Abstract
Micropollutants are increasingly prevalent in the aquatic environment. A major part of these originates from wastewater treatment plants since traditional treatment technologies do not remove micropollutants sufficiently. Moving bed biofilm reactors (MBBRs), however, have been shown to aid in micropollutant removal when applied to conventional wastewater treatment as a polishing step. Here, we used Total RNA sequencing to investigate both the active microbial community and functional dynamics of MBBR biofilms when these were exposed to increasing micropollutant concentrations over time. Concurrently, we conducted batch culture experiments using biofilm carriers from the MBBRs to assess micropollutant degradation potential. Our study showed that biofilm eukaryotes, in particular protozoa, were negatively influenced by micropollutant exposure, in contrast to prokaryotes that increased in relative abundance. Further, we found several functional genes that were differentially expressed between the MBBR with added micropollutants and the control. These include genes involved in aromatic and xenobiotic compound degradation. Moreover, the biofilm carrier batch experiment showed vastly different alterations in benzotriazole and diclofenac degradation following the increased micropollutant concentrations in the MBBR. Ultimately, this study provides essential insights into the microbial community and functional dynamics of MBBRs and how an increased load of micropollutants influences these dynamics.
Keywords: biodegradation, biofilms, MBBR, metatranscriptome, pharmaceuticals, wastewater
Shifts in the relative community composition, activity, and gene expression of moving bed biofilm reactor microbiomes upon exposure to increasing concentrations of micropollutants revealed by total RNA sequencing and degradation studies.
Introduction
Due to a growing world population, chemical applications including pharmaceuticals consumption have significantly increased over the last few decades (aus der Beek et al. 2016). This has considerably facilitated the spread of bioactive micropollutants into aquatic environments at concentrations ranging from pg L−1 to µg L−1 (Benner et al. 2013). The presence of pharmaceuticals and other micropollutants in the environment is of concern due to their intended design; to induce certain metabolic, enzymatic, or cell-signaling mechanisms (Ebele et al. 2017). Even at low concentrations, they have been linked to adverse ecological effects due to both acute and chronic exposure in non-target aquatic organisms (Huerta-Fontela et al. 2011, aus der Beek et al. 2016, Ebele et al. 2017). Several studies have addressed the occurrence, fate, and ecotoxicological impact of pharmaceuticals in the aquatic environment (e.g. Fent et al. 2006, Santos et al. 2010, aus der Beek et al. 2016, Wang et al. 2021).
The main discharge of micropollutants into the aquatic environment is via wastewater treatment plant (WWTP) discharge (aus der Beek et al. 2016, Besha et al. 2017, Ebele et al. 2017). Due to the physiochemical properties of pharmaceuticals (e.g. polarity, hydrophobicity, and type of functional groups), they typically pass through WWTPs unaltered or only partially degraded (di Biase et al. 2019). While most conventional treatments lack the ability to completely remove pharmaceuticals and other micropollutants, biofilm-based technologies, more specifically moving bed biofilm reactors (MBBRs), have shown promising removal of micropollutants from wastewater (Falås et al. 2012, Casas et al. 2015, Svendsen et al. 2020). The main functional components of MBBRs are small plastic carriers suspended in the wastewater. These carriers have a high surface area, which facilitates growth of microbial biofilms. The aerobic MBBR tank, which houses the floating carriers, receives wastewater as well as a stream of constant aeration. This allows the biofilms to receive nutrients and for the carriers to remain in perpetual motion. Biofilms are complex heterogeneous micro-ecosystems inhabited by both prokaryotic and eukaryotic microorganisms (Burmølle et al. 2014, Flemming et al. 2016, di Biase et al. 2019, Sadiq et al. 2022). The biofilm matrix provides enhanced tolerance to the associated bacteria, which may lead to enhanced survival and activity in the presence of pollutants (Flemming and Wingender 2010, Muhammad et al. 2020). These dynamic environments are the key factor allowing for the functionality of MBBRs, including nutrient and micropollutant removal efficiency (Torresi et al. 2016).
Previous MBBR microbiome studies have generally utilized DNA sequencing based methods, which provide information on the organisms present (Abu Bakar et al. 2020, Liang et al. 2021), and for metagenomes additionally the potential functions (Zhu et al. 2022, Wang et al. 2023). However, these methods only predict potential functional activity in the MBBR microbial community. Total RNA sequencing, on the other hand, allows for investigation of the functional gene expression and of the active microbial community composition across all domains of life, simultaneously. Other studies have used this technique in other environments such as agricultural, forest (Bang-Andreasen et al. 2019) and permafrost soils (Schostag et al. 2019, Scheel et al. 2023) as well as cave ice (Mondini et al. 2022). These studies identified key dynamics of their respective micro-ecosystems, including elucidation of the potential ecosystem role of microeukaryotes in those environments. Sato et al. (2019) conducted a metatranscriptomics study on the microbial community of activated sludge in a membrane bioreactor involved in degradation of heavy oil. This study identified key functional genes related to the biodegradation of heavy oils present in activated sludge. However, to our knowledge, Total RNA sequencing of MBBR systems has never been conducted specifically, nor in relation to micropollutant contamination and degradation.
Microbial communities present in other biological water treatment systems have been reported to adapt to certain micropollutants after continuous exposure resulting in improved degradation potential (Castronovo et al. 2017, Tisler and Zwiener 2019). However, both Tisler and Zwiener (2019) and Castronovo et al. (2017) focused on quantifying the biodegradation of micropollutants. Yet, the microbial community involved in the degradation of the respective micropollutants were not characterized.
In this study, we aimed to investigate how micropollutants (benzotriazole, carbamazepine, diclofenac, iohexol, and venlafaxine) affected the active microbial community, the gene expression, and the biodegradation potential of MBBR carrier biofilms. We expect the eukaryotic community in the biofilm to be negatively impacted by the micropollutant exposure, since most of the compounds are pharmaceuticals, and hence designed to induce effects in eukaryotes. On the contrary, prokaryotic organisms have previously shown to increase the gene expression of specific degrader genes when exposed to a micropollutant (Bælum et al. 2008). We therefore present the three interlinked hypotheses that MBBR exposure to higher micropollutant concentrations will 1) impact the active community composition on the carriers, especially by reducing the relative abundance of eukaryotic taxa, 2) lead to an increase in gene expression for genes involved in xenobiotic biodegradation, and 3) ultimately lead to increasing micropollutant biodegradation potential. To investigate this, two MBBRs were run in parallel, one of which received wastewater spiked with incrementally increasing micropollutant concentrations, while the other acted as a control receiving unspiked wastewater. We applied Total RNA metatranscriptomics to analyze the active microbial community in the biofilm on MBBR carriers exposed to different concentrations of micropollutants. We further integrated it with chemical analyses of micropollutant removal in a series of batch cultures inoculated with the biofilm laden carriers. This integration of analytical chemistry, microbiology, and metatranscriptomics is the first to provide functional insight into community dynamics and differential gene expression in a micropollutant exposed MBBR system. This will further provide insight into the community dynamics, across all domains of life, of a MBBR system subjected to increasing micropollutant concentrations, thus laying the foundation for optimizing towards a more efficient micropollutant removal by MBBR systems in the future.
Material and methods
Wastewater and chemicals
We collected raw and effluent wastewater from Bjergmarken WWTP, located in Roskilde, Denmark. Bjergmarken is a conventional WWTP with activated sludge treatment including biological oxygen demand removal, nitrification, and denitrification, and operates a complex biological phosphate removal within the activated sludge system. However, the plant does not operate any advanced treatment processes for the removal of micropollutants. Wastewater was collected every two weeks and subsequently stored at 4°C until use.
This study included five target compounds: benzotriazole, carbamazepine, diclofenac, iohexol, and venlafaxine. These micropollutants are all found in the wastewater effluent of the Bjergmarken WWTP with mean concentrations of 0.26 µg carbamazepine L−1, 0.52 µg venlafaxine L−1, 0.71 µg diclofenac L−1, 12.4 µg iohexol L−1, and 2.53 µg benzotriazole L−1. Benzotriazole is mostly used as corrosion inhibitor. The remaining four are all pharmaceuticals. All chemicals applied were at least of analytical purity (≥95%). Table S1 provides a list that includes CAS numbers, structures, and chemical suppliers for the micropollutants applied in this study. Solvents (HPLC-MS grade water, methanol (Lichrosolv®) and formic acid were obtained from Merck (Darmstadt, Germany).
Pre-treatment of the reactors
We ran two parallel one-liter laboratory-scale moving bed biofilm reactors during the four-month experiment (Fig. S1). Both reactors received constant aeration and had an inlet flow rate of 2.8 ml min−1. Each reactor initially contained 150 carriers (AnoxKaldnes K5; Lund, Sweden). Both reactors were subjected to a pre-treatment phase where they received raw wastewater for several weeks to establish a mature biofilm on the carriers. After the pre-treatment period, the carriers were distributed evenly between the two reactors. The reactors then received an intermediate inlet of wastewater (24 h raw wastewater, 48 h effluent wastewater) for three weeks to obtain similar conditions prior to experimental start.
