Abstract
Recognition of ubiquitin (Ub) is often mediated by small Ub binding domains such as the Ubiquitin Interacting Motif (UIM). Most Ub binding events are low affinity interactions, and designing stronger binders for Ub can be challenging. We here report the design of a short crosslinked coiled coil (CC) which is conformationally and chemically stable, and which can act as a scaffold to present the key binding residues from known UIM sequences. Doing so gives rise to a hybrid CC peptide that reconciles the important features of both UIM and CC sequences. We show by fluorescence polarization assays that this crosslinked ‘CC–UIM’ peptide exhibits enhanced binding to Ub compared to the original UIM sequence. Furthermore, we report a crystal structure of this peptide in complex with Ub. These studies show that preorganization of a small number of important binding residues onto a stable helical scaffold can be a successful strategy for binder design.
Mapping of key binding residues onto a designed coiled-coil peptide scaffold results in an enhanced-affinity binder of the protein Ubiquitin.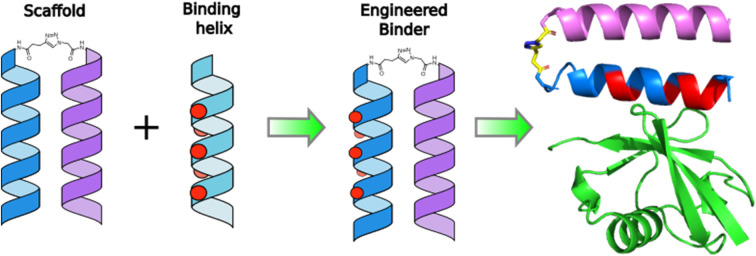
Introduction
Protein–protein interactions (PPIs) underpin many biochemical processes, and often take place over large, dynamic binding surfaces that make targeting them with designed binders challenging.1–4 In some cases it is possible to identify ‘hot-spot’ residues that provide the majority of the binding energy between interacting protein chains. α-helices are commonly found to mediate contacts between proteins, and have therefore been used as a starting point for the design of peptides that exhibit hot-spot residues and therefore mimic PPIs.2,5,6 Enhanced binding to protein partners has been targeted by stabilization of α-helical structures,7 using conformationally constrained peptides8–12 or α-helical coiled coil formation.13–17
α-helical coiled coils (CCs) are characterized by the heptad repeat, a pattern of hydrophobic and polar residues, conventionally labelled abcdefg, in which residues a and d are hydrophobic, with the other positions being occupied by polar residues. This pattern gives rise to an amphipathic α helix, which in aqueous media will tend to associate to minimize solvophobic interactions with these hydrophobic side chains. Sequence to structure relationships have been explored for CCs, and it is possible to control partner selection and oligomerization state by careful choice of peptide sequence.18–20 Because of their structural regularity and preorganization of the α helix, CCs have been used as a template for mimicking PPIs by patterning hot-spot residues onto the outward-facing surface of one helix. Related helical systems have been generated using methods that include N- and C-terminal crosslinking and side-chain crosslinking.14,15,21–23 These helical binders have been used as mimics of PPIs such as NEMO:vFLIP21 and p53:MDM2.23
Ubiquitin (Ub) is a short 76 residue protein that can be post-translationally attached to other proteins, normally via a lysine side chain or the protein N-terminus. Ubiquitination of proteins is vital for a range of cellular functions, including cell division, protein quality control, gene expression, DNA repair and the transport of proteins.24–27 The recognition of Ub is a complex process, and several small Ub binding domains (UBDs) have been characterised.24,28 Most UBDs bind to the Ile44–His68–Val70 binding groove of Ub, often by contact exclusively or primarily via a single α helix. One of the most well characterized UBDs is the ubiquitin interacting motif (UIM). UIMs bind Ub through a single α-helix, which makes contact with the hydrophobic β-sheet binding pocket of Ub.29 UIMs are characterized by their AxxxS motif, in which alanine and serine residues are spaced 4 positions apart. Binding affinities of UIMs to Ub have been reported in the low millimolar to high micromolar range.29,30 Most UBDs bind with similarly weak affinities. There is considerable variation in sequence composition between UIM sequences, which is likely to modulate the Ub binding affinity. The ability to control Ub affinity in designed Ub binding peptides could therefore be of significant utility in understanding both Ub binding and its downstream consequences.
