Abstract
The p53-induced mouse wig-1 gene encodes a Cys2His2-type zinc finger protein of unknown function. The zinc fingers in wig-1 are connected by long (56–75) amino acid linkers. This distribution of zinc finger domains resembles that of the previously described double-stranded (ds)RNA-binding proteins dsRBP-ZFa and JAZ. Ectopically expressed FLAG-tagged mouse wig-1 protein localized to nuclei and in some cells to nucleoli, whereas GFP-tagged mouse wig-1 localized primarily to nucleoli. Electrophoretic mobility shift assay using a recombinant GST–wig-1 fusion protein showed that wig-1 preferentially binds dsRNA rather than single-stranded RNA or dsDNA. A set of deletion/truncation mutants of wig-1 was tested to determine the dsRNA-binding domain(s) or region(s) in wig-1 that is involved in the stabilization of wig-1–dsRNA complexes in vitro. This revealed that the first zinc finger in wig-1 is essential for binding to dsRNA, whereas zinc fingers 2 and 3 are dispensable. wig-1 protein expressed in mammalian cells also showed a high affinity for dsRNA. wig-1 represents the first confirmed p53-induced gene that encodes a dsRNA-binding protein. This suggests that dsRNA binding plays a role in the p53-dependent stress response.
INTRODUCTION
The tumor suppressor p53 is an unstable protein expressed at low levels in most cells and tissues under normal conditions. After DNA damage and other types of cellular stress, the levels of p53 increase through protein stabilization, leading to the initiation of p53-dependent biological responses, including cell cycle arrest and apoptosis (1,2). p53 exerts its function mainly through transcriptional regulation of specific target genes. Transactivation of p21, GADD45 and 14-3-3 σ triggers G1 and G2 cell cycle arrest whereas transactivation of, for example, Bax, Fas, Noxa and PUMA is important for induction of p53-dependent apoptosis (3,4). We have previously identified a novel p53-regulated mouse gene designated wig-1 (for wild-type p53-induced gene 1) (5). wig-1 encodes a Cys2His2-type zinc finger protein of unknown function. A rat homolog, PAG608, was independently identified by Israeli et al. (6) and we have recently cloned and characterized human wig-1 (7). Mouse wig-1 is highly homologous to the rat and human proteins, sharing 97.9 and 87% amino acid sequence identity, respectively. Rat wig-1 (PAG608) has weak pro-apoptotic activity when overexpressed in human tumor cells (6). Human wig-1 can suppress cell growth by 25–30% in a colony formation assay (7).
Double-stranded (ds)RNA structures can originate in many biological processes, e.g. viral infection, or during the normal course of gene expression (8). Proteins binding to dsRNA have a wide variety of biological roles (9). For example, the dsRNA-binding protein Staufen participates in the intracellular transport of specific mRNAs (10) whereas the dsRNA-specific adenosine deaminase family proteins ADAR1 and ADAR2 are involved in site-specific pre-mRNA editing (11). Moreover, binding of the RNA-dependent protein kinase PKR to dsRNA can trigger a conformational change that activates this enzyme. Activated PKR has been implicated in the inhibition of protein synthesis in response to viral infection (12). The vast majority of dsRNA-binding proteins share an ∼70 amino acid sequence called the dsRNA-binding motif (dsRBM) responsible for the binding to RNA duplexes. Binding of the dsRBM to dsRNA is highly specific and apparently independent of the RNA sequence (13,14).
Zinc finger proteins of the Cys2His2 type represent a large class of proteins that have been assumed to function by recognition of specific DNA sequences. However, proteins of this type can also interact with single-stranded (ss)RNA, DNA–RNA hybrids, dsRNA and other proteins (15–17). Recently, several dsRNA-binding zinc finger proteins lacking the dsRBM have been described (16). This category of zinc finger proteins is characterized by long linkers (more than eight residues) between the zinc fingers and inter-histidine distances of five residues in the zinc fingers, in contrast to the three to four residues in most other zinc finger proteins. One member of this category is the Xenopus dsRBP-ZFa protein (18,19) which has linker regions of 34–44 residues separating the zinc fingers. The linker regions lack the conserved linker sequence (TGEKP) connecting adjacent zinc fingers that is common in Cys2His2 zinc finger proteins (20,21). Another dsRNA-binding zinc finger protein, JAZ, is widely expressed in mouse and human tissues (22). The zinc fingers in JAZ are homologous to the corresponding zinc fingers in dsRBP-ZFa. The linker regions in JAZ are also long but show no sequence similarity to the dsRBP-ZFa linkers. Forced expression of JAZ has been shown to induce apoptosis in NIH 3T3 cells.
