Abstract
The general transcription factor TFIID consists of the TATA-binding protein (TBP) and multiple TBP-associated factors (TAFs). We previously identified two distinct WD repeat-containing TAFs, spTAF72 and spTAF73, in the fission yeast Schizosaccharomyces pombe. Here we report the identification of another S.pombe TAF, spTAF50, which is the S.pombe homolog of histone H4-like TAFs such as human TAF80, Drosophila TAF60 and Saccharomyces cerevisiae TAF60. spTAF50 was identified in a two-hybrid screen as a protein that interacts with the C-terminal WD repeat-containing region of spTAF72. Gene disruption revealed that spTAF50 is essential for cell viability. In vitro, spTAF50 bound to spTAF72 but less efficiently to spTAF73. In S.pombe cells, spTAF50 was detected as a protein with an apparent molecular mass of ∼50 kDa. Immunoprecipitation experiments demonstrated that spTAF50 is present in both the TFIID and SAGA-like complexes as in the case of spTAF72. These results indicate that the C-terminal region of spTAF72, which largely consists of WD repeats, interacts with spTAF50 in the TFIID and SAGA-like complexes, suggesting a role for the WD repeat domain in the interaction between TAFs.
INTRODUCTION
TFIID, one of the general transcription factors required for accurate transcription initiation by RNA polymerase II in vitro (1), comprises the TATA-binding protein (TBP) and a set of TBP-associated factors (TAFs) (2–4). TAFs have been identified from human, Drosophila and Saccharomyces cerevisiae (termed hTAF, dTAF and yTAF, respectively) and found to be well conserved throughout evolution.
Some TAFs have distinct structural motifs. Most notable is the histone fold, a motif involved in heterodimerization of the core histones. For example, hTAF80, dTAF60 and yTAF60 show limited sequence homology to histone H4 and are referred to as histone H4-like TAF. In S.cerevisiae, 9 out of 14 TAFs contain a histone fold motif (5), which is likely to mediate many TAF–TAF interactions within the TFIID complex.
Another sequence motif in TAFs is the WD repeat, which has been found in proteins with a wide variety of cellular functions and implicated in protein–protein interactions (6). The WD repeat domain of the β subunit of a heterotrimeric G protein, the only WD repeat protein whose structure is known, forms a propeller-like structure (7), and this structure is assumed to be common to other WD repeat proteins. The WD repeats are found in hTAF100, dTAF80 and yTAF90. In the case of hTAF100, it has been reported that the WD repeat domain is not required for its incorporation into the TFIID complex (8). Thus, the function of the WD repeats of these TAFs in the assembly of the TFIID complex or transcription regulation remains unknown.
Both histone-like TAFs and WD repeat-containing TAFs are also present in histone acetyltransferase complexes distinct from TFIID (9–11). For instance, yTAF60 and yTAF90 are shared by the S.cerevisiae SAGA (Spt-Ada-Gcn5 acetyltransferase) complex. hTAF80 and hTAF100 are shared by TFTC (TBP-free TAF-containing complex), whereas related proteins, PAF65α and PAF65β, are present in the PCAF (p300/CBP-associated factor) complex. It has recently been reported that a dTAF80-related protein is encoded by the Drosophila gene cannonball, which is involved in spermatogenesis (12). Interestingly, mutations in cannonball cause a block at the G2/M transition of meiosis I (13).
Saccharomyces cerevisiae mutants have been used to study in vivo functions of TAFs. A taf90ts mutant shows a distinct cell cycle phenotype, G2/M arrest (14). Genome-wide expression analysis has identified a set of genes whose expression depends on yTAF90 (15). However, the presence of yTAF90 both in TFIID and SAGA makes it difficult to distinguish between the function of yTAF90 in TFIID and that in SAGA.
We have been studying TAFs using the fission yeast Schizosaccharomyces pombe as another model organism to which genetic methods are applicable. We previously identified two S.pombe TAFs, spTAF72 and spTAF73, both of which contain the WD repeats (16). spTAF72 and spTAF73 have non-redundant functions because each of them is essential for cell viability. spTAF72 is more similar in amino acid sequence to yTAF90 than spTAF73 is and is shared by the TFIID and SAGA-like complexes as in the case of yTAF90, whereas spTAF73 is present only in TFIID. Interestingly, overexpression of spTAF72 or spTAF73 suppresses the cell cycle arrest during mitosis caused by mutations in genes involved in ubiquitin-dependent proteolysis, suggesting a role for these WD repeat-containing TAFs in the expression of genes involved in progression through the M phase of the cell cycle (16).
