Abstract
The steady-state amounts of mitochondrial transcripts and transcription proteins were analyzed during mtDNA depletion and subsequent repletion to gain insight into the regulation of human mitochondrial gene expression. As documented previously, HeLa cells depleted of mtDNA via treatment with ethidium bromide (EB) were found to contain reduced steady-state levels of the mitochondrial transcription factor h-mtTFA. When partially mtDNA-depleted cells were cultured in the absence of EB, h-mtTFA recovered to normal levels at a significantly slower rate than mtDNA. Human mtRNA polymerase exhibited a similar depletion–repletion profile, suggesting that the mitochondrial transcription machinery is coordinately regulated in response to changes in mtDNA copy number. Newly synthesized mitochondrial transcripts were detected early in the recovery phase, despite the fact that mtDNA, h-mtTFA and h-mtRNA polymerase were simultaneously depleted. Although delayed relative to mtDNA, the amounts of h-mtTFA and h-mtRNA polymerase sharply increased during the later stages of the recovery phase, which was accompanied by accelerated rates of transcription and mtDNA replication. Altogether, these data indicate that when mtDNA copy number is low, it is beneficial to prevent accumulation of mitochondrial transcription proteins. In addition, h-mtTFA and h-mtRNA polymerase are either normally present in excess of the amount required for transcription or their activity is up-regulated to ensure continued expression and transcription-dependent replication of the mitochondrial genome during mtDNA-depleted states.
INTRODUCTION
The human mitochondrial genome is a 16.6 kb double-stranded circular DNA molecule that encodes 13 essential protein components of the mitochondrial oxidative phosphorylation complexes and is present in cells at 100–10 000 copies/cell (1). Mutations in mitochondrial DNA (mtDNA) cause human disease, as do mutations in nuclear genes that impact mitochondrial respiration capacity and gene expression (2,3). While important factors involved in mtDNA expression and replication in humans have been identified, how these processes are regulated remains largely undetermined. An understanding of these fundamental processes is required in order to decipher the complexities of human mitochondrial genetics and disease.
Expression and replication of mtDNA are initiated from a regulatory site in the molecule called the D-loop region that contains an origin of replication (OH) and the transcription promoters for each mtDNA strand (4). Mitochondrial transcripts are polycistronic and hence require a large number of RNA processing events to yield the mature RNA species for translation. In addition, RNA processing is required for initiation of mtDNA replication. Specifically, transcripts initiated at the Light-strand promoter (LSP) form an RNA–DNA hybrid at OH that is processed to generate the RNA primers utilized by mitochondrial DNA polymerase (pol γ) to begin DNA synthesis (5). Thus, the mitochondrial transcription machinery has a dual role in gene expression and mtDNA replication.
In humans, three proteins are known to be required for transcription initiation in mitochondria, human mitochondrial RNA (h-mtRNA) polymerase (6), the high-mobility-group box transcription factor, h-mtTFA (7,8) and the recently identified transcription factor h-mtTFB (9). Human mtTFA has unique DNA-binding properties and can activate transcription through its ability to bind upstream of mtDNA promoters (10). Based on these characteristics, mtTFA has been postulated to regulate transcription and mtDNA copy number in vivo, a supposition that is supported by several lines of evidence. First, disruption of the Tfam gene, encoding mtTFA, results in major cellular dysfunction and embryonic lethality in mice resulting from mtDNA depletion and loss of oxidative phosphorylation capacity (11,12). Second, h-mtTFA levels are responsive to the amount of mtDNA in cells. For example, it is present in low amounts in cells from patients exhibiting mtDNA depletion and in rho° cells lacking mtDNA (13–15). Third, differences in mitochondrial transcriptional activity (16) and mtDNA synthesis (17) correlate with the relative amounts of mtTFA.
In some organisms, mtTFA is a very abundant DNA-binding protein in mitochondria. For example, in Saccharomyces cerevisiae it is estimated to be present at levels capable of binding mtDNA once every 15 bp (18), a concentration predicted to be inhibitory with regard to transcription (19). Similarly, Xenopus laevis mtTFA is also found at very elevated levels during oocyte development (20). These considerations suggest that mtTFA can also serve as a DNA-packaging protein in mitochondria. However, the abundance of h-mtTFA in cultured human KB cells is estimated to be substantially lower at approximately 15 copies per genome (or approximately one molecule per every 1000 bp of mtDNA), which is a ratio predicted to be stimulatory for transcription initiation based on in vitro studies (19). The relatively low abundance of h-mtTFA may reflect optimization of its role in transcriptional regulation as opposed to a DNA-packaging function in human mitochondria.