The structural similarity between wig-1, dsRBP-ZFa and JAZ led us to analyze the intracellular localization and dsRNA-binding activity of wig-1. We found that wig-1 is nuclear and binds dsRNA with high affinity. In addition, we have found that the first zinc finger domain in wig-1 is required for efficient dsRNA binding and that wig-1 protein produced in mammalian cells also binds dsRNA.
MATERIALS AND METHODS
Cell culture
NIH 3T3 mouse fibroblasts and U2OS human osteosarcoma cells were grown in Iscove’s modified Dulbecco’s medium containing 10% fetal calf serum, 2 mM l-glutamine and 40 µg/ml gentamicin (Invitrogen BV, The Netherlands).
Immunofluorescence staining
The mouse wig-1 open reading frame (ORF) with BclI and EcoRI flanking sites introduced by polymerase chain reaction (PCR) was subcloned into the BamHI and EcoRI sites in the pCMVTag2B vector (Stratagene, CA) or in the BglII and EcoRI sites in the pEGFP-C1 vector (Clontech, CA). NIH 3T3 (100 000 cells/well) or U2OS (150 000 cells/well) cells were grown on coverslips in 6-well plates and transiently transfected using Lipofectamine 2000 (Invitrogen BV) according to the manufacturer’s instructions. Twenty-four hours after transfection cells were washed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 30 min and the green fluorescence signal from the green flourescent protein (GFP) fusion protein was analyzed using an inverted fluorescence microscope. Cells tranfected with FLAG-tagged constructs were also permeabilized with 0.2% NP-40 for 5 min and incubated in BSA blocking buffer (2% BSA, 5% glycerol, 0.2% Tween-20, 0.1% NaN3) for 1 h. FLAG–wig-1 proteins were detected using anti-FLAG M2 antibody (Stratagene, CA) at a dilution of 1:1000. FITC-conjugated rabbit anti-mouse Ig antibody (DAKO, Copenhagen, Denmark) diluted 1:40 was used as secondary antibody. Cells were washed in PBS, mounted and analyzed with a fluorescence microscope.
Expression and purification of recombinant proteins
To produce a glutathione S-transferase (GST)–wig-1 fusion protein, the wig-1 ORF with BclI and EcoRI flanking sites was subcloned into the BamHI and EcoRI sites of the pGEX-2T vector (Amersham Pharmacia Biotech, Uppsala, Sweden). The fusion protein was expressed in Escherichia coli BL21 codon plus (Stratagene, CA) and purified as described (23). Briefly, cells were induced with 0.3 mM IPTG for 3 h at 37°C and harvested. The cell pellet was resuspended in 1/25 of the bacterial culture volume in STE buffer (10 mM Tris–HCl pH 8, 1 mM EDTA, 150 mM NaCl). Lysozyme was added to a final concentration of 0.1 mg/ml and the cells were incubated for 15 min on ice. Just before sonication 10 mM DTT and 1.5% sarkosyl were added. Cell lysates were cleared by centrifugation at 15 000 r.p.m. for 30 min and incubated with glutathione–Sepharose beads (Amersham Pharmacia Biotech) overnight at 4°C. Beads were washed three times with 5 vol PBS containing 0.1% Tween-20. Fusion proteins were eluted at 4°C for 3 h in elution buffer (20 mM reduced glutathione in 50 mM Tris–HCl pH 8, 50 mM KCl, 1 mM DTT, 0.1% Triton X-100). All proteins were stored at –80°C in elution buffer containing 20% glycerol. Expression and purification of the deletion/truncation mutant proteins were performed as described for the full-length protein. Yield and purity of all recombinant proteins were assessed by 10% SDS–PAGE and Coomassie staining. Protein concentrations were determined using a BSA standard.