Experimental design of the MBBR systems
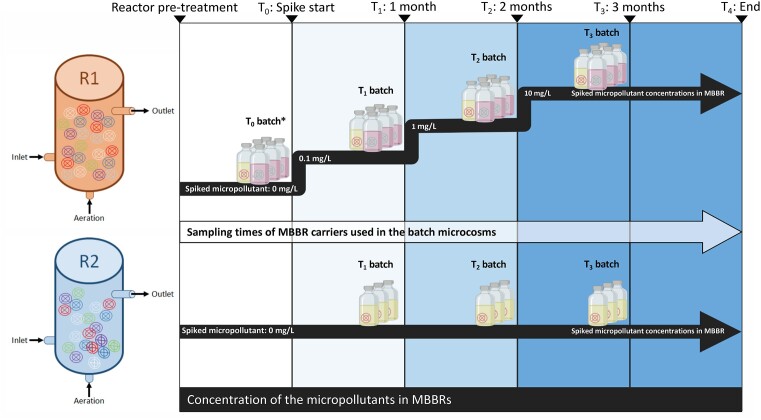
The overall adaptation experiment consisted of two sub-experiments, which we conducted simultaneously over the course of four months (Fig. 1). The first was a long-term study using Total RNA metatranscriptomics to analyze the dynamics of the active microbiome on the carriers in two MBBR systems. Of the two MBBRs, one (R1) was spiked with monthly increasing micropollutant concentrations in the feed wastewater over a three-month period, from 0.1 mg L−1 to 1 mg L−1, and finally ending at 10 mg L−1 for the last two months. The second MBBR system (R2) acted as a control and was supplied with unaltered wastewater. Both systems were under an intermittent feeding strategy; 24 h of raw wastewater, and 48 h of effluent wastewater. The MBBRs were incubated in the dark at approximately 20°C.
Figure 1.
Experimental design of the study. Two MBBRs R1 (spiked) and R2 (unspiked control) were continuously supplied with wastewater. The wastewater supplied to R1 was spiked with a mix of five micropollutants in monthly increasing concentrations; from 0.1 mg L−1 to 10 mg L−1 (held for two months). Whereas R2 acted as the control receiving the wastewater unspiked. Each month carriers from R1 and R2 were sampled for 1) Total RNA analysis and 2) inoculation in batch cultures. The batch cultures were conducted in serum bottles in triplicate, which contained a mix of micropollutants at 10 mg L−1 of each. The yellow bottles represent the treatments inoculated with biofilm carriers housing active microbiomes, and the pink bottles represent the treatment where biofilm carriers were sterilized (controls). *T0 batch microcosms were created using carriers taken from the pre-treatment reactors immediately prior to all the carriers being distributed evenly between the two reactors R1 and R2.
Microbial biofilm community and gene expression
We sampled carriers monthly in triplicates from each of the two reactors R1 and R2. These were sampled from the MBBRs just before beginning the next increase in spiked concentration. The biofilm carriers (Figure S1) were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction as described below.
RNA extraction, library building, and sequencing
The individual carriers were cut in half with sterilized scissors immediately after removing them from −80°C, and the two halves were then added directly to an extraction tube kept on ice. Total RNA was extracted from the carriers using the RNeasy Powersoil Total RNA Kit (Qiagen) with phenol:chloroform:isoamyl alcohol in a ratio of 25:24:1 (ThermoFisher Scientific) according to the manufacturer's instructions, except that the RNA was eluted in a final volume of 50 µl instead of 100 µl. DNase treatment was performed on 20 µL RNA subsamples using DNase Max Kit (Qiagen), and the DNA removal was following verified by qPCR. The integrity of the purified RNA was determined on Tapestation 4150 (Agilent Technologies, Santa Clara, CA, USA) using the RNA buffers and screentapes, while concentration was measured on a Qubit 4 fluorometer (Invitrogen, Eugene, Oregon, US) with the Qubit RNA HS Assay Kit.
RNA sequencing libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England BioLabs, Ipswich, MA, USA) without prior mRNA enrichment or rRNA depletion in combination with the NEBNext Multiplex Oligos for Illumina, according to the manufacturer's protocol (section 4). The fragment size of the resulting cDNA libraries was verified by Tapestation and DNA concentrations were measured on Qubit.
The indexed cDNA libraries were equimolarly pooled and sequenced on a NextSeq 500 platform (Illumina Inc., San Diego, USA) with the high-output 300 cycles v2.5 reagent kit, resulting in 151 bp paired-end reads. Raw sequences were submitted to SRA under the project accession number: PRJNA1046284.
Bioinformatic processing, community, and gene expression analysis
A total of 850 million paired-end reads were obtained and processed through our automated Total-RNA bioinformatics pipeline (https://zenodo.org/badge/latestdoi/546561474 v1.1.0). Briefly, adapters, poly-A tails, and unassigned nucleotides were trimmed using trim-galore (https://github.com/FelixKrueger/TrimGalore) which relies on cutadapt (Martin 2011). All the parameters and versions of the programs used are available on GitHub (https://github.com/AU-ENVS-Bioinformatics/TotalRNA-Snakemake—commit 6a199a9). Quality-filtered reads were subsequently sorted into small subunit (SSU) rRNA (used for taxonomic analysis), large subunit (LSU) rRNA (not used), or non-rRNA sequences (to be used for functional gene analysis) using SortMeRNA v2 (Kopylova et al. 2012).
rRNA
First, the reads were co-assembled into full-length SSU rRNA contigs using MetaRib (Xue et al. 2020). Contigs were taxonomically classified using CREST 4 (Lanzén et al. 2012) against the SILVA v.138 database (Quast et al. 2013) and PR2 database (Guillou et al. 2013). The SSU rRNA reads from each sample were mapped back to the rRNA contigs using BWA MEM (Li and Durbin 2009) and SAMtools (Danecek et al. 2021) under default settings for consistency across datasets (for BWA: 2 mismatches in the seed, 3 mismatches in the whole read, no gapped alignment). This resulted in a count-table of taxonomically annotated read counts across the samples from which relative abundances were calculated per contig per sample.
The Eukaryota were grouped into macroeukaryotes (Archaeplastida, Fungi and Metazoa) and Protozoa. We manually assigned rRNA contigs as Protozoa if annotated as Amoebozoa, SAR, Euglenozoa, Foraminifera, and Heterolobosa (Geisen et al. 2015) as well as Endomyxa, Telonemia, Malawimonadidae, and Choanoflagellida. The community analysis was performed using the R software v.4.2.2 in R studio (RStudio Team 2020, R Core Team 2022), and the packages phyloseq v.1.42.0 (McMurdie and Holmes 2013), tidyr v.1.3.0 (Wickham et al. 2019), and vegan v.2.6.4 (https://github.com/vegandevs/vegan). For rRNA contigs, alpha diversity was calculated using the Shannon diversity (H) index, which takes into consideration the number of unique contigs and their abundance per sample. The significance of the correlation of changes in community composition with spiking and time point was tested using the ANOVA function (PERMANOVA, 999 permutations) on Bray–Curtis dissimilarities. Consecutive Tukey's HSD post-hoc tests supplied significance of the difference between control and spiked samples, as well as across time points.
mRNA
The mRNA (non-ribosomal) reads from all samples were co-assembled into contigs using the Trinity RNA assembler (Grabherr et al. 2011). Non-coding RNA contigs were filtered out by aligning the contigs to the RFam database v.14.0 (Kalvari et al. 2021) using Infernal aligner (Nawrocki and Eddy 2013). The input sequences used for the non-ribosomal RNA assembly were then mapped back to the coding mRNA contigs using the BWA mapper. We normalized the contigs by removing those with relative expression lower than 1 out of the number of sequences in the dataset with least number of sequences, i.e. e=1 retaining only contigs with sum > 1 when divided by sum(Minimum Reads) (Anwar et al. 2019, Bang-Andreasen et al. 2019). Using diamond (Buchfink et al. 2021), the filtered contigs were then annotated against the KEGG BRITE (Kanehisa et al. 2022) and NCycDB (Tu et al. 2018) databases, which are the hierarchical database of Kyoto Encyclopedia of Genes and Genomes (KEGG) classifications and a nitrogen-cycling database (NCycDB), respectively. Full code with all options and documentation is available in the form of Jupyter Notebooks (https://zenodo.org/doi/10.5281/zenodo.10254965 v0.0.2).
Significantly differentially expressed mRNA contigs were obtained using the DESeq2 module of the SARTools v.1.7.3 (Varet et al. 2016). These functional analyses were conducted by pairwise comparisons of gene transcription levels between the spiked and control samples at the various time points. The T0 time point was not included as samples were taken before the micropollutants were first spiked into the system. We used a significance level of 0.05 for all analyses.
