Abstract
Background
The mortality risk of co-infections/secondary infections (CoI/ScI) is under-reported in patients with non-critical COVID-19, leading to the under-management of CoI/ScI and publication bias in the medical literature. We aimed to investigate the association between CoI/ScI and mortality in patients hospitalised with mild-to-severe COVID-19.
Methods
We conducted a retrospective cohort study at a COVID-19 treatment hospital in Vietnam and collected all eligible medical records, with CoI/ScI status as the exposure (non-CoI/ScI and CoI/ScI, with the latter including nature of pathogen [bacterial, fungal, or bacterial + fungal] and multidrug-resistance pathogen [no MDRp or ≥ 1 MDRp]). The outcome was all-cause mortality, defined as in-hospital death by all causes or being discharged under critical illness. We used time-dependent analysis to report rates of mortality with 95% confidence intervals (95% CI, Poisson regression) and hazard ratios (HR) with 95% CI (Cox proportional hazards regression with Holm’s method for multiplicity control).
Results
We followed 1466 patients (median age 61, 56.4% being female) for a median of 9 days. We recorded 387 (26.4%) deaths (95/144 [66.0%] in the CoI/ScI group and 292/1322 [22.1%] in the non-CoI/ScI group). Adjusted mortality rates (per 100 person-days) of the CoI/ScI (6.4, 95% CI 5.3 to 7.8), including bacterial (8.0, 95% CI 7.2 to 8.9), no MDRp (5.9, 95% CI 4.8 to 7.4), and ≥ 1 MDRp (9.0, 95% CI 8.2 to 10.0) groups were higher than that of the non-CoI/ScI group (2.0, 95% CI 1.8 to 2.2). These corresponded to higher risks of mortality in the overall CoI/ScI (HR 3.27, 95% CI 2.58 to 4.13, adjusted p < 0.001), bacterial CoI/ScI (HR 3.79, 95% CI 2.97 to 4.83, adjusted p < 0.001), no MDRp CoI/ScI (HR 3.13, 95% CI 2.42 to 4.05, adjusted p < 0.001), and ≥ 1 MDRp CoI/ScI group (HR 3.89, 95% CI 2.44 to 6.21, adjusted p < 0.001). We could not attain reliable estimates for fungal and bacterial + fungal CoI/ScI.
Conclusion
Compared with the non-CoI/ScI group, patients with CoI/ScI had a significantly higher risk of all-cause mortality, regardless of resistance status. More evidence is needed to confirm the mortality risks in patients with fungal or bacterial + fungal CoI/ScI.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-09866-0.
Keywords: Co-infection, Secondary infection, COVID-19, Mortality, Vietnam
Background
Co-infections and secondary infections (CoI/ScI) have emerged as important concerns in patients with Coronavirus Disease 2019 (COVID-19) [1]. CoI/ScI in COVID-19 can manifest in various ways, including bacterial pneumonia, bloodstream infections, urinary tract infections, or ventilator-associated infections. These conditions not only complicate the clinical management of COVID-19 but also contribute to increased morbidity, mortality, and prolonged hospital stays [2–8]. The consequences can be even more tremendous due to the increasing prevalence of drug-resistant pathogens [9].
However, current evidence primarily focuses on critically ill patients [3–8]. Thus, the risks in non-critical patients are still under-reported, leading to an incomplete awareness of CoI/ScI in COVID-19. A possible explanation behind this phenomenon is the insignificant association between CoI/ScI and poor outcomes in less severe sub-populations, which caused a publication bias during evidence synthesis. The main issue that many studies encountered was a delay between the time patients entered the study and the time they were assigned to exposed or unexposed groups. Bias would be likely if this delay was not properly analysed [10], which could under- or overestimate the effects of CoI/ScI on patient outcomes. To address this evidence gap, we aimed to investigate whether CoI/ScI were associated with poorer outcomes in patients hospitalised with mild-to-severe COVID-19.
Methods
Design and setting
Nhan Dan Gia Dinh (NDGD) Hospital is a tertiary hospital in Vietnam that served as a COVID-19 treatment facility during the 2021 outbreaks in Ho Chi Minh City, Vietnam. A retrospective cohort study was conducted at the main facility of NDGD Hospital, collecting medical records of hospitalised patients from July to October of 2021. We reported our study following the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement (Supplementary Checklist, available in the Supplementary File).