In principle, any factor required for transcription or replication of mtDNA could be involved in regulation of mtDNA copy number (21), including mtRNA polymerase, transcription factors, the subunits of DNA pol γ and other replication proteins such as single-strand DNA-binding protein. In fact, several of these factors have been shown to influence, or be influenced by, mtDNA amount or integrity in vivo (22–24). In the current study, we examined the role of the mitochondrial transcription machinery in mtDNA regulation by analyzing expression of human mtRNA polymerase and h-mtTFA in HeLa cells under conditions of mtDNA depletion and during repopulation from a mtDNA-depleted state.
MATERIALS AND METHODS
Cell lines, growth conditions and mtDNA depletion and repletion
The HeLa Tet-On cell line used in all of the experiments was obtained from Clonetech and grown in DMEM supplemented with 10% fetal bovine serum and G418 (100 µg/ml). The HeLa rho° cell line was generated by extended treatment of these cells with ethidium bromide (EB) as described (25). The mtDNA depletion–repletion protocol used in this study is as follows. HeLa Tet-On cells were grown as described above in the presence of EB (50 ng/ml), pyruvate (100 µg/ml) and uridine (50 µg/ml). After a 6 day drug treatment cells were grown in the same medium without EB to allow repopulation of mtDNA over the next ∼12 days. This scenario was chosen because treatment with EB for >6 days resulted in extensive cell death (data not shown).
Western immunoblot analysis
Protein samples were separated by SDS–PAGE using Bio-Rad Criterion pre-cast gels (12% for h-mtTFA or 7.5% for h-mtRNA polymerase) and were then prepared for immunoblot analysis as described (26). The polyclonal h-mtRNA polymerase antibody (raised against a synthesized peptide N-VNLEPSDVPQDVY-C) was obtained from Multiple Peptide Systems. The h-mtTFA antibody was obtained from Dr David Clayton (Stanford University) and the tubulin antibody was purchased from Neomarkers. Signals were generated using Pierce Supersignal Reagent and quantified using a Bio-Rad Flour S Max Bioimager. A range of protein samples was routinely analyzed to ensure the resulting signals were in a linear detection range (data not shown). The signals from h-mtRNA polymerase and h-mtTFA were subsequently normalized to those of tubulin.
Quantitative Southern analysis of mtDNA
Total DNA from HeLa cell samples was isolated using the DNeasy Tissue kit (Qiagen). DNA samples (5 µg) were digested to completion with NcoI and separated by electrophoresis through a 0.8% agarose gel. DNA was transferred to a nylon membrane as described (26) and hybridized with the indicated radiolabeled DNA probes using rapid-hyb buffer (Amersham) as described by the manufacturer. The ND2 (mtDNA) and 28S rDNA (nuclear DNA) probes were made by random-primed labeling using Ready-to-Go DNA labeling beads (Amersham) and [α-32P]dCTP (New England Nuclear) as described by the manufacturer. PCR products corresponding to portions of the ND2 and 28S rDNA genes were cloned into pGEMT (Promega) and used as templates for the labeling reactions. Primers for synthesis of the ND2 probe template were 5′-GGCCCAACCCGTCATCTAC-3′ and 5′-GAGTGTGGGGAGGAATGGGG-3′ and for the 28S rDNA probe template were 5′-GCCTAGCAGCCGACTTAGAACTGG-3′ and 5′-GGCCTTTCATTATTCTACACCTC-3′.
The ND2 probe hybridized to a 4.2 kb mtDNA restriction fragment and the 28S rDNA hybridized to a 2.6 kb nuclear DNA fragment. The same membranes were probed first for ND2, then stripped and probed with the 28S rDNA probe. Quantitative data were obtained by analyzing multiple DNA sample concentrations to ensure the resulting signals were in a linear detection range. Signals were quantified using a FUJIX Bas 1000 phosphoimager. The ND2 signal was normalized to the 28S rDNA signal and the data plotted in Figure 1C are expressed relative to this ratio obtained in cells prior to treatment with EB. The data presented are from three independent experiments.
