Abstract
The genomes of three Pyrococcus species, P.abyssi, P.furiosus and P.horikoshii, were compared at the DNA level, taking advantage of our identification of their replication origins. Three types of rearrangements have been identified: (i) inversion and translation across the replication axis (origin/terminus), (ii) inversion and translocation restricted to a replichore (the half chromosome divided by the replication axis) and (iii) apparent mobility of long clusters of repeated sequences. Rearrangements restricted within a replichore were more common between P.furiosus and the two other Pyrococcus species than between P.horikoshii and P.abyssi. A strong correlation was found between 23 homologous insertion sequence elements, present only in P.furiosus, and recombined segment boundaries, suggesting that transposition events have been a major cause of genomic disruption in this species. Moreover, gene orientation bias was much more disrupted than strand composition biases in fragments that switched their orientation within a replichore upon recombination. This allowed us to conclude that one reversion and one translation occurred in P.abyssi after its divergence from P.horikoshii, and that a smaller segment has specifically recombined in P.furiosus. Whereas a majority of genes are transcribed in the same direction as DNA replication in P.horikoshii and P.abyssi, the colinearity of transcription and replication is only maintained for highly transcribed genes in P.furiosus. We discuss the implications of genomic rearrangements on gene orientation and composition biases, and their consequences on sequence evolution.
INTRODUCTION
Comparative genomics has, up to now, mostly focused either on the comparison of distantly related organisms, aiming to reconstruct global phylogenetic trees (1–5), or on pairwise comparisons of closely related pathogenic bacteria with distinct phenotypes in order to identify genes involved in different tissue tropism and disease expression (6,7). Genomes of closely related species showed a limited number of recombination events between pairs of Mycoplasma species, Helicobacter pylori and Chlamydia isolates, whereas many rearrangements were identified inside pairs of different Pyrococcus, Mycobacterium and Chlamydia species (6–13; see 8 for a review on genome rearrangements). These rearrangements were so extensive in the Mycobacterium and Pyrococcus species that only short regions of gene order conservation (synteny) were identified between genomes. In the case of the two Pyrococcus species, P.furiosus and P.horikoshii, Robb and colleagues suggested that the genomes of these hyperthermophiles were especially prone to DNA recombination, possibly in relation to their ability to withstand high doses of ionizing radiation (9,14). However, comparative analyses of bacterial and archaeal genomic physical maps have already indicated a similar level of chromosomal rearrangement in closely related species or even strains (15–17), indicating that genome plasticity might be the rule in prokaryotic genome evolution.
Pairwise comparison of Helicobacter isolates, and Chlamydia and Mycobacterium species, led Tillier and Collins (12) to suggest that genome rearrangement occurred mainly via replication-directed translocation. This originates from the observation that many non-colinear genes between the two closely related genomes were inverted and translocated across an axis defined by the origin and terminus of replication (oriC and terC), which separates the chromosome into two ‘replichores’, as already documented in bacteria (see, for example, 18–20). Comparison of the Helicobacter isolates examined so far also suggested that insertion elements and restriction–modification genes are another important driving force behind recombination in these strains (6), whereas comparison of the Chlamydia genomes (7) revealed that some major rearrangements also occurred at pathogenicity-linked ‘plasticity zones’. A variety of other mechanisms have been proposed to explain fragment shuffling, such as homologous recombination between GC-rich repetitive sequences of the RepMP family in Mycoplasma species (21), or recombination between rRNA operons, insertion sequences (IS) or other repeated elements in Escherichia coli and other bacteria (16,22,23).
Several recombination mechanisms have also been proposed to explain chromosomal rearrangements in the Pyrococcus strains. Chinen et al., comparing P.horikoshii and Pyrococcus abyssi (24), suggested that two chromosomal rearrangements, and possibly more, could be linked to restriction–modification-bearing mobile elements. However, the actual function and mobility of these genes remains to be ascertained in Pyrococcus. Likewise, Lecompte et al. (25) concluded from comparison of the three Pyrococcus species that rearrangements originated from site-specific as well as from non-homologous recombinations, based on tRNA gene targeted integration/deletion and IS element transposition, respectively, a situation reminiscent of what has been found in Helicobacter.
As highlighted above, knowledge of the replication mode and oriC localization is of particular importance in understanding inter-species genome rearrangements. We have recently been able to identify both in silico and experimentally the replication origin of the archaeon P.abyssi (26), and pairwise comparison of P.abyssi and P.horikoshii has already indicated that the replication terminus is a hot spot for recombination in the Pyrococcus genus, as in Bacteria (25,26). Moreover, in a preliminary investigation, we and others (27,28) pointed out that most rearranged segments in the three Pyrococcus species fall into two different categories, namely replication driven, symmetrical cross-replichore recombination, and intra-replichore rearrangements often linked to IS element transposition.
