Abstract
Neurofilament light chain (NfL) levels in circulation have been established as a sensitive biomarker of neuro-axonal damage across a range of neurodegenerative disorders. Elucidation of the genetic architecture of blood NfL levels could provide new insights into molecular mechanisms underlying neurodegenerative disorders. In this meta-analysis of genome-wide association studies (GWAS) of blood NfL levels from eleven cohorts of European ancestry, we identify two genome-wide significant loci at 16p12 (UMOD) and 17q24 (SLC39A11). We observe association of three loci at 1q43 (FMN2), 12q14, and 12q21 with blood NfL levels in the meta-analysis of African-American ancestry. In the trans-ethnic meta-analysis, we identify three additional genome-wide significant loci at 1p32 (FGGY), 6q14 (TBX18), and 4q21. In the post-GWAS analyses, we observe the association of higher NfL polygenic risk score with increased plasma levels of total-tau, Aβ-40, Aβ-42, and higher incidence of Alzheimer’s disease in the Rotterdam Study. Furthermore, Mendelian randomization analysis results suggest that a lower kidney function could cause higher blood NfL levels. This study uncovers multiple genetic loci of blood NfL levels, highlighting the genes related to molecular mechanism of neurodegeneration.
Subject terms: Medical genomics, Diagnostic markers, Neurodegenerative diseases
An ancestry-specific and trans-ethnic meta-analysis of Genome-wide Association Study of blood NfL levels identified multiple loci related to neurodegeneration and potential causal association between reduced kidney function and higher blood NfL levels.
Introduction
Blood levels of the neurofilament light chain (NfL) have emerged as a robust biomarker of neuro-axonal injury and are increased in a range of neurodegenerative disorders including Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease, amyotrophic lateral sclerosis (ALS), and multiple sclerosis1. NfL proteins are expressed in the cytoplasm of neurons where they confer structural stability to the cytoskeleton of neurons1,2. Under normal physiological conditions, NfL proteins are continuously released from the axoplasm into circulation in an age- dependent manner3, whereas neuro-axonal damage has been associated with increased release of NfL in the neuronal extracellular space3,4. NfL proteins may diffuse into the cerebrospinal fluid (CSF) and circulation5. Central nervous system origin of the blood NfL levels is also supported by earlier studies demonstrating a strong correlation between CSF and blood NfL levels3,6. The advent of highly sensitive assays enables quantification of blood NfL levels5, thereby facilitating its clinical implementation as a biomarker of neuro-axonal injury and neurodegeneration7,8. Identifying the genetic basis of the NfL in blood could therefore provide a better understanding of the biological pathways underlying axonal damage and facilitate identification of shared molecular mechanisms contributing to neuronal loss across neurodegenerative disorders.
Previously, three modest size genome-wide association studies (GWAS) were performed to identify the genetic variants associated with plasma and CSF levels of NfL in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort9–11. Hong et al. reported the genome-wide significant association of variant rs1548884 within the TMEM106B gene with CSF levels of NfL11. Moreover, genetic variants near the ADAMTS1 gene have been identified as being associated with CSF NfL levels9, and blood based studies have shown sub-threshold associations of the LUXP2 and GABRB2 genes with plasma NfL levels10. To uncover the underlying genetic factors of blood NfL levels, studies with substantially larger sample sizes across different ancestries are warranted.
In the current study, we therefore performed a GWAS meta-analysis based on the findings from 11 cohorts of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium including people from both European and African-American ancestry. Furthermore, we performed a range of post-GWAS investigations, including expression quantitative trait loci (eQTLs) lookups, colocalization, pathway enrichment analysis, linkage disequilibrium score (LDSC) regression, and Mendelian Randomization (MR) analyses. Based on the identified genetic variants of NfL, we calculated polygenic risk scores (PRS) and assessed their association with the incidence of AD, and other AD-related endo-phenotypes using individual levels data from the Rotterdam Study cohort. In the current study, we identified two loci (SLC39A11 and UMOD) in European and three loci (including FMN2) in African ancestry participants. Furthermore, we detected three loci (FGGY, RN7SKP48, and TBX18) in the trans-ethnic meta-analysis. PRS analyses based on European ancestry revealed significant associations with incident AD, and AD blood biomarkers (amyloid beta (Aβ)-40, Aβ-42, and total-tau). Moreover, our MR analysis demonstrated a potentially causal association between decreased kidney function and increased blood NfL levels.
Results
Our ancestry-specific GWAS meta-analysis of circulating levels of NfL was based on 11 different cohorts of European (N = 18,532) and three cohorts of African-American ancestry (N = 1142), Supplementary Table 1. The Rotterdam Study and the Rhineland study were the major contributors (>40%) to the total sample size. Participants of cohorts of European ancestry had diverse age ranges, varying from a mean age of 51 years (standard deviation [SD] = 3.2) in the Coronary Artery Risk Development in Young Adults (CARDIA) of European-American ancestry to a mean age of 85.3 years (SD = 6.7) in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. The female proportion varied from 0% in the Vietnam Era Twin Study of Aging (VESTA) cohort to 63% in the Cardiovascular Health Study (CHS) of European-American ancestry cohort. Among the three cohorts of African-American ancestry, the Atherosclerosis Risk in Communities (ARIC) cohort contributed the largest number of participants, while the Cardiovascular Health Study (CHS) participants were older (mean = 76.3 years [SD = 4.93]) compared to the other two cohorts (mean ages of 61.5 [SD = 4.5] and 48.9 [SD = 3.5] years in the ARIC and CARDIA cohort, respectively). Moreover, the CARDIA cohort of African-American ancestry had the lowest percentage of female participants (56.7%). Overall mean NfL levels in all participating cohorts were significantly positively correlated with mean age (Pearson’s r = 0.81, P = 2.06 × 10–4).
GWAS meta-analysis findings
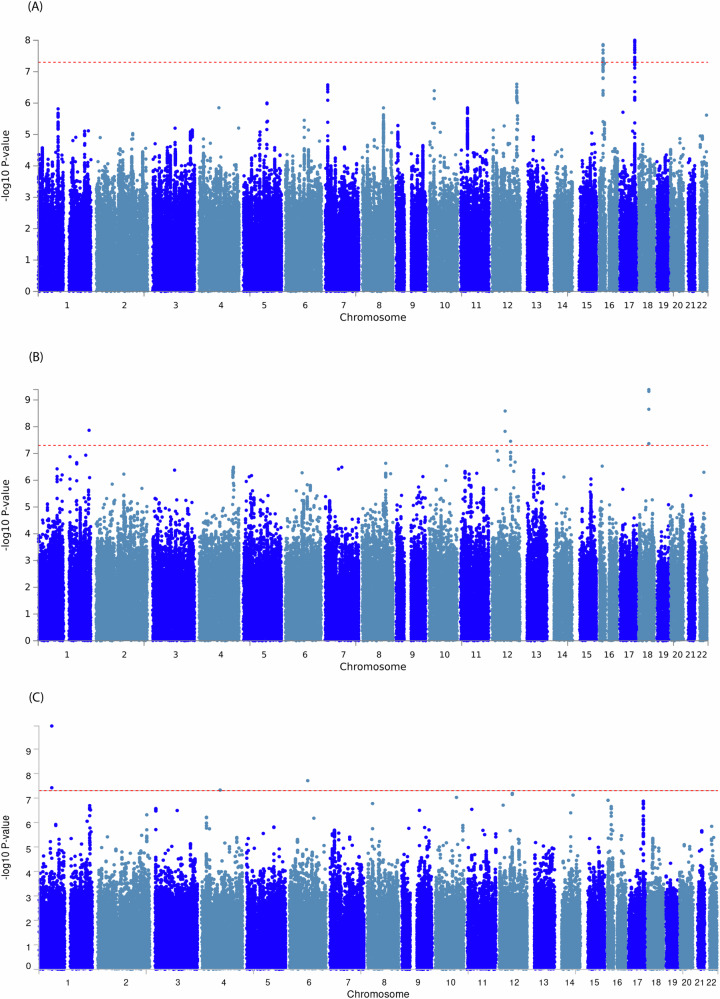
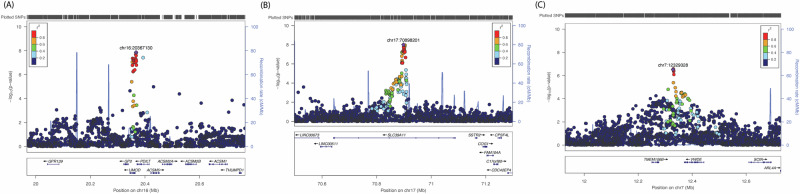
The European ancestry GWAS meta-analysis identified 26 genome-wide significant single nucleotide polymorphisms (SNPs) within two loci led by two individually significant SNPs (Supplementary Table 2). Manhattan plot and Quantile–Quantile (Q-Q) plot of the meta-analysis summary statistics are provided in Fig. 1A and Supplementary Fig. 1A. The first locus at 16p12 was mapped to the UMOD and PDILT genes, tagged by 39 SNPs with p values < 0.05 (Supplementary Data 1). This locus was led by two genetic variants reaching genome-wide significance, including rs7203642-A (effect = 0.041, standard error [SE] = 0.007, P = 1.37 × 10–8) in the intronic region of the UMOD gene and rs77924615-A (effect = –0.041, SE = 0.007, P = 3.77 × 10–8) within the intronic region of the PDILT gene. A regional plot for the 16p12 locus (Fig. 2A) shows that both variants are also in high linkage disequilibrium (LD) with each other. Therefore, we defined this locus based on the rs7203642 variant of the UMOD gene, with the A-allele of rs7203642 associated with increased blood NfL levels. The second locus at chromosome 17q24 was tagged by 117 SNPs with a p value < 0.05 (Supplementary Data 1) and was mapped to the SLC39A11 gene. At the 17q24 locus, the A-allele of the lead intronic variant rs12051560 (effect = 0.033, SE = 0.006, P = 9.94 × 10–9) was associated with increased blood NfL levels (Fig. 2B regional plot). The UMOD and SLC39A11 signals were largely consistent throughout the cohorts included in the European ancestry based meta-analysis (Supplementary Fig. 2A, 3A). We also identified three additional suggestive loci (7p21, 10p12.1 and 12q24.22) at p values < 5 × 10–7 (Supplementary Data 2). One of these suggestive loci at 7p21 and tagged by an intergenic variant, was mapped to the TMEM106B, VWDE and SCIN genes (rs3902479-T; effect = 0.0303, SE = 0.059, P = 3.05 × 10–7) (regional plot is shown in Fig. 2C). Based on LSDC software, the SNP- heritability (h2) of blood NfL levels was 0.12, meaning that the genotyped variants can explain about 12% of the variation of NfL levels in blood.
