (Figures S10A and S10B), as described for PDAC cell lines and indicating that GEM+sTRAIL MSC can target both tumoral and CAF compartments in PDAC.
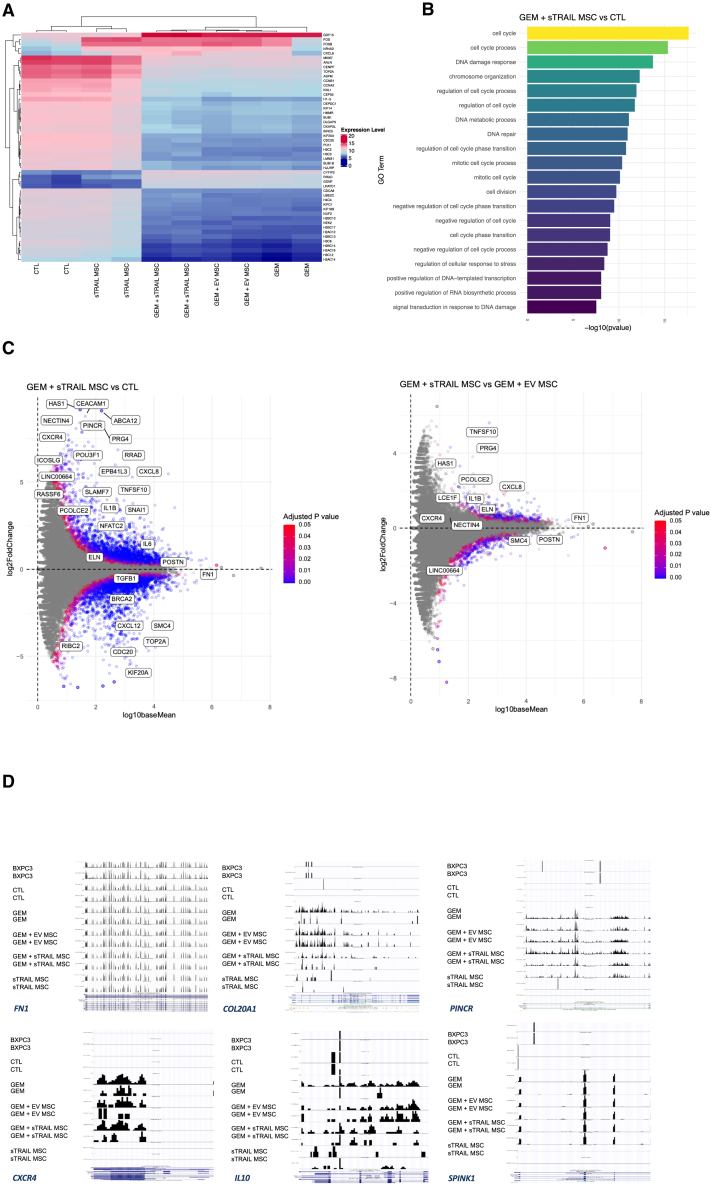
To further evaluate the impact of GEM+sTRAIL combo on CAF gene expression profile, RNA-seq was introduced. CAFs were cultured with GEM+sTRAIL MSC given as single agents or in combo. We also examined the effects of GEM with EV MSC. Post-treatment transcriptome analyses indicated that GEM+sTRAIL MSC combo downregulated CAF genes involved in cell-cycle checkpoints, histones, DNA replication, and chromosomal organization, such as MKI67, CDC20, SMC4, TOP2A, KIF20A, and BRCA2, with similar results obtained from GEM alone (Figures 7A and 7B and Data S1).
Figure 7.
GEM and sTRAIL MSC combo alters CAF transcriptome
(A) Heatmap shows a similar pattern of gene expression between GEM, GEM+EV MSC, and GEM+sTRAIL MSC.
(B) The most significant biological processes downregulated in combo treatment versus control using clusterProfiler Gene Ontology enrichment analysis.
(C) GEM+sTRAIL MSC treatment (left) shows downregulation of cell cycle and mitosis-related genes such as SMC4, TOP2A, KIF20A, and CDC20 and overexpression of CXCL8, NFATC2, and SNAI1 while expressions of FN1, POSTN, and TGFB1 do not show significant changes; GEM+sTRAIL MSC versus GEM-EV (right) shows overexpression of TNFSF10 and CXCL8.
(D) UCSC snapshot of different treatments on CAFs; BxPC-3 as a PDAC tumoral control cell line. Overexpression of COL20A1, CXCR4, IL10, PINCR, and SPINK1 in GEM, GEM-EV, and GEM+sTRAIL MSC treatment.
Considering our prevalence of myCAF, we further explored whether the GEM+sTRAIL MSC combo may alter myCAF-specific genes expression. However, no significant changes in key myCAF-related genes such as FN1, POSTN, and THY1 were observed. Very interestingly, a perturbation of genes contributing to ECM synthesis and organization emerged, with an increase in PCOLCE2, LRRC15, and COL20A1 and a decrease in COL1A1, COL27A1, and ACTA2 GEM+sTRAIL (Data S1). Moreover, the GEM+sTRAIL MSC combo upregulated epithelial-to-mesenchymal transition-related genes,56,57,58,59,60 such as SNAI1 and lncRNA PINCR, alongside ABCA12, SPINK1, and CEACAM (Figures 7C and 7D and Data S1). Finally, we report an induced immune response profile in CAFs after GEM+sTRAIL MSC combo with upregulation of IL6, IL10, IL33, HAS1, CXCL8, ICOSLG, IL1RN, NFATC2, and CXCR4, and a downregulation of CXCL12 and IL37 after (Figures 7C and 7D and Data S1). Collectively, these data suggest that the GEM+sTRAIL combo has an impact on PDAC tumor microenvironment (TME) in vitro, additionally triggering the activation of immune-related gene programs whose significance would require further experiments.
Having observed in RNA-seq data an ECM dysregulation and accounting that desmoplastic matrix a key PDAC feature,9 we investigated whether ECM could be generated in our human 3D PDAC avatars and how the GEM+sTRAIL MSC combo treatment may affect the quantity or quality of this ECM. An additional 3D PDAC model was then established in VITVO using MIA PaCa-2 and CAF, with the goal to quantify FN1 and COL1A1, as key ECM components61 (Figures S11A–S11E). Immunofluorescence of repopulated VITVO matrix demonstrated the presence of abundant well-organized bundles of FN1 in the CTL, GEM alone, sTRAIL MSC alone, and GEM+EV MSC groups (Figure S11A). Dissimilarly, in GEM+sTRAIL MSC combo FN1 resulted to be less dense and with a poorer organized structure (Figures S11A and S11B). In addition, DAPI nuclear staining highlighted the presence of small, condensed, and pyknotic nuclei in 3D PDAC avatars after GEM+sTRAIL MSC combo due tumoral and stromal cell apoptosis cells, as demonstrated by live imaging (Figure S11B, inset, white arrow). Quantification of DAPI- and FN1-positive area revealed their significant reduction after GEM+sTRAIL MSC combo compared to single agents or control groups (Figures S11C and S11D). COL1A1, tested in 3D PDAC avatars supernatants, was decreased after GEM or sTRAIL MSC, as single-agent treatments (Figure S11E). Nevertheless, GEM+sTRAIL MSC combo considerably enhanced this effect, decreasing COL1A1 levels by 2- or 3-fold compared to GEM or sTRAIL MSC alone, respectively (Figure S11E). These data indicate that the impact of GEM+sTRAIL MSC combo also reaches key ECM components next to the observed anti-cancer and anti-CAF actions.
Discussion
Despite the pro-apoptotic TRAIL being considered a promising anti-cancer agent,15,16 tumors revealed resistance by solo TRAIL treatment.62,63 Recently, combinatorial approaches were introduced to overcome TRAIL resistance, with pre-clinical and clinical studies also combining GEM and rhTRAIL in PDAC and in other cancers.41,64,65 There rhTRAIL increased GEM uptake improving efficacy versus GEM alone.66 Interestingly, GEM with agonistic TRAIL-R1 and TRAIL-R2 antibodies was tolerated in patients and was followed by partial responses.67,68
We introduced an approach using GEM combined with a multimeric form of sTRAIL delivered by MSC36 with the aim to target both tumoral and stromal component in PDAC. sTRAIL MSC represents a promising anti-cancer therapy, for tumor tropism and constant sTRAIL production, enabling a more stable bioavailability.31,36,68 Importantly, co-treatment with GEM did not impact sTRAIL release by MSC, and a very relevant proportion of sTRAIL MSC remained alive after 72 h with high GEM concentrations. Curiously, the treatment with high doses of GEM increased sTRAIL release, presumably due to a low level of MSC cytotoxicity followed by sTRAIL release into supernatant.
We targeted a panel of lines representing PDAC heterogeneity.9 WT BxPC-3 showed a better sensitivity to GEM and sTRAIL compared to MIA PaCa-2, as reported.36,69,70,71 The cellular mechanism by which MSCs expressing TRAIL variants kill tumor cells, based on the interaction with TRAIL receptors, has been demonstrated in our previous studies.28,36 Here, by the introduction of a TRAIL-neutralizing antibody to prevent the sTRAIL association with their receptors DR4 and DR5, we confirmed that PDAC mortality is specifically due to the presence of sTRAIL in CM in a TRAIL-receptor-dependent manner. Interestingly, the previously established TRAIL-resistant BxPC-3 clone37 did not display any difference in response to GEM treatment compared to WT cells. Moreover, in all considered cell lines, independently from the sensitivity levels to either GEM or sTRAIL, a synergistic effect of the combinatory approach both in 2D and 3D models was observed versus solo treatments. Notably, in 2D cultures, we obtained a higher apoptotic combinatory effect on WT BxPC-3 and MIA PaCa-2 compared to what was reported with the same GEM concentrations and rhTRAIL,64 confirming the greater impact of multimeric sTRAIL versus rhTRAIL.36 We further challenged GEM+sTRAIL MSC in a 3D avatar model with PDAC cells and their CAF to demonstrate how the combo could generate an antitumor effect in TRAIL-sensitive PDAC, also reverting TRAIL resistance on both tumor and CAF cells.
