Abstract
Our study employed molecular dynamics (MD) simulations to assess the binding affinity between short peptides derived from the tumor-associated antigen glypican 3 (GPC3) and the major histocompatibility complex (MHC) molecule HLA-A*11:01 in hepatocellular carcinoma. We aimed to improve the reliability of in silico predictions of peptide-MHC interactions, which are crucial for developing targeted cancer therapies. We used five algorithms to discover four peptides (TTDHLKFSK, VINTTDHLK, KLIMTQVSK, and STIHDSIQY), demonstrating the substantial potential for HLA-A11:01 presentation. The Anchored Peptide-MHC Ensemble Generator (APE-Gen) was used to create the initial structure of the peptide-MHC complex. This was followed by a 200 ns molecular dynamics (MD) simulation using AMBER22, which verified the precise positioning of the peptides in the binding groove of HLA-A*11:01, specifically at the A and F pockets. Notably, the 2nd residue, which serves as a critical anchor within the 2nd pocket, played a pivotal role in stabilising the binding interactions.VINTTDHLK (ΔGSIE = −14.46 ± 0.53 kcal/mol and ΔGMM/GBSA = −30.79 ± 0.49 kcal/mol) and STIHDSIQY (ΔGSIE and ΔGMM/GBSA = −14.55 ± 0.16 and −23.21 ± 2.23 kcal/mol) exhibited the most effective binding potential among the examined peptides, as indicated by both their binding free energies and its binding affinity on the T2 cell line (VINTTDHLK: IC50 = 0.45 nM; STIHDSIQY: IC50 = 0.35 nM). The remarkable concordance between in silico and in vitro binding affinity results was of particular significance, indicating that MD simulation is a potent instrument capable of bolstering confidence in in silico peptide predictions. By employing MD simulation as a method, our study provides a promising avenue for improving the prediction of potential peptide-MHC interactions, thereby facilitating the development of more effective and targeted cancer therapies.
Keywords: Molecular dynamics simulation, Binding ability, Predicted glypican-3 peptides, Human leukocyte antigen-A*11:01, Hepatocellular carcinoma
1. Introduction
Human adaptive cellular immunity relies on recognizing antigens presented on the major histocompatibility complex (MHC) [1,2]. Typically, these antigens are located on nucleated and antigen-presenting cells [1,2]. The interaction between the immunogenic peptide bound to MHC and the T-cell receptor on T lymphocytes initiates T-cell activation and differentiation [1]. Based on their antigen processing characteristics and T-cell activation function, MHC molecules are divided into two classes [1]. MHC class-I presents endogenous antigens to CD8+ cytotoxic T lymphocytes (CTLs), whereas MHC class-II processes and presents exogenous antigens taken up by endocytosis to CD4+ helper T lymphocytes. Foreign proteins undergo processing by the immunoproteasome, producing a short peptide suitable for binding within the cavity of the MHC molecule in the endoplasmic reticulum [1]. This process loads the antigen onto the MHC molecule. The Ag-binding domain of the MHC comprises a beta-sheet (β-sheet) as the base and two alpha (α)-helices positioned above the -sheet to form a cleft for the short peptide. MHC class-I molecules are derived from a single heavy chain, while MHC class-II molecules comprise two chains (α-chain and β-chain) [3,4]. Beta-2-microglobulin (β2M) is required to stabilize MHC class-I, whereas dimerization of the two chains stabilizes MHC class-II [3]. MHC molecules encoded by the human leukocyte antigen (HLA) gene complex play a crucial role in disease and immune defense by discriminating between self and non-self-antigens. MHC class-I is determined by six genes (HLA-A, -B, -C, -D, -E, -F, and -G), whereas MHC class-II is determined by five genes (HLA-DR, -DQ, -DP, DM, and -DO) [5]. The high polymorphism of HLA genes results in further classification, such as HLA-A*11:01, HLA-B*15:02, and HLA-DR1 (https://www.ebi.ac.uk/ipd/imgt/hla/stats.html), resulting in MHC molecules that are unique to each individual and affect the conformation of the binding groove and determine the loaded peptide [1].