Fig. 1. Manhattan plots for the meta-analysis of genome-wide association study (GWAS) of the blood levels of neurofilament light (NfL).
Manhattan plot based on GWAS meta-analysis of the European ancestry (A), African- American ancestry (B), and Trans-ethnic participants (C). The Observed associations of all tested genetic variants on autosomal chromosomes (X-axis) are displayed as –log10(P values) on the Y-axis. The red dotted horizontal line indicates a genome-wide significant association (P value < 5 × 10–8) with NfL levels in blood.
Fig. 2. Loci identified in the European ancestry.
Regional plot for two genome-wide significant loci in the UMOD (A), and SLC39A11 (B), and suggestive locus near TMEM106B/VWDE genes (C) identified in the meta- analysis of neurofilament light (NfL) genome-wide association study (GWAS) in European ancestry. The genetic variants are denoted as colored circles with their P values (-log10) on left Y-axis and genomic location is based on build 37 on X-axis. Lead SNPs (purple diamond) are marked with their genomic location. Recombination rates are plotted on right Y-axis to represent the local linkage disequilibrium (LD) structure. The LD between the genetic variants is provided with a color scale, ranging from blue (r2 = 0) to red (r2 = 1). LD calculations are based on 1000 genome, European ancestry.
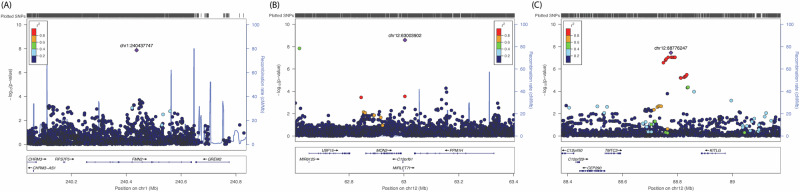
The Manhattan plot and Q-Q plots for GWAS meta-analysis of African-American ancestry are provided in Fig. 1B and Supplementary Fig. 1B. In the GWAS meta-analysis of African-American cohorts (Supplementary Table 2), we identified three independent genome-wide significant loci at chromosomes 1q43, 12q14, and 12q21 (Supplementary Data 3). An intronic variant inside the FMN2 gene (rs1026417-C, effect = –0.433, SE = 0.076, P = 1.36 × 10–8) was associated with decreased levels of NfL in circulation, while two genetic variants at 12q14 (rs17098087-C, effect = 0.440, SE = 0.074, P = 2.59 × 10–9) and 12q21 (rs73423978-T, effect = 0.332, SE = 0.060, P = 3.50 × 10–8) were associated with increased levels of NfL in blood (regional plots shown in Fig. 3). There were 12 suggestive loci (P < 5 × 10–7) in African-American ancestry (Supplementary Data 2). We have provided information about Combined Annotation Dependent Depletion (CADD) score, Regulome Database (RDB) annotation, and chromatin state information for all SNPs inside the observed genetic loci for both European and African- American ancestry using the Functional Mapping and Annotation (FUMA) in Supplementary Data 1, 3–5 and Supplementary Figs. 2–6.
Fig. 3. Loci identified in the African-American ancestry.
Regional plot for three loci in FMN2 (A), intergenic region at 12q14 (B) and 12q21 (C) identified in the meta-analysis of neurofilament light (NfL) genome-wide association study (GWAS) in African-American ancestry. The genetic variants are denoted as colored circles with their P values (-log10) on the left Y-axis and genomic location is based on build 37 on the X-axis. Lead SNPs (purple diamond) are marked with their genomic location. Recombination rates are plotted on right Y-axis to represent the local linkage disequilibrium (LD) structure. The LD between the genetic variants is provided with a color scale, ranging from blue (r2 = 0) to red (r2 = 1). LD calculations are based on 1000 genome, African ancestry.
Trans-ethnic meta-analysis
In the trans-ethnic GWAS meta-analysis of European and African-American ancestry, we identified three loci showing evidence of association with plasma NfL levels at genome-wide significance (Supplementary Table 3). The Manhattan plot and Q-Q plots for trans-ethnic meta-analysis are provided in Fig. 1C and Supplementary Fig. 1C. The first locus, 1p32, was mapped to the FGGY gene led by rs11583796 (P = 1.14 × 10–10, PANS-HET = 3.23 × 10-12), and the second locus at 6q14 was found near the TBX18 gene (rs58152294: P = 1.95 × 10–8, PANS-HET = 1.05 × 10–9). We identified 16 additional suggestive loci (P < 5 × 10–7), one of which (8p21) mapped to the NEFM gene (P = 1.69 × 10–7, PANS-HET = 1.5 × 10–4) (Supplementary Data 6 and Supplementary Fig. 7). SNPs identified in ancestry-specific meta-analysis were also observed in trans-ethnic meta-analysis including FMN2 (P = 3.08 × 10–7, PANS-HET = 3.32 × 10–8), UMOD (P = 2.25 × 10–7, PANS-HET = 0.122), and SLC39A11 (P = 1.34 × 10–7, PANS-HET = 0.03891). Annotation information of SNPs identified in trans-ethnic meta-analysis is provided in Supplementary Data 7 and 8.
Conditional analysis on kidney function and Alzheimer’s disease
In the European ancestry meta-analysis, the locus at 16p12 (UMOD gene) is also a known locus for kidney function12. Kidney function was not included as a covariate in the GWAS of blood NfL levels by the participating cohorts, although it may have a role in blood protein clearance and thus may confound genetic associations of blood protein levels. To investigate whether genetic variants associated with kidney function may have confounded the associations between the identified SNPs and blood NfL levels, we conducted a conditional analysis by conditioning the observed genetic association effect size estimates on the estimated kidney glomerular filtration rate (eGFR)12 associated genetic variants in European ancestry using mtCOJO13. Results of this conditional analysis showed that one of the lead genetic variants at 17q24 (rs12051560-A, Effect = 0.033, SE = 0.006, P = 9.13 × 10–9) is independent of kidney function (Supplementary Data 9). However, the second variant inside the intronic region of UMOD gene became less significant (rs7203642-A, effect = 0.034, SE = 0.007, P = 3.51 × 10–6) upon conditioning the meta-analysis on kidney function. Since some of the participating cohorts included in the meta-analysis also included AD patients, we also conditioned the observed effect size estimates of blood NfL levels in European ancestry meta-analysis on AD18 associated genetic variants using mtCOJO (Supplementary Data 9). Results showed that both top genetic loci remained significant after adjusting the summary statistics of NfL levels in European ancestry for AD associated variants (rs12051560-A, Effect = 0.034, SE = 0.006, P = 7.75 × 10–9; rs7203642-A, effect = –0.041, SE = 0.007, P = 1.69 × 10–8).