Figure 1.
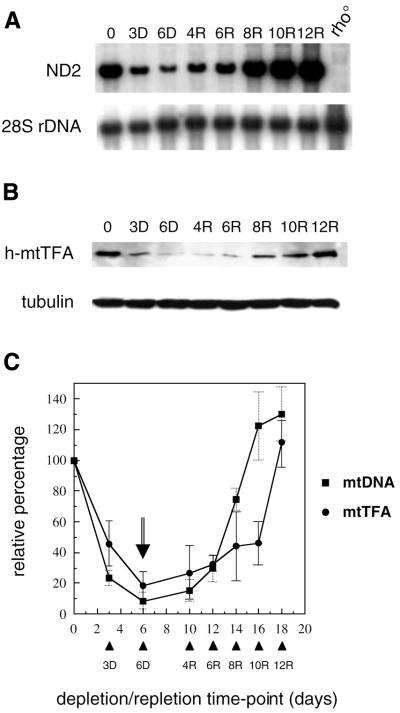
Delayed accumulation of h-mtTFA after mtDNA depletion in HeLa cells. (A) Southern analysis of nuclear DNA (28S rDNA gene) and mtDNA (ND2 gene) from cells partially depleted and then repopulated with mtDNA. Shown is the analysis of total DNA samples from HeLa Tet-On cells prior to treatment with EB (time 0); cells depleted of mtDNA with EB for 3 (3D) and 6 days (6D); and cells subsequently grown in the absence of EB (mtDNA recovery) for 4 (4R), 6 (6R), 8 (8R), 10 (10R) and 12 days (12R). Also shown is total DNA from a corresponding rho° cell line that is devoid of mtDNA. (B) Immunoblot analysis of h-mtTFA and tubulin in total protein (20 µg) from the samples described in (A) above. (C) Quantitative analysis of data from (A) and (B) above (see Materials and Methods). Plotted on the ordinate is the relative percentage (amount relative to time 0) of h-mtTFA (circles) or mtDNA (squares) at each time point during the depletion–repletion experiment [numbers directly below the graph indicate the total time of the experiment in days; specific time points in the depletion–repletion experiment are indicated by arrowheads and labeled as in (A) above]. The point in the experiment when cells were removed from EB is indicated by the arrow. Error bars (solid, h-mtTFA; dashed, mtDNA) indicate ± one standard deviation from the mean of three independent experiments.
RNA analysis
Total RNA was extracted from HeLa cells using the RNeasy kit (Qiagen) after treatment with RNAlater (Ambion) and quantitated by measuring the OD260 of the samples. Northern analysis was performed as described (27) using total HeLa cell RNA (10 µg) and randomly 32P-labeled probes generated from PCR products corresponding to the mitochondrial ND2 or ND6 genes and the nuclear β-actin gene using Ready-To-Go labeling beads (Amersham/Pharmacia). RNase protection assays were performed as described (26) using total HeLa cell RNA (10 µg). The RNA probe corresponded to a 534 nt RNA transcript that overlapped a 5′-region of human mtRNA polymerase mRNA. After hybridization and RNase treatment, a protected fragment of 448 nt remained, which was analyzed by denaturing PAGE and autoradiography. A range of RNA concentrations were analyzed to ensure that the conditions used in this assay were linear over ≥10-fold range (data not shown). EB staining revealed that the amounts of total cell RNA loaded in each lane did not vary by >2-fold.
RESULTS
Delayed accumulation of h-mtTFA during recovery of HeLa cells from a mtDNA-depleted state
Treatment of cultured vertebrate cells with low amounts of EB results in rapid and selective depletion of mtDNA (28). We established a protocol that allowed depletion of mtDNA in HeLa cells after growth in medium containing EB for 6 days, followed by subsequent growth in the absence of drug to allow repopulation of the cells with mtDNA. Under these conditions, mtDNA was depleted to ∼10% of normal after 6 days of growth in EB (Fig. 1). When these cells were subsequently cultured in the absence of drug, mtDNA was fully repopulated after 10–12 days, but exhibited a 4–6 day lag phase before rapid mtDNA replication was observed (Fig. 1).