This study focuses on MHC class-I because of its essential function in activating CTLs that are responsible for destroying infected or cancerous cells. The MHC class-I binding groove is divided into six pockets (A to F) that adhere to the immunogenic peptide [6]. Both extremities of the groove are closed by conserved tyrosine residues, limiting the loaded immunogenic peptide to 8–10 residues [7]. Identifying the proper peptide for MHC loading remains a challenging task. Advanced predictive tools have been created to address this issue. These techniques utilize intricate algorithms and data from the crystallographic structures of different peptide-MHC (pMHC) complexes gathered from various sources [8]. Among these tools, motif-based prediction systems stand out as a prominent technique. They rely on the binding propensity of specific amino acids to precisely defined positions on the MHC molecule, known as anchor positions [8,9].
The SYFPEITHI database employs an algorithmic model that considers the anchor positions and factors, such as unusual and auxiliary anchors within a given peptide [10]. This exhaustive methodology accurately predicts T-cell epitopes [10]. Significant progress has been made in recent years by incorporating quantitative metrics into the predictive framework. KD, EC50, and stability metrics have been developed to capture the affinity properties of peptides. Combining these metrics yields a comprehensive matrix that captures the relative significance of each position in the peptide [9]. These vast datasets are currently used to train machine learning models, employing advanced techniques such as artificial neural networks (ANN) and support vector machines [9]. This innovative method identifies potential binding interactions between specific amino acid positions in a given peptide and an MHC molecule and quantifies the strength of these interactions. A variety of cutting-edge prediction tools employing these quantitative metrics are currently available. Among these, RankPep [11], NetMHC 4.0 [12], and NetCTL [13] occupy prominent positions. These tools provide a robust method for determining the likelihood of a peptide's binding to an MHC molecule by incorporating diverse factors into their predictive models. It is essential to note that not all top-ranked predicted peptides successfully bind to the desired HLA or influence T-cell stimulation [14]. In vitro, functional investigations, such as binding assays with transporter associated with antigen processing (TAP)-deficient T2 cell lines expressing specific HLAs, cytokine response testing, and cytolytic activity assays, are required to confirm successful T-cell stimulation. However, these in vitro investigations are both time-consuming and costly to conduct. Molecular dynamics (MD) simulations provide an alternative method for predicting potential peptides for MHC loading, thereby reducing the number of peptides requiring in vitro functional testing [15]. While in vitro testing is still necessary for definitive results, MD simulations can expedite the procedure significantly [15]. To date, however, data and information regarding the use of MD simulation to predict pMHCs are limited.
Before conducting in vitro functional testing, this study seeks to evaluate the dynamic behavior and atomic-level binding affinity of predicted peptides loaded onto MHC molecules using MD simulation. This investigation employed the tumor-associated antigen glypican-3 (GPC3) in hepatocellular carcinoma (HCC) as a model. This study used the complete translated amino acid sequence of GPC3 to determine the optimal nonamer peptide for loading onto the HLA-A*11:01 molecule, the predominant HLA subtype in most world populations [16]. Prediction algorithms are utilized, including SYFPEITHI, IEDB-MHCI, Rankpep, NetMHC 4.0, and NetCTL. The post-MD simulation evaluated and analyzed the intermolecular hydrogen bonds, free energy of decomposition, and binding affinity. In addition to assessing the binding properties of predicted peptides to HLA-A*11:01 presented on T2 cells, the focus was predominantly on peptide binding within the MHC groove. Through extensive in silico examination of the binding affinity of predicted peptides to MHC molecules and subsequent in vitro binding testing, this study aimed to increase the precision and efficiency of identifying optimum peptides for MHC loading.
2. Materials and methods
2.1. Ethics consideration
The MD simulations were performed in silico, and the binding affinity testing was conducted in vitro using a transporter associated with antigen processing TAP-deficient T2 cell line. No experiments in this study involved either humans or animals. The protocol for this study was approved by the Siriraj Institutional Review Board (SIRB) of the Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok, Thailand (COA no. 055/2022).