Gene enrichment analysis
Gene enrichment analysis of both European (number of genes = 18,718) and African- American (number of genes = 17,370) ancestry-based meta-analysis showed enrichment of several Gene Ontology (GO) terms, though they did not pass the Bonferroni-adjusted thresholds for multiple testing (Supplementary Data 10). We also did not find an overlap in the top ten curated GO terms in the ancestry-specific enrichment analysis. Yet, the top GO terms enriched in European ancestry meta-analysis included GO molecular function beta-2 adrenergic receptor binding (P = 1.89 × 10–5), GO biological process glycerolipid catabolic process (P = 2.97 × 10–5), germ cell proliferation (P = 6.85 × 10–5), and canonical wnt signaling pathway (P = 5.57 × 10–5). In addition, genes were enriched in curated gene sets including sharma pilocytic astrocytoma location dn, reactome foxo mediated transcription of cell death genes, and pid p38 alpha beta downstream pathway. In African-America ancestry meta-analysis findings were enriched for GO biological process astrocyte differentiation (P = 1.38 × 10–4), compartment pattern specification (P = 1.85 × 10–4) and GO astrocyte development (P = 9.38 × 10–4). In the trans-ethnic meta-analysis, we identified significant association of GO term CUL3 Ring Ubiquiti ligase complex (P = 1.66 × 10–6). We also observed enrichment of GO biological process neurofilament cytoskeleton organization (P = 1.25 × 10–4), but this association was not significant after Bonferroni correction.
eQTL analysis for the identified genetic variants
The eQTL analysis findings related to SNPs located within genome-wide significant and suggestive loci in the European, African-American, and the trans-ethnic ancestry are presented in Supplementary Data tables 1, 4, 5, 8. Notably, the effect allele of the lead genetic variant on chromosome 17, rs12051560-A, was associated with decreased expression of the SSTR2 gene in cerebellar hemispheres (Normalized Effect Size [NES] = -0.28, P = 1.66 × 10–7) and the cerebellum (NES = –0.26, P = 7.20 × 10–6). Multiple suggestive loci at 7p21 in European ancestry showed significant association with the expression of the VWDE gene in various brain tissues (Supplementary Data 4). In the trans-ethnic meta-analysis, seven intergenic SNPs within suggestive loci (near NEFM) also acted as eQTL for the expression levels of the NEFM gene in the basal ganglia (Supplementary Data 8)
Genetic correlation of NfL with neurological traits
In the LD regression analysis based on the results of European ancestry meta-analysis (Supplementary Data 11), we observed no genetic correlation with any neurological or neurology-derived traits. The genetic correlation coefficients were in positive direction for AD, T-tau, Aβ-40, Aβ-42, and Aβ-ratio. As a sensitivity test, we also repeated the LD regression analysis using the European ancestry NfL summary statistics conditioned on kidney function (Supplementary Data 11), but the results remained similar to the original unadjusted summary statistics. Furthermore, colocalization analysis of two loci in individuals of European ancestry did not demonstrate a significant posterior probability of colocalization with neurological traits (Supplementary Data 12).
Polygenic risk score analysis in the Rotterdam Study
We further corroborated the results of the LD regression analyses, which were based on summary statistics, by deriving a PRS based on individual-level data (Supplementary Data 13). In the Rotterdam Study cohort, the PRS based genome-wide significant threshold (PRS Threshold = 5 × 10–8) in European ancestry participants showed strong associations with plasma levels of total tau (effect size = 0.836, False discovery rate (FDR) = 6.81 × 10–4), Aβ-40 (effect size = 0.550, FDR = 1.24 × 10–4), and Aβ-42 (effect size = 0.638, FDR = 9.33 × 10–4). Association analysis of PRS score based on higher p value thresholds (PRS Threshold 1.0 × 10–4; FDR = 1.50 × 10–3 ; PRS Threshold 0.001; FDR = 1.60 × 10–3 ; PRS Threshold 0.05; FDR = 2.32 × 10–5) showed significant association with AD incidence, as well as with imaging phenotypes including total brain volume (PRS Threshold 1 × 10–6; FDR = 3.30 × 10–2), and total white matter lesions (PRS Threshold 0.05; FDR = 1.34 × 10–3).
Relation of identified single genetic variants with neurological traits
We also performed look-ups of the two identified genetic variants associated with blood NfL levels in European ancestry participants using AD14,15, PD, and other GWAS (meta-analysis) summary statistics included in our LD regression analyses (Supplementary Data 14).
Neither of the two genetic variants showed an association with AD or PD. The genetic variant inside UMOD (rs7203642-A) gene showed weak evidence of association (Bonferroni correction [0.05/12] <4.16 × 10–3) with Aβ-40 (β = 0.039, P = 1.37 × 10–2), Aβ-42 (β = 0.033, P = 4.43 × 10–2) and significant association with total-tau (β = 0.028, P = 6.21 × 10–4). The associations of the variant within the SLC39A11 gene (rs12051560-A) with total-tau (β = 0.012, P = 7.30 × 10–2) and total brain volume (P = 3.25 × 10–2) were not significant after multiple testing correction. Of the single genetic variants associated with blood NfL levels, which were assessed in relation to CSF levels of Aβ-42, phosphorylated tau (p-tau), total-tau in the MEMENTO cohort (Supplementary Data 15), only one genetic variant rs12051560-A (SLC39A11) was related to CSF levels of total tau (β = 0.138, P = 4.96 × 10–2).
Two-sample MR
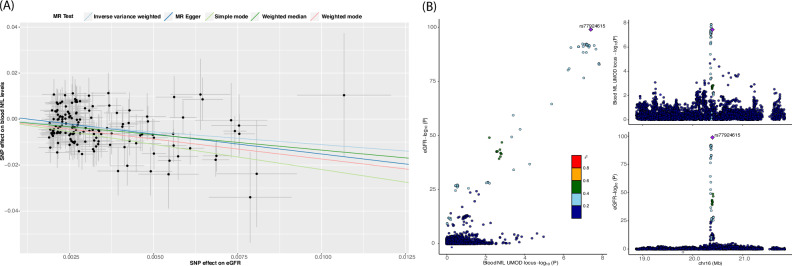
In the forward MR (the effect of kidney function on blood NfL levels) analysis, we observed a potential causal association between kidney function and blood NfL levels based on the inverse variance weighted (IVW) method (β = –1.045, P = 6.19 × 10–8). MR estimates from other robust methods also showed consistent results, including MR-Egger (β = –1.912, P = 1.29 × 10–4), weighted median (β = –1.32, P = 4.31 × 10-7), simple mode (β = –2.13, P = 6.84 × 10–3), and weighted mode (β = –1.75, P = 5.73 × 10–3) (Supplementary Data 16). However, the significant Cochran Q statistic for inverse variance weighted method (P = 7.0 × 10–4) and the MR-Egger method (P = 3.98 × 10–4) indicated heterogeneity in our MR estimates. No evidence of directional horizontal pleiotropy was observed based on the Egger intercept of 0.003 (p value = 0.054). Among the 168 instrumental variables, we identified 15 outliers using the radial MR method that contributed to the significant heterogeneity. Sensitivity analysis after excluding the outliers from the instrumental variables showed consistent significant results among all MR methods (Fig. 4A) with no evidence of heterogeneity or horizontal pleiotropy in estimates (Q-PIVW = 0.733 and Q-PMR-Egger = 0.709, PEgger intercerpt = 0.147). MR-PRESSO results also showed significant MR estimates after removal of outliers (β = –0.977, P = 1.19 × 10–7). In the reverse MR (the effect of blood NfL levels on kidney function), we did not find evidence for causality based on the inverse variance weighted MR method (β = –0.104, P = 1.02 × 10–1).
Fig. 4. Mendelian Randomization and Colocalization analysis.
Scatter plot of instrumental variable (SNPs) effect size estimates of kidney function (eGFR) on NfL in two sample Mendelian Randomization analysis (A), and Colocalization plot between UMOD locus and eGFR (kidney function) in European ancestry (B).
Colocalization analysis between NfL loci and kidney function in European ancestry
Colocalization analysis revealed that the NfL locus within the UMOD gene and kidney function (eGFR) share a single causal variant with posterior probabilities of the single shared causal variant (PP:H4) > 0.819 (Fig. 4B, Supplementary Data 12), whereas the other locus near SLC39A11 showed no evidence of colocalization of causal variants between both traits (PP H4: 1.21 × 10–4) (Supplementary Fig. 8).
Discussion
In the ancestry-specific GWAS meta-analyses, we identified two genome-wide significant loci (16p12 and 17q24) associated with blood NfL levels within the UMOD and SLC39A11 genes, and three suggestive loci, one (7p21) mapped to the TMEM106B gene, among European ancestry participants. Additionally, we identified three genome-wide significant loci (1q43, 12q14, and 12q21) in participants from the African-American ancestry. Further, in the trans-ethnic meta-analysis, we identified three more loci (1p32, 4q21, 6q14) mapped to the FGGY, RN7SKP48, TBX18 genes, and 16 suggestive loci, one (8p21) located near to the NEFM gene. Evaluation of the genetic correlation of the blood NfL levels (European ancestry) with neurological traits demonstrated no significant genetic correlation. However, a PRS based on European ancestry was associated with plasma levels of Aβ-42, Aβ-40, total-tau, and the incidence of AD in the Rotterdam Study cohort. MR and colocalization analyses demonstrated evidence for a causal association between lower kidney function and higher blood NfL levels in European ancestry.