To then better elucidate the mechanism of this synergy, we show that the combo enhances p38 MAPK phosphorylation compared to single agents, provoking an MD which ultimately leads to apoptosis.72,73,74 Focusing Bcl-2, we additionally confirm this anti-apoptotic player to be involved in our strategy, further suggesting how selective Bcl-2 inhibitors may be here introduced alone or with GEM.75 Previously, we demonstrated that sT-resistant-BxPC-3 line upregulates the anti-apopotic Bcl-xL, cFLIP, XIAP, nuclear factor κB (NF-κB)1, and NF-κB2 and downregulates the pro-apoptotic BID and BAK that can be counteracted by Taxol, a known anti-PDAC agent.37 In this study, we did not directly assess the action of the MSC sTRAIL and GEM on those pathways. However, the observed pro-apoptotic effect of the combo in sT-resistant-BxPC-3 may indirectly suggests that GEM, similarly to Taxol, empowers sTRAIL even in resistant settings, prompting additional investigations in complex combinatory strategies with sTRAIL, Taxol, and GEM.
After the in vitro encouraging results, we challenged the combo in two orthotopic models to better mimic the PDAC clinical scenario compared to subcutis. Thus, we selected two distinct PDAC lines having different aggressiveness and responses to treatment: WT BxPC-3 retaining a high responsiveness to GEM or sTRAIL and MIA PaCa-2 having an aggressive phenotype with poor sensitivity to GEM or sTRAIL combined with a relevant metastatic potential.45 US imaging provided crucial information concerning tumor composition, enabling US-guided injections and real-time monitoring of cells distribution. Up to seven days after injection, sTRAIL MSC remained detectable in tumors as viable, metabolically active cells. The MIA PaCa-2 model also allowed histopathological evidence that the combo led to tumor architecture shredding with reduction of CK8/18 expression, while a significant increase of tumor necrosis emerged in the BxPC-3 model. Remarkably, in the MIA PaCa-2 model, we registered splenic metastases abrogation in mice treated with GEM+sTRAIL MSC, thus suggesting an increased therapeutic profile of the combo in controlling the metastatic disease. Histology on collected WT BxPC-3 specimens treated by GEM+sTRAIL MSC revealed tumor-extensive necrosis without affecting the murine desmoplastic reaction. We considered this probably due to the large amount of PMS infiltrating the damaged PDAC tissue. Thus, we tested the impact of the combo on isolated PMS cells, originally showing their resistance to GEM+sTRAIL MSC therapy. Given these data, we hypothesize that PMS survival can partially limit the anti-PDAC potential of GEM+sTRAIL MSC in murine models.
Efforts have been recently focused in developing anti-stroma treatments for PDAC. However, given the limited benefits of those strategies in clinics, it has been suggested that a simple stromal depletion could favor PDAC progression rather than inhibition.76 Therefore, we believe that combinatorial approaches able to target both malignant and stromal cells are needed. With this aim, we isolated several human primary CAF lines confirming the typical CAF phenotype and the high expression of functional DR5.77,78 This result parallels our histology findings indicating that human PDAC stromal cells stain positive for DR5, being negative for DR4.4 Additionally, human fibroblasts in vitro reportedly express moderate levels of DR5 but not DR4.79 Through transcriptome analysis, we also have confirmed the expression of myCAFs and iCAFs markers as significant subpopulations within our cultured CAF, underlining their capacity in ECM production and immunomodulatory activity.
Thus, introducing a 3D PDAC avatar to recreate the complex PDAC architecture, we demonstrated that GEM+sTRAIL MSC can significantly counteract tumor cells even in co-culture with CAF. To validate pre-clinical approaches, in vitro 3D models have limitations but also retain specific advantages when relevant in vivo complexity has to be mimicked, as in this case. Recently, the Food and Drug Administration (FDA) endorsed 3D models as relevant for faster clinical translation,80 and, in this case, a 3D model allowed to mimic the in vitro ECM deposition from PDAC, investigating the impact of a gene therapy combo on desmoplastic reaction. In the 3D PDAC avatar model, we demonstrated that GEM+sTRAIL MSC significantly reduces FN1 and COL1A1, as a key player in PDAC ECM. While this requires further investigations, we presume this may be linked to the impact of GEM+sTRAIL MSC on both tumor and CAF with a reduction of ECM protein deposition after treatments, indicating the potential of our chemo and gene combination in reducing desmoplastic reaction while killing PDAC cells. These data coupled with RNA-seq analysis of CAF indicate that the combo suppresses DNA replication and mitotic division, triggering cell-cycle arrest, ultimately revealing TME remodeling and immunomodulatory shifts.
Focusing on CAF targeting, we additionally showed that, despite their high DR5 expression, all isolated CAF samples were resistant to sTRAIL alone, even at the highest tested concentration (2,500 pg/mL). Szegezdi et al. showed that human fibroblasts are not sensitive to rhTRAIL apoptotic induction.78 It was additionally reported that full-length transmembrane TRAIL cannot induce pancreatic stellate cell death within PDAC microenvironment, unlike with nearby tumor cells.81 However, no other studies have reported data on the cytotoxic effect of multimeric sTRAIL complexes on PDAC-derived CAF. Subsequently, we investigated GEM+sTRAIL efficacy against primary CAF, registering a synergistic pro-apoptotic impact of combined treatment versus either sTRAIL or GEM alone. Aligning with our results, evidence suggests that CAF and normal fibroblasts do not display much sensitivity to GEM alone and that CAF contributes to GEM resistance in PDAC.82,83
The present work demonstrates the therapeutic potential of combining an sTRAIL gene therapy approach with a standard of care for PDAC treatment, as reported using Nab-PTX as a synergizing agent with sTRAIL MSC.37 The combinatory approach was able to strongly potentiate in vitro single-drug treatment in PDAC cell lines, and the synergistic effect was further demonstrated in vivo using two PDAC orthotopic models. Although further investigation is needed, these encouraging findings support the synergistic effect of GEM+sTRAIL on PDAC CAF, suggesting how clinical investigation should consider this proposed therapeutic regimen for integrated therapy toward both tumor and stromal components in PDAC.
Limitations of the study
We demonstrated that GEM+sTRAIL MSC represents a possible therapeutic option to target both stromal and tumoral compartments in PDAC. While very promising, some aspects of our strategy will need to be further investigated. Starting from molecular insights, it has been reported that an active p38 MAPK provokes the loss of mitochondrial membrane potential through the phosphorylation of Bcl-2.42,43 Although we here demonstrated that the combo enhances p38 MAPK phosphorylation compared to single agents, more studies shall be demanded to fully identify the players involved in MD in mitochondria after GEM+sTRAIL treatment. On route of administration, we here report two orthotopic PDAC models in which modified MSCs were locally delivered by US-guided injection, also considering phase 1/2 clinical trials in PDAC where the anti-cancer agents have been intratumorally injected to maximize drug bioavailability.84 Both our xenografts demonstrated safety and feasibility of intratumorally implanted gene-modified MSC with systemically delivered chemotherapy. Albeit these pre-clinical data suggest a possible clinical transferability of the approach, further optimization may foresee a systemic injection that, once validated, could be a more accessible and less invasive approach.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-human TRAIL | Peprotech | 500-P135; RRID:AB_147781 |
| PE mouse anti-human p38 MAPK (pT180/pY182) | Becton Dickinson | 562065 |
| Alexa Fluor® 647 mouse anti-human Bcl-2 | BD Pharmingen | 563600 |
| Alexa Fluor™ 647 mouse IgG1 κ Isotype control | BD Pharmingen | 570232 |
| PE mouse anti-human CD73 | Miltenyi Biotec | REA804 |
| APC mouse anti-human CD90 | BD Pharmingen | 561971 |
| FITC mouse anti-human CD105 | BD Pharmingen | 561443 |
| FITC mouse anti-human CD45 | BD Pharmingen | 560976 |
| PE mouse anti-human CD45 | eBioscience | 12-9326-42 |
| FITC mouse anti-human HLA-DR | BD Pharmingen | 555560 |
| BD Via-Probe™ Cell Viability Solution | BD Pharmingen | 555816 |
| PE mouse anti-human CD261 (DR4, TRAIL-R1) | BioLegend | 307205; RRID:AB_314669 |
| PE mouse anti-human CD263 (TRAIL-R3, DcR1) | BioLegend | 307005; RRID:AB_2287506 |
| APC mouse anti-human CD262 (DR5, TRAIL-R2) | BioLegend | 307407; RRID:AB_2204813 |