2.2. Epitope prediction
HLA-A*11:01-restricted epitopes from the GPC3 protein (NM_004484) were identified using five prediction tools with different algorithms, including SYFPEITHI (http://www.syfpeithi.de) [10], IEDB-MHCI (http://tool.immuneepitope.org/mhci) [17], Rankpep (http://imed.med.ucm.es/Tools/rankpep.html) [11], NetMHC 4.0 (http://www.cbs.dtu.dk/services/NetMHC 4.0) [12], and NetCTL (http://www.cbs.dtu.dk/services/NetCTL) [13]. The top five GPC3 candidate epitope sequences (nonamers) were selected based on the highest score and rank of binding percentage. Peptides that were consensus-predicted by at least three algorithms were chosen for inclusion in the MD simulation.
2.3. Structure preparation
The initial step in preparing the pMHC structures for MD simulation was using the Anchored Peptide-MHC Ensemble Generator (APE-Gen) [18]. We followed the instructions at https://github.com/KavrakiLab/hla-arena to integrate APE-Gen into the HLA-Arena environment [19]. We implemented the program using Jupyter Notebook and Docker, ensuring that all necessary protocols were followed for seamless execution.
To validate the reliability of the structures generated by APE-Gen, we subjected three X-ray crystallography-resolving structures from the Protein Data Bank (PDB) to re-docking. This process involved re-docking the respective peptides and the MHC molecule using the APE-Gen algorithm. Specifically, we examined the structures 1X7Q, 6JOZ, and 7S8R, along with their corresponding peptides: KTFPPTEPK, ATIGTAMYK, and SALEWIKNK.
Subsequently, we generated the pMHC complexes that comprised the four predicted GPC3 peptides listed in Table 1. Additionally, we included the well-known peptide FVGFFTDV, which was recognized for its strong binding to HLA-A*02:01 [20] as a negative HLA-A*11:01 control. The generation process was facilitated by APE-Gen, with the reference structure for the MHC molecule being 1X7Q [21]. This approach ensured consistency and enabled direct comparisons throughout the study.
Table 1.
Summary of the top 5 predicted GPC3 peptides that could be successfully loaded onto HLA-Aa11:01 from each of the five prediction tools used for in silico analysis. Only peptides predicted by at least 3 prediction tools were selected for MD simulation.
| Peptides | Positiona | Prediction tools |
||||
|---|---|---|---|---|---|---|
| SYFPEITHI | IEDB-MHCI | NetMHC 4.0 | NetCTL | Rankpep | ||
| TTDHLKFSK | 242–250 | ✓ | ✓ | ✓ | ✓ | ✓ |
| VINTTDHLK | 239–247 | ✓ | ✓ | ✓ | ||
| KLIMTQVSK | 213–221 | ✓ | ✓ | ✓ | ||
| STIHDSIQY | 325–333 | ✓ | ✓ | ✓ | ||
| KNYTNAMFK | 123–131 | ✓ | ✓ | |||
| LQSASMELK | 93–101 | ✓ | ||||
| KLKSFISFY | 394–402 | ✓ | ||||
| PVVSQIIDK | 547–565 | ✓ | ||||
| VSKSLQVTR | 219–227 | ✓ | ||||
| VSQIIDKLK | 459–467 | ✓ | ||||
| RTMSMPKGR | 474–482 | ✓ | ||||
| NQFNLHELK | 443–451 | ✓ | ||||
| IVVRHAKNY | 117–125 | ✓ | ||||
| LLRTMSMPK | 472–480 | ✓ | ||||
Abbreviations: GPC3, glypican-3; HLA-A*11:01, human leukocyte antigen-A*11:01; MD, molecular dynamics.
NM_004484 was used to designate the amino acid position.