The locus at 17q24 (SLC39A11) was associated with blood NfL levels at the genome-wide level of significance in European ancestry. Several lines of evidence from earlier studies indicate the role of the SLC39A11 gene in neurodegeneration. The SLC39A11 gene plays a role in zinc homeostasis16 and has been associated with ALS17,18 (P = 8.11x10-6). The KEGG pathway database (map05010 and map05012) queries indicated a role of the SLC39A11 gene in the AD- as well as PD-related pathways19. The SLC39A11 gene is expressed in the brain based on data from the human protein atlas20 and the genotype-tissue expression (GTEx) database, but apart from the SLC39A11 variants (Supplementary Data 1) that acted as an eQTL for the SSTR2 gene in cerebellar tissue, no other variants in UMOD locus were identified as eQTLs in the GTEx database. Another interesting observation that links the SLC39A11 polymorphisms to neuro-axonal injury is that the rs12051560-A (SLC39A11) associated with decreased expression of the somatostatin receptor 2 (SSTR2) gene in the cerebellum and the cerebellar hemispheres based on the queries of the GTEx eQTL database. Decreased expression of SSTR2 gene was linked to axonal degeneration of noradrenergic projections in SSTR2–/– mice studies21. Earlier studies also implicate the SSTR2 gene in neurodegeneration under ischemia22, and in hypoxia-induced neuronal cell death23.
Another locus for blood NfL levels in European ancestry participants was identified in the 16p12.3 region, which is tagged by two lead genetic variants located inside the UMOD and PDILT genes, which have previously been associated with kidney function12. Conditional analysis on kidney function showed that the association of our lead SNP rs7203642 became less significant (P = 3.51 × 10–6). The UMOD findings are in line with results from earlier studies reporting an association between decreased kidney function and increased blood NfL levels, which may be due to aging, cardiovascular risk factors, and diabetes mellitus24–27. The observation of higher blood levels of NfL in children with chronic kidney disease28 contradicts the role of cardiovascular diseases and diabetes as the sole drivers of blood NfL levels. Nevertheless, kidney function has been associated with cognitive decline, brain atrophy, and white matter abnormalities29,30 which suggest a direct role of kidney function in determining blood NfL levels. Our MR analysis robustly demonstrated a potential causal relationship between decreased kidney function and increased blood NfL levels, which remained consistent across different MR methods. Among the two loci identified in European ancestry (UMOD and SLC39A11), only the UMOD locus exhibited significant colocalization with kidney function (eGFR), reinforcing the UMOD gene’s role as a shared genetic driver of blood NfL levels and kidney function. The exact mechanism through which genetic variants in the UMOD and blood levels of NfL are related requires further investigation. However, the key role of UMOD in kidney function may be instrumental in understanding its link to neurodegeneration. The UMOD gene encodes for the uromodulin protein and mutations in this gene have been associated with hyperuricemia and tubulointerstitial nephritis31. One of the primary reasons for UMOD-related kidney disease is the accumulation of misfolded UMOD protein inside the endoplasmic reticulum (ER)32 and thereby generation of ER stress which results in cell death and inflammation33. Hyperglycemia and ER stress related pathways may trigger the generation of advanced glycation end products (AGE), which have been associated with neurodegeneration33,34. Second, the lead genetic variants of identified locus (i.e., rs77924615) was found inside the PDILT gene, which belongs to the protein-disulfide isomerase (PDI) family of proteins35. PDI proteins play an important role in protein folding in the ER and their dysfunction may lead to diseases involving the accumulation of misfolded proteins, which is also a hallmark of neurodegenerative diseases such as AD and PD12.
One of the suggestive loci of blood NfL levels in European ancestry meta-analysis was located near the TMEM106B gene, which is known to influence CSF NfL levels11. The identification of a common gene influencing NfL levels in both blood and CSF aligns with the reported correlation between plasma and CSF NfL levels36. An early reported protein quantitative trait locus (pQTL) of the TMEM106B protein37 (rs1548884-C, effect size = –0.26, SE = 0.019, P = 7.23e–42) also showed an association with blood NfL levels in our meta-analysis (rs1548884-C, effect size = –0.0209, SE = 0.0068, P = 3.34 × 10–4), suggesting genetic pleiotropy or shared biological pathways that warrants further investigation. To gain further insight, we performed a two-sample MR analysis to evaluate the causal association between blood TMEM106B and NfL levels, which showed a significant link between increased TMEM106B protein levels with increased NfL levels (wald ratio: effect size = 0.08, P = 3.14 × 10–4). The TMEM106B gene is a known locus for neurodegenerative diseases, with reported implications in sites of onset, motor functions, and cognition in ALS38 and AD39. It also acts as a modifier of frontotemporal dementia (FTD) risk40. Moreover, the TMEM106B gene contributes to the pathophysiology of frontotemporal lobar degeneration (FTLD) by interacting with the progranulin protein and altering lysosomal activity41.
We identified a locus in the trans-ethnic meta-analysis in the FGGY gene, which has been previously associated with sporadic ALS42,43, but was not replicated in subsequent studies44. In our meta-analysis, the direction of the association was not consistent across different cohorts and the lead SNP showed highly significant ancestral heterogeneity (P = 3.23 × 10–12). The FGGY gene codes for a family of carbohydrate kinases45, which have a role in energy metabolism and glycolysis46. The identification of the FGGY locus is relevant due to the importance of NfL levels as specific markers of ALS disease progression and survival47.
We also identified a suggestive association of blood NfL levels with a locus mapped to the NEFM gene, which encodes the neurofilament medium chain protein that plays a role in axonal radial growth and stability of neurofilament network48. The direction of effect size estimates of the lead variant (rs196876) was consistent over all 14 cohorts from two ethnicities, which makes this locus a plausible target for future investigations. This locus is also supported by the observation that 7 SNPs within the locus have shown association (eQTLs) with NFEM expression in the basal ganglia (Supplementary Data 8). The GTex database showed expression of NEFM predominantly in brain tissues and earlier studies have reported the association of NEFM expression dysregulation with ALS49. An earlier study also linked a point mutation within the NEFM gene to early onset PD50, which did not emerge in large genetic studies.
NfL PRS at the genome-wide significant threshold was associated with plasma levels of t-tau, Aβ-40, and Aβ-42 in the Rotterdam Study. Notably, PRS analyses constructed using more lenient p value thresholds (i.e., 1 × 10–4, 1 × 10–3, and 0.05) were significantly associated with the incidence of AD. Although single SNP lookups and genetic correlation analyses using summary-level data did not show associations between NfL and Aβ-40 or Aβ-42, the association of PRS at different p value thresholds suggest a shared biological mechanism underlying these core AD biomarkers. Additionally, the broader set of NfL-associated variants better captured AD risk, potentially indicating the presence of neurodegeneration related loci that did not reach genome-wide significance in the meta-analysis such as TMEM106B. PRS based association findings, together with the observed association of the variant inside the UMOD gene (P = 6.21 × 10–4) with blood total-tau levels, supports the notion that genetic determinants of blood NfL levels are likely linked to central neurodegeneration.
In the participants from the African-American ancestry, one of the three loci was located near the FMN2 gene (rs1026417-C). FMN2 is a coding gene involved in the cytoskeleton assembly, which makes it an important discovery since NfL is a cytoskeleton protein released into extracellular space as a result of neuro-axonal damage7. FMN2 gene is highly expressed in the brain and involved in synaptic plasticity and memory formation51. Our observation was concordant with several studies that reported the association of the FMN2 gene with cognition52, ALS53, intellectual development disorder54 and neuropsychiatry traits55. All these interlinked pieces of evidence support the role of the FMN2 gene in determining the blood NfL levels in various neurological diseases in African-American ancestry. None of the genetic variants identified in the African-American ancestry meta-analysis showed association with blood NfL levels in the European ancestry.
Next, in the pathway-enrichment analysis based on the European ancestry, we did not identify significant enrichment of GO biological processes for our observed genes after multiple testing correction. However, GO: canonical wnt signaling pathway (P = 5.57 × 10–5) and GO: regulation of wnt signaling pathway (P = 1.43 × 10–4) were among the top pathways that were enriched for 307 and 334, respectively, of the putative genes identified in our study. Wnt signaling is one of the most crucial pathways involved in brain development and involves several genes associated with neurodegenerative diseases such as AD and PD56. Furthermore, the ‘beta 2 adrenergic receptor’ binding GO molecular process ranked first in our analysis (P = 1.89 × 10–5). Interestingly, blocking the beta 2 adrenergic receptors is found to be an effective approach in PD to reduce neuroinflammation and degeneration of dopaminergic neurons57,58. In the African-American ancestry, in GO biological processes, astrocyte differentiation (P = 1.38 × 10–4) and astrocyte development (P = 3.38 × 10–4) were the most notable terms enriched in the GWAS, which reiterates the role of astrocytes in neurodegeneration59. In the trans-ethnic pathway-enrichment analysis, GO cellular component term CUL3 ring ubiquitin ligase complex emerged, which has been linked to regulation of neurofilaments60,61. GO biological process neurofilament cytoskeleton organization (P = 1.25 × 10–4) showed enrichment for 8 genes.
Our study represents the largest GWAS to uncover the genetic determinants of NfL levels in blood. Our GWAS sample included 11 different cohorts of both European and African-American ancestry, which is also the main strength of our study. The genetic variant inside UMOD gene (rs7203642) identified in our study not only highlight the importance of kidney function in neurodegeneration but also indicates that kidney function should be taken into account when assessing blood-based protein biomarkers and specifically NfL. This study has also limitations. A few participating cohorts also included AD patients in the GWAS. Although, we adjusted the analyses for case-control status, and also performed a conditional analysis based on European GWAS meta-analysis summary statistics, a more sensitive approach would be to consider a stratified GWAS based on a dementia-free population for future NfL GWAS. The small sample size of African-American cohorts was a major limitation of the trans-ethnic meta-analysis.