We screened all medical records of patients with COVID-19 in the study timeframe for eligibility. Records were selected if the patients: (1) were ≥ 18 years old; (2) were hospitalised with COVID-19; and (3) had mild-to-severe illness (classified by the clinical spectrum of the National Institute of Health [11]). During the study timeframe, COVID-19 hospitalisation was considered for patients with moderate-to-severe COVID-19 or patients with mild COVID-19 and other reasons requiring hospital-level care. We excluded records of patients who: (1) were pregnant or breastfeeding; (2) were moderately or severely immunocompromised (immunosuppressive medications, moderate or severe primary immunodeficiency, advanced or untreated human immunodeficiency virus infection, active cancer treatment, or white blood cell (WBC) count < 4 × 109/L); (3) had chronic kidney disease stage 4–5; (4) had hepatic impairment (Child–Pugh class B or C); or (5) were discharged or transferred within 3 days of hospitalisation.
Exposures
The primary exposure was presence of CoI/ScI. Based on the clinical symptoms and organ-specific microbial cultures, patients with COVID-19 were classified as having (1) CoI/ScI or (2) no CoI/ScI (non-CoI/ScI). As COVID-19 outbreaks caused severe shortages of medical equipment in Vietnam, we only focused on the following types of specimens: blood, peritoneal fluid, sputum, urine, and wound drainage. If contaminated cultures were suspected, another sample would be ordered to detect the pathogens. For patients with persistently contaminated or negative microbial cultures, at least 3 positive clinical symptoms/signs for ≥ 24 h ((1) fever; (2) increased WBC; (3) increased C-reactive protein; (4) increased procalcitonin; and (5) organ-specific manifestations) were required to confirm CoI/ScI diagnosis. For patients with negative clinical symptoms/signs but positive microbial cultures, another valid positive result at the same site would imply CoI/ScI. Due to the limitations in laboratory services, we only focused on bacterial and fungal CoI/ScI.
We also investigated 2 CoI/ScI-based secondary exposures, which were type of CoI/ScI and resistance status. Type of CoI/ScI was classified based on the nature of causative pathogens (bacterial, fungal, or bacterial plus fungal). Resistance status was classified as having (1) no multidrug-resistance (MDR) pathogen (MDRp) or (2) ≥ 1 MDRp. The definition of MDR was issued by the European Centre for Disease Prevention and Control [12], which was non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories available for testing. The reference for these secondary exposures was the non-CoI/ScI group.
The in vitro antimicrobial susceptibility testing for MDR detection was conducted using the automated VITEK 2 system (bioMérieux Inc., France). Results were reported as minimal inhibitory concentration (MIC) for each tested antimicrobial agent. Breakpoints for susceptibility status were based on the M100 document from the Clinical Laboratory Standards Institute [13]. Specifically, for Acinetobacter baumannii complex, susceptibility testing of colistin (or polymyxin E) could not be conducted due to the lack of broth microdilution panels amid COVID-19 outbreaks.
Outcome
The outcome of interest was all-cause mortality. This was defined as in-hospital mortality or being discharged under critical illness, obtained by screening patient medical records. We analysed this as a survival endpoint. Survival time was calculated from admission (for non-CoI/ScI group) or secondary infection (for CoI/ScI group) to mortality date or censored by the end of follow-up. We followed patients from 1 July 2021 to 31 October 2021.
Covariates
We included the following covariates as potential confounders: age (years), sex (female/male), body mass index (BMI, kg/m2), comorbidities (yes/no), and disease severity (mild/moderate/severe). We decided, a priori, that some factors, such as biological markers, were not adjusted to avoid overadjustment bias [14].
Statistical analysis
We screened all medical records (n = 1923) and included all eligible ones for analysis (n = 1466). Records with missing data (exposure, covariates, or outcome) were excluded from the analysis. We used descriptive analysis to summarise the cohort characteristics, where median (interquartile range, IQR) or frequency (percentage) were presented as appropriate. For most patients, we only observed CoI/ScI after they had been followed. This created an immortal time where no mortality event would be observed in the CoI/ScI group. If this immortal time was misclassified or excluded in the analysis, bias would be introduced into the study [10]. Thus, we used the time-dependent approach as the primary analysis to clarify and mitigate this immortal time bias [15], in addition to conducting an exploratory time-fixed analysis.
Rates of all-cause mortality per 100 person-days with 95% confidence intervals (95% CI) were calculated using Poisson regression. We reported the effects of the exposure on time-to-mortality as hazard ratios (HR, from Cox proportional hazards regression) with the 95% CI. The proportional hazard assumption of Cox proportional hazards regression was checked using Schoenfeld residuals. This assumption was satisfied in all multivariable time-dependent models. Kaplan–Meier curves (in time-fixed analysis) and Simon–Makuch curves (in time-dependent analysis) were also provided to visualise the unadjusted mortality risk over time. To avoid inflated type I error rate, we used the Holm method to control for multiplicity in the time-dependent analysis [16].