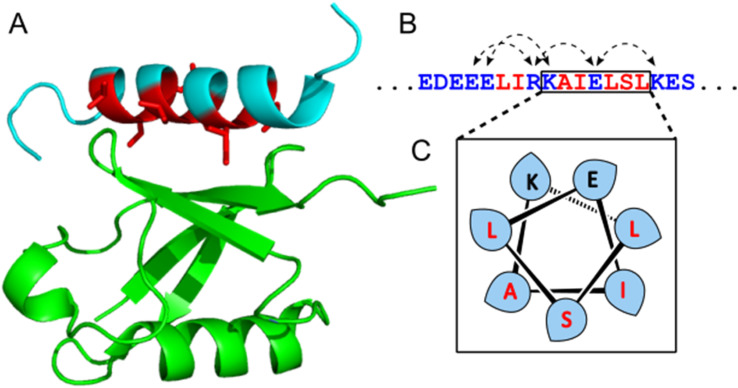
The Vps27 UIM-1 peptide
The UIM-1 sequence of Vsp27 was chosen as a starting point, due to it having both an NMR structure (1q0w) and a reported dissociation constant of 227 μmol l−1. The solution structure of the Ub:UIM-1 complex shows that the binding region of the peptide is α-helical. UIM-1 binds through a continuous stripe of hydrophobic residues, centered around the Ala266 and Ser270 residues in the core binding region of UIM-1 covering residues 257–274. Overall, UIM-1 binds through six hydrophobic residues and a buried serine, which all make contact with the well-defined Ile44–His68–Val70 binding region of Ub. Computational alanine scanning experiments indicate that the central hydrophobic residues Leu262, Ile263, Ile267, Leu269 and Leu271 constitute the main binding surface of the peptide. The bound state of the peptide also exhibits a series of i − i + 4 salt bridges across the ‘back’ face of the helix. These interactions may contribute to Ub binding by stabilizing the helical form of the peptide. In general terms, it is likely that preorganization of the binding residues exhibited by UIMs could contribute to Ub binding by offsetting the entropic cost of helix formation. We therefore reasoned that a stable helical scaffold could be designed such that a UIM sequence could be patterned onto the outer surface of a preorganized helix, giving rise to enhanced Ub binding properties. To this end we first designed a stabilized α-helical coiled coil peptide.
Results and discussion
Design of a short crosslinked helical dimer
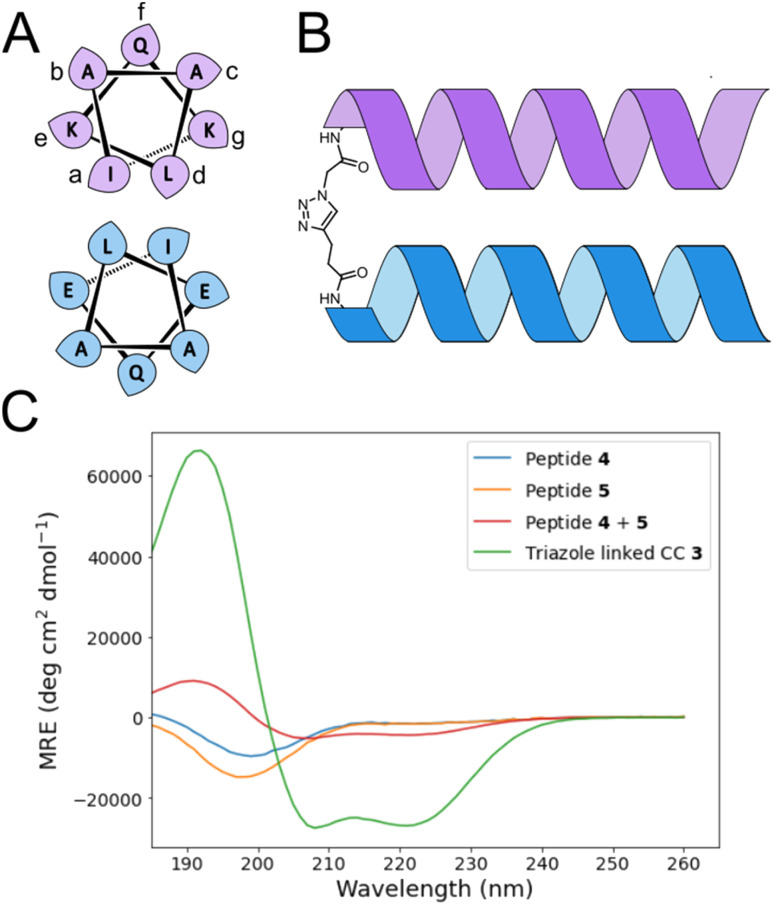
α-helical coiled coils are a common protein structural motif, and have been the subject of numerous engineering studies. CC folding is primarily mediated by the masking of hydrophobic residues. Because of this the conformational stability of CCs varies as a function of helix length, among other factors, and CCs shorter than three heptad repeats (21 residues, six helical turns), which would mask only six or fewer hydrophobic residues, are not commonly observed. We reasoned that a short, conformationally stable CC structure could be achieved by covalently connecting two peptides with a linker that is of the right size to stabilize CC formation. We based our design on known design principles for the formation of CC heterodimers, as described by Thomas et al.18 Briefly, Ile and Leu residues were placed at a and d positions and complementary salt bridges were programmed by one peptide having Glu residues at the e and g positions while the other sequence has Lys residues at these positions. The remainder of the heptad positions were populated with helix-favoring Ala residues, as well as Gln for solubility and Tyr as a UV chromophore. In order to covalently link the peptides we selected an eight-atom linker that could be installed via a triazole ligation. The length of the linker was chosen to approximately match the approximate separation of 6–7 Å between amino acid α-carbon atoms at a d register position.16 It was further anticipated that the hydrophobic nature of this linker could stabilize the folded CC structure by partially masking the adjacent Ile residues from unfavorable interactions with water.31 The linker was placed at the N-terminus so that its precursor azide/alkyne functionality could be installed easily under standard solid phase peptide synthesis conditions. These considerations gave rise to sequences 1 and 2 (Table 1). These peptides were synthesized and crosslinked under mild conditions (Fig. S1†) to yield peptide 3. Acetyl capped versions of the CC sequences, peptides 4 and 5 were made for reference purposes.