| APC mouse anti-human TRAILR4/TNFRSF10D/DcR2 | R&D Systems | FAB633A; RRID:AB_2044736 |
| Rabbit anti-human Fibronectin | ABCAM | ab2413; RRID:AB_2262874 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 594 | Invitrogen | A21207 |
| Mouse anti-human Cytokeratin 7 (CK-7) | Roche | 790–4462; RRID:AB_2861319 |
| Mouse anti-human Cytokeratin 8 (CK-8) | Roche | 760–4344 |
| Rabbit anti-human fibronectin | Abcam | AB2413; RRID:AB_2262874 |
| Donkey anti-rabbit antibody Alexa Fluor Dye 594 | Invitrogen | A21207 |
| Bacterial and virus strains | ||
| Precision LentiORF Bcl-2 transcript variant alpha (NM_000633.3) | Horizon Discovery | FE6OHS5899 |
| Lentiviral particles for Firefly Luciferase | GeneCopoeia | LPP-HLUC-Lv105-025-C |
| Biological samples | ||
| Human PDAC tissues were collected from four patients (ID: PZ1, 2, 3, 4) undergoing surgery | Complex Structure of Hepato-Bilio-Pancreatic Oncological Surgery and Liver Transplant Surgery (University Hospital of Modena), |
N/A |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 Medium | Thermo Fisher Scientific | 11875101 |
| FBS - Fetal Bovine Serum | Thermo Fisher Scientific | A5256701 |
| L-Glutamine | Euroclone | ECB3000D/1 |
| Penicillin-Streptomicin | Sigma-Aldrich | P4333 |
| DMEM medium | Thermo Fisher Scientific | 41966–052 |
| Horse Serum, New Zealand origin | Thermo Fisher Scientific | 16050130 |
| MEM α medium | Thermo Fisher Scientific | 22561–054 |
| Human platelet lysate | Macopharma | BC0190030 |
| Phosphate-buffered saline (DPBS, 1X), Dulbecco’s formula | Thermo Fisher Scientific | 14190–169 |
| Ciprofloxacin 2 mg/mL | Fresenius Kabi | N/A |
| Heparin | Sigma-Aldrich | H3149 |
| Gemcitabina 40 mg/mL | Sandoz S.p.A | N/A |
| Luciferin substrate | Promega | E1605 |
| Trypsin 0.05% EDTA 0.02% in PBS | Eurolone | ECB3052D |
| Matrigel matrix | Corning | 356234 |
| Polybrene Infection/Transfection Reagent | Sigma-Aldrich | TR-1003 |
| BASESCOPE PROBE BA-TNFSF10-modified-1zz-st | Biotechne | 822411 |
| Blocking buffer | Roche | 10057177103 |
| Newborn Bovine Calf Serum, US Origin | Euroclone | CHA30118D |
| Triton™ X-100 | Sigma-Aldrich | X100 |
| Fluoroshield™ with DAPI | Sigma-Aldrich | Sigma-Aldrich |
| Critical commercial assays | ||
| CellTiter 96® AQueous One Solution Cell Proliferation Assay | Promega | G3582 |
| RNeasy Mini Kit | Qiagen | 74104 |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1621 |
| Fast SYBR® green master mix | Thermo Fisher Scientific | 4385612 |
| MitoStatus TMRE | BD Pharmingen | 564696 |
| FIX & PERM Cell Fixation & Cell Permeabilization Kit | Thermo Fisher Scientific | GAS004 |
| Cytofix/Cytoperm™ Fixation/Permeabilization Kit | BD Bioscience | 554714 |
| CellTrace™ CFSE Cell Proliferation Kit | Invitrogen | C34554 |
| QIAseq Fast Select RNA Removal kit | Qiagen | 333180 |
| QIAseq stranded total RNA library kit | Qiagen | 180753 |
| Agilent High Sensitivity DNA Kit | Agilent | 5067–4626 |
| Orange CMRA Dye and CellTrace | Invitrogen | C34551 |
| CellTracke Deep Red dye | Invitrogen | C34565 |
| Human Pro-Collagen I alpha 1 ELISA Kit - Quantikine | Biotechne | DPCA00 |
| Deposited data | ||
| Raw data of RNA-seq | BioProject NCBI | PRJNA1124875 |
| Differential Gene Expression, R script | Data are deposited in Zenodo | https://zenodo.org/records/12104006 |
| Experimental models: Cell lines | ||
| Human PDAC line: BxPC-3 | Interlab Cell Line Collection, ICLC | N/A |
| Human PDAC line: sTRAIL resistant BxPC-3 | Ref. 37 | N/A |
| Human PDAC line: MIA PaCa-2 | ATCC | CRL-1420 |
| Experimental models: Organisms/strains | ||
| Mouse: NOD.CB17-Prkdcscid/J | Charles River (Jax mice strain) | STRAIN: 001303 RRID:IMSR_JAX:001303Info |
| Oligonucleotides | ||
| Primers for ACTB | F:ACCTTCTACAATGAGCTGCG R:CCTGGATAGCAACGTACATGG |
|
| Primers for MIK67 | F:GTCGTGTCTCAAGATCTAGCTTC R: GTCATCTGCGGTACTGTCTTC |
|
| Primers for GAPDH | F: ACATCGCTCAGACACCATG R: TGTAGTTGAGGTCAATGAAGGG |
|
| Software and algorithms | ||
| Living Image software (v.4.3.1) | Perkin Elmer | http://caliperls.com/support/software-downloads.html |
| BD FACSDiva™ Software | Becton Dickinson | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Vevo Lab 3.1.0 software | FUJIFILM VisualSonics Inc | https://www.visualsonics.com/product/software/vevo-lab |
| FastQC Version 0.12.0 | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic version 0.39 | Usadellab | http://www.usadellab.org/cms/?page=trimmomatic |
| HISAT2 version 2.1.0 | The University of Texas Southwestern Medical Center | http://daehwankimlab.github.io/hisat2/ |
| HTSeqCount 1.99.2 | Stanford University and EMBL Heidelberg | https://htseq.readthedocs.io/en/release_0.11.1/count.html |
| DESeq2 | Bioconductor | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| ClusterProfiler | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| StringTie v2.2.1 | The Center for Computational Biology at Johns Hopkins University | https://ccb.jhu.edu/software/stringtie/ |
| SAMtools 1.10 | Genome Research Limited | http://www.htslib.org/ |
| bamCoverage 3.5.3 | Max Planck Institute for Immunobiology and Epigenetics, Freiburg | https://deeptools.readthedocs.io/en/develop/content/tools/bamCoverage.html |
| UCSC Genome Browser | https://genome.ucsc.edu/ | |
| ImageJ | ImageJ | https://imagej.net/ij/download.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Giulia Grisendi (giulia.grisendi@unimore.it).
Materials availability
This study did not generate new materials.
Data and code availability
-
•
RNA-seq data have been deposited at NCBI and are publicly available as of the date of publication. Accession number is listed in the key resources table (accession number: PRJNA1124875). Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
R custom script used in this study has been deposited at Zenodo (https://zenodo.org/records/12104006) and is publicly available as of the date of publication. DOIs are also listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Experimental model and study participant details
Animal husbandry
Adult (six to eight weeks old) male and female NOD.CB17-Prkdcscid/J mice (Charles River, Lecco, Italy) were kept in accordance with guidelines at the University of Modena and Reggio Emilia Animal Facility in a 12-h light/dark cycle (lights on from 6:00 a.m. to 6:00 p.m.) with food and water ad libitum. All animal procedures were approved by the National Ministry of Health and the local Institutional Animal Care and Use Committee (502/2018-PR. 02/07/2018).
Cell lines
Three human PDAC cell lines (wild-type BxPC-3, sTRAIL resistant BxPC-3, and MIA PaCa-2) were used. Wild-type BxPC-3 (WT BxPC-3; Interlab Cell Line Collection, ICLC, Genova, Italy) and sTRAIL-resistant BxPC-3 [37] (sT-resistant BxPC-3) were cultured in RPMI 1640 (Gibco-Life Technologies, Grand Island, NY, USA) with 10% heat-inactivated fetal bovine serum (FBS; Gibco-Life Technologies), 1% L-glutamine (Euroclone, Milan, Italy), and 1% penicillin/streptomycin (Sigma-Aldrich, Saint Louis, MO, USA). MIA PaCa-2 (ATCC, LGC Standards S.r.l., Milan, Italy) were maintained in DMEM (Gibco), 10% FBS, 2.5% horse serum (Euroclone), 1% pen/strep, and 1% L-glutamine. Authentication of PDAC cell lines was performed by the Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures (GmbH, Braunschweig, Germany).
MSC were isolated from lipoaspirate specimens of individuals undergoing aesthetic liposuction after approval from a local ethics committee28,29,36. Cells were maintained in α-Mem (Gibco) supplemented with 2.5% human platelet lysate (Macopharma, Tourcoing, France), 1% L-glutamine, 0.5% ciprofloxacin (Fresenius Kabi Italia S.r.l., Verona, Italy), and 1 IU/mL heparin (Sigma Aldrich). MSC transduced with a lentiviral vector coding for sTRAIL gene (sTRAIL MSC) or empty vectors (EV MSC) were obtained as described as a control36.
Method details
Luciferase transduction of PDAC lines
PDAC cell lines were engineered to express a luciferase protein (Lentiviral particles for Firefly Luciferase, GeneCopoeia, MD, USA) per manufacturer instructions. Photon emissions from luciferase-positive (Luc+) PDAC cells were measured via IVIS Lumina XRMS (PerkinElmer, Waltham, Massachusetts, USA) with Living Image software (v.4.3.1, PerkinElmer).
Cytotoxicity assays
sTRAIL and GEM dose response
PDAC cell lines were seeded in 96-well plates at 5000 cells/well (Corning, New York, USA) and treated the next day with increasing concentrations of GEM (Sandoz, Canada) (0.5–2000 μM) for 24 h. PDAC cell viability was assessed via Cell Titer AQueous96 One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) and quantified via GloMax Discover Microplate Reader (Promega). Similarly, sTRAIL cytotoxic impact was assessed by treating PDAC cells with conditioned medium (CM) collected from engineered MSC containing increasing concentrations of sTRAIL (50–2500 pg/mL) for 24 h. CM from EV MSC was collected and used as a control. Tumor cell viability was evaluated as above.