Using this mtDNA depletion–repletion protocol, we analyzed the expression of h-mtTFA in response to changes in mtDNA copy number. During depletion, the amount of h-mtTFA decreased in concert with mtDNA (Fig. 1B) and remained at low levels during the lag phase before mtDNA replication was induced. However, in the later stages of the mtDNA recovery phase, h-mtTFA was re-established toward normal levels at a rate significantly slower than that of mtDNA. This relative delay was most obvious at day 16 of the experiment (10 days of recovery, 10R), at which point mtDNA was fully re-established while h-mtTFA remained at ∼40% of normal (Fig. 1C). Over the next 2 days the steady-state amounts of h-mtTFA were re-established more quickly and eventually reached normal levels.
Reduced amounts of h-mtRNA polymerase in mtDNA-depleted and rho° HeLa cells
Using an antibody raised against a h-mtRNA polymerase peptide (see Materials and Methods), we were able to detect a protein of ∼140 kDa in total cell extracts prepared from HeLa cells by western immunoblot analysis (Fig. 2A, lane 1). We observed a specific increase in cross reactivity to the 140 kDa species in whole cell extracts from cells that were transiently transfected with a human mtRNA polymerase over-expression plasmid (Fig. 2A, lane 2) or in enriched mitochondrial fractions from non-transfected cells (Fig. 2A, lane 3), confirming that the antibody recognizes h-mtRNA polymerase.
Figure 2.
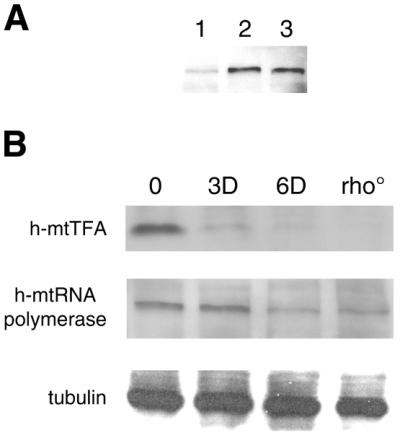
Human mtRNA polymerase is down-regulated in partially mtDNA-depleted and rho° HeLa cells. (A) Detection of human mtRNA polymerase in HeLa cell extracts by immunoblot analysis of the following protein samples (10 µg each): total protein from HeLa Tet-On cells (lane 1); total protein from cells transiently transfected with a human mtRNA polymerase over-expression plasmid (lane 2); protein from a mitochondria-enriched fraction from non-transfected cells (lane 3). (B) Immunoblot analysis of rho° HeLa cells and mtDNA-containing (rho+) HeLa cells prior to EB treatment (0) and after 3 (3D) and 6 days (6D) of drug treatment. Blots were probed using antibodies against h-mtRNA polymerase, h-mtTFA and tubulin (normalization control).
Next, whole cell extracts from parental, mtDNA-containing (rho+) HeLa cells before and after 3 or 6 days of EB treatment were probed for h-mtRNA polymerase by western immunoblot analysis, as was a corresponding rho° cell line. Similar to h-mtTFA, the steady-state amount of h-mtRNA polymerase was reduced in cells depleted of mtDNA by the EB treatment and in the corresponding rho° cell line (Fig. 2B). However, the depletion of h-mtRNA polymerase occurs more slowly and to a lesser degree than that observed with h-mtTFA. For example, h-mtTFA was significantly reduced at 3 and 6 days of EB treatment, while the levels of h-mtRNA polymerase were not significantly reduced until 6 days of EB treatment. Likewise, h-mtTFA was barely detectable in the rho° cells as reported previously (13), while h-mtRNA polymerase was present at significant but reduced levels (∼70% of normal, based on quantitative western analysis; see Materials and Methods). Though we were unable to reliably measure the amounts of h-mtRNA polymerase in EB-treated cell lines (due to a low signal-to-noise ratio with this antibody), it is clear that it was decreased to an even greater extent in the 6-day EB-treated cells than it was in rho° cells (Fig. 2B).