In conclusion, we identified two unique loci associated with blood NfL levels in participants from European ancestry, three loci in African, and three unique loci in a trans-ethnic meta-analysis. Further, we validated a known locus (near TMEM106B gene) of CSF NfL levels, which is also implicated in ALS, FTD, and AD. Our findings highlight the role of the UMOD gene in linking reduced kidney function to increased blood NfL levels.
Methods
Study populations
The current study includes 18532 participants of European and 1142 participants of African- American ancestry from 11 different cohorts of the CHARGE consortium including: the Rotterdam Study (RS-I and RS-II, N = 4119), the Rhineland Study (N = 4019), the MEMENTO cohort (N = 2195), the Framingham Heart Study (FHS, N = 2048), the BiDirect study (N = 1899), the CHS (African-American N = 273, European-American N = 1396), the ARIC (African-American N = 823, European American N = 742), the VESTA (N = 828), the ADNI (N = 578), the CARDIA (African-American N = 128, European-American N = 343) Study, and the Austrian Stroke Prevention Family Study (ASPS-Fam, N = 287). Prior to participation, each participant gave written, informed consent. A detailed description of each of the participating cohorts, their genotyping information, and the quantification of NfL is described in the supplementary materials. General demographic information is provided in Supplementary Table 1.
NfL quantification
Different protocols were adopted by participating cohorts for sample preparation, plasma or serum extraction, and NfL quantification. Methodological details concerning NfL quantification are provided in the cohort descriptions included in the supplement (Supplementary Data 17). In summary, the Rotterdam Study used the single molecule array (Simoa) HD-1 analyzer platform, the Rhineland study used the Quanterix Simoa NF-light assay (103186), the FHS, ARIC, and CARDIA cohorts used the Quanterix 4-Plex, the MEMENTO cohort used Simoa NF-light kit on a Quanterix H1 analyzer, BiDirect Study profiled NFL on Simoa HDX analyzer, ADNI cohort used simoa HD-1 analyzer, VESTA cohort used single analyte assays using the Quanterix Simoa HD-1 platform, CHS used the Simoa Human Neurology 4-Plex A assay and the ASPS-fam used Simoa HDX analyzer.
Genotyping and imputation
The participating cohorts genotyped their samples employing various genotyping kits and imputed using either 1000 Genomes (1Kg)62 or the Haplotype Reference Consortium (HRC)63 panels. Detailed descriptions of the genotyping and imputation methods are provided in cohort description (Supplementary Data 17).
GWAS
Each participating cohort performed genome-wide association of SNP and plasma or serum levels of NfL using an additive model. Blood levels of NfL were log2 transformed before conducting the GWAS and analyses were adjusted for age, sex, study-specific covariates (including batch, study sites, case-control status (if applicable)), and genetic principal components to account for population structure and family relatedness. A post-GWAS quality control was performed on summary statistics of each study using the EasyQC software64. We excluded SNVs with low imputation quality scores (INFO score or r2 < 0.3), low frequency (minor allele count <5 or minor allele frequency < 0.01), and variants that were available in less than 30 participants for each cohort. In order to identify ancestry-specific genetic variants, we performed an ancestry- stratified GWAS meta-analysis for three cohorts of African-American ancestry and 11 cohorts of European ancestry separately, using METAL65 with inverse variance weighted average score to account for population heterogeneity and genomic inflation. In the European ancestry GWAS meta-analysis, we retained only 7,058,703 (~7 million) genetic variants that were present in at least two major cohorts (i.e., the Rotterdam Study and the Rhineland Study) of a total of 11 cohorts accounting for more than 40% of the total number of European ancestry participants. We also performed conditional analysis using GCTA mtCOJO13 to identify genome-wide significant variants independent of kidney function12 and Alzheimer’s disease14. Due to sample sizes of the three participating African-American ancestry, we only retained 8,381,611 (~8 million) genetic variants that were present in all three cohorts of African ancestry (i.e., ARIC-AA, CHS-AA, and CARDIA-AA). Moreover, we excluded variants with heterogeneity I2 values greater than 0.75 in the ancestry-specific meta-analysis. To perform the trans-ethnic meta-analysis based on European and African-American based GWAS summary data, we used MR-MEGA66 software (--pc 12), where we corrected the results of each cohort for genomic inflation. We only retained SNPs present in three large European cohorts (Rotterdam Study I, II and Rhineland study) as well as in three African-American cohorts (4,833,685 SNPs). We also filtered out variants with significant residual heterogeneity (P < 5×10-8).
Functional mapping and annotation
To perform functional mapping, and annotation of GWAS summary statistics of NfL, we used the FUMA platform version 1.3.8 which is designed to prioritize and aid in the interpretation of GWAS findings67,68. To identify independent genome-wide significant SNPs, we used r2 = 0.2 and P value < 5 × 10–8. Using FUMA, we defined the lead SNPs as independent of each other at r2 = 0.1 within a 500 kb region in the 1000 Genome Phase 3 reference panel. The individual lead SNPs were mapped based on the default 10 kb distance between SNPs and genes. The ancestry-specific GWAS and trans-ethnic GWAS meta-analysis NfL loci were visualized using Manhattan plots and regional plots using FUMA and Locus Zoom69 (using the 1000 Genomes reference panel for estimating LD), respectively. We used LD score (LDSC) regression software70 to estimate blood NfL heritability based on GWAS summary statistics. Reference LD scores were computed based on the 1000 Genomes Phase 3 reference panel.
Pathway enrichment analysis and functional analysis
Gene-based and gene-set enrichment analyses, which quantify the association of individual mapped genes with NfL levels and sets of genes with GO terms, respectively, were performed using MAGMA (version v1.0.8)71 as implemented in FUMA (version 1.3.7). The gene-based analysis was performed based on 18,718 protein-coding genes, setting the level of statistical significance at a Bonferroni-adjusted threshold of P value = 2.671 × 10–6 ( = 0.05/18718). For MAGMA gene-set analysis, gene-set p value is computed using the gene-based P value for 4728 curated gene sets (including canonical pathways) and 6,166 GO terms obtained from MsigDB v5.2, and a FDR was used to correct for multiple testing.
Similarly, tissue-specific gene expression analysis was also performed using MAGMA as integrated in FUMA. Further, we explored the effects of genetic variants identified in our GWAS on the expression levels of other genes by querying the GTEx72 database (version 8) for all SNPs included in identified loci using FUMA (in blood and brain tissue). Functional consequences for the SNPs were obtained by querying different databases, including ANNOVAR categories, CADD scores and RegulomeDB scores. ANNOVAR annotates the functional consequences of SNPs on genes (for example, intron, exon, and intergenic). CADD scores predict how deleterious the effect of a SNP may be, with scores above the 12.37 threshold flagged as potentially pathogenic. The RegulomeDB score is a categorical score based on information from eQTLs and chromatin marks, ranging from 1a to 7, with lower scores indicating an increased likelihood of having a regulatory function.
LD score regression analysis
To quantify the genetic correlation between blood NfL levels and other neurological traits and
biomarkers of neurodegeneration, we performed LD regression analysis. We obtained GWAS summary statistics for AD14,39, PD15, Huntington’s disease73, ALS74, Aβ-42, Aβ-4075, total-tau76, and brain imaging markers (total hippocampal volume77, total brain volume78, and total white matter lesions79) from the GWAS catalog80. We performed LD score regression analysis using the LDSC tool70 based on the European ancestry 1000 Genomes (phase 3) LD reference panel. Details of the GWAS studies used for LD regression and their base heritability estimates are provided in Supplementary Data 18.
PRS association with AD biomarkers
We calculated PRS based on NfL associated SNPs applying different p value thresholds (i.e., 5 × 10–8, 1 × 10–7, 1 × 10-6, 1 × 10–5, 1 × 10-4, 1 × 10–3, and 0.05) using PRSice-281. PRS was calculated in the Rotterdam Study participants by summing the number of effect alleles weighted by their effect size estimates obtained from our meta-analysis based on European ancestry. In the PRS calculation, we retained variants with MAF > 0.01 in the target population and performed clumping using an r2 value of 0.1. We used the Cox-proportional-hazard models to check the association of PRS with the incidence of AD, adjusted for age at baseline and sex. Moreover, we performed multiple linear regression analyses to assess the association of NfL PRS with plasma levels of Aβ-40, Aβ-42, Aβ-40/Aβ-42 ratio, total tau, as well as with magnetic resonance imaging (MRI) markers of neurodegeneration including total hippocampal volume, total brain volume, and total white matter lesions in the Rotterdam Study cohort. All linear regression analyses were adjusted for age, sex, batch (in case of biomarkers), and additionally for intracranial volume for MRI traits.
Look up of lead variants into previous GWASs of neurological traits
To evaluate the association of the most significant genetic variants with the two common neurodegenerative diseases AD and PD, we used the most recent GWAS meta-analyses of AD14 and PD15 and reported the results for each genetic variant. Additionally, we performed lookups for single variants in GWASs of traits used for LD regression analysis. Bonferroni correction was used for multiple comparisons adjustment.