Peptides used in this study. Coiled coil heptad registers and UIM numbering are shown where relevant. Full details are given in Table S1.
| Number | Description | Sequence & register |
|---|---|---|
| 1 | E-helix, alkyne |

|
| 2 | K-helix, azide |
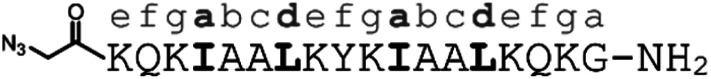
|
| 3 | Reference CC |
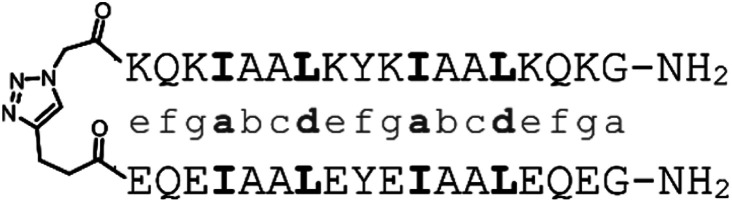
|
| 4 | E-helix, capped |
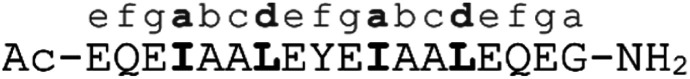
|
| 5 | K-helix, capped |
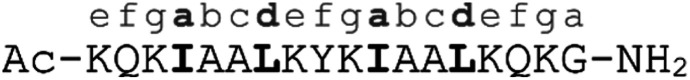
|
| 6 | CC–UIM E-helix |

|
| 7 | CC–UIM K-helix, unlabeled |
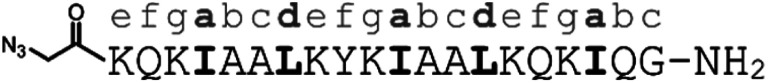
|
| 8 | CC–UIM K-helix, fluorescein labelled |

|
| 9 | CC–UIM, unlabeled |

|
| 10 | CC–UIM, fluorescein labelled |

|
| 11 | UIM reference, unlabeled |

|
| 12 | UIM reference, fluorescein labelled |

|
The effect of the triazole crosslink on folding was investigated by circular dichroism (CD). In isolation, peptides 4 and 5 showed CD spectra indicative of random coil formation. A 1 : 1 mixture of peptides 4 and 5 exhibited a CD signal that indicates a very slight degree of α-helix formation (Fig. 2C), suggesting that without crosslinking, these peptides are too short to form a CC. The triazole-linked peptide 3, however, exhibited a CD signal that indicates this peptide is predominantly in the α-helical conformation in aqueous solution. Peptide 3 was found to be chemically stable over a 30 day time period when stored at pH 7 in water at room temperature (Fig. S2†). Thermal melting CD spectroscopy experiments showed that peptide 3 was stable at moderate temperatures. Heating to temperatures exceeding 60 °C (Fig. S3 and S4†) resulted in visible aggregation of the peptide and a CD spectrum with a single broad minimum at ∼220 nm, suggesting that insoluble β-sheet structures had formed. No evidence of the formation of β-structures was observed on prolonged storage of peptide 3 at 5 °C. Peptide 3 is therefore a chemically and conformationally stable minimal CC, and we investigated its use as a scaffold for the display of UIM-derived binding residues in order to develop engineered Ub binding peptides.
Fig. 2. (A) Helical wheel showing sequence composition and register for coiled coil design. Leaf shapes indicate the direction at which amino acid side chains project from the helix. (B) Schematic of triazole cross-linking unit. (C) CD spectra of peptides 3, 4 and 5.