Batch microcosm cultures
The second sub-experiment comprised of a series of batch incubations with carriers from the MBBRs (R1 and R2) conducted monthly for four months. These microcosms were designed to provide a snapshot of the degradation potential of the microbial community on the carriers at a given time point. We sampled single carriers from the MBBRs immediately before the subsequent increase in spiked concentration and used these as inoculum in the batch incubations. Weekly samples were taken for chemical analysis to measure the concentrations of micropollutants in these batch cultures.
Media
Minimal salt media (MSN) was used as the media for the microcosms (Sørensen and Aamand 2003). MSN consisted of KH2PO4 (1.36 g/L), Na2HPO4·2H2O (1.78 g/L), MgSO4·7H2O (0.05 g/L), (NH4)2SO4 (0.238 g/L), MgSO4·7H2O (0.05 g/L), CaCl2·2H2O (0.01 g/L), H3BO4 (2.86 mg/L), MnSO4·H2O (1.54 mg/L), CuSO4·5H2O (0.04 mg/L), ZnCl2 (0.021 mg/L), CoCl2·6H2O (0.041 mg/L), Na2MoO4·2H2O (0.025 mg/L). 1 M NaOH was added to MSN before autoclaving to adjust pH to 7.1. The MSN media was spiked with the micropollutant mix to a starting concentration of 10 mg L−1 of each of the compounds.
Setup, inoculation, and sampling
Microcosms were prepared in triplicate using single biofilm laden carriers as inoculum in 30 ml sterilized serum bottles containing 15 ml MSN media with the micropollutants as the sole carbon source. Sterilized controls were created in the same manner, except the carriers were sterilized by autoclaving three times in succession prior to inoculation. The microcosms were incubated in the dark at 20°C on an orbital shaker at 120 r/m. Aliquots of the liquid were sampled every two weeks. The samples were stored at −20°C until chemical analysis. After sampling, 1 ml of fresh, spiked media was added to replace the volume taken out.
Analytical methods
All chemical analyses were performed by high performance liquid chromatography with tandem mass spectrometry (HPLC-MS/MS) utilizing electron spray ionization in positive mode (ESI (+)). An Ultimate 3000 dual gradient low pressure mixing HPLC-system (Dionex, Sunnyvale, CA, USA) coupled to an API 4000 triple-quadrupole-MS (AB Sciex, Framingham, MA, USA) was used, and all analytes were tracked and quantified using multi reaction monitoring (MRM; provided in Table S2). The separation was performed at 20°C using a Phenomenex Synergy Polar RP column (150×2 mm, 4 µm) with a multi-step gradient elution of 0.2% formic acid in water (A) and methanol containing 0.2% formic acid (B); 0–1.5 min 0% B, 1.5–3 min 0–10% B, 3–13 min 10–50% B, 13–18 min 50–60% B, 18–28 min 60–100% B, 28–30 min 100% B, 31–32 in 100–0% B at a constant flow of 250 µl/min. The injection volume was 100 µl. Data was collected and processed with analyst software version 1.5.1 (AB Sciex, Framingham, MA, USA). The quantification was done using an 8 point-calibration curve ranging from 0.05 to 300 µg L−1 linked to an internal standard (IS). Limit of detection (LOD), limit of quantification (LOQ) and different IS used are provided in Table S3 and Table S4. The difference in normalized micropollutant concentration means between the spiked and control batch culture endpoints were determined using unpaired t-tests in the program Prism v.5.04.
Results
Sequencing output results
An average (± standard error of the mean) of 32.7 ± 1.1 million reads per sample (forward and reverse) were obtained from Total RNA sequencing. After quality trimming and sorting, the total number of reads was reduced to 12.5 ± 0.5 million SSU rRNA reads and 1.1 ± 0.1 million unaligned mRNA reads, while the remaining reads belong to the LSU rRNA fraction.
Overall community composition
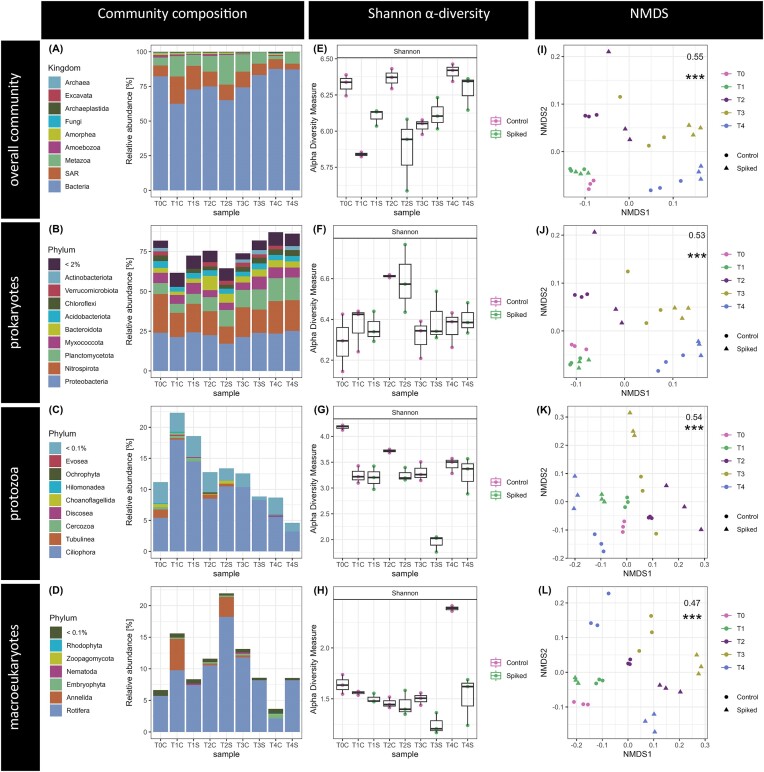
A total of 2515 full-length rRNA contigs were assembled across all samples. These contigs were annotated to 99.9% for kingdom, 99.2% for phylum, 97.3% class, 89.4% order, 69.1% family, 29.4% genus, and 1% for species level. The overall community was represented in relative abundance by 2 350 prokaryotic rRNA contigs (76.3% of counts) and 165 eukaryotic rRNA contigs (23.7% of counts) (Fig. 2A). Of the latter, more than half were represented by protozoa (132 rRNA contigs, 12.6% of counts), 12 metazoan contigs (10.5%), and four Archaeplastida contigs (0.3%) and 15 fungal rRNA contigs (0.3%).
Figure 2.
Active microbial communities of the MBBR biofilm carriers shown as microbial community composition, Shannon alpha diversity, and beta diversity of the experimental groups (C—Control, S—Spike for the time points T0-T4). All figures are given for the total community (A, E, I) and subsets with prokaryotic (B, F, J), protozoan (C, G, K) and macroeukaryotes (D, H, L) rRNA contigs. Bar plots (A-D) show the relative mean abundance for varying taxonomic orders of the microbial communities. Shannon alpha diversity (E-H) is given as boxplots with whiskers indicating standard deviation across triplicates per experimental group. Nonmetric multidimensional scaling ordination plots (I-L) were performed on rRNA contig abundances per sample and include the PERMANOVA R2 and p-value when testing the “timepoint” as factor.
Prokaryotic community
Overall prokaryotic relative abundance ranged from 62.1% (at T1 control) to 87.7% (at T4 control). The prokaryotic community composition within both reactors remained relatively constant at the phylum level across all sampling time points, although most phyla increased in relative abundance again towards the end of the experiment (T4). The dominant phyla were Proteobacteria, Nitrospirota, and Planctomycetota, followed by the predatory Myxococcota (Fig. 2B). The most abundant phyla Proteobacteria was independent of time and spiking, dominated by the order Burkholderiales with an average relative abundance of 11.2% across all samples. The most abundant family within this order was Comamonadaceae. Nitrospirota relative abundance peaked at T0, decreased towards T2 and then increased for the remainder of the experiment. This phylum was dominated by the overall most abundant order Nitrospirales (12 out of 20 most expressed contigs) with an average 16.3% relative abundance across samples. Despite an overall decrease at T2 and T3, this taxon was independent of spiking, and dominated by the Nitrospira genus (5.1% relative abundance).
Eukaryotic Community
On average, 93% of the protozoan relative abundance was assigned to the kingdom SAR and within that most represented by the Ciliophora phylum (Fig. 2C). Here, the Oligohymenophorea class accounted for 8.0% of the overall community relative abundance and was among the overall most abundant rRNA contigs.
The overall relative abundance of Fungi was low, while other non-protozoan eukaryotes were mainly represented by Metazoa (10.5% relative abundance), particularly the Rotifer order Adinetida (9.1%) (Fig. 2D).