As an attempt to understand the role of the WD repeat domain in spTAF72, we screened for proteins that interact with the C-terminal WD repeat-containing region of spTAF72 using the yeast two-hybrid system. We identified another S.pombe TAF, spTAF50, a homolog of histone H4-like TAFs. spTAF50 was associated with spTAF72 in both the TFIID and SAGA-like complexes.
MATERIALS AND METHODS
Yeast strains
Saccharomyces cerevisiae and S.pombe strains used in this study are listed in Table 1. Standard methods were used for molecular genetic analysis of S.cerevisiae (17,18) and S.pombe (19,20).
Table 1. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| S.cerevisiae | ||
| AH109 | MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ | Clontech |
| LYS2::GAL1-HIS3 GAL2-ADE2 URA3::MEL1-lacZ | ||
| S.pombe | ||
| JY741 | h– ade6-M216 leu1 ura4-D18 | M. Yamamoto |
| JY746 | h+ ade6-M210 leu1 ura4-D18 | M. Yamamoto |
| HMP10 | h–/h+ ade6-M216/ade6-M210 leu1/leu1 ura4-D18/ura4-D18 | (31) |
| HMP47 | h– ade6-M216 leu1 ura4-D18 gcn53xHA–ura4+ | (16) |
| HMP48 | h– ade6-M216 leu1 ura4-D18 gcn53xHA–ura4+ taf73FLAG | (16) |
| HMP49 | h– ade6-M216 leu1 ura4-D18 gcn53xHA–ura4+ taf72FLAG | (16) |
| HMP54 | h– ade6-M216 leu1 ura4-D18 gcn53xHA | This study |
| HMP55 | h– ade6-M216 leu1 ura4-D18 gcn53xHA taf73FLAG | This study |
| HMP56 | h– ade6-M216 leu1 ura4-D18 gcn53xHA taf72FLAG | This study |
| HMP57 | h– ade6-M216 leu1 ura4-D18 taf50Myc–ura4+ | This study |
| HMP58 | h– ade6-M216 leu1 ura4-D18 gcn53xHA taf50Myc–ura4+ | This study |
| HMP59 | h– ade6-M216 leu1 ura4-D18 gcn53xHA taf73FLAG taf50Myc–ura4+ | This study |
| HMP60 | h– ade6-M216 leu1 ura4-D18 gcn53xHA taf72FLAG taf50Myc–ura4+ | This study |
HMP10 is a diploid constructed by a cross between JY741 and JY746 and all the other HMP strains derived from JY741.
Two-hybrid screen
A Matchmaker Gal4-based two-hybrid system (Clontech) was used to identify proteins that interact with the C-terminal region (amino acids 280–643) of spTAF72. The corresponding taf72 cDNA was amplified by PCR, cloned into the EcoRI/BamHI site of the Gal4 DNA binding domain (BD) vector pGBKT7 and used to transform the S.cerevisiae strain AH109, which contains HIS3, ADE2, MEL1 and lacZ reporter genes. Expression of Gal4 BD-TAF72 protein was verified by immunoblotting with anti-c-Myc antibody. The AH109 strain carrying the bait plasmid was then transformed with the Matchmaker S.pombe cDNA library constructed in the Gal4 activation domain (AD) vector pGADGH (Clontech). Screening of ∼2.5 × 105 Trp+Leu+ colonies for a His+Ade+Mel+ phenotype yielded 11 positive clones, two of which contained the SPCC16C4.18c gene.
Disruption of the taf50 gene
The taf50 gene was disrupted by the method of Shortle et al. (21) using pBluescript (Stratagene) carrying the S.pombe ura4+ marker and a portion of the taf50 gene lacking both the 5′ and 3′ ends, which corresponds to amino acids 83–177. Integration of this plasmid into the chromosomal taf50 locus would result in two truncated taf50 genes, one encoding amino acids 1–177 and the other encoding amino acids 83–643, both of which are presumed to be non-functional. For such integration, the plasmid was linearized at the MscI site within the taf50 gene and used to transform a diploid S.pombe strain. The disruption of one chromosomal copy of the taf50 gene was confirmed by PCR prior to tetrad dissection.