GEM+sTRAIL combinatory approach in 2D
PDAC cells were seeded in 12-well plates at 21000 cells/well (Day 1) and incubated the following day (Day 2) with 10 μM GEM (WT BxPC-3 and sT-resistant BxPC-3) or 100 μM (MIA PaCa-2). At Day 3, PDAC cell lines were treated for 24 h more with CM from sTRAIL MSC containing 250 (WT BxPC-3), 500 (MIA PaCa-2), or 300 (sT-resistant BxPC-3) pg/mL sTRAIL. The dose of GEM and sTRAIL required for each cell line to generate a synergistic effect has been calculated according to the Combination Index formula CI=(CA,X/ICx,A) + (CB,X/ICX,B).85 Death rate was assessed using FACSAria III (Becton Dickinson - BD, Franklin Lakes, New Jersey, USA) following Propidium Iodide (PI) staining.
GEM+sTRAIL MSC combinatory assay in 3D
Luc+ PDAC cells were seeded in VITVO bioreactor (EIR Biotherapies srl, Mirandola, Italy) on Day 1 [38]. On Day 2, cells were treated with GEM (10 μM for WT BxPC-3 and sT-resistant BxPC-3, 100 μM for MIA PaCa-2) or not conditioned medium for 24 h. On Day 3, PDAC cells were co-cultured up to 72 h with sTRAIL MSC at different Effector:Target (E:T) ratios (WT BxPC-3 and MIA PaCa-2 E:T = 1:10 and 1:30; sT-resistant BxPC-3 E:T = 1:1 and 1:10). Luciferin substrate (Promega) was added into co-culture at 10 mg/mL. Tumor cell mortality was quantified based on decrease of Luciferase signal via GloMax Discover Microplate Reader. VITVO loaded with PDAC cells alone were used as controls.
TRAIL blocking assay
For inhibition studies, WT BxPC-3 were treated with conditioned media from sTRAIL MSC containing different concentrations (0.6; 0.8; 1.6 μg/mL) of anti-human TRAIL antibody (PeproTech, London UK). Unconditioned medium was used as negative control whereas conditioned media from sTRAIL MSC was used as positive control. The anti-TRAIL antibody prevents the sTRAIL association to its functional receptors DR4 and DR5. BxPC-3 viability was quantified after 24h of treatment by FACS based on PI staining.
CellTiter 96 AQueous one solution cell proliferation assay
For this experiment, BxPC-3 were seeded at 5000 cells/well in a 96-well plate and treated with GEM and sTRAIL CM alone or in combination. To assess cell viability, 20μL of CellTiter 96 AQueous One Solution Reagent (Promega) were added to each well at 12, 24, and 48 h after treatment. The plate was incubated in a humidified, 5% CO2 atmosphere for 1 h, after which absorbance at 490nm was measured. Background absorbance was eliminated by subtracting values from medium-only wells.
Colony formation assay
To analyze colony formation, BxPC-3 were seeded at a clonal density of 200 cells/cm2 in a T-25 flask and observed daily. After fifteen days the samples were washed with PBS 1X and fixed with cooled absolute methanol for 2 min. After washing with water, they were stained with 0.4% crystal violet aqueous solution (Sigma) for 5 min. Subsequently, the cells were washed with water to remove the excess crystal violet and air-dried for a couple of hours. Colonies with more than 50 cells were counted and the efficacy of the colony formation was calculated using the formula: E% = (number of clones/cells seeded) x 100.
Real-time PCR
BxPC-3 and CAF cells were seeded in 6-well plates at a culture density of 15000 cells/cm2 and 10000 cells/cm2 respectively. At the time points of 12, 24, and 48 h after treatment with GEM+sTRAIL CM, alone or in combo, the cells were detached using trypsin (trypsin 0.05% EDTA 0.02% in PBS) and harvested as a dry pellet at −80°C. RNA was extracted using RNeasy Mini Kit (Qiagen) following the manufacturer’s instructions and quantified using NanoDrop OneC (Thermo Fisher Scientific, Waltham, MA). For random-primed cDNA synthesis 1 μg of RNA was used by RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. We performed Real-Time PCR using fast SYBR green master mix (Applied Biosystems by Thermo Fisher Scientific) and normalized with ACTB and GAPDH. Quantitative PCR was performed by Step One Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific), and the analysis was conducted using the ΔΔCt method. Primers list: ACTB (forward: 5′-ACCTTCTACAATGAGCTGCG-3’; reverse: 5′-CCTGGATAGCAACGTACATGG-3′), MKI67 (forward: 5′-GTCGTGTCTCAAGATCTAGCTTC-3'; reverse: 5′-GTCATCTGCGGTACTGTCTTC-3′), GAPDH (forward: 5′-ACATCGCTCAGACACCATG-3′; reverse: 5′-TGTAGTTGAGGTCAATGAAGGG-3′).
Mitochondrial depolarization (MD) assay
Cells were seeded into a 12-multiwell plate, at different culture density: BxPC-3 at 6000 cells/cm2, MIA PaCa-2 at 12000 cells/cm2 and CAF at 10000 cells/cm2. Cells were pre-treated with GEM at different concentrations: MIA PaCa-2 at 100 μM, BxPC-3 and CAF 10 μM. After 24 h, CM containing sTRAIL (250 pg/mL for BxPC-3, 500 pg/mL for MIA Paca-2 and 1000 pg/mL for CAF) was added, and cells were cultured for an additional 12 h. MD was evaluated by MitoStatus TMRE (BD) staining. Samples were acquired by FACSAria III and data were analyzed (FACS Diva BD).
Analysis of p38 phosphorylation
BxPC-3 cells were seeded in 6-well plates at 25000 cells/cm2 and incubated the following day (Day 2) with 10 μM GEM. At Day 3, PDAC cell were treated for an additional 24 h with CM from sTRAIL MSC containing 250 pg/mL sTRAIL. After 4 h cells were collected, treated with FIX & PERM Cell Fixation and Cell Permeabilization Kit (Thermo Scientific) according to manufacturer instruction and stained with PE Anti-p38 MAPK (pT180/pY182; BD) or with appropriate isotype control. Analyses were performed with FACSAria III (BD). Collected data were elaborated by FACS Diva software (BD).
Generation of Bcl-2 overexpressing BxPC-3 line
Precision LentiORF Bcl-2 transcript variant alpha (NM_000633.3) w/Stop Codon viral particles (Horizon Discovery, Cambridge, UK) were used to produce a stable Bcl-2 expression in BxPC-3 line, with GFP as a gene reporter. Lentiviral particles with the same vector backbone, not expressing Bcl-2, were used as control. Cells, seeded at 20.000/cm2 the day before, were infected once with a multiplicity of infection (MOI) of 10 adding 6 μg/mL of polybrene. The percentage of infected cells was monitored by GFP-based flow cytometry. Bcl-2 expression in transduced BxPC-3 was assessed by FACS. Briefly, after fixation and permeabilization (BDCytofix/Cytoperm, BD), WT BxPC-3, BxPC-3 over-expressing Bcl-2 (Bcl-2 BxPC-3) and BxPC-3 infected with lentiviral control particles (LC BxPC-3) were stained with Alexa Fluor 647 Mouse Anti-Human Bcl-2, (BD) and isotype control (Alexa Fluor 647 Mouse IgG1, k isotype CTRL, BD). Analyses were performed with FACSAria III. Collected data were elaborated by FACS Diva software.
Gemcitabine dose response in wild-type and engineered MSC
Wild-type or sTRAIL MSC were seeded in 96-well plates at 2500 cells/well (Corning). Cells were treated with increasing concentrations of GEM (0.5–2000 μM) for 72 h. MSC viability was assessed via Cell Titer AQueous96 One Solution Cell Proliferation Assay and GloMax Discover Microplate Reader.
ELISA
Wild-type or sTRAIL MSC were cultured with/without GEM (10 or 100 μM) for 72 h. CM was collected and filtered through a 0.22-μm filter (Euroclone). sTRAIL levels from MSC supernatants were measured using a Quantikine Human TRAIL/TNFSF10 kit per manufacturer instructions (Bio-Techne, Minneapolis, MN, USA).
In vivo PDAC orthotopic models and their histology
After approval by the National Ministry of Health and the local Institutional Animal Care and Use Committee (502/2018-PR. 02/07/2018), an orthotopic mouse model was implemented to assess the therapeutic impact of the GEM+sTRAIL MSC combinatory approach. Briefly, 1 × 106 WT BxPC-3-Luc+ or MIA PaCa-2 -Luc+ cells in 50 μL of 1:1 Matrigel (Corning) and PBS 1X (Thermo Fisher Scientific, Waltham, MA) suspension were injected into the pancreas of NOD-SCID mice by surgical procedure, as reported.86,87 Tumor engraftment was evaluated by bioluminescence (BLI) 7 days after injection by the IVIS Lumina XRMS system. Tumors were monitored weekly using the ultrasound (US) Vevo 2100 Imaging System (FUJIFILM VisualSonics Inc., Bothell, WA, USA). Tumors were let to grow and, before treatment, mice were randomized into four groups by tumor volume. GEM (50 mg/kg for BxPC-3-Luc+ or 100 mg/kg for MIA PaCa-2 -Luc+) was administered intraperitoneally (i.p.) twice, 7 days apart, followed by an ultrasound-guided intratumor (i.t.) injection of sTRAIL MSC (E:T = 1:30 or 1:10) suspended in 15 μL PBS at Day 33. sTRAIL MSC were labeled using XenoLight DiR (8 μM; PerkinElmer), and their biodistribution inside the pancreas was monitored up to 7 days after injection. For tumor evaluation, tissues were quantified using Vevo Lab 3.1.0 software (FUJIFILM VisualSonics Inc.) generating six consecutive US sections and tumor volume and necrosis percentage was calculated in each tumor slices as follows: necrotic tissue (mm3)/total tumor tissue (mm3) x 100. Formalin-fixed, paraffin-embedded (FFPE) tumor sections and spleen tissue were also evaluated by hematoxylin-and-eosin staining (H&E, Carlo Erba) and immunohistochemistry analysis of Cytokeratin 7 (CK-7), Cytokeratin 8-18 (CK-8-18) (VENTANA-Roche, Oro Valley, Arizona, US) as previously reported.36
Cytotoxicity of GEM and sTRAIL in pancreatic murine stroma
A WT BxPC-3 tumor grown in vivo was collected at sacrifice and digested as described above. Pancreatic murine stroma (PMS) cells were isolated and cultured in DMEM low glucose medium (Euroclone) with 10% heat-inactivated FBS, 1% L-glutamine, and 1% penicillin/streptomycin. After isolation, murine stromal cells were plated in a 96-well plate at 2500 cells/well. Cells were pre-treated the next day with 10 μM GEM for 24 h and subsequently treated for 24 h with conditioned media containing 1000 pg/mL sTRAIL collected from sTRAIL MSC. Cell viability was assessed via CellTiter-Glo assay and registered via GloMax Discover Microplate Reader.