Analyses were conducted with R (version 4.2.1, R Foundation for Statistical Computing, Vienna, Austria). We used the survival package for Cox proportional hazards regression and glm function for Poisson regression. All statistical hypotheses were tested with a family-wise error rate of 5% (Holm correction).
Ethics approval
The study was approved by the Ethics Committee of NDGD Hospital, under approval number 61–2021/NDGD-HDDD. As only relevant information was collected without revealing the patient’s identity, patient consent was not required, as declared by the Ethics Committee.
Results
Patient and microbial characteristics
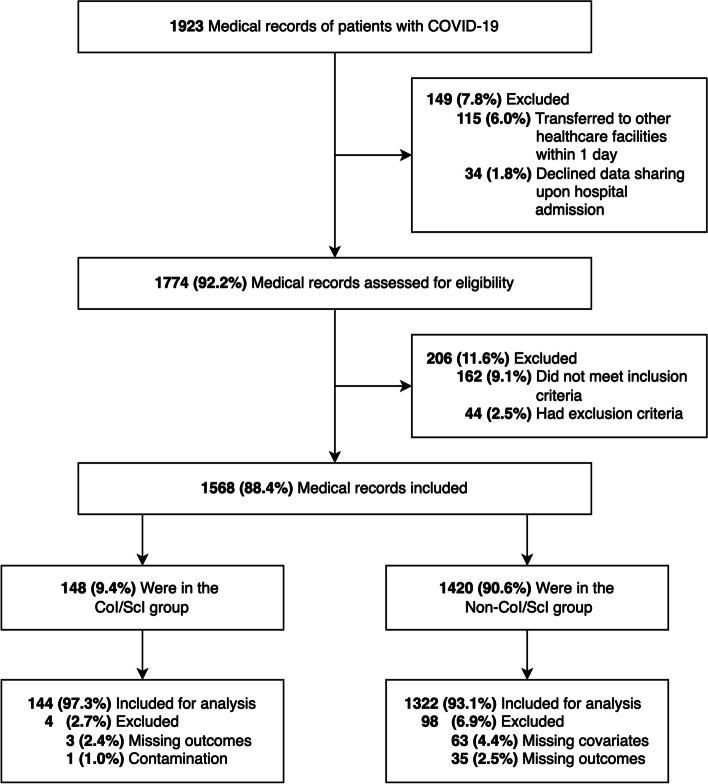
Patient characteristics of the included records (144 in the CoI/ScI group, 1322 in the non-CoI/ScI group, Fig. 1) were comparable to that of the excluded group, except for disease severity (Table S1, Supplementary File). Among 1466 patients (median age 61 (IQR 48–72), 56.4% being female), the proportion of obesity (BMI ≥ 25 kg/m2 [17]) was 25.1%. Nearly half of the study sample reported having chronic comorbidities (38.7% with type 2 diabetes, 35.3% with hypertension). Over 20.0% of the patients had severe illness at hospital admission. Corticosteroids (dexamethasone or methylprednisolone) were given to all patients with moderate-to-severe COVID-19. No patients received antimicrobial agents within 1 month prior to hospitalisation. Baseline details of the CoI/ScI and non-CoI/ScI groups are summarised in Table 1.
Fig. 1.
Flowchart of the study. Abbreviations: CoI/ScI: co-infection/secondary infection; COVID-19: Coronavirus Disease 2019. Note: We could not get access to data of patients who were transferred to other healthcare facilities within 1 day or who declined data sharing for research purposes upon hospital admission. There was a concern about contamination in 1 patient, where 2 consecutive cultures were suspected to be contaminated. Thus, we excluded this case, as well as other records with missing data (covariates or outcomes), from analysis
Table 1.