2.4. Molecular dynamics (MD) simulation
All pMHC complexes, including the structures retrieved from PDB and their re-docking with APE-Gen for reliable evaluation, along with the negative control peptide and the four HLA-A*11:01 loaded with the top four peptides, were subjected to independent triplicate MD simulations with random seeding using the AMBER 22 software package [22] (The Amber Project, San Francisco, CA, USA) and an ff19SB force field [23]. The missing atoms were added using the LEaP module [24]. In all pMHC complexes, the protonation state of all possible charged residues (arginine, lysine, histidine, aspartate, and glutamate) was assigned a pH of 7.0 by the PROPKA server [25]. Total charges with a negative value for the pMHC complex were randomly neutralized using Na+ counterions. Individual complexes were then solvated using TIP3P [26] water molecules, resulting in approximately 19,000 atoms for the entire model. The simulation box dimensions used for all systems were 97 × 97 × 82 Å3. A periodic boundary condition was used with an isothermal-isobaric ensemble and a simulation time step of 2 fs (fs). All energy minimizations and MD simulations were performed using the SANDER module of AMBER 22. All bonds and angles specific to hydrogen atoms were constrained using the SHAKE algorithm [27]. Long-range electrostatic interactions were treated using the particle mesh Ewald method, and the non-bonded interactions with a cutoff distance of 12 Å were identified [28]. All MD simulations were run with a 12 Å residue-based cutoff for non-bonded interactions. The particle mesh Ewald method was applied to treat long-range electrostatic interactions adequately [28]. Each system was subjected to the four stages of a restrained MD simulation at 298 K (K) with force constants of 10, 7.5, 5, and 2.5 kcal mol−1 Å2 for 500 ps (ps) in each stage. These subsequent steps permitted the peptide to adopt its geometry and orientation from the initial model to fit better within the peptide-binding groove. The constraints were then entirely removed, and wholly unrestrained MD simulations were performed until 200 ns. The convergences of energies and root mean square displacement (RMSD) were used to verify the stability of the systems. The MD trajectories were collected every 0.1 ps during the production phase for further analysis of the hydrogen bond (H-bond), B-factor, decomposition free energy (ΔG), and other energy components.
2.5. Binding of predicted peptides with HLA-A*11:01 expressing TAP-deficient T2 cell lines
Four predicted peptides from this study and a nucleocapsid of a SARS-CoV peptide loaded onto HLA-A*11:01 revealed from the crystalizing structure of references structure (KTFPPTEPK) and the negative control peptide (FVGFFTDV) were synthesized by GenScript (Piscataway, NJ, USA). Different concentrations of peptides were incubated with transporter associated with antigen processing (TAP)-deficient T2 cell line overexpressing HLA-A*11:01 (kindly provided by Prof. Dr. Maria Grazia Masucci of the Department of Cell and Molecular Biology, Karolinska Institutet, Stockholm, Sweden) in the presence of 3 μg/ml of beta-2 microglobulin (β2M) (Merck Millipore, Burlington, MA, USA). The cultivation was completed in Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10 % fetal bovine serum (Gibco; Thermo Fisher Scientific), 50 units of penicillin (Gibco; Thermo Fisher Scientific), and 50 μg of streptomycin (Gibco; Thermo Fisher Scientific). The incubation of peptides with T2 cells was conducted in a cell culture incubator (model BB150; Thermo Fisher Scientific) with 5 % carbon dioxide (CO2) at 37o Celsius (C) for 24 h.
Cells were collected, washed once with 1X phosphate-buffered saline (PBS), and then stained with anti-HLA-ABC conjugated with fluorescein isothiocyanate (FITC) (clone W6/32) (eBioscience™; Thermo Fisher Scientific) for 30 min. The stained cells were then washed twice with 1x PBS. After washing, the buffer was discarded, and the cell pellets were resuspended using 1 % paraformaldehyde in PBS. The cell suspensions were subjected to flow cytometry using a BD Accuri™ C6 system (BD Biosciences, Franklin Lakes, NJ, USA) to evaluate the fluorescence signal of the stained cells. The flow cytometry results were analyzed using FlowJo™ v10.0 software (FlowJo, LLC, Ashland, OR, USA).
2.6. Statistical analysis
Raw data were recorded and analyzed using GraphPad Prism version 9.5.1 (GraphPad Software, Inc., San Diego, CA, USA). The mean ± standard error of the mean (SEM) of data collected from at least three independent experiments was calculated. Statistical comparisons were performed using one-way analysis of variance (ANOVA) with Tukey's correction for analytical hypothesis testing in multiple comparisons. A p-value of less than 0.05 was regarded as being statistically significant.
2.7. Data availability
The trajectory and topology files required for the MD simulation, the files necessary for evaluating the free energy changes (ΔGSIE and ΔGMM/GBSA), and the additional datasets utilized and examined in the present investigation has been included in the Supplement Material.