Basescope assay
For the detection of sTRAIL MSC in explanted tumor samples, BaseScope Assay (ACD Bio-Techne, USA) has been introduced as in situ hybridization tool to visualize single RNA target molecules in slide-mounted FFPE samples. By a specific probe, targeting the RNA transcript of the TRAIL transgene, and a hybridization-based signal amplification system, TRAIL mRNA transcript inside sTRAIL MSC was searched as distinct dots of red chromogen.
Isolation and immunophenotypic characterization of primary human CAF
Human PDAC tissues were collected from four patients (ID: PZ1, 2, 3, 4) undergoing surgery at the Complex Structure of Hepato-Bilio-Pancreatic Oncological Surgery and Liver Transplant Surgery (University Hospital of Modena), after informed consent. Human PDAC samples were digested through mechanical and enzymatic dissociation via Tumor Dissociation Kit and the gentleMACS Octo Dissociator (MACS; Miltenyi Biotec, Auburn, CA, USA), following manufacturer instructions. Digested cells were cultured in minimal essential medium with α-MEM containing 2.5% platelet lysate, 1% glutamine, 0.5% ciprofloxacin, and 0.2% heparin for 10 days to selectively isolate four primary CAF cell lines. Immunophenotypic characterization of primary human CAF was performed via FACS. Cells were stained using phycoerythrin (PE)-conjugated anti-CD73 (Miltenyi Biotec), allophycocyanin (APC)-conjugated anti-CD90 (BD Biosciences, San Jose, CA, USA), fluorescein isothiocyanate (FITC)-conjugated anti-CD105 (BD Biosciences), FITC-conjugated anti-CD45 (BD Biosciences), PE-conjugated anti-EPCAM (eBioscience, San Diego, CA, USA), FITC-conjugated anti-HLA-DR (eBioscience), and BD Via ProbeTM Cell Viability Solution (7-Aminoactinomycin D [7-AAD]; BD Pharmingen, San Diego, CA, USA). To assess TRAIL receptor expression, CAF were tested for PE-conjugated anti-DR4 (BioLegend, San Diego, CA, USA), APC-conjugated anti-DR5 (BioLegend), PE-conjugated anti-DcR1 (BioLegend), and APC-conjugated anti-DcR2 (R&D Systems, Minneapolis, MN, USA). Isotype control antibodies were used for all cell types and antigens analyzed. Samples were acquired by FACSAria III and analyzed via BD FACS Diva software.
sTRAIL MSC and CAF co-culture with sorting
MSC sTRAIL or EV were labeled with carboxyfluorescein succinimidyl ester (CellTrace CFSE Cell Proliferation Kit, Invitrogen) according to manufacturer instructions. CAF were seeded at a density of 8000 cells/cm2 and treated by GEM (100 μM) + sTRAIL MSC (E:T 1:10) alone or in combination for 24 h. Untreated CAF or CAF treated with GEM+EV MSC were used as controls. After 24 h of co-cultures, CAF were separated from CFSE-labeled MSC by fluorescent-activating cell sorting using FACSAria III for RNA-seq analyses.
CAF characterization by RNA-seq
RNA extraction from CAF was performed by an RNA extraction kit (RNeasy Plus Mini Kit, QIAGEN, Hilden, Germany) following manufacturer instructions and quantified using NanoDrop OneC (Thermo Fisher Scientific, Waltham, MA). Short-read, stranded RNA sequencing (RNA-seq) on primary CAF alone, and CAF treated with GEM, with sTRAIL MSCs alone, with GEM+EV MSC or GEM+sTRAIL MSC were performed. All RNA-seq experiments were performed with 2 biological replicates for each condition. RNA-seq libraries were generated using QIAGEN QIAseq Fast Select RNA Removal kit and QIAGEN QIAseq stranded total RNA library kit (QIAGEN, Hilden, Germany). Specifically, for library generation 500ng of total RNA (RIN >8) was used as a starting material. Reverse transcription, second strand synthesis, end-repair, A-addition and adapters ligation were performed as reported by manufacturer’s instructions. Quality control (QC) of the libraries was performed using Agilent High Sensitivity DNA kit 1000 (Agilent, Santa Clara, California, US). RNA-seq was performed using the Illumina NextSeq 500 Instrument and the NextSeq High Output kit v.2.5 paired-end flowcell yielding between 60 and 70 million paired-end reads for each sample. To ensure the quality and reliability of data, FASTQ files were pre-processed with FastQC Version 0.12.0 and trimmed using Trimmomatic version 0.39. Subsequently, pre-processed reads were aligned to the human reference genome GRCh38 using HISAT2 version 2.1.0. We utilized SAMtools version 1.10 and HTSeqCount version 1.99.2 for the generation of BAM files and count files, respectively. Additionally, bamCoverage version 3.5.3 was employed for generating BigWig files, and the UCSC Genome Browser was used to visualize coverage plots. Differentially expressed genes and gene ontology were calculated using the DESeq2 and clusterProfiler (enrichGO) packages respectively in R 4.3.1. For transcript assembly and abundance estimation, we utilized StringTie v2.2.1. using the GENCODE GTF file release 36. The -A option in StringTie was used to generate gene-level abundance files, providing TPM (Transcripts Per Million) values as a measure of transcript abundance. TPM values visualization as log2(TPM+1) was conducted in R. The raw data of this research were deposited in NCBI and are available under the accession number PRJNA1124875.
Cytotoxicity studies on 3D PDAC avatar
Luc+ WT BxPC-3 or MIA PaCa-2 cells and primary CAF cells were loaded in VITVO bioreactor to recreate in vitro an artificial tissue mimicking PDAC microenvironment, here defined as PDAC avatar. The realistic tumor/stroma ratio, observed in patient-derived PDAC samples, was reproduced in VITVO by seeding 55% of tumor cells and 45% of stromal elements.
3D PDAC avatars were treated with GEM (10 μM for WT BxPC-3 and 100 μM for MIA PaCa-2) or not conditioned medium for 24 h. The next day, PDAC avatars were co-cultured up to 72 h with sTRAIL MSC at different E:T ratios (1:10 or 1:30). Luciferin substrate (Promega) was added into co-culture at 10 mg/mL. Tumor cell mortality was quantified based on the decrease of Luciferase signal via GloMax Discover Microplate Reader. VITVO loaded with PDAC cells and CAF alone were used as controls.
Live imaging on a 3D PDAC avatar
In an additional PDAC avatar model, MIA PaCa-2 and primary CAF were labeled respectively with Orange CMRA Dye and CellTrace CFSE (both from Invitrogen, Thermo Fisher, Eugene, Oregon, US) then, cells were seeded in VITVO bioreactor (198000 cells/VITVO for MIA PaCa-2 and 162000 cells/VITVO for CAF) in DMEM supplemented with 1% L-glutamine, 1% penicillin/streptomycin, 10% FBS, and 2.5% horse serum. On Day 2, cells were treated with GEM 100 μM for 24 h and culture medium of controls was replaced. On Day 3, sTRAIL MSC or EV MSC, labeled with CellTracker Deep Red dye (Thermo Fisher), were added into VITVO bioreactor at 1:10 E:T ratio. Cell labeling was performed according to the manufacturer instructions. Live images were taken by confocal microscope NIKON A1 (Nikon Europe B.V, Netherlands) after 24 h from MSC loading.
Primary human CAF viability assays
The isolated primary PDAC CAF were tested for sensitivity to sTRAIL, alone or with GEM, using a CellTiter-Glo Luminescent Cell Viability assay. Cells were seeded in a 96-well plate at 2500 cells/well in cell culture medium. Cells were next treated with conditioned media from sTRAIL MSC containing increasing doses of sTRAIL (50–2500 pg/mL) or were pre-treated with GEM (10 μM) for 24 h and incubated with sTRAIL (1000 pg/mL) for 24 h. CAF viability was assessed via CellTiter-Glo reagent and quantified via GloMax Discover Microplate Reader.