Colocalization analysis
We performed colocalization analysis to evaluate whether the loci discovered in the European ancestry meta-analysis are colocalized with neurological traits considered for lookups. The variants within ±1.5 megabases (Mb) of the lead SNPs in European ancestry were used for the colocalization analysis. We utilized the ‘coloc.abf ‘ function from the COLOC82 R package to test the posterior probabilities (PP) between SNPs associated with NfL and various phenotypes under the following hypotheses: (PP:H0) neither trait has a genetic association in the region; (PP:H1/PP:H2) only one trait has a genetic association in the region; (PP:H3) both traits are associated but with different causal variants; and (PP:H4) both traits are associated and share a single causal variant. The prior probability for H4, set at 1.0 × 10–6, was used to determine the likelihood that a random variant is causative for both NfL and the tested phenotypes. We used locusComparer83 R package to visualize the colocalization results.
Mendelian Randomization
To evaluate the causality of the association between kidney function and blood NfL levels in individuals of European ancestry, we performed two-sample MR analysis using the TwosampleMR84 package. We used inverse-variance weighted regression as the primary MR method, followed by other complementary methods robust to instrumental variable bias due to horizontal pleiotropy, including weighted median85 and MR-Egger86 regression. Exposure instrumental variables were defined as genome-wide significant (P < 5×10-8), independent (r2 = 0.001) within a window of 10,000 kb. To assess the robustness of our MR results to various assumptions related to instrumental variables, we performed several sensitivity analyses, including (i) the Cochran Q statistics to estimate the pleiotropy of causal estimates, (ii) MR-Egger intercept to identify horizontal pleiotropy, and (iii) radial MR87 and MR-PRESSO to detect outliers in instrumental variables and MR estimates after removing outliers, respectively. Finally, we performed a reverse MR to exclude the possibility of reverse causation.
Statistics and reproducibility
Baseline characteristics of each cohort were assessed and summarized in Supplementary Table 1. Statistical and genetic analyses were conducted using various command line tools including EasyQC64, Plink 1.9 (version 1.9, https://www.coggenomics.org/plink/), and R (version 4.0.4, https://www.r-project.org/) statistical environment. Meta-analysis of GWAS summary statistics was performed using METAL65 software (latest version released on 2011-3-25, https://csg.sph.umich.edu/abecasis/metal/download/) and MR-MEGA (version 0.2, https://genomics.ut.ee/en/tools). We used the web-based tool FUMA67 (version 1.3.8 and 1.5.2, https://fuma.ctglab. nl/) to perform LD pruning, lead loci detection, eQTL (GTEX v8, https://gtexportal.org/home/) analysis, and other functional analyses. Locuzoom was used to plot regional plots for genetic loci (http://locuszoom.org/). For PRS calculation, we used PRSice-2 (version v2.3.3, https://github.com/choishingwan/PRSice) software while MR and colocalization analyses were performed using TwoSampleMR (version 0.5.7, (https://mrcieu.github.io/TwoSampleMR/ articles/introduction.html) and coloc (version 5.2.3, https://github.com/chr1swallace/coloc) R packages. Details of tools used by each participating cohort for performing GWAS are provided in supplementary Data 17.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
Rotterdam Study cohort: The Rotterdam Study is supported by the Erasmus MC University Medical Center and Erasmus University Rotterdam, the Netherlands Organization for Scientific Research (NWO), the Netherlands Organization for Health Research and Development (ZonMW), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry of Health, Welfare and Sport, The European Commission (DGXII), the Netherlands Genomics Initiative (NGI), and the Municipality of Rotterdam. Cardiovascular Health Study (CHS) cohorts: This CHS research was supported by NHLBI contracts HHSN268201200036C, HSN268200800007C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086;, HHSN268200960009C and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and U01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629, R01AG033193, R01AG053325, and K24AG065525 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS- NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Rhineland Study: The Rhineland Study is funded by the German Center for Neurodegenerative Diseases (DZNE). Additional support was provided by the German Federal Ministry of Education and Research (BMBF) through the Diet-Body-Brain Competence Cluster in Nutrition Research (grant numbers 01EA1410C and FKZ: 01EA1809C) and grant [FKZ: 01KX2230] with the title “PreBeDem - Mit Prävention und Behandlung gegen Demenz”; the Helmholtz Association under (ExNet-0008-Phase2-3) and the 2023 Innovation Pool; the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) under Germany’s Excellence Strategy (DFG) – EXC2151 – 390873048 and SFB 1454; and the Alzheimer Forschung Initiative e.V. (#22017). NAA was partly supported by an Alzheimer’s Association Research Grant (Award Number: AARG‐19‐ 616534) and a European Research Council grant (#101041677). The BiDirect Study: The BiDirect Study is supported by grants (01ER0816, 01ER1506) of the German Ministry of Research and Education (BMBF) to the University of Muenster. Laboratory NFL analysis was performed at the University Hospital Basel and in addition supported by a grant of the Swiss National Science Foundation. The Framingham Heart Study (FHS): This work was supported by the National Heart, Lung and Blood Institute’s Framingham Heart Study Contract No. N01-HC 25195 and No. HHSN268201500001I, and by grants from the National Institute of Aging (R01s AG033193, AG008122, AG054076, AG033040, AG049607, AG05U01-AG049505), and the National Heart, Lung and Blood Institute (R01 HL093029, HL096917). The laboratory work for this investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, and by NIH contract N01-HC-25195. The analytical component of this project was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, and the Center for Information Technology, National Institutes of Health. This research is also funded from multiple grants including P30 AG066546, R01 AG059421, UF1 NS125513, and UH3NS100605. MEMENTO cohort: The MEMENTO cohort was sponsored by the Fondation Plan Alzheimer (Alzheimer Plan 2008– 2012). This work was also supported by the following: CIC 1401-EC, Bordeaux University Hospital, Inserm, and the University of Bordeaux. Genome-wide genotyping of MEMENTO was funded by a grant (EADB) from the EU Joint Program - Neurodegenerative Disease Research. S.D. is supported by a grant overseen by the French National Research Agency (ANR) as part of the “Investment for the Future Program” ANR-18-RHUS-0002, by the Precision and Global Vascular Brain Health Institute (VBHI) funded by the France 2030 IHU3 initiative ANR-23-IAHU-0001, by European Union’s Horizon 2020 research and innovation program under grant agreement No 640643 and 754517, by the Prix Burrus – Fondation pour la Recherche Médicale, and the Prix NRJ-neurosciences Académie des Sciences. AM is supported by ANR-23-CE12-0029-01 and Fondation Vaincre Alzheimer generic grant - OPE-2023-0031. Computations were performed on the Bordeaux Bioinformatics Center (CBiB) and the CREDIM computer resources, University of Bordeaux (funding provided to S.D. by the Fondation Claude Pompidou). The VETSA study was supported by Grants R03 AG065643, R01 AG050595, and R01 AG076838, K01 AG063805, and K24 AG046373 from the National Institute on Aging. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIA/NIH, or the VA. The U.S. Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of the VET Registry, including: Department of Defense; National Personnel Records Center, National Archives and Records Administration; Internal Revenue Service; National Opinion Research Center; National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. This material was, in part, the result of work supported with resources of the VA San Diego Center of Excellence for Stress and Mental Health Healthcare System. Most importantly, the VETSA co-authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families as well as the contributions of many staff members and students. ASPS-Fam: The Medical University of Graz and the Steiermärkische Krankenanstaltengesellschaft support the databank of the ASPS-Fam. The research reported in this article was funded by the Austrian Science Fund (FWF) grant numbers PI904, P20545-P05 and P13180 and supported by the Austrian National Bank Anniversary Fund, P15435 and the Austrian Ministry of Science under the aegis of the EU Joint Program-Neurodegenerative Disease Research (JPND)- www.jpnd.eu. ADNI: Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under Contract nos. (75N92022D00001, 75N92022D00002, 75N92022D00003, 75N92022D00004, 75N92022D00005). The ARIC Neurocognitive Study is supported by U01HL096812, U01HL096814, U01HL096899, U01HL096902, and U01HL096917 from the NIH (NHLBI, NINDS, NIA and NIDCD). Funding was also supported by R01HL087641 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. The authors thank the staff and participants of the ARIC study for their important contributions. This research was also supported from UH3-NS100605 and U01-AG052409 to M.F. The Coronary Artery Risk Development in Young Adults Study (CARDIA) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with the University of Alabama at Birmingham (HHSN268201800005I & HHSN268201800007I), Northwestern University (HHSN268201800003I), University of Minnesota (HHSN268201800006I), and Kaiser Foundation Research Institute (HHSN268201800004I). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging. Genotyping was funded as part of the NHLBI Candidate-gene Association Resource (N01-HC-65226) and the NHGRI Gene Environment Association Studies (GENEVA) (U01-HG004729, U01- HG04424, and U01-HG004446). This research was also supported from UH3-NS100605 and U01-AG052409 to M.F.