Expression of recombinant proteins and in vitro interaction assays
taf50 cDNA was amplified by PCR, cloned into the NdeI/XhoI site of pET-21b (Novagen) and used to transform the Escherichia coli strain BL21(DE3) to produce spTAF50 protein with a histidine tag at the C-terminus. The taf72 or taf73 cDNA encoding amino acids 302–643 or 327–642, respectively, was amplified by PCR, cloned into the EcoRI/SalI site of pGEX-5X-1 (Amersham Pharmacia Biotech) and used to transform the E.coli strain DH5α to produce the C-terminal region of spTAF72 or spTAF73 fused to glutathione-S-transferase (GST). Because the His-TAF50, GST-TAF72 and GST-TAF73 proteins were found to be in insoluble fractions after induction with 1 mM IPTG for 3 h at 37°C, the inclusion bodies were purified with the B-PER reagent (Pierce) and dissolved in buffer B (6 M urea, 50 mM Tris–HCl pH 8.0, 0.1 M KCl, 1 mM EDTA, 10 mM DTT, 20% glycerol). The denatured proteins were then renatured by dialysis against buffer C (buffer B lacking urea). Dialyzed samples were cleared by centrifugation, mixed in pairs and incubated with glutathione agarose beads (Pierce). After washing, bound proteins were eluted with glutathione and analyzed by immunoblotting with mouse anti-GST and anti-His monoclonal antibodies (BAbCO/Covance Research Products).
Construction of S.pombe strains expressing epitope-tagged spTAF50
Schizosaccharomyces pombe strains that express Myc-tagged spTAF50 protein were constructed by the same method used for the generation of the FLAG-TAF72, FLAG-TAF73 and HA-Gcn5 strains (16). Briefly, a DNA fragment consisting of a 5′ non-coding region and a 5′ portion of the taf50 gene with a Myc epitope sequence immediately after the initiation codon was amplified by PCR using primers with overlapping extension and cloned into the SacI/BamHI site of pBluescript carrying the ura4+ marker. The plasmid was linearized at the MscI site within the taf50 gene and used to transform wild-type, gcn5HA, gcn5HA taf72FLAG and gcn5HA taf73FLAG strains to construct strains expressing spTAF50 with a Myc epitope at its N-terminus. Correct integration was confirmed by PCR.
Preparation of S.pombe whole-cell extracts and immunoprecipitation
Preparation of S.pombe whole-cell extracts and immunoprecipitation with anti-FLAG antibody were carried out as described previously (16). Rat anti-HA monoclonal antibody (3F10) coupled-agarose beads (Roche Molecular Biochemicals) were used for immunoprecipitation with anti-HA antibody. Mouse anti-c-Myc and anti-HA antibodies (BAbCO/Covance Research Products) and mouse anti-FLAG monoclonal antibody (Sigma) were used for detection of Myc-TAF50, HA-Gcn5, FLAG-TAF72 and FLAG-TAF73 proteins. Rabbit polyclonal antibodies raised against S.pombe TAF111/130, PTR6 and TBP were provided by Tetsuro Kokubo (Yokohama City University).
RESULTS
Isolation of histone H4-like TAF as an spTAF72-interacting protein
We previously identified S.pombe TAF72 protein as a component of both the TFIID and SAGA-like complexes (16). spTAF72, like its homologs spTAF73, hTAF100, dTAF80 and yTAF90, contains WD repeat motifs in the C-terminal half. To gain an insight into the function of the WD repeat domain of spTAF72, we carried out a two-hybrid screen using a Gal4 BD fusion to the C-terminal region (amino acids 280–643) of spTAF72, which largely consists of WD repeat motifs, as a bait (Fig. 1A). The Gal4 BD-TAF72 protein did not activate reporter gene expression on its own (Fig. 1B).
Figure 1.