Extracellular matrix analysis on 3D PDAC avatar
ex vivo generated extracellular matrix (ECM) was tested by a combination of protein assays: on one side testing by IHC fibronectin 1 (FN1), a key ECM protein for PDAC, and on the other measuring the levels of collagen 1A1 (COL1A1) in 3D co-culture supernatant. MIA PaCa-2 and CAF were loaded in VITVO and treated by GEM (100 μM) and sTRAIL MSC (E:T ratios 1:10) alone or in combination, as described above. MIA PaCa-2 and CAF co-culture untreated or treated with GEM+EV MSC were used as controls. Twenty-four hours after loading with MSCs (EV or sTRAIL), VITVO 3D matrices were fixed in formaldehyde (4% Histo-Line, Milan, Italy) and treated at room temperature (RT) with a blocking buffer solution composed of 100 mM maleic acid, 150 mM NaCl, blocking reagent (Roche Diagnostic GmbH, Mannheim, Germany) and newborn calf serum (Euroclone, Milan, Italy). Prior to labeling, cells were permeabilized with 0.1% Triton X-100 (Sigma Aldrich) for 3 min on ice and then, washed with 1X PBS solution. To detect fibronectin1 (FN1), the 3D ECM was first labeled by an anti-fibronectin antibody (AB2413, diluted 1:100 with 0.1% BSA, Abcam, Cambridge, UK) for 1 h at room temperature. After washing with 1X PBS solution, a second labeling step was performed using an anti-rabbit antibody (Alexa Fluor Dye 594, Donkey, IGG, Invitrogen A21207, diluted 1:700 with 0.1% BSA) for 1 h at room temperature. The anti-rabbit labeling step was conducted in the dark to prevent photo-bleaching of the fluorophore. Finally, VITVO 3D matrices were mounted on slides using Fluoroshield with a DAPI histology mounting medium (Sigma-Aldrich) and analyzed using a microscope (Axio Zoom V16 Zeiss, Oberkochen, Germany) with z stack acquisition settings (Zen 2012 blue edition by Carl Zeiss Microscopy GmbH). The images acquired from each sample (N = 10) were analyzed for both FN1 and DAPI using ImageJ software (NIH, Washington DC, US) Positive areas for FN1 or DAPI were computed from each image, and then the mean and standard deviation were calculated for each sample. In addition, supernatants from VITVO were collected and collagen1A1 (COL1A1) was measured using a Quantikine Human Pro-Collagen I alpha 1 ELISA (Bio-Techne, Minneapolis, MN, USA) according to manufacturer instructions.
Quantification and statistical analysis
Data were expressed as mean values ±standard error of the mean (SEM). An unpaired 2-tailed Student’s t test was used (p ≤ 0.05 as statistically significant). Chi-square test was employed for tumor necrosis quantification by US. For MIA PaCA-2 in vivo model, a repeated measures linear mixed model analysis was carried out to assess whether the average daily variation from baseline in volume was different amongst treatment arms.
Acknowledgments
This study was supported in parts by AIRC 2022 grant 27850; Ministero Istruzione Università Ricerca (MIUR), Project “Dipartimenti Eccellenti MIUR 2017 e 2022”; and Rigenerand srl, Associazione Nastro Viola, and the European Union NextGenerationEU through the MIUR PNRR - M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to M. Dominici and G.G. We are also in debt to patients who generously donated samples for CAF isolation.
Author contributions
G.C., M. Dall’Ora, and G.G.: conception and design of the study, data collection, drafting the manuscript, data analysis, interpretation, and data finalization. A.M., V.M., C.C., and C.S.: data collection, data analysis, and interpretation. G.G., M. Dall’Ora, G.S., and A.D.: in vivo data collection. T.P., E.V., F.L., and J.V.: microscopy studies. M. Farshchian, M. Ferracin, and R.R.: RNA sequencing and analysis. A.S., P.M., B.C., C.M., and L.R.B.: procurement of biological samples. F.D.B., A.E., and E.M.H.: provision of study materials and final approval of the manuscript. F.B. and R.C.C.: statistical data analysis. M. Dominici: conception or design of the work, drafting and revising the manuscript, data analysis, interpretation, and data finalization. All authors approved the article.
Declaration of interests
M. Dominici and G.G. hold patents in the field of cell and gene therapy. EIR Biotherapies srl holds patents related to the presented technologies. M. Dall’Ora and O.C. are employees of EVOTEC Modena Srl.
Published: August 20, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101685.
Contributor Information
Giulia Grisendi, Email: giulia.grisendi@unimore.it.
Massimo Dominici, Email: massimo.dominici@unimore.it.
Supplemental information
This file contains non-filtered DESeq2 results for six pair-wise comparisons of all conditions. The columns across all sheets are defined as follows: baseMean, the average of the normalized counts for all samples; log2FoldChange, the log2 fold change between the two conditions being compared; lfcSE, the standard error of the log2 fold change; stat, the test statistic for the comparison; p value, the p value for the test statistic; padj, the adjusted p value after correcting for multiple testing using the Benjamini- Hochberg method.
References
- 1.Malvezzi M., Bertuccio P., Levi F., La Vecchia C., Negri E. European cancer mortality predictions for the year 2014. Ann. Oncol. 2014;25:1650–1656. doi: 10.1093/annonc/mdu138. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., Gavin A., Visser O., Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer. 2018;103:356–387. doi: 10.1016/j.ejca.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet (London, England) 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 4.Dall'Ora M., Rovesti G., Reggiani Bonetti L., Casari G., Banchelli F., Fabbiani L., Veronesi E., Petrachi T., Magistri P., Di Benedetto F., et al. TRAIL receptors are expressed in both malignant and stromal cells in pancreatic ductal adenocarcinoma. Am. J. Cancer Res. 2021;11:4500–4514. [PMC free article] [PubMed] [Google Scholar]
- 5.Burris H.A., Andersen J., Green M.R., Rothenberg M.L., Modiano M.R., Cripps M.C., Portenoy R.K., Storniolo A.M., Tarassoff P., et al. Moore M.J. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 6.Conroy T., Desseigne F., Ychou M., Bouché O., Guimbaud R., Bécouarn Y., Adenis A., Raoul J.L., Gourgou-Bourgade S., de la Fouchardière C., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 7.Moore M.J., Goldstein D., Hamm J., Figer A., Hecht J.R., Gallinger S., Au H.J., Murawa P., Walde D., Wolff R.A., et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 8.Pezzilli R., Fabbri D., Imbrogno A. Pancreatic ductal adenocarcinoma screening: new perspectives. World J. Gastroenterol. 2012;18:4973–4977. doi: 10.3748/wjg.v18.i36.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Principe D.R., Underwood P.W., Korc M., Trevino J.G., Munshi H.G., Rana A. The Current Treatment Paradigm for Pancreatic Ductal Adenocarcinoma and Barriers to Therapeutic Efficacy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.688377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amrutkar M., Gladhaug I.P. Pancreatic Cancer Chemoresistance to Gemcitabine. Cancers. 2017;9:157. doi: 10.3390/cancers9110157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng S., Pöttler M., Lan B., Grützmann R., Pilarsky C., Yang H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019;20:4504. doi: 10.3390/ijms20184504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C., Shi S., Meng Q., Liang D., Ji S., Zhang B., Qin Y., Xu J., Ni Q., Yu X. Do anti-stroma therapies improve extrinsic resistance to increase the efficacy of gemcitabine in pancreatic cancer? Cell. Mol. Life Sci. 2018;75:1001–1012. doi: 10.1007/s00018-017-2678-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neesse A., Algül H., Tuveson D.A., Gress T.M. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut. 2015;64:1476–1484. doi: 10.1136/gutjnl-2015-309304. [DOI] [PubMed] [Google Scholar]
- 14.Ni Y., Zhou X., Yang J., Shi H., Li H., Zhao X., Ma X. The Role of Tumor-Stroma Interactions in Drug Resistance Within Tumor Microenvironment. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.637675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashkenazi A. Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat. Rev. Cancer. 2002;2:420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 16.Wiley S.R., Schooley K., Smolak P.J., Din W.S., Huang C.P., Nicholl J.K., Sutherland G.R., Smith T.D., Rauch C., Smith C.A. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–682. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- 17.Bodmer J.L., Holler N., Reynard S., Vinciguerra P., Schneider P., Juo P., Blenis J., Tschopp J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 18.Pitti R.M., Marsters S.A., Ruppert S., Donahue C.J., Moore A., Ashkenazi A. Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J. Biol. Chem. 1996;271:12687–12690. doi: 10.1074/jbc.271.22.12687. [DOI] [PubMed] [Google Scholar]
- 19.LeBlanc H.N., Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10:66–75. doi: 10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 20.Daniels R.A., Turley H., Kimberley F.C., Liu X.S., Mongkolsapaya J., Ch'En P., Xu X.N., Jin B.Q., Pezzella F., Screaton G.R. Expression of TRAIL and TRAIL receptors in normal and malignant tissues. Cell Res. 2005;15:430–438. doi: 10.1038/sj.cr.7290311. [DOI] [PubMed] [Google Scholar]
- 21.Koornstra J.J., de Jong S., Hollema H., de Vries E.G.E., Kleibeuker J.H. Changes in apoptosis during the development of colorectal cancer: a systematic review of the literature. Crit. Rev. Oncol. Hematol. 2003;45:37–53. doi: 10.1016/s1040-8428(01)00228-1. [DOI] [PubMed] [Google Scholar]