Generation of wig-1 deletion mutants
Mutant constructs were made by fusing the corresponding PCR fragments with adequate restriction enzyme sites followed by cloning of the fragments into the pGEM-T-Easy vector (Promega, WI). The absence of PCR-induced mutations was verified by DNA sequencing using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, MA) and analyzed on an ABI Prism 310 Genetic Analyser (Perkin Elmer, MA). The fused PCR fragments were subcloned into the BamHI and EcoRI sites of the pCMVTag2B or pGEX-2T vector. Detailed cloning strategy and primer sequence information will be provided upon request.
Electrophoretic mobility shift assay (EMSA)
A protocol described for JAZ (22) was used with some modifications. A 172 bp dsDNA probe was prepared by PCR using the pBluescript II KS vector (Stratagene) as template containing the T7 and T3 promoters (T7 primer sequence, 5′-GTAATACGACTCACTATAGGGCGAATTG-3′; T3 primer sequence, 5′-AATTAACCCTCACTAAAGGG-3′). This fragment was gel purified and used as a DNA competitor or radiolabeled probe after shrimp alkaline phosphatase treatment (Roche Diagnostics, IN) and 5′-end-labeling using T4 polynucleotide kinase (Invitrogen BV) and [γ-32P]ATP (Amersham Pharmacia Biotech). For generation of the 160 nt ssRNA probe the 172 bp dsDNA fragment was in vitro transcribed using T7 and T3 polymerase (MAXIscript In Vitro Transcription Kit; Ambion, TX) in the presence of [α-32P]UTP (Amersham Pharmacia Biotech). The MEGAshortscript In Vitro Transcription Kit (Ambion, TX) was used to prepare large amounts of ssRNA for the competition experiments. To make the 130 bp dsRNA the 172 bp DNA fragments generated by PCR using plasmids pBluescript II KS and pBluescript II SK and the T3 and T7 primers were in vitro transcribed using T7 polymerase and the MEGAshortscript In Vitro Transcription Kit. The complementary strands were mixed in FBB binding buffer [80% deionized formamide, 40 mM 1,4-piperazinediethanesulfonic acid (PIPES) pH 6.7, 400 mM NaCl, 1 mM EDTA], incubated at 65°C for 5 min in a water bath and cooled slowly to room temperature. The dsRNA probe was treated with alkaline phosphatase and 5′-end-labeled using the KinaseMax labeling kit (Ambion) according to the manufacturer’s instructions. The dsDNA and dsRNA probes were purified on a native 5% polyacrylamide gel and eluted in 5 mM MgCl2 (DNA probe) or in DEPC-treated water (dsRNA probe). The ssRNA probe was gel purified in an 8 M urea/5% acrylamide denaturing gel and eluted in DEPC-treated water.
Aliquots of 50 (dsRNA gel shifts) or 100 ng (ssRNA and dsDNA gel shifts) protein were incubated with ∼0.5 ng nucleic acid probes in 20 µl of binding buffer (10% glycerol, 20 mM HEPES pH 7.4, 20 mM KCl, 1 mM MgCl2, 5 mM DTT, 10 µM ZnCl2, 100 µg/ml BSA, 0.1% NP-40) for 30 min at room temperature. Samples were loaded on a 4% native polyacrylamide gel, fixed in 10% acetic acid/10% ethanol for 15 min and vacuum dried for analysis by autoradiography.
Poly(I·C) bead binding assays
FLAG-tagged full-length wig-1 and a mutant wig-1 protein lacking the first zinc finger were transiently transfected into NIH 3T3 cells (150 000 cells/well) growing in 6-well plates using Lipofectamine 2000 (Invitrogen BV) as described above. After 48 h, cells were lysed in lysis buffer (50 mM Tris–HCl pH 7.4, 0.15 M NaCl, 1% Triton-X100, 10 mM phenylmethylsulfonyl flouride). Aliquots of cell lysates (50 µg) were incubated with 50 µl of poly(I·C)–agarose beads (Amersham Pharmacia Biotech), in the absence or presence of poly(C) or poly(I·C) (Amersham Pharmacia Biotech) for 2 h at 4°C. The beads were washed in lysis buffer three times before boiling in SDS–PAGE sample buffer. The eluted samples and 50 µg extract were loaded on a 10% SDS polyacrylamide gel followed by immunoblotting using anti-FLAG M2 antibody (Stratagene, CA) diluted 1:2000 and horseradish peroxidase-conjugated secondary anti-mouse Ig antibody (Amersham Pharmacia Biotech) diluted 1:5000.