Baseline characteristics of the study sample
| Characteristics | CoI/ScIa |
Non-CoI/ScI (n = 1322) |
Overall (n = 1466) |
|||||
|---|---|---|---|---|---|---|---|---|
| Type of CoI/ScI | Resistant status |
Total (n = 144) |
||||||
|
Bacterial (n = 129) |
Fungal (n = 2) |
Bacterial + fungal (n = 13) |
No MDRpb (n = 112) |
≥ 1 MDRpb (n = 32) |
||||
| Sexc | ||||||||
| Female | 68 (52.7) | 1 (50.0) | 7 (53.8) | 57 (50.9) | 19 (59.4) | 76 (52.8) | 751 (56.8) | 827 (56.4) |
| Male | 61 (47.3) | 1 (50.0) | 6 (46.2) | 55 (49.1) | 13 (40.6) | 68 (47.2) | 571 (43.2) | 639 (43.6) |
| Age (years)d | 66 (56–77) | 46 (41.5–50.5) | 64 (63–74) | 64 (55–76.3) | 67 (62.5–76.3) | 66 (55.8–76.3) | 60 (47–71.8) | 61 (48–72) |
| Age categoryc | ||||||||
| < 65 years | 59 (45.7) | 2 (100.0) | 7 (53.8) | 61 (54.5) | 11 (34.4) | 68 (47.2) | 783 (59.2) | 851 (58.0) |
| ≥ 65 years | 70 (54.3) | 0 (0.0) | 6 (46.2) | 51 (45.5) | 21 (65.6) | 76 (52.8) | 539 (40.8) | 615 (42.0) |
| BMI (kg/m2)d | 21.5 (19.9–23.5) | 22.9 (21.3–24.5) | 21.0 (19.1–23.8) | 21.6 (20.0–23.6) | 22.1 (20.3–23.8) | 21.3 (19.8–23.4) | 21.4 (19.6–23.2) | 21.5 (19.7–23.2) |
| Chronic comorbiditiesc | ||||||||
| Yes | 35 (27.1) | 2 (100.0) | 9 (69.2) | 28 (25.0) | 18 (56.3) | 46 (31.9) | 347 (26.2) | 393 (26.8) |
| No | 94 (72.9) | 0 (0.0) | 4 (30.8) | 84 (75.0) | 14 (43.8) | 98 (68.1) | 975 (73.8) | 1073 (73.2) |
| Disease severityc | ||||||||
| Mild | 18 (14.0) | 0 (0.0) | 0 (0.0) | 13 (11.6) | 5 (15.6) | 18 (12.5) | 109 (8.2) | 127 (8.7) |
| Moderate | 100 (77.5) | 0 (0.0) | 3 (23.1) | 85 (75.9) | 18 (56.3) | 103 (71.5) | 1128 (85.3) | 1231 (84.0) |
| Severe | 11 (8.5) | 2 (100.0) | 10 (76.9) | 14 (12.5) | 9 (28.1) | 23 (16.0) | 85 (6.4) | 108 (7.4) |
Abbreviations: BMI body mass index, CoI/ScI co-infection/secondary infection, MDRp multidrug-resistant pathogen, Non-CoI/ScI no co-infection/secondary infection
aCoI/ScI was defined as either: (1) valid positive microbial culture and ≥ 1 clinical symptoms/signs (fever, increased white blood cell, increased C-reactive protein, increased procalcitonin); (2) 2 valid positive microbial cultures at the same site; or (3) negative/contaminated microbial culture and ≥ 3 clinical symptoms/signs for ≥ 24 h
bMDR was defined as non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories available for testing
cPresented as n (%). Percentages may not equate to 100 due to rounding
dPresented as median with interquartile range (Q1–Q3)
Most secondary infections occurred in the 2 intensive care units (ICU). We recorded 87 cases (60.4%) with pneumonia, 40 cases (27.8%) with bloodstream infections, 12 cases (8.3%) with urinary tract infections, and 5 cases (3.5%) with other types of infection. The following pathogens were cultured and identified: Acinetobacter baumannii, Burkholderia cepacia, Candida spp., Enterobacter spp., Enterococcus spp., Escherichia coli, Klebsiella pneumoniae, Morganella morganii, Providencia rettgeri, Pseudomonas aeruginosa, Salmonella spp., Staphylococcus aureus, and Stenotrophomonas maltophilia. The alarming rates of MDR were observed in Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa (Figure S1–S3, Supplementary File). Cases with fungal infection were all candidaemia. The medians (IQR) MIC of the most clinically important pathogens are summarised in Table S2–S4 (Supplementary File). Treatment of CoI/ScI was empirical therapy (initial) and pathogen-specific therapy (after culturing and susceptibility testing).
Mortality
During a median follow-up of 9 days (IQR 6 to 13), 387 (26.4%) deaths were recorded (95 (66.0%) in the CoI/ScI group and 292 (22.1%) in the non-CoI/ScI group, Table 2). In the CoI/ScI group, the proportion of mortality was higher in the ICUs (73.7%) than in non-ICU departments (56.5%). Adjusted mortality rates of the CoI/ScI group or CoI/ScI-based exposures were higher than that of the non-CoI/ScI group (Table 2), except in patients with bacterial and fungal infection (2.0 versus 1.6 per 100 person-days). Presence of MDRp was associated with a higher mortality rate compared to non-MDRp (adjusted rates: 9.0 versus 5.9 per 100 person-days, Table 2). In the CoI/ScI group, more deaths were observed in patients infected by Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, and methicillin-resistant Staphylococcus aureus.
Table 2.