- 22.van Dijk M., Halpin-McCormick A., Sessler T., Samali A., Szegezdi E. Resistance to TRAIL in non-transformed cells is due to multiple redundant pathways. Cell Death Dis. 2013;4:e702. doi: 10.1038/cddis.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashkenazi A., Pai R.C., Fong S., Leung S., Lawrence D.A., Marsters S.A., Blackie C., Chang L., McMurtrey A.E., Hebert A., et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong S.H.M., Kong W.Y., Fang C.M., Loh H.S., Chuah L.H., Abdullah S., Ngai S.C. The TRAIL to cancer therapy: Hindrances and potential solutions. Crit. Rev. Oncol. Hematol. 2019;143:81–94. doi: 10.1016/j.critrevonc.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Kelley S.K., Harris L.A., Xie D., Deforge L., Totpal K., Bussiere J., Fox J.A. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J. Pharmacol. Exp. Ther. 2001;299:31–38. [PubMed] [Google Scholar]
- 26.Ashkenazi A., Holland P., Eckhardt S.G. Ligand-based targeting of apoptosis in cancer: the potential of recombinant human apoptosis ligand 2/Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL) J. Clin. Oncol. 2008;26:3621–3630. doi: 10.1200/JCO.2007.15.7198. [DOI] [PubMed] [Google Scholar]
- 27.Herbst R.S., Eckhardt S.G., Kurzrock R., Ebbinghaus S., O'Dwyer P.J., Gordon M.S., Novotny W., Goldwasser M.A., Tohnya T.M., Lum B.L., et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J. Clin. Oncol. 2010;28:2839–2846. doi: 10.1200/JCO.2009.25.1991. [DOI] [PubMed] [Google Scholar]
- 28.Grisendi G., Bussolari R., Cafarelli L., Petak I., Rasini V., Veronesi E., De Santis G., Spano C., Tagliazzucchi M., Barti-Juhasz H., et al. Adipose-derived mesenchymal stem cells as stable source of tumor necrosis factor-related apoptosis-inducing ligand delivery for cancer therapy. Cancer Res. 2010;70:3718–3729. doi: 10.1158/0008-5472.CAN-09-1865. [DOI] [PubMed] [Google Scholar]
- 29.Grisendi G., Spano C., D'souza N., Rasini V., Veronesi E., Prapa M., Petrachi T., Piccinno S., Rossignoli F., Burns J.S., et al. Mesenchymal progenitors expressing TRAIL induce apoptosis in sarcomas. Stem cells (Dayton, Ohio) 2015;33:859–869. doi: 10.1002/stem.1903. [DOI] [PubMed] [Google Scholar]
- 30.Golinelli G., Grisendi G., Spano C., Dominici M. Surrounding pancreatic adenocarcinoma by killer mesenchymal stromal/stem cells. Hum. Gene Ther. 2014;25:406–407. doi: 10.1089/hum.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golinelli G., Mastrolia I., Aramini B., Masciale V., Pinelli M., Pacchioni L., Casari G., Dall'Ora M., Soares M.B.P., Damasceno P.K.F., et al. Arming Mesenchymal Stromal/Stem Cells Against Cancer: Has the Time Come? Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.529921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan Z., Kolluri K.K., Sage E.K., Gowers K.H.C., Janes S.M. Mesenchymal stromal cell delivery of full-length tumor necrosis factor-related apoptosis-inducing ligand is superior to soluble type for cancer therapy. Cytotherapy. 2015;17:885–896. doi: 10.1016/j.jcyt.2015.03.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sage E.K., Kolluri K.K., McNulty K., Lourenco S.D.S., Kalber T.L., Ordidge K.L., Davies D., Gary Lee Y.C., Giangreco A., Janes S.M. Systemic but not topical TRAIL-expressing mesenchymal stem cells reduce tumour growth in malignant mesothelioma. Thorax. 2014;69:638–647. doi: 10.1136/thoraxjnl-2013-204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reagan M.R., Seib F.P., McMillin D.W., Sage E.K., Mitsiades C.S., Janes S.M., Ghobrial I.M., Kaplan D.L. Stem Cell Implants for Cancer Therapy: TRAIL-Expressing Mesenchymal Stem Cells Target Cancer Cells In Situ. J. Breast Cancer. 2012;15:273–282. doi: 10.4048/jbc.2012.15.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasportas L.S., Kasmieh R., Wakimoto H., Hingtgen S., van de Water J.A.J.M., Mohapatra G., Figueiredo J.L., Martuza R.L., Weissleder R., Shah K. Assessment of therapeutic efficacy and fate of engineered human mesenchymal stem cells for cancer therapy. Proc. Natl. Acad. Sci. USA. 2009;106:4822–4827. doi: 10.1073/pnas.0806647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spano C., Grisendi G., Golinelli G., Rossignoli F., Prapa M., Bestagno M., Candini O., Petrachi T., Recchia A., Miselli F., et al. Soluble TRAIL Armed Human MSC As Gene Therapy For Pancreatic Cancer. Sci. Rep. 2019;9:1788. doi: 10.1038/s41598-018-37433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossignoli F., Spano C., Grisendi G., Foppiani E.M., Golinelli G., Mastrolia I., Bestagno M., Candini O., Petrachi T., Recchia A., et al. MSC-Delivered Soluble TRAIL and Paclitaxel as Novel Combinatory Treatment for Pancreatic Adenocarcinoma. Theranostics. 2019;9:436–448. doi: 10.7150/thno.27576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Candini O., Grisendi G., Foppiani E.M., Brogli M., Aramini B., Masciale V., Spano C., Petrachi T., Veronesi E., Conte P., et al. A Novel 3D In Vitro Platform for Pre-Clinical Investigations in Drug Testing, Gene Therapy, and Immuno-oncology. Sci. Rep. 2019;9:7154. doi: 10.1038/s41598-019-43613-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki S., Okada M., Shibuya K., Seino M., Sato A., Takeda H., Seino S., Yoshioka T., Kitanaka C. JNK suppression of chemotherapeutic agents-induced ROS confers chemoresistance on pancreatic cancer stem cells. Oncotarget. 2015;6:458–470. doi: 10.18632/oncotarget.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S.H., Li D.L., Yang F., Wu Z., Zhao Y.Y., Jiang Y. Gemcitabine-induced pancreatic cancer cell death is associated with MST1/cyclophilin D mitochondrial complexation. Biochimie. 2014;103:71–79. doi: 10.1016/j.biochi.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhao B., Li L., Cui K., Wang C.L., Wang A.L., Sun Z.Q., Zhang B., Zhou W.Y., Niu Z.X., Tian H., et al. Mechanisms of TRAIL and gemcitabine induction of pancreatic cancer cell apoptosis. Asian Pac. J. Cancer Prev. 2011;12:2675–2678. [PubMed] [Google Scholar]
- 42.Lee M.W., Park S.C., Yang Y.G., Yim S.O., Chae H.S., Bach J.H., Lee H.J., Kim K.Y., Lee W.B., Kim S.S. The involvement of reactive oxygen species (ROS) and p38 mitogen-activated protein (MAP) kinase in TRAIL/Apo2L-induced apoptosis. FEBS Lett. 2002;512:313–318. doi: 10.1016/s0014-5793(02)02225-1. [DOI] [PubMed] [Google Scholar]
- 43.Koizumi K., Tanno S., Nakano Y., Habiro A., Izawa T., Mizukami Y., Okumura T., Kohgo Y. Activation of p38 mitogen-activated protein kinase is necessary for gemcitabine-induced cytotoxicity in human pancreatic cancer cells. Anticancer Res. 2005;25:3347–3353. [PubMed] [Google Scholar]
- 44.Shamas-Din A., Kale J., Leber B., Andrews D.W. Mechanisms of action of Bcl-2 family proteins. Cold Spring Harb. Perspect. Biol. 2013;5:a008714. doi: 10.1101/cshperspect.a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rouffiac V., Bouquet C., Lassau N., Opolon P., Koscielny S., Peronneau P., Perricaudet M., Roche A. Validation of a new method for quantifying in vivo murine tumor necrosis by sonography. Invest. Radiol. 2004;39:350–356. doi: 10.1097/01.rli.0000124457.99229.bb. [DOI] [PubMed] [Google Scholar]
- 46.Yu Q., Qiu Y., Chen X., Wang X., Mei L., Wu H., Liu K., Liu Y., Li M., Zhang Z., He Q. Chemotherapy priming of the Pancreatic Tumor Microenvironment Promotes Delivery and Anti-Metastasis Efficacy of Intravenous Low-Molecular-Weight Heparin-Coated Lipid-siRNA Complex. Theranostics. 2019;9:355–368. doi: 10.7150/thno.29137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito K., Iioka H., Maruyama S., Sumardika I.W., Sakaguchi M., Kondo E. PODXL1 promotes metastasis of the pancreatic ductal adenocarcinoma by activating the C5aR/C5a axis from the tumor microenvironment. Neoplasia. 2019;21:1121–1132. doi: 10.1016/j.neo.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang D., Ma Q., Wang Z., Zhang M., Guo K., Wang F., Wu E. β2-adrenoceptor blockage induces G1/S phase arrest and apoptosis in pancreatic cancer cells via Ras/Akt/NFκB pathway. Mol. Cancer. 2011;10:146. doi: 10.1186/1476-4598-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamitani N., Nakamae I., Yoneda-Kato N., Kato J.Y., Sho M. Preclinical evaluation of pentagamavunone-1 as monotherapy and combination therapy for pancreatic cancer in multiple xenograft models. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-26863-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neureiter D., Zopf S., Dimmler A., Stintzing S., Hahn E.G., Kirchner T., Herold C., Ocker M. Different capabilities of morphological pattern formation and its association with the expression of differentiation markers in a xenograft model of human pancreatic cancer cell lines. Pancreatology. 2005;5:387–397. doi: 10.1159/000086539. [DOI] [PubMed] [Google Scholar]
- 51.Grisendi G., Bussolari R., Veronesi E., Piccinno S., Burns J.S., De Santis G., Loschi P., Pignatti M., Di Benedetto F., Ballarin R., et al. Understanding tumor-stroma interplays for targeted therapies by armed mesenchymal stromal progenitors: the Mesenkillers. Am. J. Cancer Res. 2011;1:787–805. [PMC free article] [PubMed] [Google Scholar]