RESULTS
Wig-1 localizes to the nucleus and nucleoli
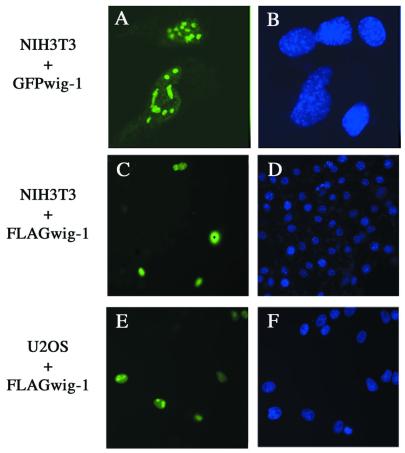
Mouse wig-1 has a potential nuclear localization signal (NLS) (residues 195–211) (5). To analyze the intracellular distribution of mouse wig-1, we transfected NIH 3T3 cells with a construct expressing a GFP–full-length mouse wig-1 fusion protein. This protein localized primarily to nucleoli (Fig. 1A). However, transfection of the same cells with FLAG-tagged full-length mouse wig-1 followed by immunostaining using anti-FLAG antibodies demonstrated that mouse wig-1 can also localize to the nucleus (Fig. 1C). Expression of the same construct in U2OS osteosarcoma cells revealed nucleolar localization in the majority of the positive cells (Fig. 1E). We have also analyzed the intracellular distribution of several FLAG-tagged deletion/truncation wig-1 mutants (Fig. 2A) in NIH 3T3 cells. All mutants were nuclear except the wig-1 mutant lacking the NLS, which was dispersed evenly in the cytoplasm (data not shown), indicating that this domain acts as a true NLS.
Figure 1.
Intracellular localization of wig-1. NIH 3T3 cells were transiently transfected with GFP-tagged (A and B) or FLAG-tagged (C and D) wig-1 expression plasmid. U2OS cells were transfected with the FLAG-tagged wig-1 expression plasmid (E and F). The green fluorescence signal from the GFP fusion construct was monitored using an inverted fluorescence microscope (A). The localization of FLAG-tagged wig-1 protein was determined by immunostaining with anti-FLAG antibody (C and E). For DNA staining, Hoechst dye 33258 was included in the BSA blocking buffer during the last antibody incubation (B, D and F). The magnification is 100× in (A) and (B) and 63× in (C)–(F).
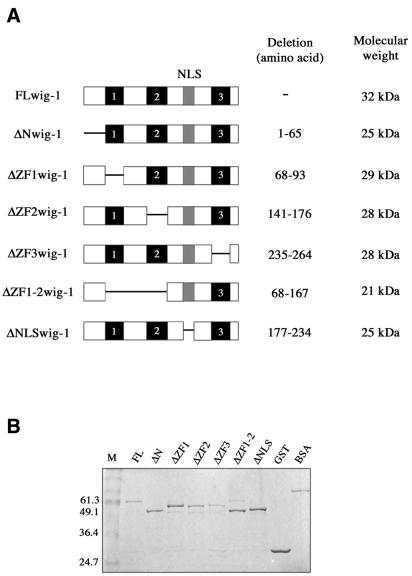
Figure 2.
wig-1 deletion mutants. (A) Schematic representation of full-length wig-1 and deletion mutants used in EMSA. (B) Purity and yield of bacterially produced GST–wig-1 proteins as estimated by SDS–PAGE and Coomassie staining.