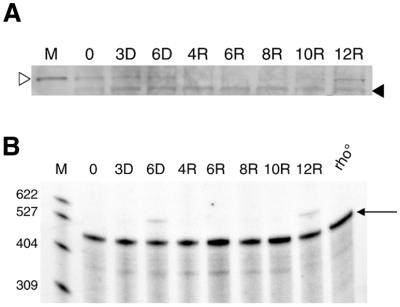
We next examined the dynamics of h-mtRNA polymerase during our mtDNA depletion–repletion protocol. Similar to h-mtTFA, the levels of h-mtRNA polymerase remained well below normal levels until the latter stages of the recovery period after mtDNA was fully repopulated (Fig. 3A). Finally, the amounts of h-mtRNA polymerase mRNA were examined during this experiment to determine if transcription of its gene in the nucleus was responsive to changes in mtDNA copy number. The steady-state amounts of h-mtRNA polymerase mRNA were found to be largely unchanged during the course of the experiment (Fig. 3B). Similar results were obtained by Moraes et al. (29) with regard to h-mtTFA and h-mtRNA polymerase mRNA levels in mtDNA-depleted human cells lines; however, our results extend this analysis to include time points during the recovery phase.
Figure 3.
Dynamics of mtRNA polymerase during mtDNA depletion–repletion in HeLa cells. (A) Western immunoblot analysis of h-mtRNA polymerase during a typical mtDNA depletion–repletion experiment. A control lane (labeled M) is included in the upper panel. This lane contains a mitochondria-enriched HeLa cell fraction, demonstrating that the band indicated by the open arrowhead is mtRNA polymerase. A cross-reacting, non-mitochondrial protein (indicated by a filled arrowhead) that is not affected by EB treatment serves as a convenient estimate of the amount of protein loaded in each lane. (B) Results of RNase protection analysis of h-mtRNA polymerase mRNA during mtDNA depletion–repletion are shown. Radiolabeled DNA size markers (M) were run with the samples and the numbers to the left of the figure indicate their length in nucleotides. The 448 nt signal (indicated with an arrow) represents the protected h-mtRNA polymerase mRNA fragment. Time points during mtDNA depletion–repletion are indicated in (A) and (B) in the same manner as in Figure 1.
Newly synthesized mitochondrial transcripts appear prior to up-regulation of mitochondrial transcription proteins and accumulate in concert with mtDNA
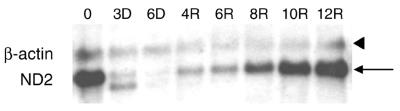
Because h-mtRNA polymerase and h-mtTFA are down-regulated in response to mtDNA depletion and are not up-regulated until relatively late in the recovery period, we analyzed mitochondrial transcription under these same circumstances. Northern analysis of total RNA using an ND2 probe (Fig. 4) revealed that mitochondrial transcription was substantially inhibited in the presence of EB. However, by 4 days of recovery in the absence of EB, significant amounts of newly synthesized mature ND2 transcripts were observed, despite the fact that mtDNA was still significantly depleted (∼10% of normal) and h-mtRNA polymerase and h-mtTFA were at their lowest concentration in the experiment (Figs 1 and 3). After 6 days of recovery, a burst of mitochondrial transcription was observed (Fig. 4) that coincided with faster rates of mtDNA replication (Fig. 1). Virtually identical results were obtained when a mitochondrial ND6 probe was used to detect mitochondrial L-strand transcripts (data not shown).
Figure 4.
Mitochondrial transcription in mtDNA-depleted cells. Northern analysis of mtDNA-encoded ND2 mRNA (indicated with an arrow) and nucleus-encoded β-actin mRNA (indicated with an arrowhead) during mtDNA depletion–repletion. A second faster-migrating RNA species (presumably a truncated ND2 transcript) was always observed in the presence of EB (most obvious in lane 3D). Time points during mtDNA depletion–repletion are indicated in the same manner as in Figure 1.
DISCUSSION
We analyzed the expression of h-mtRNA polymerase and h-mtTFA under conditions of mtDNA depletion and repopulation to gain insight into the regulation of mitochondrial transcription and mtDNA replication in human cells. Since its original documentation (13–15), whether down-regulation of h-mtTFA in mtDNA depleted cells occurs at the level of gene expression or is the result of passive instability of the protein when it is not bound to mtDNA (21) has remained unclear. Our results have shed light on this issue by demonstrating that h-mtRNA polymerase exhibits a similar fate as h-mtTFA in rho° cells and during mtDNA depletion (Fig. 2), suggesting that these two transcription proteins are coordinately down-regulated when mtDNA copy number is reduced. It is worth noting that down-regulation is not a general phenomenon exhibited by all mitochondrial regulatory factors in mtDNA-depleted cells. For example, accumulation of human mtDNA polymerase (pol γ) is unaffected by mtDNA depletion (14). While the fact that both h-mtRNA polymerase and h-mtTFA exhibit this response does not eliminate passive protein instability as an explanation, it does appear to favor the idea that cells have evolved regulatory mechanisms to coordinately down-regulate transcription proteins when mtDNA copy number is below normal. If regulation is indeed at the level of gene expression, our results with mtRNA polymerase (Fig. 3B), as well as those of Moraes et al. (29), indicate that it is most likely at the post-transcriptional level.