Author contributions
S.A., S.S., N.A.A., M.G. and M.Ar.I. Study design and drafting of the manuscript. S.A and M.A.I. writing and editing. S.A., M.A.I., A.M., R.W., M.H.R., J.C.B., M.F., G.R., E.H., M.W.L., WT.L.Jr., R.X., V.B., T.H.M., L.L., M.K., J.K., R.A.R., G.C., C.D., L.D., M.J.L., K.M., W.S.K., C.E.F., R.S., S.D., M.M.B., K.B., Q.Y., S.S., N.A.A., M.G. and M.Ar.I. Statistical analysis, interpretation of data and revision of the manuscript. J.C.B., M.F., T.H.M., L.L., C.D., L.D., M.J.L., K.M., W.S.K., C.E.F., R.S., S.D., M.M.B., K.B., Q.Y., S.S., N.A.A., M.G. and M.Ar.I. Acquisition of the data.
Peer review
Peer review information
Communications Biology thanks Lauren Byrne and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Melanie Bahlo and Benjamin Bessieres.
Data availability
All data generated during this study are included in this published article and its supplementary information/data files. Summary statistics of the GWAS is made available publicly in GWAS catalog (GCP000993). We used publicly available data in this manuscript, including data from GTEx version 8 (https://gtexportal.org/home/) and publicly available GWAS summary statistics for total hippocampal volume (https://www.ebi.ac.uk/gwas/publications/30279459), total brain volume (https://www.nature.com/articles/s41588-019-0516-6#Sec22), Alzheimer’s disease (https://www.ebi.ac.uk/gwas/publications/30820047,https://www.ebi.ac.uk/gwas/publications/35379992), Parkinson’s disease (https://www.ebi.ac.uk/gwas/studies/GCST90043734, https://www.ebi.ac.uk/gwas/studies/GCST009324), white matter lesions (https://www.ebi.ac.uk/gwas/publications/33293549), t-tau (https://www.ebi.ac.uk/gwas/publications/35396452), amyotrophic lateral sclerosis (https://www.projectmine.com/research/download-data/), Huntington’s disease (https://datadryad.org/stash/dataset/doi:10.5061%2Fdryad.5d4s2r8), amyloid beta 40, 42 and ratio (https://alz-journals.onlinelibrary.wiley.com/doi/epdf/10.1002/alz.12333).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shahzad Ahmad, Mohammad Aslam Imtiaz.
These authors jointly supervised this work: N. Ahmad Aziz, Mohsen Ghanbari, M. Arfan Ikram.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06804-3.
References
- 1.Gaetani, L. et al. Neurofilament light chain as a biomarker in neurological disorders. J. Neurol. Neurosurg. Psychiatry90, 870–881 (2019). 10.1136/jnnp-2018-320106 [DOI] [PubMed] [Google Scholar]
- 2.Zetterberg, H. Neurofilament light: A dynamic cross-disease fluid biomarker for neurodegeneration. Neuron91, 1–3 (2016). 10.1016/j.neuron.2016.06.030 [DOI] [PubMed] [Google Scholar]
- 3.Disanto, G. et al. Serum Neurofilament light: A biomarker of neuronal damage in multiple sclerosis. Ann. Neurol.81, 857–870 (2017). 10.1002/ana.24954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil, M. et al. Serum neurofilament light levels in normal aging and their association with morphologic brain changes. Nat. Commun.11, 812 (2020). 10.1038/s41467-020-14612-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders - towards clinical application. Nat. Rev. Neurol.20, 269–287 (2024). 10.1038/s41582-024-00955-x [DOI] [PubMed] [Google Scholar]
- 6.Alagaratnam, J. et al. Correlation between cerebrospinal fluid and plasma neurofilament light protein in treated HIV infection: results from the COBRA study. J. Neurovirol28, 54–63 (2022). 10.1007/s13365-021-01026-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol.14, 577–589 (2018). 10.1038/s41582-018-0058-z [DOI] [PubMed] [Google Scholar]
- 8.Bacioglu, M. et al. Neurofilament light chain in blood and CSF as marker of disease progression in mouse models and in neurodegenerative diseases. Neuron91, 56–66 (2016). 10.1016/j.neuron.2016.05.018 [DOI] [PubMed] [Google Scholar]
- 9.Niu, L. D. et al. Genome-wide association study of cerebrospinal fluid neurofilament light levels in non-demented elders. Ann. Transl. Med. 7, 657 (2019). 10.21037/atm.2019.10.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, J. Q. et al. Genome-wide association study identifies two loci influencing plasma neurofilament light levels. BMC Med. Genomics11, 47 (2018). 10.1186/s12920-018-0364-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, S. et al. TMEM106B and CPOX are genetic determinants of cerebrospinal fluid Alzheimer’s disease biomarker levels. Alzheimers Dement17, 1628–1640 (2021). 10.1002/alz.12330 [DOI] [PubMed] [Google Scholar]
- 12.Wuttke, M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet.51, 957–972 (2019). 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet.44, 369–375 (2012). 10.1038/ng.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet.51, 414–430 (2019). 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol.18, 1091–1102 (2019). 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu, Y. et al. Characterization of the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J. Nutr. Biochem24, 1697–1708 (2013). 10.1016/j.jnutbio.2013.02.010 [DOI] [PubMed] [Google Scholar]
- 17.Landers, J. E. et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA106, 9004–9009 (2009). 10.1073/pnas.0812937106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie, T. et al. Genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol. Aging35, 1778.e1779–1778.e1723 (2014). 10.1016/j.neurobiolaging.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science347, 1260419 (2015). 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 21.Ádori, C. et al. Critical role of somatostatin receptor 2 in the vulnerability of the central noradrenergic system: New aspects on Alzheimer’s disease. Acta Neuropathol.129, 541–563 (2015). 10.1007/s00401-015-1394-3 [DOI] [PubMed] [Google Scholar]
- 22.Stumm, R. K. et al. Somatostatin receptor 2 is activated in cortical neurons and contributes to neurodegeneration after focal ischemia. J. Neurosci.24, 11404–11415 (2004). 10.1523/JNEUROSCI.3834-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, D. et al. Involvement of mitochondrial K+ release and cellular efflux in ischemic and apoptotic neuronal death. J. Neurochem86, 966–979 (2003). 10.1046/j.1471-4159.2003.01913.x [DOI] [PubMed] [Google Scholar]
- 24.Korley, F. K. et al. Serum NfL (neurofilament light chain) levels and incident stroke in adults with diabetes mellitus. Stroke50, 1669–1675 (2019). 10.1161/STROKEAHA.119.024941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akamine, S. et al. Renal function is associated with blood neurofilament light chain level in older adults. Sci. Rep.10, 20350 (2020). 10.1038/s41598-020-76990-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barro, C., Chitnis, T. & Weiner, H. L. Blood neurofilament light: a critical review of its application to neurologic disease. Ann. Clin. Transl. Neurol.7, 2508–2523 (2020). 10.1002/acn3.51234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garzone, D. et al. Neurofilament light chain and retinal layers’ determinants and association: A population-based study. Ann. Clin. Transl. Neurol.9, 564–569 (2022). 10.1002/acn3.51522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Plas, E. et al. Associations between neurofilament light-chain protein, brain structure, and chronic kidney disease. Pediatr. Res91, 1735–1740 (2022). 10.1038/s41390-021-01649-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheppach, J. B. et al. Albuminuria and estimated GFR as risk factors for dementia in midlife and older age: Findings from the ARIC study. Am. J. Kidney Dis.76, 775–783 (2020). 10.1053/j.ajkd.2020.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheppach, J. B. et al. Association of kidney function measures with signs of neurodegeneration and small vessel disease on brain magnetic resonance imaging: The atherosclerosis risk in communities (ARIC) study. Am. J. Kidney Dis.81, 261–269.e261 (2023). 10.1053/j.ajkd.2022.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adam, J. et al. Endoplasmic reticulum stress in UMOD-related kidney disease: A human pathologic study. Am. J. Kidney Dis.59, 117–121 (2012). 10.1053/j.ajkd.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 32.Williams, S. E. et al. Uromodulin mutations causing familial juvenile hyperuricaemic nephropathy lead to protein maturation defects and retention in the endoplasmic reticulum. Hum. Mol. Genet18, 2963–2974 (2009). 10.1093/hmg/ddp235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, K. & Kaufman, R. J. From endoplasmic-reticulum stress to the inflammatory response. Nature454, 455–462 (2008). 10.1038/nature07203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piperi, C., Adamopoulos, C., Dalagiorgou, G., Diamanti-Kandarakis, E. & Papavassiliou, A. G. Crosstalk between advanced glycation and endoplasmic reticulum stress: emerging therapeutic targeting for metabolic diseases. J. Clin. Endocrinol. Metab.97, 2231–2242 (2012). 10.1210/jc.2011-3408 [DOI] [PubMed] [Google Scholar]
- 35.Li, H. et al. Crystal and solution structures of human protein-disulfide isomerase-like protein of the testis (PDILT) provide insight into its chaperone activity. J. Biol. Chem.293, 1192–1202 (2018). 10.1074/jbc.M117.797290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preische, O. et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat. Med.25, 277–283 (2019). 10.1038/s41591-018-0304-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gudjonsson, A. et al. A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat. Commun.13, 480 (2022). 10.1038/s41467-021-27850-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manini, A. et al. TMEM106B acts as a modifier of cognitive and motor functions in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 23, 10.3390/ijms23169276 (2022). [DOI] [PMC free article] [PubMed]