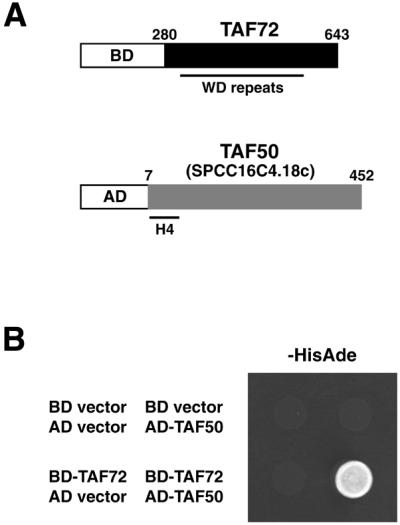
Two-hybrid screen for spTAF72-interacting protein. (A) The bait plasmid (upper) encodes a Gal4 BD fusion of the C-terminal portion of spTAF72, which contains WD-repeat motifs. An isolated clone (lower) encodes a Gal4 AD fusion of a nearly entire region of a predicted protein, SPCC16C4.18c (referred to as spTAF50), which has homology to histone H4-like TAFs. (B) AH109 cells carrying the indicated pairs of plasmids were spotted onto synthetic complete medium lacking tryptophan, leucine, histidine and adenine and allowed to grow at 30°C for 2 days. Growth on this medium implies interaction. The cells carrying the BD-TAF72 and AD-TAF50 plasmids (lower right) also showed a Mel+ phenotype as judged by blue color on plates containing 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (data not shown).
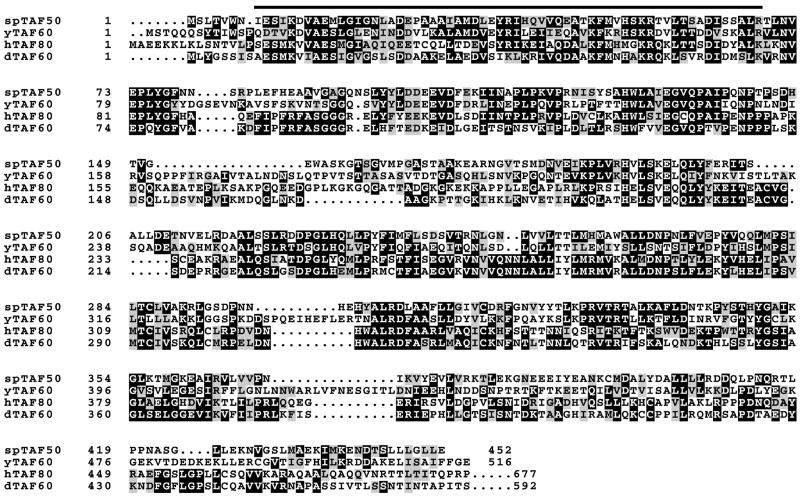
The bait plasmid was used to screen an S.pombe cDNA library constructed in a Gal4 AD plasmid. Of ∼2.5 × 105 transformants screened, 11 showed a His+Ade+Mel+ phenotype, which indicates the simultaneous expression of the reporter genes. Sequencing revealed that two identical clones encode a Gal4 AD fusion of a nearly entire region (lacking only the first six residues) of SPCC16C4.18c (Fig. 1A), a 452-amino acid protein predicted from the S.pombe genome sequencing project by the Sanger Centre, UK (DDBJ/EMBL/GenBank accession number CAA20756). The clone by itself did not activate the reporter expression (Fig. 1B), confirming the interaction between Gal4 BD-TAF72 and Gal4 AD-SPCC16C4.18c. A comparison of our cDNA sequence with the SPCC16C.18c genomic sequence confirmed the presence of a predicted 158-bp intron in the N-terminal proximal region (nucleotides 133–290). This gene encodes a protein with homology to histone H4-like TAFs, such as hTAF80, dTAF60 and yTAF60 (Fig. 2). We refer to this gene as taf50 for its calculated molecular mass of 50 kDa and its product as spTAF50. In accordance with the predicted molecular mass, apparent molecular mass of spTAF50 in SDS–PAGE was found to be ∼50 kDa (see below). spTAF50 is 39% identical to yTAF60, 24% identical to dTAF60 and 22% identical to hTAF80. spTAF50, like yTAF60, lacks the C-terminal extension present in both hTAF80 and dTAF60 (188 and 122 residues, respectively).
Figure 2.