Ub-binding CC based on crosslinked CC-scaffold
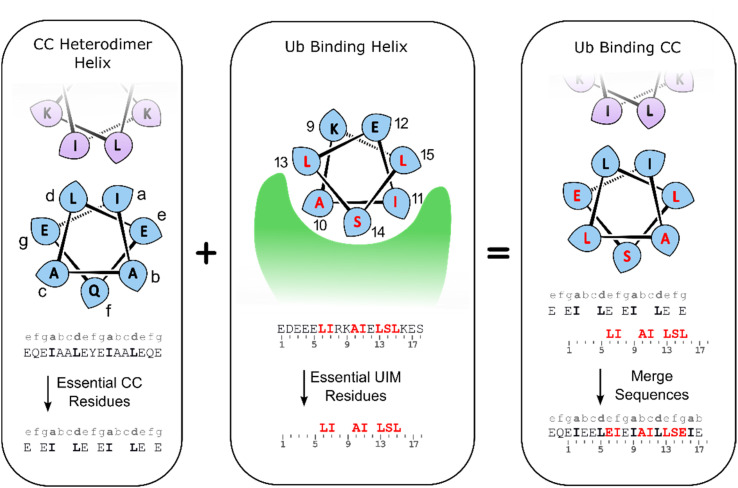
Alignment of the UIM-1 sequence with that of the CC scaffold 3 allowed us to propose a merged sequence containing key features of both the CC and UIM. The core residues of the UIM-1 sequence can be placed at the b, c and f positions, away from the g, a, d and e positions that define the helix–helix interaction surface of the CC. We chose to place the Ser14 residue at the C-terminal of the two available f positions of the CC. This necessitated increasing the length of the CC by two residues, incorporating an additional hydrophobic a position, but allowed us to preserve the negatively charged Glu residues at the N-terminal 1, 3, and 4 positions of the UIM-1 sequence. A cluster of negatively charged residues at these positions is a common sequence feature of UIMs, and may contribute to binding by interaction with the positively charged C-terminal tail of Ub.32 The positively charged auxiliary strand was also extended by two residues at its C-terminus. There is partial overlap between the UIM-1 sequence and the CC motif at the e and g positions corresponding to UIM-1 residues 6, 13 and 15. In the UIM-1 sequence these are all Leu residues, whereas in the CC sequence they are all Glu. UIM sequences commonly feature negatively charged residues, so we examined the degree to which Glu substitutions were tolerated in the original UIM-1 sequence. We tested the L6E and L15E variations of UIM-1 (Fig. S5†), and found that both variations resulted in slightly weaker Ub binding, despite minimal changes in secondary structure (Fig. S6†). We reasoned that the inclusion of two Glu residues into the Ub binding interface presented by the CC represented a suitable compromise between the two sequence patterns, and would avoid making the CC overly hydrophobic. We therefore specified the peripheral residues 6 and 15 to be Glu, and the more central residue 13 to be Leu (Fig. 3).
Fig. 3. Sequence design for CC–UIM peptides.
These considerations gave rise to peptide 6, a CC–UIM binding strand, peptide 7, a CC–UIM auxiliary strand, and peptide 8, a CC–UIM auxiliary strand with a fluorescein label. A triazole formation between 6 and 7 gave rise to the unlabeled CC–UIM 9, and reaction of 6 and 8 yielded the fluorescein labelled CC–UIM 10. The native UIM-1 peptide sequences was synthesized in unlabeled form (peptide 11), and with a fluorescein label appended to the side chain of a C-terminal Lys residue, giving peptide 12.
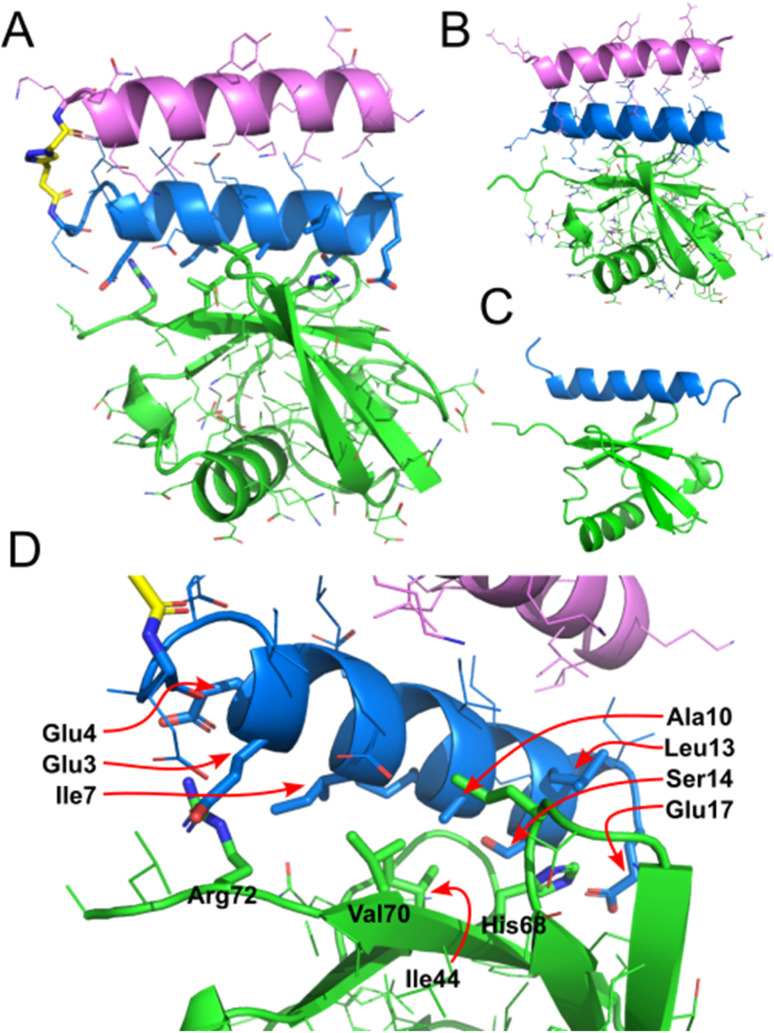
As a preliminary test of the resulting CC–UIM peptide sequences, the complex between Ub and the peptide components of 9 was modelled using AlphaFold Multimer (Fig. 5B).33 This model omits the triazole linker between the two helices comprising the CC, but nevertheless predicts the intended UIM-like complex between the CC and Ub. This suggests that it is possible to reconcile the design requirements for CC formation and Ub binding within a short peptide sequence.