Community Changes with Varying Pollutant Concentrations
The overall average Shannon index (Fig. 2E) per sample was 6.14 ± 0.22 (SD). For prokaryotes, Shannon diversity was also overall high and showed no differences over time nor treatment. While eukaryotic Shannon diversity decreased in spiked samples compared to controls from time point T2 onwards. This trend was mirrored in protozoan alpha diversity as well (Fig. 2F-H).
No significant differences in community composition between the spiked and the control biocarrier samples were found for the total community or any of the sub-communities, except for the protozoa (ANOVA, Table 1; Fig. 2K). Yet, there were observed significant differences between time points (ANOVA, Table 1) with approximately half of the variance (R2 ∼0.5 each) across samples correlated with the time points (Fig. 2I-L). Tukey tests revealed no significant differences for the total microbial community composition between specific time points, while prokaryotic beta diversity significantly varied between T2 and T4 and protozoan beta diversity between T1 and T3. The macroeukaryotic community composition significantly varied between T4 compared to T0 and T3.
Table 1.
Permutational multivariate analysis results based on the mean Bray–Curtis dissimilarities per Time point and Control-Spike experimental group depicted for total community, prokaryotic, protozoan and fungal/metazoan rRNA contigs. P-values < 0.05 were consider ed significant with * P < 0.05, **P < 0.01, and ***P < 0.005. Nonsignificant (NS) categories and pair-wise contrasts in time points were removed for the Control-Spike test.
| Total | Prokaryotes | Protozoa | Other Eukaryotes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F.Model | R2 | P | F.Model | R2 | P | F.Model | R2 | P | F.Model | R2 | P | |
| Control-Spike | 1.88 | 0.073 | NS | 1.84 | 0.071 | NS | 3.92 | 0.140 | ** | 1.47 | 0.058 | NS |
| Time point | 6.13 | 0.539 | *** | 5.68 | 0.520 | *** | 5.93 | 0.530 | *** | 4.40 | 0.456 | *** |
| T4-T0 | NS | NS | NS | * | ||||||||
| T3-T1 | NS | NS | * | NS | ||||||||
| T4-T2 | NS | * | NS | NS | ||||||||
| T4-T3 | NS | NS | NS | * | ||||||||
Functional gene annotation and expression
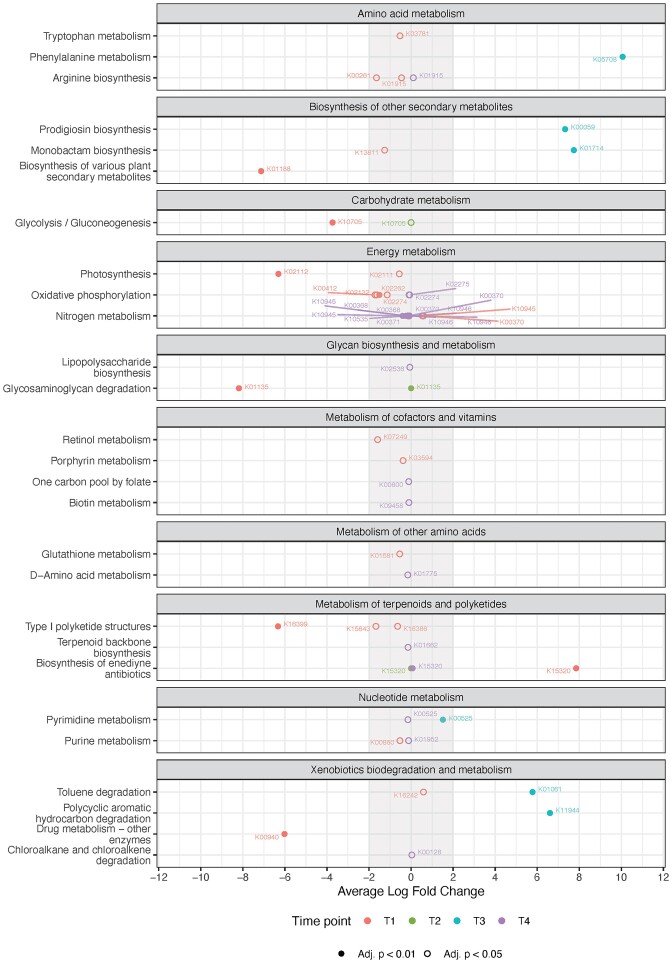
Of the 28 925 mRNA contigs, 28 441 KEGG Brite annotations were assigned as well as 4 981 alignments with the NCyc database. Cumulatively amongst all time points, 50 BRITE and 48 NCycDB annotated mRNA genes were determined to be differentially expressed between the spike and control MBBR biocarriers over time (Fig. 3 and Figure S1).
Figure 3.
Differential gene expression between the microbiomes of the carriers taken from the spiked and the control MBBRs at the different timepoints (T1- T4). mRNA contigs were annotated using BRITE database and tested with SARTools for differential expression between carriers from the control and spiked MBBR. Annotated contigs—labelled with unique Entry ID (K-number) for the KEGG functional ortholog number—are shown per overall expressed pathway categories based on their log fold changes with filled and empty dots indicating adjusted P-values < 0.01 and < 0.05, respectively.
BRITE annotated genes
The 2459 unique KEGG IDs spanned 11 BRITE pathways (of the KEGG Ontology), of which the most expressed pathways included Energy, Amino acid, and Cofactor and vitamin metabolism as well as Xenobiotics degradation. All BRITE pathway categories were less expressed at T1 and T2 compared to the other experimental time points, except the Metabolism of lipids and other amino acids. For BRITE, time point T1 exhibited the highest total number of differentially expressed functional genes (21 down- and 4 up-regulated; 0.1 mg L−1), followed by T4 (20 down- and 3 up-regulated; 10 mg L−1), then T3 (6 up-regulated; 10 mg L−1) and lastly T2 (3 up-regulated; 1 mg L−1).
Overall, within the energy metabolism category, nitrogen metabolism was the most expressed pathway category, especially represented by counts annotated to nitrate reductase, nitrite oxidoreductases, (alpha)-subunit (K00370). For energy metabolism, especially mRNA contigs annotated within photosynthesis, oxidative phosphorylation, and nitrogen metabolism were differentially expressed, although mostly with comparatively low overall logFC values (Fig. 3).
Notably, the relative abundance of contigs annotated within xenobiotic degradation and metabolism was highest at T4 and mainly characterized by annotations within nitrotoluene degradation, such as pyruvate ferrodoxin oxidoreductases (alpha) subunits (K00169). Xenobiotic degradation and metabolism annotated contigs that were differentially expressed included both toluene and polycyclic aromatic hydrocarbon degradation counts that were upregulated at T3 between control and spiked MBBR biocarriers samples, while one contig (K00940) related to drug metabolism was downregulated at T1 spiked samples (Fig. 3).
NCycDB annotated genes
The mRNA contigs annotated to the nitrogen-cycling database (NCycDB) were most expressed in the pathways: Denitrification (average 42.9% of all counts), organic degradation and synthesis, and nitrification. Denitrification was most characterized by contigs annotated as nirK, nirS and nosZ. Organic degradation and synthesis overall were more expressed at T1 and T2 and mainly dominated by counts annotated as dgh, as well as nmo. Nitrification pathways included nxrB annotated contigs with lower overall abundance at T2, and amoA/B/C annotations.
These nitrogen cycling genes also included significantly differentially expressed contigs, with overall downregulation of organic degradation and denitrification contigs from control to spiked MBBR biocarriers at T1, as opposed to nitrification nxrB gene upregulation for spiked compared to control samples at T1 (Figure S2). Time point T4 exhibited the highest total number of differentially expressed functional genes (17 down- and 11 up-regulated; 10 mg L−1), followed by T1 (17 down- and 6 up-regulated; 0.1 mg L−1), while both T2 and T3 had each one up-regulated contig.
Batch culture chemical analysis
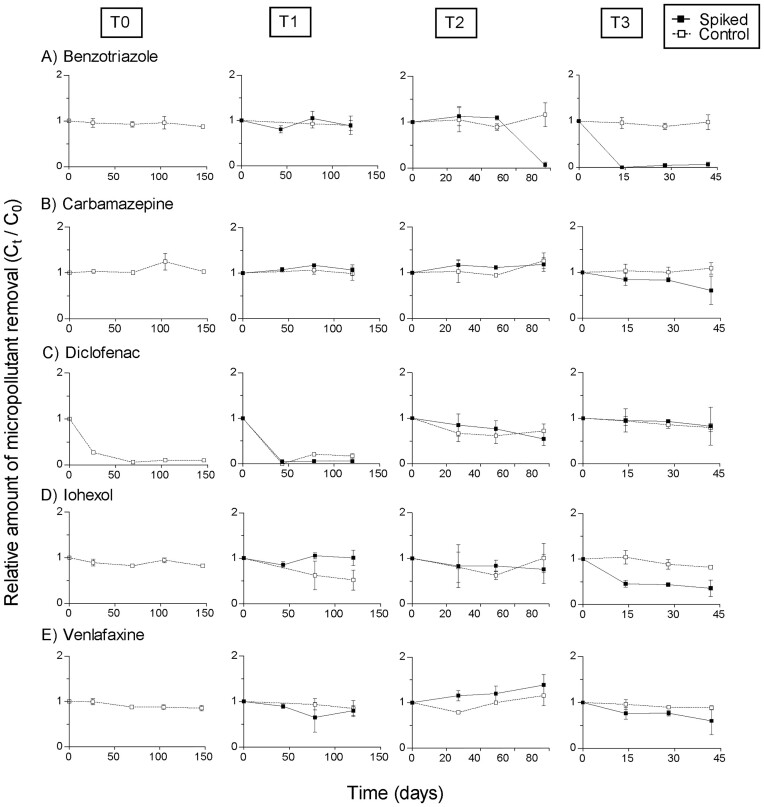
The relative micropollutant concentrations in the batch cultures of the various time points as a result of degradation by the biocarrier microbial communities are shown in Fig. 4. T-tests were conducted on the endpoints of each batch culture experiment to compare if there was a significant difference in the relative concentration for each micropollutant between batches with biocarriers from the spiked and control MBBR (Table S5).