Our identification of the replication origins in three closely related Pyrococcus species (P.abyssi, P.horikoshii and P.furiosus) (26) allowed us to analyze in more detail the different types of chromosomal rearrangements that have occurred between their three genomes. The observation of a clear correlation between modifications of strand composition biases and the different kinds of rearrangements within the chromosome replichores allowed us to gain insights into the mechanism of genome evolution in the archaeal domain.
MATERIALS AND METHODS
Genome sequences
Pyrococcus abyssi and P.horikoshii complete nucleotide sequences and annotations were retrieved from GenBank at http://www.ncbi.nlm.nih.gov/Genomes/. Accession numbers are AL096836 and BA000001 for P.abyssi and P.horikoshii, respectively.
Pyrococcus furiosus complete nucleotide sequence and annotation is as provided by the Utah Genome Center at http://www.genome.utah.edu/sequence.html. The unpublished sequence annotation is credited to R. B. Weiss, D. M. Dunn, F. T. Robb and J. R. Brown.
Whole genome alignments, repeated sequences analysis
Inter-genome comparisons were performed by pairwise BLASTN alignments (29) with the following settings: default alignment parameters (gap penalties: existence, 5; extension, 2) were used without filtering and with an exclusion threshold of E < 10–05 (expectation value). Genome sequences were chopped into 100 nt segments, and subsequently blasted against the two other genomes. Sequences having the best high-scoring segment pair were used to plot the alignment diagram, retaining 100 nt segments showing 100% identity over at least 25 nt within both matched genomes. Control comparisons were done by aligning protein sequence sets from each genome with a 60% identity threshold, showing plot diagrams almost identical to those obtained by nucleic alignments.
Intra-genome comparison for long clusters of tandem repeats (LCTRs) was performed as above, except that 10 000 nt segments were used.
Sequence composition analysis
Strand composition analyses were performed essentially as described (30,31). Briefly, a skew is defined as the ratio of one oligonucleotide (word skew) to its reverse complement over a fixed length window (here one-fiftieth of genome length). Informative words, i.e. strongly biased, are used [such as G over C skew (G/C skew) or the Chi octamer GCTGGTGG of E.coli (32)]. For all three Pyrococcus species, the GGTT tetramer has a strong bias and a well-defined singularity (26) and thereafter was chosen as a reference. The skew was computed over the entire genome sequence by shifting the window by 1/240th of genome length increments. For gene orientation bias, the ratio of the number of genes (as annotated in public databases) in each orientation within the window was used in place of oligonucleotides. For ‘AT3’ skew, the ratio of A to T nucleotides at the third codon position only was computed. Cumulative skews (30) were used in this study, i.e. skew values were integrated from the starting point in the sequence as given in the database.
Codon usage bias was measured by the codon adaptation index (CAI) method (33). The CAI was calibrated on ribosomal proteins, whose sequence should be encoded by the most highly expressed genes.
RESULTS AND DISCUSSION
Chromosomal rearrangements among the three Pyrococcus genomes
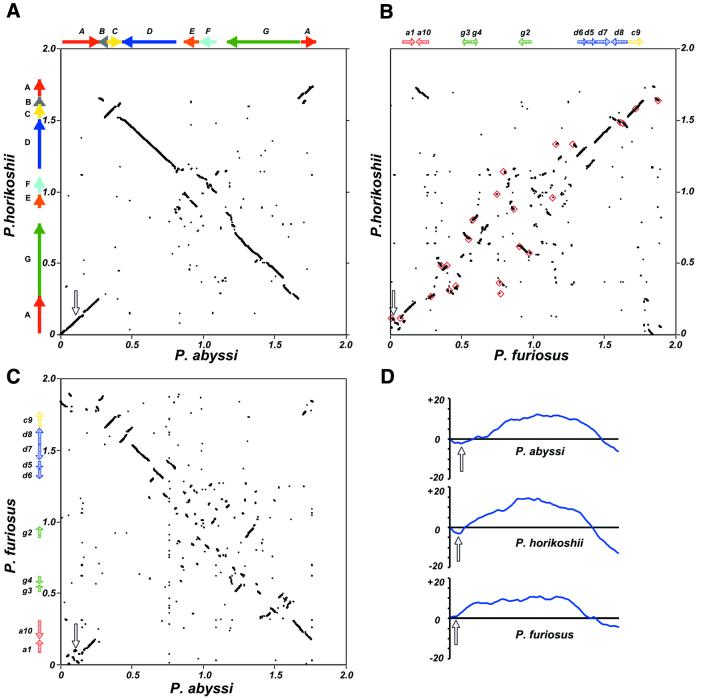
Whole Pyrococcus genome alignments (Fig. 1) showed that the extent of rearrangement that occurred after the three species diverged is much lower between P.abyssi and P.horikoshii than between these two species and P.furiosus. Our analysis showed that there are three large and four small chromosomal segments (segments A–G; Fig 1A) between P.abyssi and P.horikoshii, ranging from 40 to 500 kb, for which nucleotide similarity is sufficient to produce continuous segments on plot diagrams and overall gene order conservation is preserved (synteny regions; see Materials and Methods), whereas 10 smaller segments of colinear gene order ranging from 40 to 120 kb existed between the three genomes (a1, a10, d5–d8, g2–g4 and c9), along with several other even smaller fragments (Fig. 1B and C).