Fig. 5. (A) Crystal structure of CC–UIM 9 bound to Ub. (B) AlphaFold structure of CC–UIM helices (as a non-crosslinked dimer) bound to Ub. (C) Literature NMR structure of UIM-1 bound to Ub (PDB 1q0w). (D) Detailed view of binding interactions, highlighting key residues.
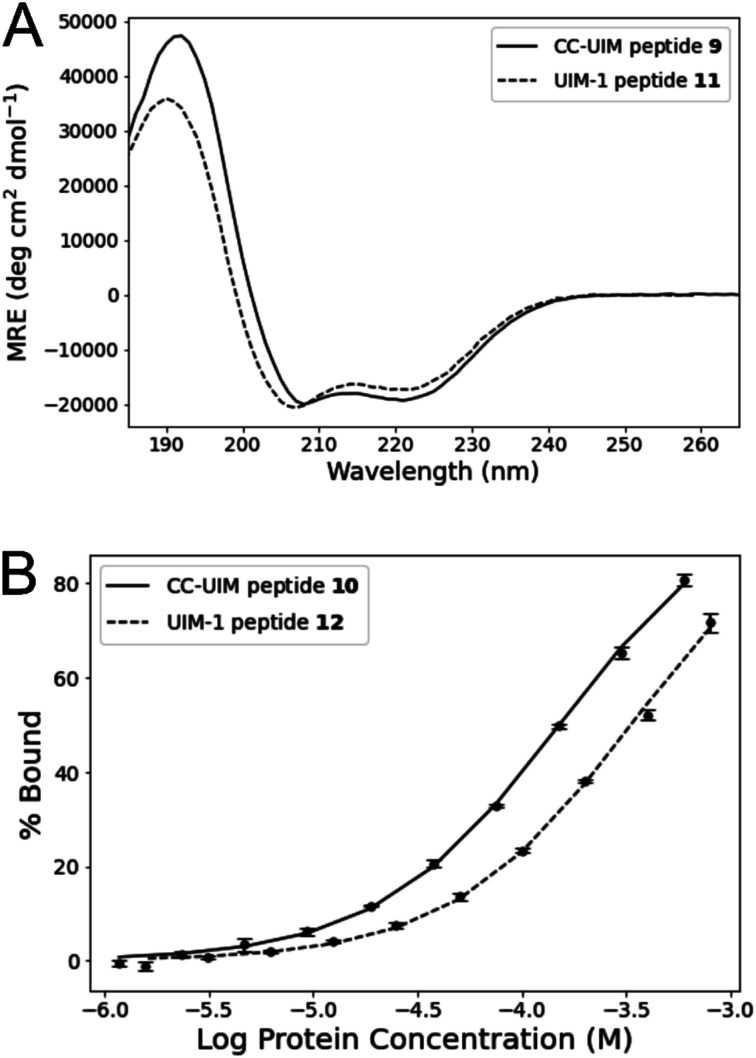
CD spectroscopy of peptides 9 and 11 showed both peptides to have a substantial degree of helical character (Fig. 4A). Comparison of the CD signals at 222 nm shows that peptide 9 is somewhat more helical than peptide 11, indicating that crosslink-mediated CC formation favors the helical form to a greater degree than the i − i + 4 salt bridges present in the original UIM-1 sequence (Fig. 1B). Moreover, peptide 9 remains helical at temperatures below 60 °C, though an eventual irreversible β-transition occurs at temperatures beyond this (Fig. S7†). The greater degree of helical character of the CC–UIM peptide implies that the binding residues are also preorganized to a greater degree.
Fig. 4. (A) CD spectroscopy of unlabeled peptides 9 and 11. (B) FP titration assay of fluorescein labelled peptides 10 and 12 against Ub.
Fig. 1. Details of the Ub:UIM-1 interaction. (A) View of NMR structure (PDB 1q0w), showing Ub in green, UIM-1 in cyan, with key binding residues in red. (B) Binding region of UIM-1, with i − i + 4 salt bridges highlighted with dashed arrows and key binding residues shown in red. (C) Helical wheel depiction of part of the binding region of UIM-1, showing disposition of key residues.
We investigate the Ub binding capacity of peptide 10 through an FP titration (Fig. 4 and S8†). Peptide 10 was maintained at a constant concentration of 50 nM, and titrated against Ub by serial dilution in a concentration range of 600 to 1.2 μM. Fitting these data yielded a Kd of 138.8 ± 5.0 μM. A comparable titration showed that the reference UIM-1 peptide 12, bound with a Kd of 311.3 ± 15.5 μM. The CC–UIM peptide 10 therefore binds more strongly than the native UIM-1 peptide, indicating that preorganization of the helical conformation does indeed contribute significantly to the energy of binding. It is notable that the enhanced binding exhibited by peptide 10 occurs despite it retaining only three of the original five hydrophobic residues. Retaining these same three hydrophobic residues in analogues of UIM-1 (Fig. S5†) results in a Kd that is an order of magnitude higher, whereas with peptide 10 tighter binding is observed. Thus the same set of interfacial residues result in different changes to affinity in these two different scaffolds. These results indicate that even a modest enhancement of helical character can play a significant role in enhancing binding affinity.