Figure 4.
The relative micropollutant concentrations in the batch cultures of the various time points as a result of degradation by the microbial communities on the biofilm carriers from the spiked and control MBBRs. The measured relative concentrations in each batch culture are shown as means with standard error bars (n=3) over time for the micropollutants: (A) Benzotriazole, (B) Carbamazepine, (C) Diclofenac, (D) Iohexol, and (E) Venlafaxine. T-tests were conducted for the endpoint concentrations of each batch culture to compare if there was a significant difference in the relative concentration for each micropollutant between the spiked and control batches (Table S5).
Benzotriazole was the only micropollutant showing a significant difference in degradation between cultures with biocarriers from the spiked and control MBBR (Fig. 4A). For the T1 batch cultures, the degradation potential of benzotriazole was not significantly different when comparing carriers taken from the spiked and the control reactors (P = 0.4336). However, the degradation of benzotriazole by biofilm carriers from T2 and T3 differed significantly (P= 0.0079 and 0.004, respectively) between the spiked and control MBBR carriers, indicating an adaptation of the microbial community towards benzotrialzole degradation when exposed to higher concentrations (T2, T3). Furthermore, with regard to the biocarriers from the spiked MBBR, degradation of benzotriazole for T3 occurred much earlier than for T2. The microbial community inoculated from T3 had almost fully removed benzotriazole after approximately 13 days, compared to approximately 79 days for the microbial community inoculated from T2.
A removal of more than 90% of diclofenac (Fig. 4C) occurred within the batch cultures inoculated with carriers from T0 and T1 within approximately 50 days. However, there is no significant difference between the diclofenac removal of the batches with biocarriers from the control and spiked MBBR of T1 (P > 0.05). For T2 and T3, the degradation potential for diclofenac was much lower than at T0 and T1. Still, the endpoint concentrations for the treatments with from the spiked and control MBBRs for T2 and T3 were not significantly different (P > 0.05).
The degradation of carbamazepine (Fig. 4B), iohexol (Fig. 4D), and venlafaxine (Fig. 4E) was not significantly different between the adapted batch cultures using biofilm laden carriers taken from the spiked and control MBBRs (all P > 0.05). Removal ranged from 0% to 50% without significant patterns. No biodegradation of any of the five micropollutants was observed in the sterilized controls (data not shown).
Discussion
Here, we present the first Total RNA study revealing the changes in the entire microbial community over time for MBBR carrier biofilms, as well as the changes in microbial gene expression and degradation potential as a result of increased micropollutant concentrations.
Prokaryotic response over time
The prokaryotic community compositions and diversity did not differ significantly between carriers from the control and spiked MBBRs. However, the relative abundance of prokaryotes increased over time in both (spiked and control) treatments, with total prokaryotic relative abundance increasing from roughly 68% at T1 to 87% at T4 across the entire dataset. Yet, prokaryotic diversity of MBBR carriers has been shown to be related to micropollutant removal efficiency (Torresi et al. 2016). The increased relative abundance is a logical outcome of the constant supply of feed wastewater throughout the experiment supplying the prokaryotes with ample organic substrate for growth. Over the course of the entire experiment, the dominant phyla were Proteobacteria, Nitrospirota, Planctomycetota, and Myxococcota (Fig. 2B). Proteobacteria are ubiquitous and abundant in most environments and have been reported as some of the most dominant taxa in other MBBR studies (Ma et al. 2013, Su et al. 2019, Liang et al. 2021). Nitrospirota are likewise found in wastewater facilities and MBBRs, where they significantly contribute to the nitrification process (Hoang et al. 2014). Planctomycetota, which have also previously been reported in wastewater systems, possess both antibiotic and multidrug resistant qualities (Cayrou et al. 2010, Kaboré et al. 2020). Furthermore, Planctomycetota has been shown to respond positively in comparison to other taxa when exposed to increased concentrations of micropollutants in surface waters (Yergeau et al. 2012). Notably, Myxococcota annotated contigs were among the most abundant in our system. This phylum consists of predatory prokaryotes—keystone taxa within many environmental systems predating on other bacteria (Petters et al. 2021, Scheel et al. 2023). We hypothesize that they have proliferated throughout the course of the experiment since competition for food has decreased following the decline in the relative abundance of protozoa.
It was not possible to pin-point prokaryotic species or genera of known or suspected micropollutant biodegradation potential that were significantly enriched on the carriers from the spiked MBBR. Since the MBBRs contained a high load of other energy sources proliferation of degraders could be less pronounced than the overall increase in prokaryotes over time in both treatments. However, we suggest that experiments with individual micropollutants as the sole carbon source are needed to elucidate the organisms involved in especially benzotriazole and diclofenac degradation.
Of the five micropollutants selected for this study, three (benzotriazole, carbamazepine, and diclofenac) have reported toxicity in prokaryotic model organisms. EC50 concentration for V. fischeri have been reported as 41 mg L−1 benzotriazole (Hem et al. 2003), > 81 mg L−1 carbamazepine and 11.5 mg L−1 diclofenac (Ferrari et al. 2003). Little to no research has been conducted on the toxicity of iohexol or venlafaxine on prokaryotic model organisms. Even though our study had a much longer duration than the above-mentioned toxicity tests, we did not observe a significant impact of the micropollutant exposure on the prokaryotic community of the MBBRs, which corresponds well with the micropollutants concentration of our study being much lower than the toxicity endpoints found in the studies above.
Protozoan response over time
Throughout the duration of this experiment, the relative abundance of protozoa decreased. At each time point, except T2, the spiked reactor had a relatively lower abundance of protozoa compared to the control reactor. Ciliates (Ciliophora) constituted the most dominant protozoan phylum throughout the entire experiment in both the control and spiked MBBRs. Ciliates are common in biofilms of various environments and play an important role in the microbial food webs mainly as predators of other microorganisms. Ciliates dominate mature biofilms in wastewater systems, where they graze upon bacteria (Eisenmann et al. 2001), which our findings support.
In the present study, the protozoan community composition was the only subset impacted by spiking with the increasing micropollutant concentrations, whereas the community composition of control samples was more stable (Fig. 2K). Still, the overall relative protozoan abundance in the control reactor also decreased with time. We suggest that further studies are needed to establish a plausible toxic effect of these specific micropollutants on protozoan taxa. To our knowledge, no studies have been reported on the toxic effects of micropollutants specifically on Ciliates in wastewater biofilms for an exposure this long in duration. Our results suggest that concentrations between 1 and 10 mg L−1 (which is about 1000 times higher concentrations than what we found in the unspiked wastewater (Table S3)) adversely impact the protozoa and cause their gradual decline in relative abundance over time.
Macroeukaryotic response over time
Fungi and Metazoa displayed no clear difference in the community composition and relative abundance of the taxa between biocarriers from the control and spiked MBBRs. However, the beta-diversity NMDS plot indicate that the communities on the carriers from the spiked MBBR at the three last time points were distinct from the others (Fig. 2L), implying a potential impact of increased pollutant concentrations specifically on macroeukaryotic community compositsion.
The most dominant Metazoa on the carriers were the Rotifers, which are common in biofilms of WWTPs (Lapinski and Tunnacliffe 2003). The relatively high abundance of Rotifers in the spiked MBBR at several time points suggests that it is unlikely that they are negatively affected by the increasing micropollutant concentrations.
At two occurrences (control T1 and spiked T2) the presence of Annelida, represented by only Rheomorpha sp., increased drastically in our reactors (Fig. 2D). Annelids are common in wastewater systems and are often associated with polluted environments (Elissen et al. 2008, Hendrickx et al. 2009). Further, Annelids have been found to feed on organic material present in the activate sludge of wastewater treatment plants, as well as on protozoa or bacteria (Elissen 2007, Elissen et al. 2008).