Mortality incidences
| Event/Total (%) | ImT-included time-fixed analysisa | ImT-excluded time-fixed analysisa | Time-dependent analysisa | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Person-days | Rate (95% CI)b | Person-days | Rate (95% CI)b | Person-days | Rate (95% CI)b | |||||
| Unadjusted | Adjustedc | Unadjusted | Adjustedc | Unadjusted | Adjustedc | |||||
| Non-CoI/ScI | 292/1322 (22.1) | 13,339 | 2.2 (1.9 to 2.5) | 2.4 (2.2 to 2.7) | 13,339 | 2.2 (1.9 to 2.5) | 2.2 (2.0 to 2.4) | 14,624 | 2.0 (1.8 to 2.2) | 2.0 (1.8 to 2.2) |
| CoI/ScId | 95/144 (66.0) | 2717 | 3.5 (2.8 to 4.3) | 1.9 (1.6 to 2.2) | 1432 | 6.6 (5.4 to 8.1) | 6.3 (5.3 to 7.7) | 1432 | 6.6 (5.4 to 8.1) | 6.4 (5.3 to 7.8) |
| Type of CoI/ScI | ||||||||||
| Bacterial | 85/129 (65.9) | 2270 | 3.7 (3.0 to 4.6) | 2.1 (1.9 to 2.4) | 1147 | 7.4 (5.9 to 9.2) | 7.9 (7.1 to 8.7) | 1147 | 7.4 (5.9 to 9.2) | 8.0 (7.2 to 8.9) |
| Fungal | 1/2 (50.0) | 28 | 3.6 (0.1 to 19.9) | 4.7 (1.6 to 14.5) | 21 | 4.8 (0.1 to 26.5) | 8.8 (1.7 to 44.6) | 21 | 4.8 (0.1 to 26.5) | 8.7 (1.7 to 45.0) |
| Bacterial + fungal | 9/13 (69.2) | 419 | 2.1 (1.0 to 4.1) | 0.7 (0.6 to 0.8) | 264 | 3.4 (1.6 to 6.5) | 1.6 (1.3 to 1.9) | 264 | 3.4 (1.6 to 6.5) | 1.6 (1.3 to 2.0) |
| Resistance statuse | ||||||||||
| No MDRp | 76/112 (67.9) | 2391 | 3.2 (2.5 to 4.0) | 1.5 (1.3 to 1.8) | 1190 | 6.4 (5.0 to 8.0) | 5.9 (4.7 to 7.2) | 1190 | 6.4 (5.0 to 8.0) | 5.9 (4.8 to 7.4) |
| ≥ 1 MDRp | 19/32 (59.4) | 326 | 5.8 (3.5 to 9.1) | 5.2 (4.7 to 5.8) | 242 | 7.9 (4.7 to 12.3) | 8.9 (5.8 to 13.5) | 242 | 7.9 (4.7 to 12.3) | 9.0 (8.2 to 10.0) |
Abbreviations: CI confidence interval, CoI/ScI co-infection/secondary infection, ImT immortal time, MDRp multidrug-resistant pathogen, Non-CoI/ScI no co-infection/secondary infection
aOutcome follow-up was from 1 July 2021 to 31 October 2021
bRates were calculated using Poisson regression and presented per 100 person-days
cAdjusted rates were controlled for sex, age, body mass index, comorbidities, and disease severity
dCoI/ScI was defined as either: (1) valid positive microbial culture and ≥ 1 clinical symptoms/signs (fever, increased white blood cell, increased C-reactive protein, increased procalcitonin); (2) 2 valid positive microbial cultures at the same site; or (3) negative/contaminated microbial culture and ≥ 3 clinical symptoms/signs for ≥ 24 h
eMDR status is defined as non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories available for testing
The mortality curves implied that deaths were more likely to occur in patients with CoI/ScI (Fig. 2). Results from the time-dependent Cox proportional hazards regression also showed a higher risk of mortality in the CoI/ScI group compared with the non-CoI/ScI group (adjusted HR = 3.27, 95% CI: 2.58 to 4.13, Holm-adjusted p < 0.001, Table 3). Similar associations were also found in subgroups of patients infected by non-MDRp (adjusted HR = 3.13, 95% CI: 2.42 to 4.05, Holm-adjusted p < 0.001) or ≥ 1 MDRp (adjusted HR = 3.89, 95% CI: 2.44 to 6.21, Holm-adjusted p < 0.001). For type of CoI/ScI, we only found significantly increased mortality risk in patients with bacterial CoI/ScI (adjusted HR = 3.79, 95% CI: 2.97 to 4.83, Holm-adjusted p < 0.001). With fungal or bacterial plus fungal CoI/ScI, we could not detect any differences due to the small subgroups.
Fig. 2.