Amino acid sequence alignment of histone H4-like TAFs. spTAF50 is aligned with yTAF60, hTAF80 and dTAF60, using the ClustalW program (29). Identical and similar residues are shaded. The C-terminal extension of hTAF80 and dTAF60, which is absent from yTAF60 and spTAF50, is not shown. The bar indicates the histone fold region based on the crystal structure of dTAF60 (30).
spTAF50 is essential for cell viability
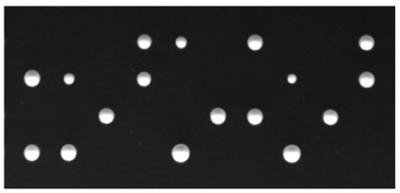
We tested whether the S.pombe taf50 gene is essential for cell viability. A diploid S.pombe strain was constructed in which one copy of taf50 was disrupted. The taf50+/Δtaf50::ura4+ cells were sporulated and subjected to tetrad analysis. As shown in Figure 3, most tetrads showed a 2+:2– segregation for spore viability, and no tetrads with more than two viable spores were observed. Importantly, all the viable spores were Ura– and thus presumed to be taf50+. Microscopic observation revealed that the Δtaf50::ura4+ spores germinated but did not divide more than twice. These results indicate that taf50, like its S.cerevisiae counterpart TAF60 (22), is an essential gene.
Figure 3.
Disruption of the taf50 gene. The taf50+/Δtaf50::ura4+ cells were sporulated on malt extract medium at 27°C for 2 days. Tetrads were dissected on yeast extract medium, and spores were grown at 30°C for 4 days. Ten tetrads are shown; four spores from each tetrad are aligned vertically.
Direct interaction of spTAF50 with spTAF72
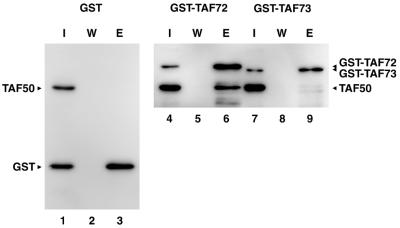
To test for direct interaction between spTAF72 and spTAF50 in vitro, we carried out GST pull-down assays with recombinant proteins expressed in E.coli. spTAF50 was produced as a His-tagged protein while the C-terminal region (amino acids 302–643) of spTAF72 was produced as a fusion to GST. The C-terminal region of spTAF72 is 22 residues shorter than that (amino acids 280–643) fused to Gal4 BD for the two-hybrid screen. The corresponding region (amino acids 327–642) of spTAF73 was also fused to GST and tested for interaction with spTAF50. When expressed, the His-TAF50, GST-TAF72 and GST-TAF73 proteins were all found to be insoluble. Accordingly, each protein was recovered in insoluble fractions and then solubilized in a denaturing buffer containing 6 M urea. After renaturing by dialysis, the proteins were mixed and incubated with glutathione agarose. Bound proteins were analyzed by immunoblotting with anti-GST and anti-His antibodies. As shown in Figure 4, TAF50 bound to GST-TAF72 (lane 6) but not to GST-TAF73 (lane 9) despite the fact that the C-terminal regions of spTAF72 and spTAF73 fused to GST are 45% identical. spTAF50 did not bind to GST (lane 3), confirming the interaction of spTAF50 with spTAF72. Similar results were obtained when the denatured proteins were mixed and then renatured by dialysis (data not shown).
Figure 4.
Direct interaction of spTAF50 with spTAF72. GST pull-down assays were carried out using GST, GST-TAF72 and GST-TAF73. Bound proteins were separated by 10% SDS–PAGE followed by immunoblotting. I, 10% of input; W, the final wash of the same volume with the elution; E, elution.
Association of spTAF50 with spTAF72 in the TFIID and SAGA-like complexes
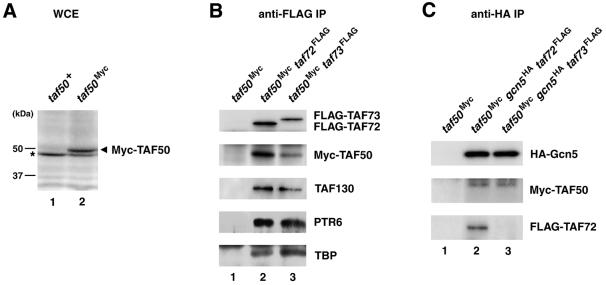
To detect spTAF50 protein in S.pombe cells, we constructed strains expressing Myc-tagged spTAF50 by replacing the chromosomal taf50+ allele by taf50Myc. In this strain, Myc-tagged spTAF50, instead of wild-type spTAF50, is expressed from the native promoter. Whole-cell extracts were prepared from the taf50Myc strain as well as a wild-type strain, which did not express any Myc-tagged protein. Immunoblotting with anti-Myc antibody identified Myc-TAF50 that was present only in the extract from the taf50Myc strain with an apparent molecular mass of ∼49 kDa (Fig. 5A), which is close to the calculated molecular mass of spTAF50.