Finally, Fungi were present in our MBBR system, but at a low relative abundance. Similarly, Zhou et al. (2018) and Ribera-Pi et al. (2020) reported fungi in MBBRs, with a community richness and diversity much lower than for the prokaryotes.
For eukaryotic model organisms, Seeland et al. (2012) reported that benzotriazole negatively affected reproduction in Daphnia galeata with EC10 of 0.97 mg L−1 in a 21-day study. Further, Ferrari et al. (2003) found that the lowest observed effect concentrations in the rotifer Brachionus calyciflorus were 0.75 mg L−1 for carbamazepine and 25.0 mg L−1 for diclofenac, while a LC50 for diclofenac of 2.00 mg L−1 have been reported for a Daphnia magna 21-day study (Du et al. 2016). Most of these effect concentrations are below the concentrations used in our experiment, and the adverse effects observed on eukaryotic organisms on the MBBR in the present study could at least partly be due to benzotriazole, carbamazepine and/or diclofenac exposure. The longest duration for the abovementioned studies was 21 days; given that our study lasted four months, it is also plausible that long-term toxicity negatively impacted the eukaryotes, especially protozoans.
Up- and down-regulated mRNA genes
Of the 50 significantly up- or downregulated genes annotated by the BRITE database, five were related to xenobiotic degradation. Of these, one was upregulated at time point T1 and four at T3 (Fig. 3). T1 samples were taken one month after the introduction of our specific micropollutants (0.1 mg L−1) into the system. The upregulation of this one gene (phenol/toluene 2-monooxygenase) could allude to the introduction of micropollutant in concentrations above ambient effluent triggered the expression of this gene (Fig. 3). The carriers for RNA analysis of T3 were taken a month after the 10 mg L−1 spiking commenced. The gene with the highest upregulation of this time point was a gene annotating for phenylalanine metabolism. Genes within this grouping are known for being involved in the degradation of xenobiotics with aromatic rings (Teufel et al. 2010). This coincides well with the batch cultures, as the spiked batch cultures taken from time point T3 were able to degrade benzotriazole from the system within 14 days. Other gene classes that were upregulated significantly at this time point fall into the classes of toluene degradation, polycyclic aromatic hydrocarbon degradation, and pyrimidine metabolism (Karpouzas and Singh 2006, Kunze et al. 2009, Lima-Morales et al. 2016).
Further, the relative abundance of contigs annotated within the xenobiotic degradation and metabolism groupings was highest at T4 and mainly characterized by annotations within nitrotoluene degradation, such as pyruvate ferredoxin oxidoreductases. However, these were not differentially expressed between spiked and control MBBR carriers, hence these genes are more likely to be involved in the degradation of other compounds than benzotriazole.
For genes involved in N cycling, many of the significantly up- or downregulated genes annotated by the NCyc database were related to nitrification or denitrification (Figure S2), which is not surprising for wastewater treating systems (Kuypers et al. 2018, Rose et al. 2021). Particularly for biofilms, oxygen-dependent nitrifiers and the anaerobic denitrifiers have been shown to coexist by occupying different layers of wastewater-associated biofilms, such that nitrification and denitrification occurred simultaneously in a membrane-aerated biofilm reactor (Terada et al. 2003).
Of particular interest, in relation to micropollutant degradation, are the amo genes, encoding for the non-specific enzyme ammonia monooxygenase. The amo genes facilitate oxidative co-metabolic breakdown of a variety of micropollutants in wastewater such as aliphatic and aromatic hydrocarbons (Helbling et al. 2012, Fischer and Majewsky 2014). In the present study, the expression of the amo genes did not follow any specific trend in regard to the spiking of the micropollutants, which is what we would expect since these genes are induced by ammonium (Sayavedra‐Soto et al. 1996) and not micropollutants. However, the expression of the amo genes fits well with the high abundance of Nitrospira that we observed and that Nitrospira has been shown to degrade certain micropollutants (Han et al. 2019). Whether any genes, including amo gene complex, in the nitrogen cycle may be involved in the degradation of the specific micropollutants investigated the present study warrants a controlled study that includes a treatment where these genes are inhibited.
Micropollutant degradation by biofilm carriers
Of all the micropollutants, we observed that only the biodegradation of benzotriazole became more efficient over time and with increased spiking concentrations (increased degradation at T2 and T3) for the biofilm carriers from the spiked MBBR compared to the control. This suggests that the microbial community and/or activity had been altered by the higher micropollutant concentrations in the spiked MBBR compared to the control, such that it achieved a higher potential for benzotriazole degradation. Herzog et al. (2013) presented similar results when acclimating activated sludge microbiota to benzotriazole in batch cultures. After a 49-day acclimation period, benzotriazole was rapidly degraded within seven days. This is similar to the results we showed in the batch cultures inoculated with carriers from time point T3, where benzotriazole was removed completely before the first time point was taken at 14 days. The long-term exposure to benzotriazole may have successfully selected for specific degraders, although these could not be pin-pointed in the present study.
Biodegradation of diclofenac has been reported in batch cultures under a variety of conditions (Tiehm et al. 2011, Langenhoff et al. 2013). These studies showed a degradation of diclofenac over the course of 3 to 12 weeks. The variability presented in these systems is likely due to the microbial community present. In our batch cultures, the increasing concentrations suggest a potentially inhibitory effect on the proliferation of the microbial community capable of degrading diclofenac, as seen by the batch cultures becoming less efficient at degrading diclofenac over time. Time points T0 and T1 were the most adept at degrading diclofenac, while the later time points T2 and T3 were incapable of removal. We observed complete removal of diclofenac from the microcosms of T0 and T1 after 40–60 days. This coincides with the reported values of the other studies mentioned above.
From the batch culture experiment, there appears to be no microbial degradation of carbamazepine (Fig. 4B). Carbamazepine is a recalcitrant micropollutant and persists in WWTPs (Joss et al. 2006). However, carbamazepine removal in the range of 2.7%–20% has been reported in other biofilm experiments (Casas et al. 2015, Ahmadi et al. 2023). Iohexol and venlafaxine showed similar results to that of carbamazepine. However, iohexol was degraded to some extent in the batch cultures of T3 (Fig. 4D), albeit the values were not significantly different between treatments.
The results confirm previous studies showing that microbial transformation of micropollutants is primarily through co-metabolism, with endogenous metabolism playing a minor role (Liang et al. 2022). It is assumed that the indigenous concentrations of micropollutants in wastewater are insufficient to serve as a primary substrate to support microbial growth. Our observations show that, even with concentrations increased 1000-fold compared to the indigenous concentrations (Table S3), most compounds could not be utilized by microorganisms as a carbon source. This study implies that a “training” phase of MBBR carriers by spiking micropollutants in the feed would not necessarily improve the overall degradation efficiency in full-scale MBBR reactors.
Lastly, we recommend that future metatranscriptomic studies of dynamic systems such as MBBRs are designed to provide increased statistical power by increasing both the number of sample replicates and if relevant the concentration range. The present study design with sampling the MBBR carriers over time somewhat accommodates for the challenges of studying these dynamic systems. However, we suggest that future in-depth studies would benefit from a reduced complexity of the systems to better elucidate the causal relationships within the systems.
Conclusion
Through the implementation of Total RNA sequencing, this is the first demonstration of fluctuations in the relative community composition, activity, and gene expression of microorganisms across all domains of life within an MBBR system. Overall, we found that biofilm eukaryotes, in particular protozoa, were negatively influenced by prolonged micropollutant exposure. The prokaryotes’ relative abundance on the other hand increased over time with increasing micropollutant concentrations. The expression of functional genes in the MBBRs provided valuable insights into microbial activity, particularly in response to increasing micropollutant concentrations. Notably, we observed a significant upregulation of functional genes related to metabolism, including those involved in aromatic and xenobiotic compound degradation, within the biofilms of the spiked MBBR compared to the control. These findings shed light on potential genes responsible for the degradation of specific micropollutants. Additionally, our biofilm carrier batch experiment revealed distinct effects on benzotriazole and diclofenac degradation efficiency due to prolonged exposure and increased micropollutant concentrations. Overall, this study enhances our understanding of the microbial community and functional dynamics in MBBRs, particularly when subjected to higher micropollutant loads.
Supplementary Material
Acknowledgements
We would like to express appreciation to laboratory technicians Tanja Begovic and Tina Thane for their assistance, as well as to Francisco Campuzano Jiménez for his assistance with creating visualizations.
Contributor Information
Joseph Donald Martin, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark; Department of Biology, University of Copenhagen, Copenhagen, Denmark, Terrestrial Ecology Section, Department of Biology, University of Copenhagen, Universitetsparken 15, 2100 Copenhagen, Denmark.
Selina Tisler, Department of Plant and Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, Frederiksberg 1871, Denmark.
Maria Scheel, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Sif Svendsen, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Muhammad Zohaib Anwar, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark; The Center for Infectious Disease Genomics and One Health, Faculty of Health Sciences, Simon Fraser University, 8888 University Dr. W, Burnaby, BC V5A 1S6, Canada.