Crystal structure of CC:Ub interaction
To investigate the binding interaction between CC–UIM and Ub, the unlabeled version of CC–UIM, peptide 9 was co-crystallized with Ub. Two crystal forms of peptide 9 bound to Ub were obtained in the Morpheus Screen condition H1. Crystal form 1 (space group C121) contains one molecule of CC–UIM:Ub complex in the asymmetric unit, while crystal form 2 (space group P1211) has two molecules of CC–UIM:Ub complex in the asymmetric unit. All of the CC–UIM:Ub complexes are of essentially the same geometry (with an RMSD between 0.30–0.55 Å), differing only in the orientation of the triazole linker (Fig. 5 and S9†).
The CC–UIM peptide 9 is observed to bind to Ub in an antiparallel manner, with the N-terminus of the CC adjacent to the C-terminal tail of the Ub. This is consistent with the mode of binding observed for in the published NMR structure of the Ub:Vps27 UIM-1 complex (PDB 1q0w).29 The crystal structure of the Ub:CC–UIM complex is geometrically very similar to that of the NMR structure, with an all-atom RMSD of 1.01 Å (Fig. S10†). Details of the Ub:CC–UIM interaction are consistent with the NMR structure: the central Ile44 of Ub aligns with the AxxxS motif formed by Ala10 and Ser14. Similarly, residue His68 of Ub interacts with the pocket formed by with Ala10, Leu13, Ser14 and Glu17 of the CC–UIM. A hydrogen bond is formed between CC–UIM Glu17 and Ub His68. The C-terminal Arg72 of Ub forms contacts with Glu3 and Glu4 of CC–UIM. Thus, the Ub:CC–UIM interaction is structurally almost identical to the Ub:UIM-1 interaction. The somewhat tighter binding exhibited by CC–UIM is therefore most likely a result of its greater structural preorganisation.
An alignment of the AlphaFold model of the Ub:CC–UIM complex with the crystal structure of the same complex has a very low RMSD of 0.26 Å (Fig. S11†). This similarity suggests that the triazole linker is consistent with the ideal folded structure of the peptide, and moreover indicates that AlphaFold Multimer has significant potential as a predictive tool for peptide protein complexes, even where chemical modifications are present. The use of machine learning based methods such as AlphaFold combined with rational design principles and experimental studies offer significant scope to engineer peptide-protein interactions.
Conclusion
In conclusion, we have designed and synthesized a crosslinked CC which, despite its small size, folds into a stable α-helical structure. We have demonstrated that it is possible to use this scaffold CC to make a structured mimic of the Vps27 UIM-1 binding sequence, and that this mimic binds to Ub with a higher affinity than the original UIM-1 peptide. The Ub binding affinity of CC–UIM is within the range of values measure for other Ub binding motifs such as the UBA domain. An X-ray crystal structure of the CC–UIM:Ub complex shows that the peptide binds through interactions similar to the parent peptide, indicating that the overall binding motif is preserved when transferred to the new CC context. In a general sense, these findings show that PPIs involving α-helices can be mimicked using a crosslinked CC system. We envision that CC–UIM peptides will be useful tools for the study of Ub binding processes, and that the CC scaffold demonstrated here could be used to target other PPIs.
Data availability
Coordinates and structure factors for the crystal structures of CC–UIM:Ub form 1 (PDB 9jf4) and form 2 (PDB 9jf3) have been deposited in the Protein Data Bank. The data and methods relating to this article are presented in the ESI.†
Author contributions
P. V. and A. R. T. designed peptides and analyzed data. P. V. synthesized peptides and carried out biophysical analysis. P. P. V. expressed and purified Ub, and set up crystal screens. P. P. V. and D. T. H. analyzed crystallographic data. A. R. T. designed the research program.
Conflicts of interest
D. T. H. is a consultant for Triana Biomedicines.
Supplementary Material
Acknowledgments
D. T. H. was supported by a Cancer Research UK core programme grant (A23278). We thank Diamond Light Source for access to beamline I04 (Proposal mx28516). We thank Dr Brian Smith (University of Glasgow, School of Molecular Biosciences) for helpful advice on biophysical analysis.