Figure 1.
Pyrococcus whole genome DNA alignments and GGTT word skew analysis. (A–C) Genome pairwise alignments as indicated besides and beneath the frames axes; each dot represents a 100 nt segment significantly matching in both genomes (see Materials and Methods). Plots of coordinate origins and sequence orientations are taken from the reference sequences. (A) Solid colored arrows indicate the large A to G synteny segments between P.horikoshii and P.abyssi and their relative orientation in each genome. (B and C) Shaded colored arrows represent the location and relative orientation of smaller segments conserved in all three genomes (a1, a10, g2–g4, d5–d8 and c9). (B) Red squares denote the locations of IS elements identified in P.furiosus. Scales are in Mb, open arrows indicate the location of oriC in each genome. (D) Cumulative GGTT skews were calculated for each Pyrococcus genome and plotted. The x-axis is the genome length, the y-axis represents the cumulative difference in arbitrary units. Open arrows indicate the location of oriC in each genome as in (A–C).
The most parsimonious explanation for large segments shuffling between P.abyssi and P.horikoshii implies that two inversions (segments A and C) and one transposition (E/F) occurred after their divergence (Fig. 2). The ~370 kb A segment is, moreover, remarkable because it contains the replication origin. As a consequence, its reversion in P.abyssi compared to P.horikoshii does not change gene orientation relative to the origin and terminus of replication axis (26), and produces the characteristic X-shaped plot diagrams (12,13) (Fig. 1A).
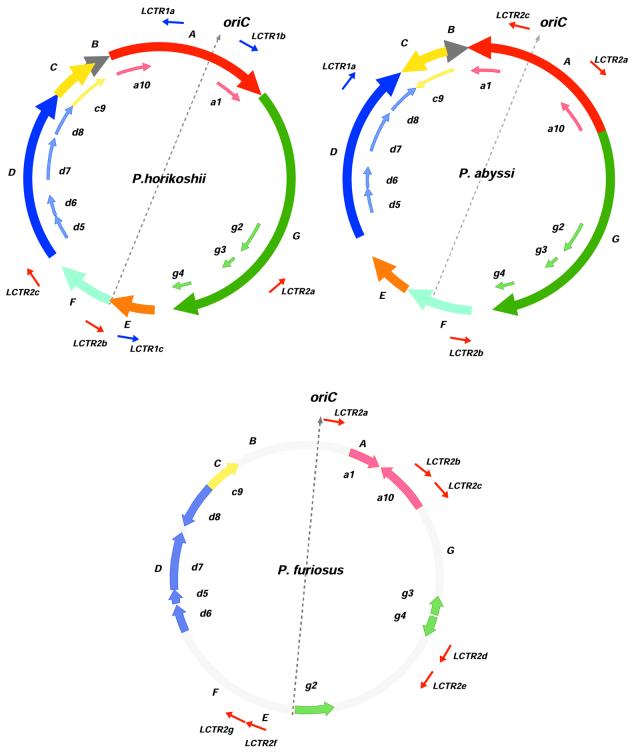
Figure 2.
A sketch of P.abyssi, P.horikoshii and P.furiosus genome organization. Location and orientation of major syntenic A to G segments between P.abyssi and P.horikoshii are represented on outer circles by solid colored arrows, along with 10 syntenic segments of P.furiosus (a1, a10, g2–g4, d5–d8 and c9) represented with shaded color arrows on inner circles. For P.furiosus (bottom) only a, g, d and c small fragments are presented in shaded arrows. For each genome, oriC, the replication origin, and the symmetrically located putative terminus of replication are connected by a gray dashed arrow, dividing the chromosome into two replichores. LCTR elements are indicated for each genome with their respective orientation.
When comparing the P.abyssi and P.horikoshii pair to P.furiosus, large A to G segments were disrupted into many pieces often <50 kb, and no synteny remained in the plot’s central region. Although substantial, these rearrangements did not alter the cumulative word skew obtained for P.furiosus, which remained quite similar to those of P.abyssi and P.horikoshii (Fig. 1D), the singularity in the P.furiosus skew curve coinciding with the genome region homologous to oriC already identified in the other Pyrococcus species.