Wig-1 preferentially binds dsRNA
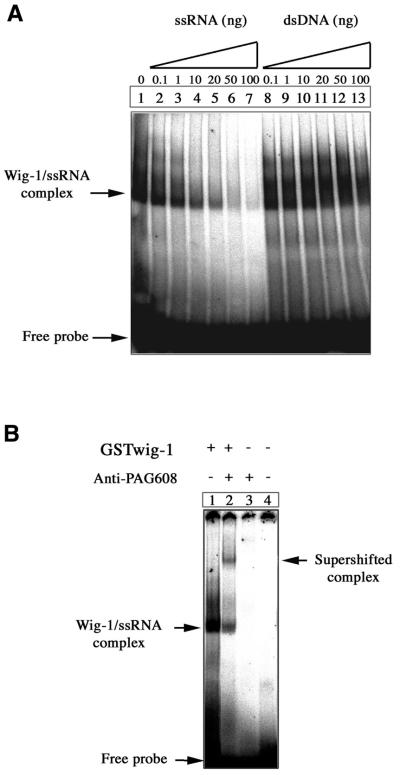
Most zinc finger proteins have been shown to bind nucleic acids or other proteins. In order to investigate the nucleic acid-binding properties of wig-1 we performed EMSAs using recombinant GST–full-length mouse wig-1 protein produced in bacteria. The protein was incubated with different types of DNA and RNA probes and subjected to native PAGE. As shown in Figure 3A, the wig-1 protein binds preferentially to ssRNA compared to dsDNA. Formation of the complex between wig-1 and the 160mer ssRNA probe (Fig. 3A, lane 1) was efficiently competed out with increasing concentrations of the same unlabeled ssRNA probe (Fig. 3A, lanes 2–7). In contrast, increasing amounts of an unlabeled 172mer dsDNA probe did not have any effect on the ability of wig-1 to bind ssRNA (Fig. 3A, lanes 8–13). No binding of wig-1 to dsDNA was observed under these experimental conditions (data not shown). To demonstrate the presence of wig-1 protein in the complex, we used anti-PAG608 antibodies that cross-react with mouse wig-1. Addition of these antibodies to the binding reaction mixture gave rise to a supershifted band presumably corresponding to a larger complex containing wig-1, anti-PAG608 antibodies and ssRNA (Fig. 3B, lane 2).
Figure 3.
EMSA showing the binding of wig-1 to ssRNA. (A) An aliquot of 100 ng bacterially produced GST–wig-1 protein was incubated with 32P-labeled 160mer ssRNA probe and analyzed by PAGE. In the competition experiments the indicated amounts of the unlabeled 160mer ssRNA probe or an unlabeled 172mer dsDNA probe were added (up to 200-fold excess over the labeled probe). (B) Addition of anti-PAG608 antibodies (lane 2) gave rise to a supershifted band, demonstrating the presence of wig-1 in the complex.
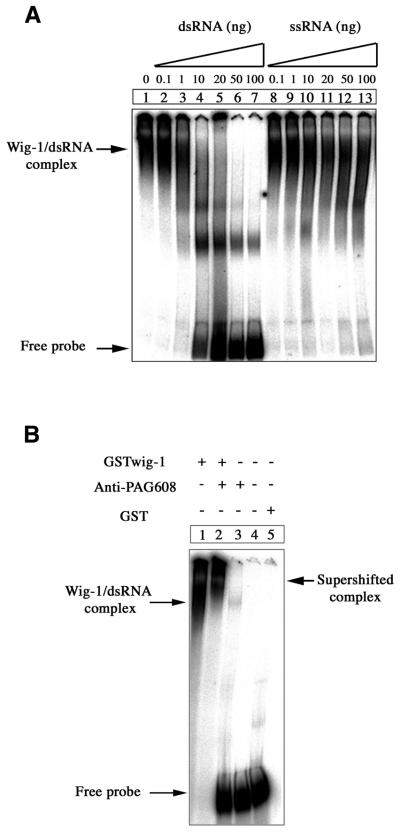
The structural homology between wig-1 and the dsRNA-binding proteins dsRBP-ZFa and JAZ (19,22) prompted us to examine whether wig-1 could bind dsRNA. For this purpose, we performed EMSAs as described above but using a 130mer dsRNA as radiolabeled probe. wig-1 bound to the dsRNA probe with higher affinity than to the 160mer ssRNA probe, since much less protein (50 ng) was needed to detect the formation of a complex similar to that formed with ssRNA (Fig. 4A, lane 1). In fact, wig-1 binding to dsRNA was observed with <10 ng protein (data not shown). Moreover, only the same unlabeled dsRNA probe (Fig. 4A, lane 2–7) but not an unlabeled ssRNA probe (Fig. 4A, lane 8–13) was able to efficiently compete out wig-1–dsRNA complexing. Again, the presence of wig-1 in the complex was confirmed by supershift with anti-PAG608 antibodies (Fig. 4B, lane 2). To rule out the possibility that the GST portion of the fusion protein participated in binding to dsRNA, GST protein alone was incubated with all probes and analysed by EMSA. No binding of GST to dsRNA (Fig. 4B, lane 5) or ssRNA or dsDNA (not shown) was detected.