- 39.Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet.54, 412–436 (2022). 10.1038/s41588-022-01024-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu, Y. et al. rs1990622 variant associates with Alzheimer’s disease and regulates TMEM106B expression in human brain tissues. BMC Med.19, 11 (2021). 10.1186/s12916-020-01883-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brady, O. A., Zheng, Y., Murphy, K., Huang, M. & Hu, F. The frontotemporal lobar degeneration risk factor, TMEM106B, regulates lysosomal morphology and function. Hum. Mol. Genet22, 685–695 (2013). 10.1093/hmg/dds475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Es, M. A. et al. Analysis of FGGY as a risk factor for sporadic amyotrophic lateral sclerosis. Amyotroph. Lateral Scler.10, 441–447 (2009). 10.3109/17482960802673042 [DOI] [PubMed] [Google Scholar]
- 43.Aberg, K. et al. Genomewide association study of movement-related adverse antipsychotic effects. Biol. Psychiatry67, 279–282 (2010). 10.1016/j.biopsych.2009.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daoud, H., Valdmanis, P. N., Dion, P. A. & Rouleau, G. A. Analysis of DPP6 and FGGY as candidate genes for amyotrophic lateral sclerosis. Amyotroph. Lateral Scler.11, 389–391 (2010). 10.3109/17482960903358857 [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., Zagnitko, O., Rodionova, I., Osterman, A. & Godzik, A. The FGGY carbohydrate kinase family: Insights into the evolution of functional specificities. PLoS Comput Biol.7, e1002318 (2011). 10.1371/journal.pcbi.1002318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunckley, T. et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N. Engl. J. Med.357, 775–788 (2007). 10.1056/NEJMoa070174 [DOI] [PubMed] [Google Scholar]
- 47.Mullard, A. NfL makes regulatory debut as neurodegenerative disease biomarker. Nat. Rev. Drug Discov.22, 431–434 (2023). 10.1038/d41573-023-00083-z [DOI] [PubMed] [Google Scholar]
- 48.Garcia, M. L. et al. NF-M is an essential target for the myelin-directed “outside-in” signaling cascade that mediates radial axonal growth. J. cell Biol.163, 1011–1020 (2003). 10.1083/jcb.200308159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campos-Melo, D., Hawley, Z. C. E. & Strong, M. J. Dysregulation of human NEFM and NEFH mRNA stability by ALS-linked miRNAs. Mol. Brain11, 43 (2018). 10.1186/s13041-018-0386-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavedan, C., Buchholtz, S., Nussbaum, R. L., Albin, R. L. & Polymeropoulos, M. H. A mutation in the human neurofilament M gene in Parkinson’s disease that suggests a role for the cytoskeleton in neuronal degeneration. Neurosci. Lett.322, 57–61 (2002). 10.1016/S0304-3940(01)02513-7 [DOI] [PubMed] [Google Scholar]
- 51.Peleg, S. et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science328, 753–756 (2010). 10.1126/science.1186088 [DOI] [PubMed] [Google Scholar]
- 52.Sherva, R. et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement10, 45–52 (2014). 10.1016/j.jalz.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cronin, S. et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum. Mol. Genet17, 768–774 (2008). 10.1093/hmg/ddm361 [DOI] [PubMed] [Google Scholar]
- 54.Agís-Balboa, R. C. et al. Formin 2 links neuropsychiatric phenotypes at young age to an increased risk for dementia. Embo j.36, 2815–2828 (2017). 10.15252/embj.201796821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, L. et al. Genome-wide DNA methylation profiles of autism spectrum disorder. Psychiatr. Genet32, 131–145 (2022). 10.1097/YPG.0000000000000314 [DOI] [PubMed] [Google Scholar]
- 56.Wexler, E. M. et al. Genome-wide analysis of a Wnt1-regulated transcriptional network implicates neurodegenerative pathways. Sci. Signal4, ra65 (2011). 10.1126/scisignal.2002282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian, L. et al. β2-adrenergic receptor activation prevents rodent dopaminergic neurotoxicity by inhibiting microglia via a novel signaling pathway. J. Immunol.186, 4443–4454 (2011). 10.4049/jimmunol.1002449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peterson, L., Ismond, K. P., Chapman, E. & Flood, P. Potential benefits of therapeutic use of β2-adrenergic receptor agonists in neuroprotection and Parkinsonμs disease. J. Immunol. Res. 2014, 103780 (2014). 10.1155/2014/103780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksanen, M. et al. Astrocyte alterations in neurodegenerative pathologies and their modeling in human induced pluripotent stem cell platforms. Cell Mol. Life Sci.76, 2739–2760 (2019). 10.1007/s00018-019-03111-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park, H. M. et al. The CRL3(gigaxonin) ubiquitin ligase-USP15 pathway governs the destruction of neurofilament proteins. Proc. Natl Acad. Sci. USA120, e2306395120 (2023). 10.1073/pnas.2306395120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dubiel, W., Dubiel, D., Wolf, D. A. & Naumann, M. Cullin 3-based ubiquitin ligases as master regulators of Mammalian cell differentiation. Trends Biochem Sci.43, 95–107 (2018). 10.1016/j.tibs.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Auton, A. et al. A global reference for human genetic variation. Nature526, 68–74 (2015). 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet.48, 1279–1283 (2016). 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Winkler, T. W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc.9, 1192–1212 (2014). 10.1038/nprot.2014.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics26, 2190–2191 (2010). 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mägi, R. et al. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet26, 3639–3650 (2017). 10.1093/hmg/ddx280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun.8, 1826 (2017). 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe, K., Umićević Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun.10, 3222 (2019). 10.1038/s41467-019-11181-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics26, 2336–2337 (2010). 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet47, 291–295 (2015). 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol.11, e1004219 (2015). 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet.45, 580–585 (2013). 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.CAG repeat not polyglutamine length determines timing of Huntington’s disease onset. Cell178, 887–900.e814 (2019). [DOI] [PMC free article] [PubMed]
- 74.van Rheenen, W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet53, 1636–1648 (2021). 10.1038/s41588-021-00973-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Damotte, V. et al. Plasma amyloid β levels are driven by genetic variants near APOE, BACE1, APP, PSEN2: A genome-wide association study in over 12,000 non-demented participants. Alzheimers Dement17, 1663–1674 (2021). 10.1002/alz.12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarnowski, C. et al. Meta-analysis of genome-wide association studies identifies ancestry-specific associations underlying circulating total tau levels. Commun. Biol.5, 336 (2022). 10.1038/s42003-022-03287-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van der Meer, D. et al. Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Mol. Psychiatry25, 3053–3065 (2020). 10.1038/s41380-018-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, B. et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet51, 1637–1644 (2019). 10.1038/s41588-019-0516-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sargurupremraj, M. et al. Cerebral small vessel disease genomics and its implications across the lifespan. Nat. Commun.11, 6285 (2020). 10.1038/s41467-020-19111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–d901 (2017). 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi, S. W. & O’Reilly, P. F. PRSice-2: Polygenic risk score software for biobank-scale data. Gigascience8, 10.1093/gigascience/giz082 (2019). [DOI] [PMC free article] [PubMed]
- 82.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet10, e1004383 (2014). 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu, B., Gloudemans, M. J., Rao, A. S., Ingelsson, E. & Montgomery, S. B. Abundant associations with gene expression complicate GWAS follow-up. Nat. Genet.51, 768–769 (2019). 10.1038/s41588-019-0404-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife7, 10.7554/eLife.34408 (2018). [DOI] [PMC free article] [PubMed]
- 85.Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol.40, 304–314 (2016). 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J. Epidemiol.44, 512–525 (2015). 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowden, J. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int. J. Epidemiol.47, 1264–1278 (2018). 10.1093/ije/dyy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
All data generated during this study are included in this published article and its supplementary information/data files. Summary statistics of the GWAS is made available publicly in GWAS catalog (GCP000993). We used publicly available data in this manuscript, including data from GTEx version 8 (https://gtexportal.org/home/) and publicly available GWAS summary statistics for total hippocampal volume (https://www.ebi.ac.uk/gwas/publications/30279459), total brain volume (https://www.nature.com/articles/s41588-019-0516-6#Sec22), Alzheimer’s disease (https://www.ebi.ac.uk/gwas/publications/30820047,https://www.ebi.ac.uk/gwas/publications/35379992), Parkinson’s disease (https://www.ebi.ac.uk/gwas/studies/GCST90043734, https://www.ebi.ac.uk/gwas/studies/GCST009324), white matter lesions (https://www.ebi.ac.uk/gwas/publications/33293549), t-tau (https://www.ebi.ac.uk/gwas/publications/35396452), amyotrophic lateral sclerosis (https://www.projectmine.com/research/download-data/), Huntington’s disease (https://datadryad.org/stash/dataset/doi:10.5061%2Fdryad.5d4s2r8), amyloid beta 40, 42 and ratio (https://alz-journals.onlinelibrary.wiley.com/doi/epdf/10.1002/alz.12333).