Segment boundaries in P.abyssi and P.horikoshii often remained difficult to ascertain even by examining pairwise nucleotide genome alignments (see Materials and Methods), and transitions between segments could not be narrowed to regions <0.2 kb at best, and up to ∼10 kb. This, in turn, hampered recognition of clear-cut features at boundaries, although some of the smaller indel borders were indeed correlated with the nearby presence of tRNA genes or restriction–modification genes (24,25).
The case of P.furiosus is somewhat different, as there is a noticeable correlation between the location of 23 homologous transposon-associated IS-like elements identified by Diruggiero et al. (34) and the segmentation pattern. Their distribution is clearly non-random (Fig. 1B), since 16 of them are located near shuffled segments boundaries, five are in completely scrambled regions and only two are found within an undisturbed segment. Since multicopy IS elements are not found in P.horikoshii and P.abyssi, the P.furiosus genome could have been invaded by these elements after its divergence from the common lineage to P.horikoshii and P.abyssi.
Surprisingly, 8 out of the 10 syntenic segments that remained between P.furiosus and the other Pyrococcus species were shuffled in P.furiosus (segments a1, a10, d5, d6, d8 and g2–g4; Fig. 2), but stay inside the same replichore in all three species (28). The origin/terminus of the replication axis therefore seems to act as an effective barrier to fragment exchange in Pyrococcus, in contrast to what has been observed for replication-related fragment shuffling in Chlamydia and Mycobacterium species.
Strand composition bias as a gauge of genome evolution
Large genomic rearrangements identified between Pyrococcus strains were investigated using strand composition bias analysis, more specifically cumulative skew analysis, a method that allowed us to identify the replication origin in Pyrococcus (26). Regular compositional skews (oligonucleotide skews, GC skews, etc.) often observed in whole genomic sequences are tightly correlated with the chromosome replication origin and terminus axis (30,35–37). Consequently, chromosomal rearrangements have different consequences on skew curves, depending on whether or not rearranged fragments change their orientation relative to replication direction. In the first case, termed non-synonymous rearrangement, a significant perturbation is expected in the curves, whereas in the second, synonymous rearrangement case, skew curve modification should be minimal. As noted above, however, skew curves of reversed segments containing the replication origin should not change, because gene orientation, hence sequence composition bias, is unchanged relative to replication direction.
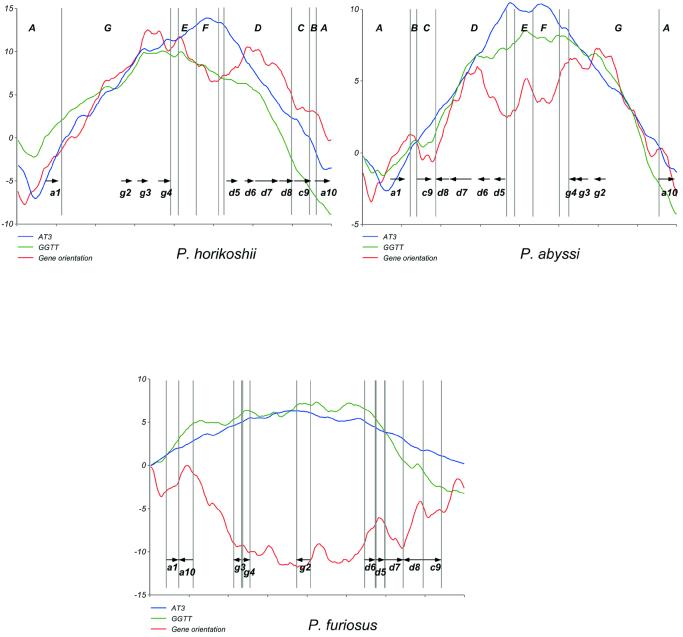
Close comparison of the three Pyrococcus species led us to focus on three segments with conserved up and downstream genomic environments, making them suitable for comparison, and having experienced non-synonymous recombinations: the C reversion and the E/F translation, both between P.horikoshii and P.abyssi (bordered by segments B/D and segments D/G, respectively), and the d8 segment reversal in P.furiosus (bordered by segments d7 and c9 in all three Pyrococcus species; see Fig. 2).
All three segments share a common pattern of differences in their skew curves (Fig. 3). Indeed, for each segment, one genome (or group of genomes) showed smooth and regular curves, whereas the other (or group) showed disturbed skew curves, with gene orientation skew always most highly perturbed. Hence C, and to a lesser extent E/F, regions displayed quite regular skews in P.horikoshii for GGTT word, gene orientation and AT at the third base of the codon (AT3; Fig. 3) but the gene orientation and GGTT skews are markedly upset in P.abyssi. The GGTT skew perturbation was, however, less pronounced for the E/F region. In the d8 segment, gene orientation skew was reversed in P.furiosus compared to GGTT and AT3 skews, whereas all skews had the same orientation in P.abyssi and P.horikoshii. The fact that these segments share equivalent genomic environments in all genomes gives us a strong argument to conclude that segments C and likely E/F were rearranged in P.abyssi and not in P.horikoshii, and that the d8 segment was rearranged in P.furiosus.