Mortality curves of the primary and secondary exposures. Abbreviations: MDRp: multidrug-resistant pathogen; CoI/ScI: co-infection/secondary infection; CoI/ScI (b): bacterial CoI/ScI; CoI/ScI (f): fungal CoI/ScI; CoI/ScI (b + f): bacterial and fungal CoI/ScI; Non-CoI/ScI: no co-infection/secondary infection. Note: Panel A, B, and C depicted the mortality curves of the CoI/ScI and non-CoI/ScI groups. Panel D, E, and F depicted the mortality curves stratified by types of CoI/ScI. Panel G, H, and I depicted the mortality curves stratified by resistance status. Mortality Kaplan–Meier curves in Panel A, D, and G were generated from the immortal time-included time-fixed analysis. Mortality Kaplan–Meier curves in Panel B, E, and H were generated from the immortal time-excluded time-fixed analysis. Mortality Simon–Makuch curves in Panel C, F, and I were generated from the time-dependent analysis. No comparisons were made among these curves, as they were for exploratory purposes. MDR status is defined as non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories available for testing
Table 3.
Associations between the exposures and mortality
| ImT-included Time-fixed Analysisa | ImT-excluded Time-fixed Analysisa | Time-dependent Analysisa | ||||
|---|---|---|---|---|---|---|
| Unadjusted HRb | Adjusted HRb,c | Unadjusted HRb | Adjusted HRb,c | Unadjusted HRb | Adjusted HRb,c,d | |
| Non-CoI/ScI | reference | reference | reference | reference | reference | reference |
| CoI/ScIe | 1.16 (0.90 to 1.49) | 1.10 (0.86 to 1.41) | 2.98 (2.35 to 3.78) | 2.90 (2.29 to 3.67) | 3.33 (2.63 to 4.21) | 3.27 (2.58 to 4.13)f |
| Type of CoI/ScI | ||||||
| Bacterial | 1.26 (0.97 to 1.63) | 1.19 (0.92 to 1.54) | 3.50 (2.75 to 4.47) | 3.38 (2.65 to 4.31) | 3.88 (3.04 to 4.95) | 3.79 (2.97 to 4.83)g |
| Fungal | 1.47 (0.21 to 10.50) | 2.50 (0.35 to 17.91) | 2.20 (0.31 to 15.69) | 4.59 (0.64 to 32.99) | 2.42 (0.34 to 17.24) | 4.92 (0.68 to 35.37)h |
| Bacterial + fungal | 0.58 (0.29 to 1.16) | 0.55 (0.28 to 1.10) | 1.08 (0.52 to 2.21) | 1.07 (0.52 to 2.19) | 1.33 (0.67 to 2.65) | 1.29 (0.65 to 2.58)i |
| Resistance statusj | ||||||
| No MDRp | 0.97 (0.73 to 1.28) | 0.95 (0.72 to 1.25) | 2.76 (2.13 to 3.58) | 2.77 (2.14 to 3.59) | 3.10 (2.39 to 4.01) | 3.13 (2.42 to 4.05)k |
| ≥ 1 MDRp | 2.73 (1.71 to 4.34) | 2.21 (1.38 to 3.52) | 4.20 (2.64 to 6.69) | 3.49 (2.19 to 5.57) | 4.59 (2.88 to 7.31) | 3.89 (2.44 to 6.21)l |
Abbreviations: CoI/ScI co-infection/secondary infection, ImT immortal time, HR hazard ratio, MDRp multidrug-resistant pathogen, Non-CoI/ScI no co-infection/secondary infection
aOutcome follow-up was from 1 July 2021 to 31 October 2021
bHRs were calculated using Cox proportional hazards regression and presented with the 95% confidence intervals
cAdjusted HRs were controlled for sex, age, body mass index, comorbidities, and disease severity
dHolm method was used to control for multiplicity, with a family-wise error rate of 5%
eCoI/ScI was defined as either: (1) valid positive microbial culture and ≥ 1 clinical symptoms/signs (fever, increased white blood cell, increased C-reactive protein, increased procalcitonin); (2) 2 valid positive microbial cultures at the same site; or (3) negative/contaminated microbial culture and ≥ 3 clinical symptoms/signs for ≥ 24 h
fUnadjusted p < 0.001; multiplicity-adjusted p < 0.001
gUnadjusted p < 0.001; multiplicity-adjusted p < 0.001
hUnadjusted p = 0.210; multiplicity-adjusted p = 0.420
iUnadjusted p = 0.885; multiplicity-adjusted p = 0.885
jMDR status is defined as non-susceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories available for testing
kUnadjusted p < 0.001; multiplicity-adjusted p < 0.001
lUnadjusted p < 0.001; multiplicity-adjusted p < 0.001
Discussion
Overall, after adjusting for potential confounders, we found a higher rate and risk of mortality in patients with CoI/ScI compared with the non-CoI/ScI group. This association is consistent across subgroups of CoI/ScI, i.e., bacterial infections and with or without resistance status). Presence of MDRp is an indicator of poorer outcomes.