Athanasios Zervas, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Flemming Ekelund, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark; Department of Biology, University of Copenhagen, Copenhagen, Denmark, Terrestrial Ecology Section, Department of Biology, University of Copenhagen, Universitetsparken 15, 2100 Copenhagen, Denmark.
Kai Bester, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Lars Hestbjerg Hansen, Department of Plant and Environmental Sciences, University of Copenhagen, Thorvaldsensvej 40, Frederiksberg 1871, Denmark.
Carsten Suhr Jacobsen, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Lea Ellegaard-Jensen, Department of Environmental Science, Aarhus University, Frederiksborgvej 399, 4000 Roskilde, Denmark.
Author contributions
Joseph Donald Martin (Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing), Selina Tisler (Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing), Maria Scheel (Data curation, Formal analysis, Software, Validation, Writing – review & editing), Sif Svendsen (Data curation, Formal analysis, Writing – review & editing), Muhammad Zohaib Anwar (Methodology, Software, Validation, Writing – review & editing), Athanasios Zervas (Data curation, Methodology, Resources, Software, Supervision, Validation, Writing – review & editing), Flemming Ekelund (Validation, Writing – review & editing), Kai Bester (Conceptualization, Resources, Writing – review & editing), Lars Hestbjerg Hansen (Conceptualization, Resources, Supervision, Writing – review & editing), Carsten Suhr Jacobsen (Conceptualization, Writing – review & editing), and Lea Ellegaard-Jensen (Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – review & editing)
Conflict of interest
None declared.
Funding
This work was supported by Aarhus University Research Foundation [starting grant AUFF-E-2017-7-21] and the BONUS CLEANWATER project. MZA was supported by the Canadian Institutes of Health Research (CIHR) and Michael Smith Health Research BC (MSHRBC) fellowships.
Data Availability
Raw sequence data generated using this experiment have been deposited in the NCBI SRA (Accession Number PRJNA1046284).
References
- Abu Bakar SNH, Abu Hasan H, Mohammad AW et al. Performance of a laboratory-scale moving bed biofilm reactor (MBBR) and its microbial diversity in palm oil mill effluent (POME) treatment. Process Saf Environ Prot. 2020;142:325–35. [Google Scholar]
- Ahmadi N, Abbasi M, Torabian A et al. Biotransformation of micropollutants in moving bed biofilm reactors under heterotrophic and autotrophic conditions. J Hazard Mater. 2023;460:132–232. [DOI] [PubMed] [Google Scholar]
- Anwar MZ, Lanzen A, Bang-Andreasen T et al. To assemble or not to resemble—a validated comparative metatranscriptomics workflow (CoMW). GigaScience. 2019;8:giz096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- aus der Beek T, Weber FA, Bergmann A et al. Pharmaceuticals in the environment-global occurrences and perspectives. Enviro Toxic and Chemistry. 2016;35:823–35. [DOI] [PubMed] [Google Scholar]
- Bælum J, Nicolaisen MH, Holben WE et al. Direct analysis of tfdA gene expression by indigenous bacteria in phenoxy acid amended agricultural soil. ISME J. 2008;2:677–87. [DOI] [PubMed] [Google Scholar]
- Bang-Andreasen T, Anwar MZ, Lanzén A et al. Total RNA sequencing reveals multilevel microbial community changes and functional responses to wood ash application in agricultural and forest soil. FEMS Microbiol Ecol. 2019;96:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner J, Helbling DE, Kohler HPE et al. Is biological treatment a viable alternative for micropollutant removal in drinking water treatment processes?. Water Res. 2013;47:5955–76. [DOI] [PubMed] [Google Scholar]
- Besha AT, Gebreyohannes AY, Tufa RA et al. Removal of emerging micropollutants by activated sludge process and membrane bioreactors and the effects of micropollutants on membrane fouling: a review. J Environ Chem Eng. 2017;5:2395–414. [Google Scholar]
- Buchfink B, Reuter K, Drost H-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18:366–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmølle M, Ren D, Bjarnsholt T et al. Interactions in multispecies biofilms: do they actually matter?. Trends Microbiol. 2014;22:84–91. [DOI] [PubMed] [Google Scholar]
- Casas ME, Chhetri RK, Ooi G et al. Biodegradation of pharmaceuticals in hospital wastewater by staged moving bed biofilm reactors (MBBR). Water Res. 2015;3:293–302. [DOI] [PubMed] [Google Scholar]
- Castronovo S, Wick A, Scheurer M et al. Biodegradation of the artificial sweetener acesulfame in biological wastewater treatment and sandfilters. Water Res. 2017;110:342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Raoult D, Drancourt M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother. 2010;65:2119–22. [DOI] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Biase A, Kowalski MS, Devlin TR et al. Moving bed biofilm reactor technology in municipal wastewater treatment: a review. J Environ Manage. 2019;247:849–66. [DOI] [PubMed] [Google Scholar]
- Du J, Mei CF, Ying GG et al. Toxicity thresholds for diclofenac, acetaminophen and ibuprofen in the water flea daphnia magna. Bull Environ Contam Toxicol. 2016;97:84–90. [DOI] [PubMed] [Google Scholar]
- Ebele AJ, Abdallah MA-E, Harrad S. Pharmaceuticals and personal care products (PPCPs) in the freshwater aquatic environment. Emerg Contam. 2017;3:1–16. [Google Scholar]
- Eisenmann H, Letsiou I, Feuchtinger A et al. Interception of small particles by flocculent structures, sessile ciliates, and the basic layer of a wastewater biofilm. Appl Environ Microb. 2001;67:4286–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elissen HJH, Peeters ETHM, Buys BR et al. Population dynamics of free-swimming Annelida in four Dutch wastewater treatment plants in relation to process characteristics. Hydrobiologia. 2008;605:131–42. [Google Scholar]
- Elissen HJH. Sludge reduction by aquatic worms in wastewater treatment: with emphasis on the potential application of lumbriculus variegatus. 2007.
- Falås P, Baillon-Dhumez A, Andersen HR et al. Suspended biofilm carrier and activated sludge removal of acidic pharmaceuticals. Water Res. 2012;46:1167–75. [DOI] [PubMed] [Google Scholar]
- Fent K, Weston AA, Caminada D. Ecotoxicology of human pharmaceuticals. Aquat Toxicol. 2006;76:122–59. [DOI] [PubMed] [Google Scholar]
- Ferrari B, Paxéus N, Giudice RL et al. Ecotoxicological impact of pharmaceuticals found in treated wastewaters: study of carbamazepine, clofibric acid, and diclofenac. Ecotoxicol Environ Saf. 2003;55:359–70. [DOI] [PubMed] [Google Scholar]
- Fischer K, Majewsky M. Cometabolic degradation of organic wastewater micropollutants by activated sludge and sludge-inherent microorganisms. Appl Microbiol Biotechnol. 2014;98:6583–97. [DOI] [PubMed] [Google Scholar]
- Flemming H-C, Wingender J, Szewzyk U et al. Biofilms: an emergent form of bacterial life. Nat Rev Micro. 2016;14:563–75. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Micro. 2010;8: 623–33. [DOI] [PubMed] [Google Scholar]
- Geisen S, Tveit AT, Clark IM et al. Metatranscriptomic census of active protists in soils. ISME J. 2015;9:2178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small Sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Yu Y, Zhou L et al. Specific micropollutant biotransformation pattern by the comammox bacterium nitrospira inopinata. Environ Sci Technol. 2019;53:8695–705. [DOI] [PubMed] [Google Scholar]
- Helbling DE, Johnson DR, Honti M et al. Micropollutant biotransformation kinetics associate with WWTP process parameters and microbial community characteristics. Environ Sci Technol. 2012;46:10579–88. [DOI] [PubMed] [Google Scholar]
- Hem LJ, Hartnik T, Roseth R et al. Photochemical degradation of benzotriazole. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2003;38:471–81. [DOI] [PubMed] [Google Scholar]
- Hendrickx TLG, Temmink H, Elissen HJH et al. Aquatic worms eating waste sludge in a continuous system. Bioresour Technol. 2009;100:4642–8. [DOI] [PubMed] [Google Scholar]
- Herzog B, Huber B, Lemmer H et al. Analysis and in situ characterization of activated sludge communities capable of benzotriazole biodegradation. Environ Sci Eur. 2013;25:1–8. [Google Scholar]
- Hoang V, Delatolla R, Abujamel T et al. Nitrifying moving bed biofilm reactor (MBBR) biofilm and biomass response to long term exposure to 1 °C. Water Res. 2014;49:215–24. [DOI] [PubMed] [Google Scholar]
- Huerta-Fontela M, Galceran MT, Ventura F. Occurrence and removal of pharmaceuticals and hormones through drinking water treatment. Water Res. 2011;45:1432–42. [DOI] [PubMed] [Google Scholar]
- Joss A, Zabczynski S, Göbel A et al. Biological degradation of pharmaceuticals in municipal wastewater treatment: proposing a classification scheme. Water Res. 2006;40:1686–96. [DOI] [PubMed] [Google Scholar]