The reason for this coordinate regulation (or instability) of mitochondrial transcription proteins is not known, but one possibility is that as mtDNA levels decline, it is necessary to prevent accumulation of these proteins in order to maintain the remaining genomes in a transcription and replication competent state. For example, it has been shown that, when in excess, h-mtTFA binds and saturates the DNA template, resulting in complete inhibition of transcription from the mitochondrial LSP in vitro (19). Thus, if h-mtTFA were not down-regulated during mtDNA depletion, in principle, the remaining mtDNA templates could become subject to this type of transcriptional inhibition. However, under this scenario, it is not clear why h-mtRNA polymerase would likewise be down-regulated. Another possibility, that might better account for the need to down-regulate h-mtRNA polymerase as well as h-mtTFA, is that these proteins may bind to other mitochondrial factors as a means to couple them to transcription or to the mtDNA template. Thus, coordinately decreasing their accumulation during mtDNA depletion, and preventing their subsequent accumulation until after mtDNA copy number is largely re-established (Figs 1–3), may prevent such interactions from occurring in a mtDNA-unbound state, a situation that could potentially impair mitochondrial gene expression. Perhaps consistent with this idea is the finding that Nam1p binds the N-terminal domain of mtRNA polymerase in yeast as a means to couple post-transcriptional gene expression events to transcription (27). In all likelihood, it is probably a combination of the above factors, and perhaps others, that have led cells to evolve mechanisms to coordinately down-regulate mitochondrial transcription proteins in response to mtDNA depletion.
Analysis of the relative kinetics of h-mtTFA and mtRNA polymerase, mtDNA and mitochondrial transcript accumulation also revealed significant new insights into the control of mitochondrial transcription and replication. First, at the earliest recovery time-point analyzed after partial mtDNA depletion, significant amounts of newly synthesized mitochondrial transcripts are present (Fig. 4), while the amounts of mtDNA, h-mtRNA polymerase and h-mtTFA remain at very low levels (Figs 1 and 3). Therefore, even when mtDNA and these two key transcription proteins are present at seemingly limiting amounts, a significant amount of transcription still occurs.
Two potential explanations for this result are worthy of discussion. The first is that h-mtTFA and h-mtRNA polymerase are normally present in excess of the amount needed for transcription. The degree to which h-mtTFA and h-mtRNA polymerase are in excess for transcription in vivo is not known. However, with regard to h-mtTFA, data from in vitro studies may provide some insight. For example, the amount of h-mtTFA in human KB cells has been estimated at approximately 15 molecules per mtDNA genome (30). At this h-mtTFA/DNA ratio, significant, albeit not maximal, levels of transcription occur from the mitochondrial LSP in vitro (19). In addition, a >10-fold drop in this ratio results in a significant reduction, if not complete lack, of transcription initiation capacity in vitro (19). Based on these considerations, a conservative estimate is that at approximately 15 molecules per genome, h-mtTFA is present in vivo at levels that are in ∼10-fold excess of that required for substantial amounts of transcription initiation from the LSP. Therefore, it is a formal possibility that a small pool of h-mtTFA and h-mtRNA polymerase that remains after mtDNA depletion is adequate to allow significant transcription to occur from the remaining mtDNA templates. In fact, down-regulation of h-mtTFA and h-mtRNA polymerase during depletion, as well as the observed lag in the accumulation of these proteins behind mtDNA during repletion (Figs 1–3), may provide a mechanism to ensure that optimal protein/mtDNA ratios for transcription are maintained. Alternatively, it is possible that compensatory mechanisms exist to enhance transcriptional efficiency when the amounts of h-mtTFA and h-mtRNA polymerase are limiting. For example, enhanced transcription efficiency due to post-translational modification of transcription proteins or the influence of other regulators of mitochondrial transcription, such as the recently identified transcription factor h-mtTFB (9), are formal possibilities that remain largely unexplored in human cells.