- 52.Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menezes S., Okail M.H., Jalil S.M.A., Kocher H.M., Cameron A.J.M. Cancer-associated fibroblasts in pancreatic cancer: new subtypes, new markers, new targets. J. Pathol. 2022;257:526–544. doi: 10.1002/path.5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brichkina A., Polo P., Sharma S.D., Visestamkul N., Lauth M. A Quick Guide to CAF Subtypes in Pancreatic Cancer. Cancers. 2023;15:2614. doi: 10.3390/cancers15092614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M., Hancock S.E., Sultani G., Wilkins B.P., Ding E., Osborne B., Quek L.E., Turner N. Snail-Overexpression Induces Epithelial-mesenchymal Transition and Metabolic Reprogramming in Human Pancreatic Ductal Adenocarcinoma and Non-tumorigenic Ductal Cells. J. Clin. Med. 2019;8:822. doi: 10.3390/jcm8060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaudhary R., Gryder B., Woods W.S., Subramanian M., Jones M.F., Li X.L., Jenkins L.M., Shabalina S.A., Mo M., Dasso M., et al. Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife. 2017;6 doi: 10.7554/eLife.23244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui J., Christin J.R., Reisz J.A., Cendali F.I., Sanawar R., Coutinho De Miranda M., D'Alessandro A., Guo W. Targeting ABCA12-controlled ceramide homeostasis inhibits breast cancer stem cell function and chemoresistance. Sci. Adv. 2023;9 doi: 10.1126/sciadv.adh1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Man K.F., Zhou L., Yu H., Lam K.H., Cheng W., Yu J., Lee T.K., Yun J.P., Guan X.Y., Liu M., Ma S. SPINK1-induced tumor plasticity provides a therapeutic window for chemotherapy in hepatocellular carcinoma. Nat. Commun. 2023;14:7863. doi: 10.1038/s41467-023-43670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gebauer F., Wicklein D., Horst J., Sundermann P., Maar H., Streichert T., Tachezy M., Izbicki J.R., Bockhorn M., Schumacher U. Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez V.M., Kearney J.F., Yeh J.J. The PDAC Extracellular Matrix: A Review of the ECM Protein Composition, Tumor Cell Interaction, and Therapeutic Strategies. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.751311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Todaro M., Lombardo Y., Francipane M.G., Alea M.P., Cammareri P., Iovino F., Di Stefano A.B., Di Bernardo C., Agrusa A., Condorelli G., et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 63.Soria J.C., Márk Z., Zatloukal P., Szima B., Albert I., Juhász E., Pujol J.L., Kozielski J., Baker N., Smethurst D., et al. Randomized phase II study of dulanermin in combination with paclitaxel, carboplatin, and bevacizumab in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:4442–4451. doi: 10.1200/JCO.2011.37.2623. [DOI] [PubMed] [Google Scholar]
- 64.Elia A., Henry-Grant R., Adiseshiah C., Marboeuf C., Buckley R.J., Clemens M.J., Mudan S., Pyronnet S. Implication of 4E-BP1 protein dephosphorylation and accumulation in pancreatic cancer cell death induced by combined gemcitabine and TRAIL. Cell Death Dis. 2017;8:3204. doi: 10.1038/s41419-017-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hylander B.L., Pitoniak R., Penetrante R.B., Gibbs J.F., Oktay D., Cheng J., Repasky E.A. The anti-tumor effect of Apo2L/TRAIL on patient pancreatic adenocarcinomas grown as xenografts in SCID mice. J. Transl. Med. 2005;3:22. doi: 10.1186/1479-5876-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hylander B.L., Sen A., Beachy S.H., Pitoniak R., Ullas S., Gibbs J.F., Qiu J., Prey J.D., Fetterly G.J., Repasky E.A. Tumor priming by Apo2L/TRAIL reduces interstitial fluid pressure and enhances efficacy of liposomal gemcitabine in a patient derived xenograft tumor model. J. Control. Release. 2015;217:160–169. doi: 10.1016/j.jconrel.2015.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oldenhuis C.N.A.M., Stegehuis J.H., Walenkamp A.M.E., de Jong S., de Vries E.G.E. Targeting TRAIL death receptors. Curr. Opin. Pharmacol. 2008;8:433–439. doi: 10.1016/j.coph.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 68.Sikic B.I., Wakelee H.A., von Mehren M., Lewis N., Calvert A.H., Plummer E.R., Fox N.L., Howard T., Jones S.F., Burris H.A., III A phase Ib study to assess the safety of lexatumumab, a human monoclonal antibody that activates TRAIL-R2, in combination with gemcitabine, pemetrexed, doxorubicin or FOLFIRI. J. Clin. Oncol. 2007;25:14006. [Google Scholar]
- 69.Razeghian E., Suksatan W., Sulaiman Rahman H., Bokov D.O., Abdelbasset W.K., Hassanzadeh A., Marofi F., Yazdanifar M., Jarahian M. Harnessing TRAIL-Induced Apoptosis Pathway for Cancer Immunotherapy and Associated Challenges. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.699746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Zhou J., Zhang L., Zheng H., Ge W., Huang Y., Yan Y., Zhou X., Zhu W., Kong Y., Ding Y., Wang W. Identification of chemoresistance-related mRNAs based on gemcitabine-resistant pancreatic cancer cell lines. Cancer Med. 2020;9:1115–1130. doi: 10.1002/cam4.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amrutkar M., Vethe N.T., Verbeke C.S., Aasrum M., Finstadsveen A.V., Sántha P., Gladhaug I.P. Differential Gemcitabine Sensitivity in Primary Human Pancreatic Cancer Cells and Paired Stellate Cells Is Driven by Heterogenous Drug Uptake and Processing. Cancers. 2020;12:3628. doi: 10.3390/cancers12123628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J., Ren Y., Gui C., Zhao M., Wu X., Mao K., Li W., Zou F. Phosphorylation of Parkin at serine 131 by p38 MAPK promotes mitochondrial dysfunction and neuronal death in mutant A53T α-synuclein model of Parkinson's disease. Cell Death Dis. 2018;9:700. doi: 10.1038/s41419-018-0722-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farley N., Pedraza-Alva G., Serrano-Gomez D., Nagaleekar V., Aronshtam A., Krahl T., Thornton T., Rincón M. p38 mitogen-activated protein kinase mediates the Fas-induced mitochondrial death pathway in CD8+ T cells. Mol. Cell Biol. 2006;26:2118–2129. doi: 10.1128/MCB.26.6.2118-2129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gräb J., Rybniker J. The Expanding Role of p38 Mitogen-Activated Protein Kinase in Programmed Host Cell Death. Microbiol. Insights. 2019;12 doi: 10.1177/1178636119864594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ploumaki I., Triantafyllou E., Koumprentziotis I.A., Karampinos K., Drougkas K., Karavolias I., Trontzas I., Kotteas E.A. Bcl-2 pathway inhibition in solid tumors: a review of clinical trials. Clin. Transl. Oncol. 2023;25:1554–1578. doi: 10.1007/s12094-022-03070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lafaro K.J., Melstrom L.G. The Paradoxical Web of Pancreatic Cancer Tumor Microenvironment. Am. J. Pathol. 2019;189:44–57. doi: 10.1016/j.ajpath.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Awaji M., Singh R.K. Cancer-Associated Fibroblasts' Functional Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancers. 2019;11:290. doi: 10.3390/cancers11030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Öhlund D., Elyada E., Tuveson D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szegezdi E., O'Reilly A., Davy Y., Vawda R., Taylor D.L., Murphy M., Samali A., Mehmet H. Stem cells are resistant to TRAIL receptor-mediated apoptosis. J. Cell Mol. Med. 2009;13:4409–4414. doi: 10.1111/j.1582-4934.2008.00522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wadman M. FDA no longer has to require animal testing for new drugs. Science (New York, N.Y.) 2023;379:127–128. doi: 10.1126/science.adg6276. [DOI] [PubMed] [Google Scholar]
- 81.Jiang H., Wang S., Zhou X., Wang L., Ye L., Zhou Z., Tang J., Liu X., Teng L., Shen Y. New path to treating pancreatic cancer: TRAIL gene delivery targeting the fibroblast-enriched tumor microenvironment. J. Control. Release. 2018;286:254–263. doi: 10.1016/j.jconrel.2018.07.047. [DOI] [PubMed] [Google Scholar]
- 82.Fang Y., Zhou W., Rong Y., Kuang T., Xu X., Wu W., Wang D., Lou W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019;383 doi: 10.1016/j.yexcr.2019.111543. [DOI] [PubMed] [Google Scholar]
- 83.Liu Y., Li Z. Cancer-associated fibroblasts promote progression and gemcitabine resistance via the SDF-1/SATB-1 pathway in pancreatic cancer. Cell Death Dis. 2018;9:1065. doi: 10.1038/s41419-018-1104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Willink C.Y., Jenniskens S.F.M., Klaassen N.J.M., Stommel M.W.J., Nijsen J.F.W. Intratumoral injection therapies for locally advanced pancreatic cancer: systematic review. BJS Open. 2023;7:7. doi: 10.1093/bjsopen/zrad052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao L., Au J.L.S., Wientjes M.G. Comparison of methods for evaluating drug-drug interaction. Front. Biosci. 2010;2:241–249. doi: 10.2741/e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Qiu W., Su G.H. Development of orthotopic pancreatic tumor mouse models. Methods Mol. Biol. 2013;980:215–223. doi: 10.1007/978-1-62703-287-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim M.P., Evans D.B., Wang H., Abbruzzese J.L., Fleming J.B., Gallick G.E. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 2009;4:1670–1680. doi: 10.1038/nprot.2009.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains non-filtered DESeq2 results for six pair-wise comparisons of all conditions. The columns across all sheets are defined as follows: baseMean, the average of the normalized counts for all samples; log2FoldChange, the log2 fold change between the two conditions being compared; lfcSE, the standard error of the log2 fold change; stat, the test statistic for the comparison; p value, the p value for the test statistic; padj, the adjusted p value after correcting for multiple testing using the Benjamini- Hochberg method.
Data Availability Statement
-
•
RNA-seq data have been deposited at NCBI and are publicly available as of the date of publication. Accession number is listed in the key resources table (accession number: PRJNA1124875). Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
R custom script used in this study has been deposited at Zenodo (https://zenodo.org/records/12104006) and is publicly available as of the date of publication. DOIs are also listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request