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc04204b
References
- Russell R. B. Alber F. Aloy P. Davis F. P. Korkin D. Pichaud M. Topf M. Sali A. A Structural Perspective on Protein–Protein Interactions. Curr. Opin. Struct. Biol. 2004;14(3):313–324. doi: 10.1016/j.sbi.2004.04.006. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Wójcik P. Berlicki Ł. Peptide-Based Inhibitors of Protein–Protein Interactions. Bioorg. Med. Chem. Lett. 2016;26(3):707–713. doi: 10.1016/j.bmcl.2015.12.084. doi: 10.1016/j.bmcl.2015.12.084. [DOI] [PubMed] [Google Scholar]
- Keskin O. Tuncbag N. Gursoy A. Predicting Protein–Protein Interactions from the Molecular to the Proteome Level. Chem. Rev. 2016;116(8):4884–4909. doi: 10.1021/acs.chemrev.5b00683. doi: 10.1021/acs.chemrev.5b00683. [DOI] [PubMed] [Google Scholar]
- Walport L. J. Low J. K. K. Matthews J. M. Mackay J. P. The Characterization of Protein Interactions – What, How and How Much? Chem. Soc. Rev. 2021;50(22):12292–12307. doi: 10.1039/D1CS00548K. doi: 10.1039/D1CS00548K. [DOI] [PubMed] [Google Scholar]
- Mabonga L. Kappo A. P. Peptidomimetics: A Synthetic Tool for Inhibiting Protein–Protein Interactions in Cancer. Int. J. Pept. Res. Ther. 2020;26(1):225–241. doi: 10.1007/s10989-019-09831-5. doi: 10.1007/s10989-019-09831-5. [DOI] [Google Scholar]
- Rezaei Araghi R. Keating A. E. Designing Helical Peptide Inhibitors of Protein–Protein Interactions. Curr. Opin. Struct. Biol. 2016;39:27–38. doi: 10.1016/j.sbi.2016.04.001. doi: 10.1016/j.sbi.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritzer J. A. Zutshi R. Cheah M. Ran F. A. Webman R. Wongjirad T. M. Schepartz A. Miniature Protein Inhibitors of the P53–hDM2 Interaction. ChemBioChem. 2006;7(1):29–31. doi: 10.1002/cbic.200500324. doi: 10.1002/cbic.200500324. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Kajino M. Inouye M. Development of a Series of Cross-Linking Agents That Effectively Stabilize α-Helical Structures in Various Short Peptides. Chem.–Eur. J. 2008;14(3):857–863. doi: 10.1002/chem.200700843. doi: 10.1002/chem.200700843. [DOI] [PubMed] [Google Scholar]
- Taylor J. W. The Synthesis and Study of Side-Chain Lactam-Bridged Peptides. Pept. Sci. 2002;66(1):49–75. doi: 10.1002/bip.10203. doi: 10.1002/bip.10203. [DOI] [PubMed] [Google Scholar]
- Haney C. M. Loch M. T. Horne W. S. Promoting Peptide α-Helix Formation with Dynamic Covalent Oxime Side-Chain Cross-Links. Chem. Commun. 2011;47(39):10915–10917. doi: 10.1039/C1CC12010G. doi: 10.1039/C1CC12010G. [DOI] [PubMed] [Google Scholar]
- Spokoyny A. M. Zou Y. Ling J. J. Yu H. Lin Y.-S. Pentelute B. L. A Perfluoroaryl-Cysteine SNAr Chemistry Approach to Unprotected Peptide Stapling. J. Am. Chem. Soc. 2013;135(16):5946–5949. doi: 10.1021/ja400119t. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assem N. Ferreira D. J. Wolan D. W. Dawson P. E. Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling. Angew. Chem., Int. Ed. 2015;54(30):8665–8668. doi: 10.1002/anie.201502607. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone N. S. A. Kros A. Boyle A. L. Modulation of Coiled-Coil Binding Strength and Fusogenicity through Peptide Stapling. Bioconjugate Chem. 2020;31(3):834–843. doi: 10.1021/acs.bioconjchem.0c00009. doi: 10.1021/acs.bioconjchem.0c00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuo M. G. Mahon A. B. Arora P. S. An Effective Strategy for Stabilizing Minimal Coiled Coil Mimetics. J. Am. Chem. Soc. 2015;137(36):11618–11621. doi: 10.1021/jacs.5b05525. doi: 10.1021/jacs.5b05525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H. Nguyen T. Arora P. Design and Synthesis of Crosslinked Helix Dimers as Protein Tertiary Structure Mimics. Curr. Protoc. 2022;2(1):e315. doi: 10.1002/cpz1.315. doi: 10.1002/cpz1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torner J. M. Arora P. S. Conformational Control in a Photoswitchable Coiled Coil. Chem. Commun. 2021;57(12):1442–1445. doi: 10.1039/D0CC08318F. doi: 10.1039/D0CC08318F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer N. Watkins A. M. Arora P. S. Protein Domain Mimics as Modulators of Protein–Protein Interactions. Acc. Chem. Res. 2017;50(6):1313–1322. doi: 10.1021/acs.accounts.7b00130. doi: 10.1021/acs.accounts.7b00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F. Boyle A. L. Burton A. J. Woolfson D. N. A Set of de Novo Designed Parallel Heterodimeric Coiled Coils with Quantified Dissociation Constants in the Micromolar to Sub-Nanomolar Regime. J. Am. Chem. Soc. 2013;135(13):5161–5166. doi: 10.1021/ja312310g. doi: 10.1021/ja312310g. [DOI] [PubMed] [Google Scholar]