Figure 5.
Presence of spTAF50 in the TFIID and SAGA-like complexes. (A) Identification of spTAF50 protein. Whole-cell extracts (WCE) prepared from wild-type (lane 1) and taf50Myc (lane 2) strains were used for immunoblotting with an anti-Myc antibody. The asterisk indicates a protein in the WCE that reacts to the anti-Myc antibody. (B) Co-immunoprecipitation of spTAF50 with spTAF72 and spTAF73. WCE prepared from taf50Myc (lane 1), taf50Myc taf72FLAG (lane 2) and taf50Myc taf73FLAG (lane 3) strains were used for immunoprecipitation (IP) with an anti-FLAG antibody, followed by immunoblotting with antibodies to FLAG, Myc, spTAF111/130, PTR6 and TBP. (C) Co-immunoprecipitation of spTAF50 and spTAF72 with Gcn5. WCE prepared from taf50Myc (lane 1), taf50Myc gcn5HA taf72FLAG (lane 2) and taf50Myc gcn5HA taf73FLAG (lane 3) were used for IP with an anti-HA antibody, followed by immunoblotting with antibodies to HA, Myc and FLAG.
To demonstrate that spTAF50, a putative TAF predicted from the amino acid sequence, is indeed a component of the S.pombe TFIID complex, we prepared whole-cell extracts from taf50Myc taf72FLAG and taf50Myc taf73FLAG strains and carried out co-immunoprecipitation experiments. As shown in Figure 5B, immunoprecipitation of spTAF72 or spTAF73 with anti-FLAG antibody revealed that spTAF50 was co-immunoprecipitated with spTAF72 and spTAF73, along with other TFIID components: spTAF111/130 (the S.pombe homolog of hTAF250/dTAF250/yTAF145), PTR6 (the S.pombe homolog of hTAF55/yTAF67) and TBP. Because spTAF73 is specific to TFIID (16), the association of spTAF50 with spTAF73 indicates that spTAF50 is a component of the TFIID complex.
In addition to TFIID, spTAF72 is present in the S.pombe SAGA-like complex, which contains the S.pombe homolog of the Gcn5 histone acetyltransferase (16). We next tested whether spTAF50 is also present in the SAGA-like complex. Immunoprecipitation of Gcn5 with anti-HA antibody revealed that spTAF50 was co-immunoprecipitated with Gcn5 (Fig. 5C), indicating that spTAF50, like spTAF72, is present both in the TFIID and SAGA-like complexes. In addition, spTAF72 but not spTAF73 was co-immunoprecipitated with Gcn5 (Fig. 5C). We showed previously that Gcn5 is co-immunoprecipitated with spTAF72 but not with spTAF73 and concluded that spTAF72 but not spTAF73 is shared by the TFIID and SAGA-like complexes (16). The results shown in Figure 5C confirm the conclusion. The S.cerevisiae SAGA complex contains Gcn5, yTAF90 and yTAF60, in addition to Spt and Ada proteins (9–11). Our results have shown that S.pombe has a similar complex containing Gcn5, spTAF72 and spTAF50, which we refer to as a SAGA-like complex.