Regardless of how, the fact that transcription continues even in the presence of very low amounts of h-mtTFA and h-mtRNA polymerase ensures that mitochondrial gene expression and the synthesis of RNA primers for mtDNA replication continues under conditions of mtDNA depletion. This situation likely accounts for the normal amounts of mitochondrial transcripts observed in muscle cells from patients with severe mtDNA depletion (31) and, likewise, may explain the results of Goto et al. (32), who observed normal mitochondrial transcription in cultured Drosophila cells despite severe reductions of h-mtTFA and mtDNA depletion caused by RNAi interference of mtTFA expression.
Finally, the fact that the appearance of newly synthesized mitochondrial transcripts after mtDNA depletion immediately precedes, and then closely mirrors, re-establishment of mtDNA copy number (Fig. 4) suggests an intimate association between transcription rate and mtDNA replication capacity. This is substantiated further by the fact that the portion of the recovery phase where mtDNA accumulates at the fastest rate (Fig. 1) also corresponds to when accumulation of h-mtTFA and h-mtRNA polymerase is most dramatic (Figs 1 and 3). Also consistent with this notion is the ability of mtTFA to stimulate 7S DNA synthesis in isolated rat mitochondria (17). Altogether, these data strongly support a transcription-primed mtDNA replication mechanism (4) operating during mtDNA repletion after transient depletion in HeLa cells.
In summary, the results of this study indicate that there is a dynamic relationship that exists between mtDNA copy number and the accumulation of two known factors that are required for transcription initiation in human mitochondria, h-mtTFA and h-mtRNA polymerase. These two proteins are coordinately regulated during mtDNA depletion and repletion, suggesting that cells strive to maintain an optimal ratio of transcription complexes/mtDNA that ensures that transcription and mtDNA replication can occur over a range of mtDNA concentrations. Understanding the precise mechanisms underlying these regulatory phenomena will help unravel the complexities of human mitochondrial genetics and disease.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Matt Rodeheffer for critically evaluating the manuscript. This work was supported by a grant (HL-59655) from the NIH awarded to G.S.S.
REFERENCES
- 1.Shadel G.S. (1999) Yeast as a model for human mtDNA replication. Am. J. Hum. Genet., 65, 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schon E.A. (2000) Mitochondrial genetics and disease. Trends Biochem. Sci., 25, 555–560. [DOI] [PubMed] [Google Scholar]
- 3.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 4.Shadel G.S. and Clayton,D.A. (1997) Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem., 66, 409–435. [DOI] [PubMed] [Google Scholar]
- 5.Lee D.Y. and Clayton,D.A. (1998) Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem., 273, 30614–30621. [DOI] [PubMed] [Google Scholar]
- 6.Tiranti V., Savoia,A., Forti,F., D’Apolito,M.F., Centra,M., Rocchi,M. and Zeviani,M. (1997) Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum. Mol. Genet., 6, 615–625. [DOI] [PubMed] [Google Scholar]
- 7.Fisher R.P. and Clayton,D.A. (1988) Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol., 8, 3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisi M.A. and Clayton,D.A. (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science, 252, 965–969. [DOI] [PubMed] [Google Scholar]
- 9.McCulloch V., Seidel-Rogol,B.L. and Shadel,G.S. (2002) A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol., 22, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher R.P., Lisowsky,T., Parisi,M.A. and Clayton,D.A. (1992) DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem., 267, 3358–3367. [PubMed] [Google Scholar]
- 11.Larsson N.G., Wang,J., Wilhelmsson,H., Oldfors,A., Rustin,P., Lewandoski,M., Barsh,G.S. and Clayton,D.A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genet., 18, 231–236. [DOI] [PubMed] [Google Scholar]
- 12.Silva J.P., Kohler,M., Graff,C., Oldfors,A., Magnuson,M.A., Berggren,P.O. and Larsson,N.G. (2000) Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nature Genet., 26, 336–340. [DOI] [PubMed] [Google Scholar]