The results of our study (overall proportion of deaths 66.0%, ICU-based mortality 73.7%) were consistent with other previous findings [8, 18–20]. However, most of these reports did not investigate the effects of fungal pathogens. Our study suggested that the proportion of fungi-associated mortality might range from 50.0% to 69.2%, which could facilitate a more accurate prognosis in patients with COVID-19. Noteworthily, despite a high proportion of deaths (69.2%), patients with bacterial-fungal CoI/ScI had the lowest adjusted mortality rate (1.6 per 100 person-days) among the subgroups. This could be partially due to the intensive care provided to patients with sequential infections, leading to prolonged length of hospitalisation. This also proposed an association between longer treatment duration and worse outcomes, although we did not have enough data to clarify this hypothesis.
No prior reports evaluated the mortality risk over time in patients with CoI/ScI and without CoI/ScI. In our study, we have addressed this gap with a time-dependent approach to control for potential immortal time bias. Physicians could apply our findings to improve timely prognosis and adequate antimicrobial coverage. Nevertheless, current evidence about the mortality risk of CoI/ScI seems to be conflicting. Similar to our result, some studies confirmed the significantly increasing mortality risk of CoI/ScI [8, 20], whereas other reported no statistical difference between CoI/ScI and non-CoI/ScI groups [18]. While there could be many factors contributing to this discrepancy, one of the most important issues was MDRp, which was reported to have an abnormally high prevalence during COVID-19 in our hospital. Type of pathogens (bacteria, fungi, or both) and disease severity might also play substantial roles, but there were not sufficient data to confirm this in our study.
To the best of our knowledge, this is one of the first studies to investigate the mortality risk of CoI/ScI in patients with COVID-19 in low-resource countries, providing new evidence to improve timely prognosis. However, there are some remaining limitations. First, due to the lack of data, we could not separate patients with CoI and ScI to investigate these exposures further. Second, the sample sizes of fungal and bacterial-fungal CoI/ScI groups were too small to draw any strong conclusion. Third, we did not have enough resources to monitor all patients for recurrent infections. However, as recurrent infections had the carry-over effects of CoI/ScI (treatment-induced resistance [21]), we anticipated that the prognosis of patients with CoI/ScI could be even worse. Finally, we only had data on the delta and omicron variants, which could limit the applicability of our findings to newer variants.
Conclusion
In a low-resource Asian setting, compared with the non-CoI/ScI group, patients with CoI/ScI had a significantly higher risk of all-cause mortality, regardless of resistance status. More evidence is needed to confirm the mortality risks in patients with fungal or bacterial-fungal CoI/ScI.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- 95% CI
95% Confidence interval
- BMI
Body mass index
- CoI
Co-infection
- COVID-19
Coronavirus Disease 2019
- HR
Hazard ratio
- ICU
Intensive care unit
- ImT
Immortal time
- IQR
Interquartile range
- MDR
Multidrug-resistance
- MDRp
Multidrug-resistance pathogen
- MIC
Minimal inhibitory concentration
- NDGD Hospital
Nhan Dan Gia Dinh Hospital
- ScI
Secondary infection
- STROBE
Strengthening the Reporting of Observational Studies in Epidemiology
- WBC
White blood cell
Authors’ contributions
Conceptualization: HTP and M-HT. Study design and methods: HTP, THT, K-HT-N, and M-HT. Data collection: HTP, THT, K-HT-N, and M-HT. Data analysis and interpretation: HTP, BKN, and M-HT. Manuscript drafting and revision: all authors. Supervision: M-HT. All authors read and agreed to the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of NDGD Hospital (under approval number 61–2021/NDGD-HDDD) and was conducted in accordance with the Declaration of Helsinki. As only relevant information was collected without revealing the patient’s identity, patient consent was not required, as declared by the Ethics Committee of NDGD Hospital.
Consent for publication
Not applicable.