DISCUSSION
We previously identified S.pombe spTAF72, spTAF73, spTAF111/130 and PTR6 as proteins associated with TBP (16). Both spTAF72 and spTAF73 are the S.pombe homologs of hTAF100/dTAF80/yTAF90, whereas spTAF111/130 and PTR6 are the homologs of hTAF250/dTAF250/yTAF145 and hTAF55/yTAF67, respectively. In this study we identified another S.pombe TAF, spTAF50, a homolog of the histone H4-like hTAF80/dTAF60/yTAF60. spTAF50 was present both in the TFIID and SAGA-like complexes. Similarly, yTAF60 is shared by TFIID and SAGA. In contrast, hTAF80 is replaced by a related protein, PAF65α, in PCAF. Our database searches did not reveal another S.pombe protein that is closely related to spTAF50, which is consistent with the fact that spTAF50 is shared by the TFIID and SAGA-like complexes.
spTAF50 was identified as an spTAF72-interacting protein. The interaction of WD repeat-containing TAF and histone H4-like TAF has been reported for the human counterparts hTAF100 and hTAF80 (23). However, the region of hTAF100 that is involved in this interaction has not been delineated. In this regard, our results showed that the C-terminal half of spTAF72 mediates the interaction with spTAF50. This region largely consists of WD repeats, suggesting that one of the functions of the WD repeats in spTAF72 and its homologs is the interaction with histone H4-like TAF, although we cannot rule out the possibility that the region outside of the WD repeats mediates the interaction. Recently, it has been reported that mutations corresponding to the C-terminal region of yTAF90 result in broad transcription defects and defects in its incorporation into TFIID and SAGA (24). Our results suggest the possibility that some of the defects caused by the mutations result from impaired interaction with the histone H4-like yTAF60.
spTAF50 bound less efficiently to spTAF73 than to spTAF72 in vitro. The difference between spTAF72 and spTAF73 in their ability to interact with spTAF50 could be useful to identify residues in the WD repeat domain that are important in the interaction with histone H4-like TAF. It has been shown that the C-terminal WD repeat-containing region of spTAF72 can be replaced by that of yTAF90 for complementation of Δtaf72 although the spTAF72–yTAF90 chimera confers temperature-sensitive growth (25). This observation suggests that the C-terminal region of yTAF90 can interact with spTAF50 to some extent. Interestingly, in regard to the C-terminal region fused to GST, spTAF72 is more similar to yTAF90 (51% identical) than to spTAF73 (45% identical) although overall spTAF72 shows similar identities to yTAF90 (46%) and spTAF73 (45%).
BD fusion of TAF often activates the reporter expression by recruitment of transcription machinery (26–28). Thus, activation by BD-TAF implies the incorporation of the TAF into the TFIID complex. The Gal4 BD fusion of the C-terminal region of spTAF72 used for our two-hybrid screen did not activate transcription. In contrast, the full-length spTAF72 fused to Gal4 BD caused weak activation (data not shown). Similar results have been reported for yTAF90 (14). These results indicate that the N-terminal region is required for incorporation of WD repeat-containing TAF into TFIID, which is consistent with the results of Dubrovskaya et al. (8).
Our results have revealed that S.pombe has TFIID and SAGA-like complexes similar to those characterized in human, Drosophila and S.cerevisiae. The S.pombe TFIID complex is unique, however, in that it contains two distinct WD repeat-containing TAFs. In contrast to hTAF100 and yTAF90, spTAF73 seems to be specific to TFIID, which provides an advantage for analyzing the in vivo function of WD repeat-containing TAF in the context of TFIID. Interestingly, spTAF72 and spTAF73 have been implicated in the expression of genes involved in progression through the M phase of the cell cycle (16). Thus, S.pombe would serve as a good model for studying the functions of TAFs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Emi Kanda for assistance. We also thank Masayuki Yamamoto for S.pombe strains and Tetsuro Kokubo for antibodies to S.pombe TBP, TAF111/130 and PTR6. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Science and Technology Corporation.