Figure 3.
Different cumulative skews for two Pyrococcus species. Skews are computed as described in Materials and Methods. The GGTT curve is given in arbitrary units, and gene orientation and AT3 skews are fitted on it for clarity, meaning that scale is not informative. Syntenic A to G and a, c, d, g segments from Figure 1 are delimited by vertical lines.
The observed uncoupling of compositional biases implies that they evolved at different rates, in agreement with the hypothesis that skews tend to settle with different kinetics (AT3 > word > gene orientation) depending on differential selective pressures, such as mutation bias at specific positions in the codon, strand-specific replication or transcription-coupled repair, or others (30,31,35). Our results also showed that the AT3 skew was less upset than the word skew (here GGTT), which was itself, as expected, less perturbed than the gene orientation skew (Fig. 3). This explains why the three Pyrococcus genomes exhibit such regular AT3 skews despite their numerous rearrangements. In particular, a restoration of the smooth gene orientation curve after non-synonymous recombinations (such as the C, E/F and d8 segments) would require all genes to be reversed back, whereas settlement of other nucleotide or word skews do not require a gene orientation switch, due to degeneracy of the genetic code.
Long clusters of tandem repeats mobility
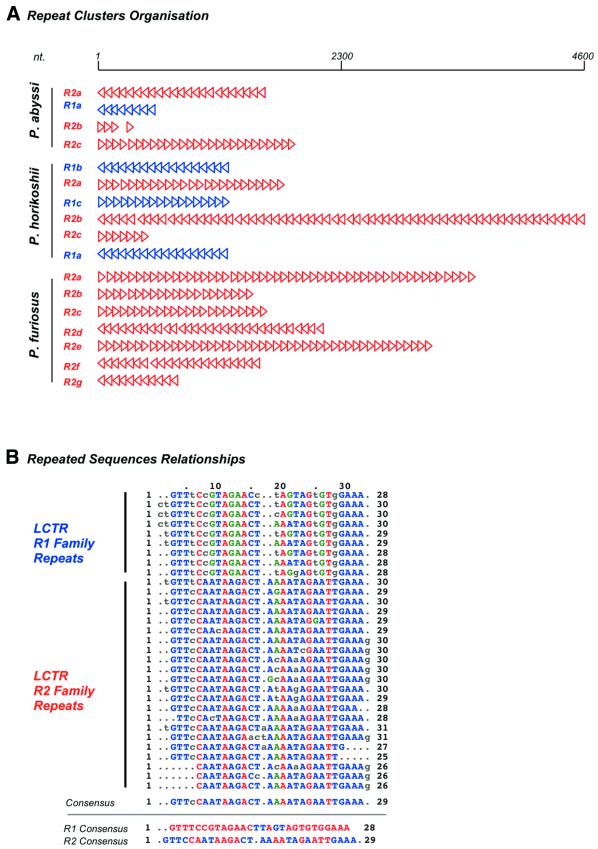
The three Pyrococcus genomes contained several copies of LCTRs also detected in other chromosomes and plasmids from archaea or some bacteria (38–41). LCTRs are non-coding regions characterized by clusters of short (~30 nt) repeated elements organized in tandem, each element being regularly spaced by unique intervening sequences ranging from 60 to 100 nt (Fig. 4). Pyrococcus short repeats split into two families of related sequences, R1 and R2, which never combine within the same repeats cluster. As shown in Figure 2, two LCTRs are symmetrically located around and roughly at the same distance from replication origins of P.abyssi and P.horikoshii, a third one being located at ∼50 kb of the putative terminus (Fig. 2). Surprisingly, although both R1 and R2 families of LCTRs are present in each genome, those flanking the replication origin and terminus of P.horikoshii are from the R1 family, whereas the P.abyssi oriC and terminus are surrounded by R2 LCTRs. Moreover, R1 family clusters found in the A and E segments in P.horikoshii are confined to segment D in P.abyssi, whereas family R2 clusters (on segments D, F and G) in P.horikoshii switch to segments A and F in P.abyssi (Fig. 2). A tentative explanation for this reciprocal shuffling between two Pyrococcus strains is that LCTRs behave like mobile elements.
Figure 4.
(A) LCTR family 1 and 2 (blue and red arrowheads, respectively) organization is represented for P.abyssi, P.horikoshii and P.furiosus. Each arrowhead represents a ~30 nt repeat unit and its orientation in the cluster, the scale is in nt. (B) Sequence characteristics of repeat units from family 1 and 2, aligned with clustal v1.8 program.