- Burkhard P. Ivaninskii S. Lustig A. Improving Coiled-Coil Stability by Optimizing Ionic Interactions. J. Mol. Biol. 2002;318(3):901–910. doi: 10.1016/S0022-2836(02)00114-6. doi: 10.1016/S0022-2836(02)00114-6. [DOI] [PubMed] [Google Scholar]
- Dong H. Hartgerink J. D. Short Homodimeric and Heterodimeric Coiled Coils. Biomacromolecules. 2006;7(3):691–695. doi: 10.1021/bm050833n. doi: 10.1021/bm050833n. [DOI] [PubMed] [Google Scholar]
- Sadek J. Wuo M. G. Rooklin D. Hauenstein A. Hong S. H. Gautam A. Wu H. Zhang Y. Cesarman E. Arora P. S. Modulation of Virus-Induced NF-κB Signaling by NEMO Coiled Coil Mimics. Nat. Commun. 2020;11(1):1786. doi: 10.1038/s41467-020-15576-3. doi: 10.1038/s41467-020-15576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara T. Kitada H. Fujiwara D. Fujii I. Macrocyclization and Labeling of Helix–Loop–Helix Peptide with Intramolecular Bis-Thioether Linkage. Pept. Sci. 2016;106(4):415–421. doi: 10.1002/bip.22826. doi: 10.1002/bip.22826. [DOI] [PubMed] [Google Scholar]
- Fujiwara D. Kitada H. Oguri M. Nishihara T. Michigami M. Shiraishi K. Yuba E. Nakase I. Im H. Cho S. Joung J. Y. Kodama S. Kono K. Ham S. Fujii I. A Cyclized Helix-Loop-Helix Peptide as a Molecular Scaffold for the Design of Inhibitors of Intracellular Protein–Protein Interactions by Epitope and Arginine Grafting. Angew. Chem., Int. Ed. 2016;55(36):10612–10615. doi: 10.1002/anie.201603230. doi: 10.1002/anie.201603230. [DOI] [PubMed] [Google Scholar]
- Hicke L. Schubert H. L. Hill C. P. Ubiquitin-Binding Domains. Nat. Rev. Mol. Cell Biol. 2005;6(8):610–621. doi: 10.1038/nrm1701. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Welchman R. L. Gordon C. Mayer R. J. Ubiquitin and Ubiquitin-like Proteins as Multifunctional Signals. Nat. Rev. Mol. Cell Biol. 2005;6(8):599–609. doi: 10.1038/nrm1700. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Damgaard R. B. The Ubiquitin System: From Cell Signalling to Disease Biology and New Therapeutic Opportunities. Cell Death Differ. 2021;28(2):423–426. doi: 10.1038/s41418-020-00703-w. doi: 10.1038/s41418-020-00703-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. Dixit V. M. Drugging the Undruggables: Exploring the Ubiquitin System for Drug Development. Cell Res. 2016;26(4):484–498. doi: 10.1038/cr.2016.31. doi: 10.1038/cr.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. Wakatsuki S. Walters K. J. Ubiquitin-Binding Domains — from Structures to Functions. Nat. Rev. Mol. Cell Biol. 2009;10(10):659–671. doi: 10.1038/nrm2767. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. A. Kang R. S. Stamenova S. D. Hicke L. Radhakrishnan I. Solution Structure of Vps27 UIM-Ubiquitin Complex Important for Endosomal Sorting and Receptor Downregulation. EMBO J. 2003;22(18):4597–4606. doi: 10.1093/emboj/cdg471. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Young P. Walters K. J. Structure of S5a Bound to Monoubiquitin Provides a Model for Polyubiquitin Recognition. J. Mol. Biol. 2005;348(3):727–739. doi: 10.1016/j.jmb.2005.03.007. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Truebestein L. Leonard T. A. Coiled-Coils: The Long and Short of It. BioEssays. 2016;38(9):903–916. doi: 10.1002/bies.201600062. doi: 10.1002/bies.201600062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrughi M. Maiani E. Aykac Fas B. Shaw G. S. Kragelund B. B. Lindorff-Larsen K. Teilum K. Invernizzi G. Papaleo E. Ubiquitin Interacting Motifs: Duality Between Structured and Disordered Motifs. Front. Mol. Biosci. 2021;8:676235. doi: 10.3389/fmolb.2021.676235. doi: 10.3389/fmolb.2021.676235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J. Evans R. Pritzel A. Green T. Figurnov M. Ronneberger O. Tunyasuvunakool K. Bates R. Žídek A. Potapenko A. Bridgland A. Meyer C. Kohl S. A. A. Ballard A. J. Cowie A. Romera-Paredes B. Nikolov S. Jain R. Adler J. Back T. Petersen S. Reiman D. Clancy E. Zielinski M. Steinegger M. Pacholska M. Berghammer T. Bodenstein S. Silver D. Vinyals O. Senior A. W. Kavukcuoglu K. Kohli P. Hassabis D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors for the crystal structures of CC–UIM:Ub form 1 (PDB 9jf4) and form 2 (PDB 9jf3) have been deposited in the Protein Data Bank. The data and methods relating to this article are presented in the ESI.†