Figure 4.
EMSA showing preferential binding of wig-1 to dsRNA. (A) A radiolabeled 130mer dsRNA probe was incubated with 50 ng bacterially produced GST–wig-1 protein. Increasing amounts of unlabeled dsRNA or ssRNA probes were used as competitors (up to 200-fold excess over the labeled probe). (B) The wig-1–dsRNA complex was supershifted by anti-PAG608 antibodies (lane 2). The GST protein was used as a control (lane 5).
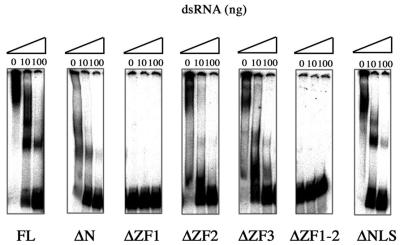
Mapping of the dsRNA-binding activity
To determine the region(s) in the wig-1 protein that is involved in dsRNA binding, we analyzed a set of deletion mutants subcloned into the pGEX-2T vector and expressed as GST fusion proteins (Fig. 2A). The purity and yield of the recombinant proteins was assessed by Coomassie staining (Fig. 2B). For the deletion studies, recombinant GST mutant proteins (50 ng) were incubated with the labeled dsRNA probe and assayed in EMSA. The wig-1 deletion mutants lacking the first zinc finger or the first and the second zinc fingers failed to bind dsRNA (Fig. 5), whereas all other mutants retained dsRNA binding with similar affinity as the full-length protein.
Figure 5.
The first zinc finger in wig-1 is necessary for binding to dsRNA. Fifty nanograms of full-length GST–wig-1 and deletion mutants were tested by EMSA for their ability to bind dsRNA. The complexes were competed out by the addition of 10 or 100 ng unlabeled dsRNA probe.
Wig-1 expressed in mammalian cells also binds dsRNA
In order to investigate the RNA-binding capacity of wig-1 protein expressed in mammalian cells, we transfected FLAG-tagged full-length wig-1 and the wig-1 mutant lacking the first zinc finger into NIH 3T3 cells. Total protein cell lysates were incubated with poly(I·C)–agarose beads. The beads were pelleted and the binding of wig-1 to poly(I·C) was analyzed by western blotting. This confirmed the results obtained with the bacterially produced recombinant wig-1 proteins. As shown in Figure 6A, full-length wig-1 expressed in NIH 3T3 cells was able to bind poly(I·C) beads and this binding could only be competed out by addition of an excess of poly(I·C), whereas the ssRNA polymer poly(C) was inefficient as a competitor. In contrast, the wig-1 deletion mutant lacking the first zinc finger could not bind the poly(I·C) beads (Fig. 6B), confirming the critical role of this zinc finger domain for dsRNA binding.
Figure 6.
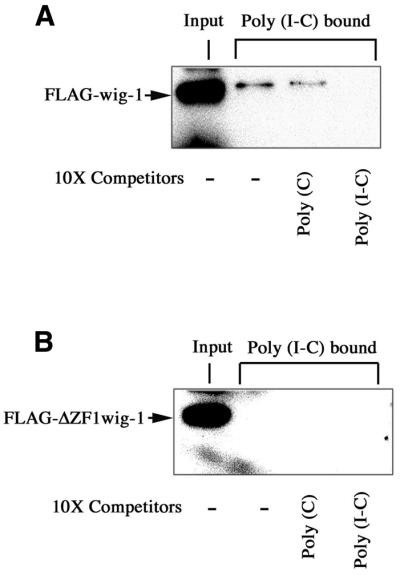
Wig-1 protein produced in mammalian cells also specifically binds dsRNA. (A) NIH 3T3 cells were transfected with the FLAG-tagged mouse wig-1 expression plasmid. After 48 h total protein cell lysates were prepared and 50 µg aliquots were incubated with poly(I·C) immobilized on agarose beads. The incubations were performed in the absence or presence of the competitor poly(C) or poly(I·C). The bound wig-1 protein was analyzed by immunoblotting using anti-FLAG antibodies. (B) As in (A) except that NIH 3T3 cells were transfected with a FLAG-tagged wig-1 expression plasmid lacking the first zinc finger domain.