Competing interests
HTP reported receiving speaking fees and travel reimbursement from Servier Vietnam Ltd and Pfizer Vietnam Ltd, grants from Servier Vietnam Ltd outside the submitted work. M-HT reported receiving travel reimbursement from Pfizer Vietnam Ltd, speaking fees and grants from Servier Vietnam Ltd outside the submitted work. The remaining authors declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–75. 10.1016/j.jinf.2020.05.046. 10.1016/j.jinf.2020.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa RLD, Lamas CDC, Simvoulidis LFN, et al. Secondary infections in a cohort of patients with COVID-19 admitted to an intensive care unit: impact of gram-negative bacterial resistance. Rev Inst Med Trop Sao Paulo. 2022;64: e6. 10.1590/s1678-9946202264006. 10.1590/s1678-9946202264006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Bruyn A, Verellen S, Bruckers L, et al. Secondary infection in COVID-19 critically ill patients: a retrospective single-center evaluation. BMC Infect Dis. 2022;22(1):207. 10.1186/s12879-022-07192-x. 10.1186/s12879-022-07192-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasselli G, Cattaneo E, Florio G. Secondary infections in critically ill patients with COVID-19. Crit Care. 2021;25(1):317. 10.1186/s13054-021-03672-9. 10.1186/s13054-021-03672-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lie KC, Shakinah S, Pasaribu A, Sinto R, Nainggolan L. Observational Study on Secondary Bacterial Infection and the Use of Antibiotics in COVID-19 Patients Treated in a Tertiary Referral Hospital. Acta Med Indones. 2022;54(2):161–9. [PubMed] [Google Scholar]
- 6.Murgia F, Fiamma M, Serra S, et al. The impact of secondary infections in COVID-19 critically ill patients. J Infect. 2022;84(6):e116–7. 10.1016/j.jinf.2022.03.017. 10.1016/j.jinf.2022.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Na YS, Baek AR, Baek MS, et al. Clinical outcomes of and risk factors for secondary infection in patients with severe COVID-19: a multicenter cohort study in South Korea. Korean J Intern Med. 2023;38(1):68–79. 10.3904/kjim.2022.084. 10.3904/kjim.2022.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santis V, Corona A, Vitale D, et al. Bacterial infections in critically ill patients with SARS-2-COVID-19 infection: results of a prospective observational multicenter study. Infection. 2022;50(1):139–48. 10.1007/s15010-021-01661-2. 10.1007/s15010-021-01661-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu S, You Y, Zhang S, et al. Multidrug-resistant infection in COVID-19 patients: A meta-analysis. J Infect. 2023;86(1):73–5. 10.1016/j.jinf.2022.10.043. 10.1016/j.jinf.2022.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suissa S. Immortal Time Bias in Pharmacoepidemiology. Am J Epidemiol. 2008;167(4):492–9. 10.1093/aje/kwm324. 10.1093/aje/kwm324 [DOI] [PubMed] [Google Scholar]
- 11.COVID-19 Treatment Guidelines Panel. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines. National Institutes of Health. Accessed 20 September 2022. Available from: https://www.covid19treatmentguidelines.nih.gov/
- 12.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. 10.1111/j.1469-0691.2011.03570.x. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 13.CLSI. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. 30th ed. Pennsylvania, USA: Clinical and Laboratory Standards Institute; 2020.
- 14.van Zwieten A, Tennant PWG, Kelly-Irving M, Blyth FM, Teixeira-Pinto A, Khalatbari-Soltani S. Avoiding overadjustment bias in social epidemiology through appropriate covariate selection: a primer. J Clin Epidemiol. 2022;149:127–36. 10.1016/j.jclinepi.2022.05.021. 10.1016/j.jclinepi.2022.05.021 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Reinikainen J, Adeleke KA, Pieterse ME, Groothuis-Oudshoorn CGM. Time-varying covariates and coefficients in Cox regression models. Ann Transl Med. 2018;6(7):121. 10.21037/atm.2018.02.12. 10.21037/atm.2018.02.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 17.World Health Organization, Regional Office for the Western Pacific. The Asia-Pacific perspective: redefining obesity and its treatment. Available from: https://apps.who.int/iris/handle/10665/206936
- 18.Kaçmaz B, Keske Ş, Sişman U, et al. COVID-19 associated bacterial infections in intensive care unit: a case control study. Sci Rep. 2023;13(1):13345. 10.1038/s41598-023-39632-2. 10.1038/s41598-023-39632-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pourajam S, Kalantari E, Talebzadeh H, et al. Secondary Bacterial Infection and Clinical Characteristics in Patients With COVID-19 Admitted to Two Intensive Care Units of an Academic Hospital in Iran During the First Wave of the Pandemic. Front Cell Infect Microbiol. 2022;12: 784130. 10.3389/fcimb.2022.784130. 10.3389/fcimb.2022.784130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafran N, Shafran I, Ben-Zvi H, et al. Secondary bacterial infection in COVID-19 patients is a stronger predictor for death compared to influenza patients. Sci Rep. 2021;11(1):12703. 10.1038/s41598-021-92220-0. 10.1038/s41598-021-92220-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q, Li D, Beiersmann C, et al. Risk factors for antibiotic resistance development in healthcare settings in China: a systematic review. Epidemiol Infect. 2021;149: e141. 10.1017/s0950268821001254. 10.1017/s0950268821001254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.