3. Results
3.1. Potential predicted peptides derived from GPC3 loaded onto HLA-A*11:01
This study aimed to identify potential peptides suitable for loading onto MHC class-I molecules with a peptide length of nine amino acids. The complete amino acid sequence of GPC3 (NM_004484) was analyzed using five web-based prediction tools: SYFPEITHI, IEDB-MHCI, Rankpep, NetMHC 4.0, and NetCTL. Specifically, HLA-A*11:01, the predominant HLA subtype in the Thai and world populations [16], was selected as the specific MHC class-I molecule for prediction. This study was primarily carried out with the Thai population to assist our population and simplify recruiting healthy volunteers for in vitro binding testing. Nevertheless, HLA-A11:01 exhibits the highest prevalence among the global population, making this study relevant worldwide. The binding groove of MHC class-I is closed and contains two conserved residues, Y84 and W167, which line the A and F pockets, respectively. Therefore, the length of the peptide repertoire that a specific MHC I molecule can effectively bind is limited. Most MHC class-I peptides consist of 8–10 amino acids, with a preference for 9-mers. Longer peptides either bind in a zig-zag orientation within the groove or protrude from the middle of the groove [29].
Consequently, the study focused on 9-mers in this study. Supplementary Table 1 presents the rankings and scores of the predicted peptides with high binding potential to HLA-A*11:01 from all five prediction tools. The predicted peptides exhibited a binding affinity with IC50 values less than 500 nM. Table 1 displays the top five peptides identified by each prediction algorithm. Only peptides predicted by at least three algorithms were selected for further MD simulation. The most highly ranked predicted peptides were TTDHLKFSK, VINTTDHLK, KLIMTQVSK, and STIHDSIQY (Table 1).
3.2. Starting structure preparation for molecular dynamics (MD) simulation of peptide-loaded MHC (pMHC)
The initial structures for MD simulations of pMHC complexes were generated using the Anchored Peptide-MHC Ensemble Generator (APE-Gen). APE-Gen efficiently produces diverse pMHC conformations by anchoring the terminal residues of peptides to specific pockets within the MHC binding site, leveraging prior structural knowledge for precise initial positioning. The methodology involves iterative loop modeling and energy minimization to optimize peptide and MHC conformations. Initially, peptide ends are aligned to the MHC binding site using a template structure, followed by peptide backbone sampling with the Random Coordinate Descent algorithm, which introduces randomization for efficient conformational exploration. Sidechains are added using PDBFixer, and energy minimization is performed with SMINA to optimize electrostatic, hydrogen bonding, solvation, and hydrophobic interactions. Multiple rounds of these steps allow APE-Gen to explore diverse conformations, starting each new round with the highest quality conformation from the previous one. APE-Gen's effectiveness is validated by reproducing native-like conformations with an average RMSD of 2.02 Å across a dataset of 535 pMHC structures, supported by successful reproduction of crystal structures, accurate generation from sequence information, and effective modeling of non-canonical peptides [18].
To evaluate the viability of the initial structures created by APE-GEN for further MD simulation, three pMHC complexes from the Protein Data Bank (PDB) were selected for analysis: 1X7Q [21], 6JOZ [30], and 7S8R [31]. X-ray crystallography with respective resolutions of 1.45, 1.35, and 2.95 Å confirmed the accuracy of these structures, respectively (Fig. 1A). Using the APE-GEN method, the HLA-specific peptides KTFPPTEPK for 1X7Q (Fig. 1B and C), SALEWIKNK for 7S8R (Fig. 1D), and ATIGTAMYK for 6JOZ (Fig. 1E) were three independently replicated re-docked. Compared to the original PDB structure, it was determined that the structure with the lowest optimized binding energy was the most suitable. Compared to the original crystal structure, the results indicated that the APE-GEN model aligns exceptionally well, particularly at the center of all three peptides (Fig. 1C–E). This discovery was further supported by the small root mean square deviation (RMSD) between the PDB database structure and the APE-Gen generated structure (Fig. 1F). APE-Gen performed a vital role in producing trustworthy starting structures of pMHC for MD simulation. Among the examined pMHC complexes, 1X7Q, which contains the peptide sequence KTFPPTEPK, displayed the lowest RMSD relative to the equivalent APE-Gen structure.