DISCUSSION
The p53-induced mouse wig-1 gene encodes a zinc finger protein with structural homology to several dsRNA-binding proteins, including dsRBP-ZFa and JAZ. We have found here that mouse wig-1, like JAZ and dsRBP-ZFa, is a nuclear protein. Like JAZ, wig-1 may also localize in nucleoli. These results are in agreement with the presence of a NLS in the mouse wig-1 protein (5) and the observed nuclear and nucleolar localization of the human and rat wig-1 (PAG608) proteins (6,7).
The structural similarity between wig-1, dsRBP-ZFa and JAZ raised the possibility that wig-1 also complexes with dsRNA. We have shown that wig-1 binds preferentially to dsRNA as compared to ssRNA or dsDNA of corresponding size. Another feature shared between wig-1, dsRBP-ZFa and JAZ is the inability to bind dsDNA under the conditions used.
dsRNA binding by proteins containing the consensus dsRBM is apparently sequence independent (8). Similarly, both dsRBP-ZFa and JAZ bind dsRNA in a sequence-independent manner (18,22). Consistent with this, binding of wig-1 to dsRNA appears to be independent of RNA sequence, since wig-1 is able to bind poly(I·C). The molecular basis of the interaction between zinc finger proteins like wig-1, dsRBP-ZFa or JAZ and dsRNA is poorly understood. We have demonstrated that deletion of the first zinc finger in wig-1 abrogates binding to dsRNA. This indicates that this region either represents the dsRNA-binding domain itself or is critical for maintaining the overall structure of the dsRNA-binding domain in the wig-1 protein. Our finding that wig-1 protein expressed in mammalian cells binds poly(I·C) and that the first zinc finger is required for this interaction suggests that wig-1 binds dsRNA also in living cells.
The biological significance of the dsRNA-binding activity of wig-1 remains unclear. However, there are several conceivable mechanisms by which a p53-induced dsRNA-binding protein could contribute to the p53-mediated biological response. We have observed that wig-1 is localized in nucleoli in some cells. This raises the possibility that wig-1 binds to ribosomal dsRNA. Such binding could interfere with ribosome biogenesis and/or protein synthesis, ultimately leading to inhibition of cell growth and possibly cell death. In this model wig-1 would act as a downstream effector of p53-dependent growth suppression. This idea is supported by our previous demonstration that human wig-1 inhibits cell growth in a colony formation assay (7).
dsRNA also exists in cells during the sequence-specific post-transcriptional gene silencing process called RNA interference or RNAi (24,25). It has been proposed that certain dsRNA molecules can guide protein complexes to target specific mRNAs for cleavage. wig-1 could be one component in such complexes accompanying the dsRNAs to their specific targets and thus participate in specific gene silencing. Another possibility is that wig-1 binds directly to specific mRNAs and thereby regulates expression of certain gene products, for example proteins that control cell growth and/or apoptosis.
Moreover, dsRNA is produced in cells during viral infection. It is possible that viral infection per se could act as a stress signal to p53, resulting in the induction of p53 target genes, including wig-1. In this situation, wig-1 could inhibit viral replication by binding viral dsRNA. wig-1 would thus serve as an antiviral protein induced by p53 to protect cells from virus infection and/or lysis.
A recently described novel p53 target gene, MCG10, encodes a protein that binds ssRNA and suppresses cell growth by inducing cell cycle arrest in G2/M and apoptosis (26). MCG10 binds poly(C) through two so-called KH domains and this activity is essential for function of the protein. However, wig-1 clearly represents a different class of p53-induced gene products since wig-1 binds to dsRNA rather than ssRNA. To our knowledge, wig-1 is the first reported dsRNA-binding protein induced by p53. Further functional studies of wig-1 and the role of its dsRNA-binding activity should provide important information about the p53 tumor suppressor pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Moshe Oren (Weizmann Institute of Science, Rehovot, Israel) for the anti-PAG608 antibodies. This work was supported by grants from the Swedish Cancer Society (Cancerfonden), Konung Gustaf V Jubilee Fund, Åke Wibergs Stiftelse and the Karolinska Institute.