REFERENCES
- 1.Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 2.Burley S.K. and Roeder,R.G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem., 65, 769–799. [DOI] [PubMed] [Google Scholar]
- 3.Green M.R. (2000) TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci., 25, 59–63. [DOI] [PubMed] [Google Scholar]
- 4.Albright S.R. and Tjian,R. (2000) TAFs revisited: more data reveal new twists and confirm old ideas. Gene, 242, 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Gangloff Y.-G., Romier,C., Thuault,S., Werten,S. and Davidson,I. (2001) The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci., 26, 250–257. [DOI] [PubMed] [Google Scholar]
- 6.Smith T.F., Gaitatzes,C., Saxena,K. and Neer,E.J. (1999) The WD repeat: common architecture for diverse functions. Trends Biochem. Sci., 24, 181–185. [DOI] [PubMed] [Google Scholar]
- 7.Neer E.J. and Smith,T. (1996) G protein heterodimers: new structures propel new questions. Cell, 84, 175–178. [DOI] [PubMed] [Google Scholar]
- 8.Dubrovskaya V., Lavigne,A.-C., Davidson,I., Acker,J., Staub,A. and Tora,L. (1996) Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIFβ (RAP30) and incorporation into the TFIID complex. EMBO J., 15, 3702–3712. [PMC free article] [PubMed] [Google Scholar]
- 9.Struhl K. and Moqtaderi,Z. (1998) The TAFs in the HAT. Cell, 94, 1–4. [DOI] [PubMed] [Google Scholar]
- 10.Bell B. and Tora,L. (1999) Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res., 246, 11–19. [DOI] [PubMed] [Google Scholar]
- 11.Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. [DOI] [PubMed] [Google Scholar]
- 12.Hiller M.A., Lin,T.-Y., Wood,C. and Fuller,M.T. (2001) Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev., 15, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin T., Visvanathan,S., Wood,C., Wilson,P.G., Wolf,N. and Fuller,M.T. (1996) Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development, 122, 1331–1341. [DOI] [PubMed] [Google Scholar]
- 14.Apone L.M., Virbasius,C.A., Reese,J.C. and Green,M.R. (1996) Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev., 10, 2368–2380. [DOI] [PubMed] [Google Scholar]
- 15.Lee T.I., Causton,H.C., Holstege,F.C.P., Shen,W.-C., Hannett,N., Jennings,E.G., Winston,F., Green,M.R. and Young,R.A. (2000) Redundant roles for the TFIID and SAGA complexes in global transcription. Nature, 405, 701–704. [DOI] [PubMed] [Google Scholar]
- 16.Mitsuzawa H., Seino,H., Yamao,F. and Ishihama,A. (2001) Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem., 276, 17117–17124. [DOI] [PubMed] [Google Scholar]
- 17.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 18.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, NY.
- 19.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 20.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, NY.
- 21.Shortle D., Haber,J.E. and Botstein,D. (1982) Lethal disruption of the yeast actin gene by integrative DNA transformation. Science, 217, 371–373. [DOI] [PubMed] [Google Scholar]
- 22.Poon D., Bai,Y., Campbell,A.M., Bjorklund,S., Kim,Y.-J., Zhou,S., Kornberg,R.D. and Weil,P.A. (1995) Identification and characterization of a TFIID-like multiprotein complex from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 92, 8224–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao Y., Guermah,M., Martinez,E., Oelgeschläger,T., Hasegawa,S., Takada,R., Yamamoto,T., Horikoshi,M. and Roeder,R.G. (1997) Specific interactions and potential functions of human TAFII100. J. Biol. Chem., 272, 6714–6721. [DOI] [PubMed] [Google Scholar]
- 24.Durso R.J., Fisher,A.K., Albright-Frey,T.J. and Reese,J.C. (2001) Analysis of TAF90 mutants displaying allele-specific and broad defects in transcription. Mol. Cell. Biol., 21, 7331–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto T. and Horikoshi,M. (1998) Defect in cytokinesis of fission yeast induced by mutation in the WD40 repeat motif of a TFIID subunit. Genes Cells, 3, 347–355. [DOI] [PubMed]
- 26.Gonzalez-Couto E., Klages,N. and Strubin,M. (1997) Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl Acad. Sci. USA, 94, 8036–8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keaveney M. and Struhl,K. (1998) Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell, 1, 917–924. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer S.M., Ranallo,R.T., Ogg,R.C. and Stargell,L.A. (2001) TFIIA interacts with TFIID via association with TATA-binding protein and TAF40. Mol. Cell. Biol., 21, 1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X., Kokubo,T., Cohen,S.L., Mirza,U.A., Hoffmann,A., Chait,B.T., Roeder,R.G., Nakatani,Y. and Burley,S.K. (1996) Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature, 380, 316–322. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai H., Mitsuzawa,H., Kimura,M. and Ishihama,A. (1999) The Rpb4 subunit of fission yeast Schizosaccharomyces pombe RNA polymerase II is essential for cell viability and similar in structure to the corresponding subunits of higher eukaryotes. Mol. Cell. Biol., 19, 7511–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]