Indeed, it has been argued that LCTRs may be involved in gene transfer (38), but analogy with some plasmid partition systems led several authors to suggest that archaeal LCTRs could be analogs of bacterial partition sequences (40,41). In good agreement with this hypothesis, the peculiar organization of LCTRs around oriC and terC observed in P.abyssi and P.horikoshii was also found in genomes of the archaeon Methanobacterium thermoautotrophicum and of the bacterium Thermotoga maritima (not shown).
However, P.furiosus does not apparently follow the same rules, as only R2 family LCTRs are present, and their overall distribution is different from the previous scheme. If our model of the recent invasion of P.furiosus by IS elements is correct, it could be hypothesized that this strain is in the process of reorganizing his LCTRs.
Replication/transcription fitting in the course of chromosome evolution
Bacterial genomes for which the location of the replication origin is known typically verify that a majority of genes are transcribed in the same direction as DNA replication (42–44). This appears also true for P.horikoshii and P.abyssi, in which ∼54% of predicted genes are transcribed in the same direction as DNA replication (Table 1). The situation in P.furiosus is strikingly different, as exemplified by the concave shape of the gene orientation skew curve (Fig. 3) and the fact that only about 45% of genes are transcribed in the same direction as replication (Table 1). This feature is unique among completely sequenced genomes until now. However, if we restrict this analysis to subclasses of most highly expressed genes (e.g. ribosomal protein genes, see top ranking CAI 10, 5 and 2% in Table 1), the codirectionality of replication and transcription is restored for the three genomes. A notable exception is P.abyssi, for which the colinearity of gene orientation and replication decreases as the sampling is narrowed from 5 down to 2% top ranking subclasses. This anomaly can be readily explained by observing that the C segment, which has switched orientation in P.abyssi (see above), contains 23 (out of a total of 64) ribosomal protein genes which are counter-replication oriented in this species, thus accounting for the decreased percentage in this subclass.
Table 1. Replication-translation colinearity in the three Pyrococcus genomes.
| Top ranking CAI (%) | P.abyssi | P.horikoshii | P.furiosus |
|---|---|---|---|
| 2 | 60.0 | 73.1 | 54.8 |
| 5 | 62.5 | 67.0 | 50.0 |
| 10 | 60.8 | 65.0 | 48.6 |
| Total | 53.8 | 54.7 | 45.3 |
Pyrococcus genes are sorted by strength of expression, as measured by the CAI. In each subclass of top-ranking genes, the percentage of genes oriented in the same direction as replication is indicated. The CAI was calibrated with ribosomal proteins, whose sequence should be encoded by the most highly expressed genes.
Though it has been argued that the colinearity of transcription and replication was under strong selective pressure in E.coli or Bacillus subtilis, this might not be such a heavy constraint in other prokaryotes. This is exemplified by P.furiosus global gene orientation and by the C segment in P.abyssi, for which a significant portion of highly expressed genes can be reversed without a major influence on cell viability. Nevertheless, selection appears to favor replication/transcription codirectionality for highly expressed genes in Archaea as in Bacteria. This is highlighted by the increased ratio of replication-oriented genes in P.furiosus for the most highly expressed fraction of them (10–5 and 2% top ranking CAI genes; Table 1). As the massive invasion of P.furiosus by IS elements may have disrupted the global gene orientation and replication colinearity, once again this strain could be in a reorganization process.
CONCLUSION
Pyrococcus genomes comparison exemplifies well the recognized major causes of chromosome instability: first, non-homologous crossovers linked to replication forks, often resulting in very large segment rearrangements across the replication axis, which leads to characteristic X shaped alignment plots (12,13) (Fig. 1). This type of event does not alter significantly the compositional organization features of the chromosome, such as gene orientation and strand oligonucleotide biases, because the resulting rearrangements are mostly synonymous. The other main recombination type is small scale segment shuffling, caused by homologous or site-targeted recombination of tRNAs, restriction–modification genes and IS transposition activity. This often leads to non-synonymous rearrangements, as opposed to the first scheme, and thus appears to promptly trigger active remodeling of the chromosome, presumably as the result of perturbations to sensitive sequence compositional parameters.
The three Pyrococcus genomes allow us to establish unambiguously the chronology of some recombination events by correlating rearrangements of syntenic segments with the corresponding modifications in sequence bias curves. The observed behavior of bias curves—with a decrease in perturbations going from the highly upset gene orientation skew, then oligonucleotide words, down to A/T at the third position in the codon triplet—are interpreted as a mechanism which favors restoration of the original curve shape. The most obvious consequence will be that the underlying sequence will have to evolve to fit its new environment (11,37,45). We suggest that this perturbation–restoration model of genome evolution can be extended to other genome elements such as LCTRs, which exhibit a quite surprising mobile-like behavior. There is growing evidence that these elements might be involved at some stage of chromosome partitioning in archaea (40,41), but this does not seem to be critical for proper cell functioning. We infer that this preferred, but not obliged, sequence organization is maintained by appropriate LCTR shuffling whenever a chromosomal rearrangement occurs. The same conclusion does indeed apply to other ‘plastic’ parameters, i.e. strand composition and gene orientation.