- 13.Larsson N.G., Oldfors,A., Holme,E. and Clayton,D.A. (1994) Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem. Biophys. Res. Commun., 200, 1374–1381. [DOI] [PubMed] [Google Scholar]
- 14.Davis A.F., Ropp,P.A., Clayton,D.A. and Copeland,W.C. (1996) Mitochondrial DNA polymerase gamma is expressed and translated in the absence of mitochondrial DNA maintenance and replication. Nucleic Acids Res., 24, 2753–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulton J., Morten,K., Freeman-Emmerson,C., Potter,C., Sewry,C., Dubowitz,V., Kidd,H., Stephenson,J., Whitehouse,W., Hansen,F.J. et al. (1994) Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum. Mol. Genet., 3, 1763–1769. [DOI] [PubMed] [Google Scholar]
- 16.Montoya J., Perez-Martos,A., Garstka,H.L. and Wiesner,R.J. (1997) Regulation of mitochondrial transcription by mitochondrial transcription factor A. Mol. Cell. Biochem., 174, 227–230. [PubMed] [Google Scholar]
- 17.Gensler S., Weber,K., Schmitt,W.E., Perez-Martos,A., Enriquez,J.A., Montoya,J. and Weisner,R.J. (2001) Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res., 29, 3657–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diffley J.F. and Stillman,B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dairaghi D.J., Shadel,G.S. and Clayton,D.A. (1995) Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim. Biophys. Acta, 1271, 127–134. [DOI] [PubMed] [Google Scholar]
- 20.Shen E.L. and Bogenhagen,D.F. (2001) Developmentally-regulated packaging of mitochondrial DNA by the HMG-box protein mtTFA during Xenopus oogenesis. Nucleic Acids Res., 29, 2822–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moraes C.T. (2001) What regulates mitochondrial DNA copy number in animal cells? Trends Genet., 17, 199–205. [DOI] [PubMed] [Google Scholar]
- 22.Lefai E., Calleja,M., Ruiz de Mena,I., Lagina,A.T.,III, Kaguni,L.S. and Garesse,R. (2000) Overexpression of the catalytic subunit of DNA polymerase gamma results in depletion of mitochondrial DNA in Drosophila melanogaster. Mol. Gen. Genet., 264, 37–46. [DOI] [PubMed] [Google Scholar]
- 23.Maier D., Farr,C.L., Poeck,B., Alahari,A., Vogel,M., Fisher,S., Kaguni,L.S. and Schneuwly,S. (2001) Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol. Biol. Cell, 12, 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultz R.A., Swoap,S.J., McDaniel,L.D., Zhang,B., Koon,E.C., Garry,D.J., Li,K. and Williams,R.S. (1998) Differential expression of mitochondrial DNA replication factors in mammalian tissues. J. Biol. Chem., 273, 3447–3451. [DOI] [PubMed] [Google Scholar]
- 25.King M.P. and Attardi,G. (1996) Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol., 264, 304–313. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J., Fritsch,E.P. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Rodeheffer M.S., Boone,B.E., Bryan,A.C. and Shadel,G.S. (2001) Nam1p, a protein involved in RNA processing and translation, is coupled to transcription through an interaction with yeast mitochondrial RNA polymerase. J. Biol. Chem., 276, 8616–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dejardins P., Frost,E. and Morais,R. (1985) Ethidium bromide-induced loss of mitochondrial DNA from primary chicken embyro fibroblasts. Mol. Cell. Biol., 5, 1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moraes C.T., Kenyon,L. and Hao,H. (1999) Mechanisms of human mitochondrial DNA maintenance: the determining role of primary sequence and length over function. Mol. Biol. Cell, 10, 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher R.P., Lisowsky,T., Breen,G.A. and Clayton,D.A. (1991) A rapid, efficient method for purifying DNA-binding proteins. Denaturation-renaturation chromatography of human and yeast mitochondrial extracts. J. Biol. Chem., 266, 9153–9160. [PubMed] [Google Scholar]
- 31.Barthelemy C., de Baulny,H.O., Diaz,J., Cheval,M.A., Franchon,P., Romero,N., Goutieres,F., Fardeau,M. and Lombes,A. (2001) Late-onset mitochondrial DNA depletion: DNA copy number, multiple deletions, and compensation. Ann. Neurol., 49, 607–617. [PubMed] [Google Scholar]
- 32.Goto A., Matsushima,Y., Kadowaki,T. and Kitagawa,Y. (2001) Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem. J., 354, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]