REFERENCES
- 1.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 2.Vousden K.H. (2000) p53: death star. Cell, 103, 691–694. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry W.S. (1998) Regulation of p53 downstream genes. Semin. Cancer Biol., 8, 345–357. [DOI] [PubMed] [Google Scholar]
- 4.Ryan K.M., Phillips,A.C. and Vousden,K.H. (2001) Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol., 13, 332–337. [DOI] [PubMed] [Google Scholar]
- 5.Varmeh-Ziaie S., Okan,I., Wang,Y., Magnusson,K.P., Warthoe,P., Strauss,M. and Wiman,K.G. (1997) Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene, 15, 2699–2704. [DOI] [PubMed] [Google Scholar]
- 6.Israeli D., Tessler,E., Haupt,Y., Elkeles,A., Wilder,S., Amson,R., Telerman,A. and Oren,M. (1997) A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes papooses. EMBO J., 16, 4384–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellborg F., Wang,Q., Méndez-Vidal,C., Asker,C., Kost-Alimova,M., Wilhelm,M., Imreh,S. and Wiman,K.G. (2001) Human wig-1, a p53-target gene that encodes a growth inhibitory zinc finger protein. Oncogene, 20, 5466–5474. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson A.W. (1996) Structure, reactivity and biology of double-stranded RNA. Prog. Nucleic Acid Res. Mol. Biol., 52, 1–65. [DOI] [PubMed] [Google Scholar]
- 9.Fierro-Monti I. and Mathews,M.B. (2000) Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci., 25, 241–246. [DOI] [PubMed] [Google Scholar]
- 10.St Johnston D., Beuchle,D. and Nusslein-Volhard,C. (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell, 66, 51–63. [DOI] [PubMed] [Google Scholar]
- 11.Bass B.L. and Weintraub,H. (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell, 55, 1089–1098. [DOI] [PubMed] [Google Scholar]
- 12.Clemens M.J. (1997) PKR—a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol., 29, 945–949. [DOI] [PubMed] [Google Scholar]
- 13.Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos A., Grunert,S., Adams,J., Micklem,D.R., Proctor,M.R., Freund,S., Bycroft,M., St Johnston,D. and Varani,G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laity J.H., Lee,B.M. and Wright,P.E. (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol., 11, 39–46. [DOI] [PubMed] [Google Scholar]
- 16.Iuchi S. (2001) Three classes of C2H2 zinc finger proteins. Cell Mol. Life Sci., 58, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackay J.P. and Crossley,M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- 18.Finerty P.J. Jr and Bass,B.L. (1997) A Xenopus zinc finger protein that specifically binds dsRNA and RNA-DNA hybrids. J. Mol. Biol., 271, 195–208. [DOI] [PubMed] [Google Scholar]
- 19.Finerty P.J. Jr and Bass,B.L. (1999) Subsets of the zinc finger motifs in dsRBP-ZFa can bind double-stranded RNA. Biochemistry, 38, 4001–4007. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct., 29, 183–212. [DOI] [PubMed] [Google Scholar]
- 21.Laity J.H., Dyson,H.J. and Wright,P.E. (2000) DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J. Mol. Biol., 295, 719–727. [DOI] [PubMed] [Google Scholar]
- 22.Yang M., May,W.S. and Ito,T. (1999) JAZ requires the double-stranded RNA-binding zinc finger motifs for nuclear localization. J. Biol. Chem., 274, 27399–27406. [DOI] [PubMed] [Google Scholar]
- 23.Frangioni J.V. and Neel,B.G. (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem., 210, 179–187. [DOI] [PubMed] [Google Scholar]
- 24.Bass B.L. (2000) Double-stranded RNA as a template for gene silencing. Cell, 101, 235–238. [DOI] [PubMed] [Google Scholar]
- 25.Carthew R.W. (2001) Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol., 13, 244–248. [DOI] [PubMed] [Google Scholar]
- 26.Zhu J. and Chen,X. (2000) MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G(2)-M. Mol. Cell. Biol., 20, 5602–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]