Thus, our results clearly establish how structural modifications of the chromosome, i.e. recombination in this study, are able to promote and/or favor sequence mutations and rearrangements such that the original organization is restored. It might be that this mechanism is one of the major driving forces in genome evolution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. Weissenbach and his staff for sequencing the complete genome of P.abyssi. This work was supported by a research grant from the ‘Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires’ of the Ministère de l’Education Nationale de la Recherche et de la Technologie.
REFERENCES
- 1.Snel B., Bork,P. and Huynen,M.A. (1999) Genome phylogeny based on gene content. Nature Genet., 21, 66–67. [DOI] [PubMed] [Google Scholar]
- 2.Fitz-Gibbon S.T. and House,C.H. (1999) Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res., 27, 4218–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tekaia F., Lazcano,A. and Dujon,B. (1999) The genomic tree as revealed from whole proteome comparisons. Genome Res., 9, 550–557. [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen J.A. (2000) Assessing evolutionary relationships among microbes from whole-genome analysis. Curr. Opin. Microbiol., 3, 475–480. [DOI] [PubMed] [Google Scholar]
- 5.Sicheritz-Ponten T. and Andersson,S.G. (2001) A phylogenomic approach to microbial evolution. Nucleic Acids Res., 29, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm R.A., Ling,L.S., Moir,D.T., King,B.L., Brown,E.D., Doig,P.C., Smith,D.R., Noonan,B., Guild,B.C., deJonge,B.L., Carmel,G., Tummino,P.J., Caruso,A., Uria-Nickelsen,M., Mills,D.M., Ives,C., Gibson,R., Merberg,D., Mills,S.D., Jiang,Q., Taylor,D.E., Vovis,G.F. and Trust,T.J. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature, 397, 176–180. [DOI] [PubMed] [Google Scholar]
- 7.Read T.D., Brunham,R.C., Shen,C., Gill,S.R., Heidelberg,J.F., White,O., Hickey,E.K., Peterson,J., Utterback,T., Berry,K., Bass,S., Linher,K., Weidman,J., Khouri,H., Craven,B., Bowman,C., Dodson,R., Gwinn,M., Nelson,W., DeBoy,R., Kolonay,J., McClarty,G., Salzberg,S.L., Eisen,J. amd Fraser,C.M. (2000) Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res., 28, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes D. (2000) Evaluating genome dynamics: the constraints on rearrangements within bacterial genomes. Genome Biol., 1, REVIEWS0006.1–0006.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeder D.L., Weiss,R.B., Dunn,D.M., Cherry,J.L., Gonzalez,J.M., DiRuggiero,J. and Robb,F.T. (1999) Divergence of the hyperthermophilic archaea Pyrococcus furiosus and P. horikoshii inferred from complete genomic sequences. Genetics, 152, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev A. (2000) Graphical genome comparison: rearrangements and replication origin of Helicobacter pylori.Trends Genet., 16, 376–378. [DOI] [PubMed] [Google Scholar]
- 11.Tillier E.R. and Collins,R.A. (2000) Replication orientation affects the rate and direction of bacterial gene evolution. J. Mol. Evol., 51, 459–463. [DOI] [PubMed] [Google Scholar]
- 12.Tillier E.R. and Collins,R.A. (2000) Genome rearrangement by replication-directed translocation. Nature Genet., 26, 195–197. [DOI] [PubMed] [Google Scholar]
- 13.Eisen J.A., Heidelberg,J.F., White,O. and Salzberg,S.L. (2000) Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol., 1, RESEARCH0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRuggiero J., Santangelo,N., Nackerdien,Z., Ravel,J. and Robb,F.T. (1997) Repair of extensive ionizing-radiation DNA damage at 95°C in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol., 179, 4643–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Garcia P., St Jean,A., Amils,R. and Charlebois,R.L. (1995) Genomic stability in the archaeae Haloferax volcanii and Haloferax mediterranei. J. Bacteriol., 177, 1405–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daveran-Mingot M.L., Campo,N., Ritzenthaler,P. and Le Bourgeois,P. (1998) A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J. Bacteriol., 180, 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens S. (1998) The diverse and dynamic structure of bacterial genomes. Annu. Rev. Genet., 32, 339–377. [DOI] [PubMed] [Google Scholar]
- 18.Rebollo J.E., Francois,V. and Louarn,J.M. (1988) Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA, 85, 9391–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segall A., Mahan,M.J. and Roth,J.R. (1988) Rearrangement of the bacterial chromosome: forbidden inversions. Science, 241, 1314–1318. [DOI] [PubMed] [Google Scholar]