- Kaboré OD, Godreuil S, Drancourt M. Planctomycetes as host-associated bacteria: a perspective that holds promise for their future isolations, by mimicking their native environmental niches in clinical microbiology laboratories. Front Cell Infect Microbiol. 2020;10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Ontiveros-Palacios N et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021;49:D192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Y et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpouzas DG, Singh BK. Microbial degradation of organophosphorus xenobiotics: metabolic pathways and molecular basis. Adv Microb Physiol. 2006;51:119–85. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–7. [DOI] [PubMed] [Google Scholar]
- Kunze M, Zerlin KF, Retzlaff A et al. Degradation of chloroaromatics by Pseudomonas putida GJ31: assembled route for chlorobenzene degradation encoded by clusters on plasmid pKW1 and the chromosome. Microbiology. 2009;155:4069–83. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Marchant HK, Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–76. [DOI] [PubMed] [Google Scholar]
- Langenhoff A, Inderfurth N, Veuskens T et al. Microbial removal of the pharmaceutical compounds ibuprofen and diclofenac from wastewater. Biomed Res Int. 2013;2013:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzén A, Jørgensen SL, Huson DH et al. CREST—classification resources for environmental sequence tags. PLoS One. 2012;7:49334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinski J, Tunnacliffe A. Reduction of suspended biomass in municipal wastewater using bdelloid rotifers. Water Res. 2003;37:2027–34. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Carvalho PN, Bester K. Effects of substrate loading on co-metabolic transformation pathways and removal rates of pharmaceuticals in biofilm reactors. Sci Total Environ. 2022;853:158607. [DOI] [PubMed] [Google Scholar]
- Liang C, de Jonge N, Carvalho PN et al. Biodegradation kinetics of organic micropollutants and microbial community dynamics in a moving bed biofilm reactor. Chem Eng J. 2021;415:128963. [Google Scholar]
- Liang D, Hu Y, Liang D et al. Bioaugmentation of moving bed biofilm reactor (MBBR) with Achromobacter JL9 for enhanced sulfamethoxazole (SMX) degradation in aquaculture wastewater. Ecotoxicol Environ Saf. 2021;207:111258. [DOI] [PubMed] [Google Scholar]
- Lima-Morales D, Jáuregui R, Camarinha-Silva A et al. Linking microbial community and catabolic gene structures during the adaptation of three contaminated soils under continuous long-term pollutant stress. Appl Environ Microb. 2016;82:2227–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Wen X, Zhao F et al. Effect of temperature variation on membrane fouling and microbial community structure in membrane bioreactor. Bioresour Technol. 2013;133:462–8. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10–2. [Google Scholar]
- McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondini A, Anwar MZ, Ellegaard-Jensen L et al. Heat shock response of the active microbiome from perennial cave ice. Front Microbiol. 2022;12:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad MH, Idris AL, Fan X et al. Beyond risk: bacterial biofilms and their regulating approaches. Front Microbiol. 2020;11:928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petters S, Groß V, Söllinger A et al. The soil microbial food web revisited: predatory myxobacteria as keystone taxa?. ISME Journal. 2021;15:2665–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribera-Pi J, Badia-Fabregat M, Arias D et al. Coagulation-flocculation and moving bed biofilm reactor as pre-treatment for water recycling in the petrochemical industry. Sci Total Environ. 2020;715:136800. [DOI] [PubMed] [Google Scholar]
- Rose A, Padovan A, Christian K et al. The diversity of nitrogen-cycling microbial genes in a waste stabilization pond reveals changes over space and time that is uncoupled to changing nitrogen chemistry. Microb Ecol. 2021;81:1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq FA, Hansen MF, Burmølle M et al. Trans-kingdom interactions in mixed biofilm communities. FEMS Microbiol Rev. 2022;46:1–20. [DOI] [PubMed] [Google Scholar]
- Santos LHMLM, Araújo AN, Fachini A et al. Ecotoxicological aspects related to the presence of pharmaceuticals in the aquatic environment. J Hazard Mater. 2010;175:45–95. [DOI] [PubMed] [Google Scholar]
- Sato Y, Hori T, Koike H et al. Transcriptome analysis of activated sludge microbiomes reveals an unexpected role of minority nitrifiers in carbon metabolism. Commun Biol. 2019;2:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto LA, Hommes NG, Russell SA et al. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol Microbiol. 1996;20:541–8. [DOI] [PubMed] [Google Scholar]
- Scheel M, Zervas A, Rijkers R et al. Abrupt permafrost thaw triggers activity of copiotrophs and microbiome predators. FEMS Microbiol Ecol. 2023;99:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schostag M, Priemé A, Jacquiod S et al. Bacterial and protozoan dynamics upon thawing and freezing of an active layer permafrost soil. ISME J. 2019;13:1345–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeland A, Oetken M, Kiss A et al. Acute and chronic toxicity of benzotriazoles to aquatic organisms. Environ Sci Pollut Res. 2012;19:1781–90. [DOI] [PubMed] [Google Scholar]
- Sørensen SR, Aamand J. Rapid mineralisation of the herbicide isoproturon in soil from a previously treated Danish agricultural field. Pest Manage Sci. 2003;59:1118–24. [DOI] [PubMed] [Google Scholar]
- Su JF, Zhang YM, Liang DH et al. Performance and microbial community of an immobilized biofilm reactor (IBR)for Mn(II)-based autotrophic and mixotrophic denitrification. Bioresour Technol. 2019;286:121407. [DOI] [PubMed] [Google Scholar]
- Svendsen SB, El-taliawy H, Carvalho PN et al. Concentration dependent degradation of pharmaceuticals in WWTP effluent by biofilm reactors. Water Res. 2020;186:116389. [DOI] [PubMed] [Google Scholar]
- Terada A, Hibiya K, Nagai J et al. Nitrogen removal characteristics and biofilm analysis of a membrane-aerated biofilm reactor applicable to high-strength nitrogenous wastewater treatment. J Biosci Bioeng. 2003;95:170–8. [DOI] [PubMed] [Google Scholar]
- Teufel R, Mascaraque V, Ismail W et al. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. P Natl Acad Sci USA. 2010;107:14390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiehm A, Schmidt N, Stieber M et al. Biodegradation of phmaceutical compounds and their occurrence in the Jordan Valley. Water Resour Manage. 2011;25:1195–203. [Google Scholar]
- Tisler S, Zwiener C. Aerobic and anaerobic formation and biodegradation of guanyl urea and other transformation products of metformin. Water Res. 2019;149:130–5. [DOI] [PubMed] [Google Scholar]
- Torresi E, Fowler SJ, Polesel F et al. Biofilm thickness influences biodiversity in nitrifying MBBRs—implications on micropollutant removal. Environ Sci Technol. 2016;50:9279–88. [DOI] [PubMed] [Google Scholar]
- Tu Q, Lin L, Cheng L et al. NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics. 2019;35:1040–8. [DOI] [PubMed] [Google Scholar]
- Varet H, Brillet-Guéguen L, Coppée JY et al. SARTools: a DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS One. 2016;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xi H, Xu L et al. Ecotoxicological effects, environmental fate and risks of pharmaceutical and personal care products in the water environment: a review. Sci Total Environ. 2021;788:147819. [DOI] [PubMed] [Google Scholar]
- Wang Q, Wu S, Chu G et al. Metagenomic analysis and microbial activity shifts reveal the effect of the carbon/nitrogen ratio on the nitrogen removal performance of a moving bed biofilm reactor treating mariculture wastewater. J Water Process Eng. 2023;51:103363. [Google Scholar]
- Wickham H, Averick M, Bryan J et al. Welcome to the Tidyverse. JOSS. 2019;4:1686. [Google Scholar]
- Xue Y, Lanzén A, Jonassen I. Reconstructing ribosomal genes from large scale total RNA meta-transcriptomic data. Bioinformatics. 2020;36:3365–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Sanschagrin S, Waiser MJ et al. Sub-inhibitory concentrations of different pharmaceutical products affect the meta-transcriptome of river biofilm communities cultivated in rotating annular reactors. Environ Microbiol Rep. 2012;4:350–9. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wang G, Wu M et al. Phenol removal performance and microbial community shift during pH shock in a moving bed biofilm reactor (MBBR). J Hazard Mater. 2018;351:71–9. [DOI] [PubMed] [Google Scholar]
- Zhu L, Yuan H, Shi Z et al. Metagenomic insights into the effects of various biocarriers on moving bed biofilm reactors for municipal wastewater treatment. Sci Total Environ. 2022;813:151904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data generated using this experiment have been deposited in the NCBI SRA (Accession Number PRJNA1046284).