- 20.Guijo M.I., Patte,J., del Mar Campos,M., Louarn,J.M. and Rebollo,J.E. (2001) Localized remodeling of the Escherichia coli chromosome: the patchwork of segments refractory and tolerant to inversion near the replication terminus. Genetics, 157, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Himmelreich R., Plagens,H., Hilbert,H., Reiner,B. and Herrmann,R. (1997) Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium.Nucleic Acids Res., 25, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segall A.M. and Roth,J.R. (1994) Approaches to half-tetrad analysis in bacteria: recombination between repeated, inverse-order chromosomal sequences. Genetics, 136, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S.L. and Sanderson,K.E. (1998) Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett., 164, 275–281. [DOI] [PubMed] [Google Scholar]
- 24.Chinen A., Uchiyama,I. and Kobayashi,I. (2000) Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene, 259, 109–121. [DOI] [PubMed] [Google Scholar]
- 25.Lecompte O., Ripp,R., Puzos-Barbe,V., Duprat,S., Heilig,R., Dietrich,J., Thierry,J.C. and Poch,O. (2001) Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res., 11, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myllykallio H., Lopez,P., Lopez-Garcia,P., Heilig,R., Saurin,W., Zivanovic,Y., Philippe,H. and Forterre,P. (2000) Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science, 288, 2212–2215. [DOI] [PubMed] [Google Scholar]
- 27.Makino S.-I. and Suzuki,M. (2001) Bacterial genomic reorganization upon DNA replication. Science, 292, 803. [DOI] [PubMed] [Google Scholar]
- 28.Zivanovic Y., Myllykallio,H. and Forterre,P. (2001) Bacterial genomic reorganization upon DNA replication. Science, 292, 803a. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoriev A. (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res., 26, 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez P. and Philippe,H. (2001) Composition strand asymmetries in prokaryotic genomes: mutational bias and biased gene orientation. C. R. Acad. Sci. III, 324, 201–208. [DOI] [PubMed] [Google Scholar]
- 32.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 33.Sharp P.M. and Li,W.H. (1987) The codon Adaptation Index–a measure of directional synonymous codon usage bias and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diruggiero J., Dunn,D., Maeder,D.L., Holley-Shanks,R., Chatard,J., Horlacher,R., Robb,F.T., Boos,W. and Weiss,R.B. (2000) Evidence of recent lateral gene transfer among hyperthermophilic archaea. Mol. Microbiol., 38, 684–693. [DOI] [PubMed] [Google Scholar]
- 35.Freeman J.M., Plasterer,T.N., Smith,T.F. and Mohr,S.C. (1998) Patterns of genome organization in bacteria. Science, 279, 1827a. [Google Scholar]
- 36.Salzberg S.L., Salzberg,A.J., Kerlavage,A.R. and Tomb,J.F. (1998) Skewed oligomers and origins of replication. Gene, 217, 57–67. [DOI] [PubMed] [Google Scholar]
- 37.Tillier E.R. and Collins,R.A. (2000) The contributions of replication orientation, gene direction and signal sequences to base-composition asymmetries in bacterial genomes. J. Mol. Evol., 50, 249–257. [DOI] [PubMed] [Google Scholar]
- 38.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A., McDonald,L., Utterback,T.R., Malek,J.A., Linher,K.D., Garrett,M.M., Stewart,A.M., Cotton,M.D., Pratt,M.S., Phillips,C.A., Richardson,D., Heidelberg,J., Sutton,G.G., Fleischmann,R.D., Eisen,J.A., Fraser,C.M. et al. (1999) Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima.Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 39.Suckow J.M. and Suzuki,M. (1999) A sequence of thirty bases that is highly repetitive in archaebacterial genomes. Proc. Jpn Acad. Ser. B, 75, 16–21. [Google Scholar]
- 40.Mojica F.J., Diez-Villasenor,C., Soria,E. and Juez,G. (2000) Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol., 36, 244–246. [DOI] [PubMed] [Google Scholar]
- 41.Mojica F.J., Ferrer,C., Juez,G. and Rodriguez-Valera,F. (1995) Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol., 117, 85–93. [DOI] [PubMed] [Google Scholar]
- 42.Schmid M.B. and Roth,J.R. (1987) Gene location affects expression level in Salmonella typhimurium. J. Bacteriol., 169, 2872–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brewer B.J. (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell, 53, 679–686. [DOI] [PubMed] [Google Scholar]
- 44.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S., Borriss,R., Boursier,L., Brans,A., Braun,M., Brignell,S.C., Bron,S., Brouillet,S., Bruschi,C.V., Caldwell,B., Capuano,V., Carter,N.M., Choi,S.K., Codani,J.J., Connerton.I.F., Danchinm,A. et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 45.Rocha E.P. and Danchin,A. (2001) Ongoing evolution of strand composition in bacterial genomes. Mol. Biol. Evol., 18, 1789–1799. [DOI] [PubMed] [Google Scholar]