Fig. 1.
The HLA-A*11:01 complex is loaded with the GPC3 peptide structure. (A), comprising a single heavy α-chain and associating with β2M. (B) defines the binding pockets on the MHC class-I groove, distinguished by colors such as blue, pink, yellow, green, purple, and red for pockets A to F, respectively. (C) showcases the peptide embedded in the MHC blind groove of 1X7Q (KTFPPTEPK), characterized by two α-helices and a β-sheet forming the binding groove base. Peptide alignments from the crystal structure and three independently generated structures using APE-Gen are color-coded, with red, blue, yellow, and green representing distinct alignments. (D) and (E) present the alignment of loaded peptides on the MHC binding groove of 7S8R (SALEWIKNK) and 6JOZ (ATIGTAMYK), respectively. (F) illustrates the average RMSD of the loaded peptides on their desired MHC binding groove, calculated from three independent experiments and presented as mean ± SEM. Statistical analysis employing one-way ANOVA with Tukey's correction demonstrates no significant differences (ns: not significantly different) among the samples.
Consequently, 1X7Q was chosen as the reference structure for APE-Gen to generate the HLA-A*11:01 complex loaded with the four predicted GPC3 peptides: TTDHLKFSK, VINTTDHLK, KLIMTQVSK, and STIHDSIQY. In addition, the GPC3 peptide FVGEFFTDV, which has been shown to bind to HLA-A*02:01 precisely [20], acted as a negative control for HLA-A*11:01 binding in this work. The construction of the pMHC with each individual predicted peptide involved the addition of missing hydrogen atoms, and minimization was performed. A water molecule was then added to the simulation box as a solvent. The pMHCs were subsequently ready for further MD analysis.
3.3. Stability of the pMHC
The MD simulations were conducted for each predicted peptide pMHC complex, with 200 ns (ns) duration in three simulation replicas with random seeding numbers. The stability of the pMHC complex was assessed by calculating the RMSD of atomic positions, considering the loaded peptides, binding grooves, and the entire HLA molecule, including the heavy α chain and β2M. Fig. 2 displays the RMSD plots for the four predicted peptide pMHC complexes, the reference structures (KTFPPTEPK), and the negative control (FVGEFFTDV). RMSD values were obtained by comparing the geometric coordinates of the MHC backbone throughout the simulation to its initial configuration (Fig. 2). The deviation of all atoms in the seven systems examined ranged from approximately 1.0 to 1.5 Å. Throughout the MD simulation, a consistent pattern of stability was observed for all six pMHC complexes. The binding groove of the MHC molecule (residue 1 to 180) and the loaded peptide, all six pMHC systems displayed highly stable fluctuations of less than 1 Å throughout the simulation. Equilibrium was attained by all six pMHC systems after 150 ns, with fluctuations of approximately 0.5 Å. Consequently, the trajectories obtained during the last 50 ns (150–200 ns) of the MD simulation were deemed representative of the production phase, and the data were recorded and collected for further analysis.
Fig. 2.
The RMSD of all atoms of peptides loaded onto an MHC molecule with three independent MD simulations. The loaded peptide movement was relatively stable when analyzing the binding groove of the MHC molecule.
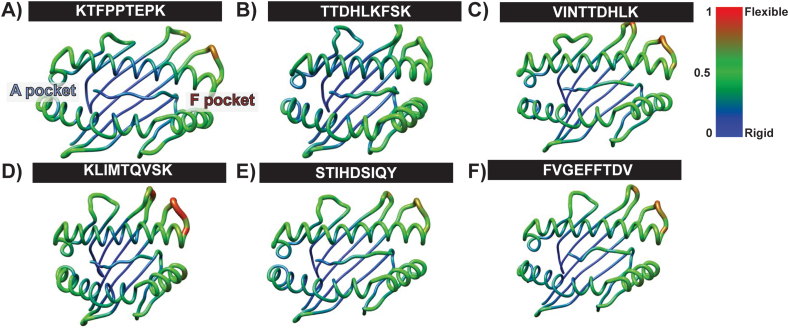
3.4. The flexibility of the loaded peptide into the MHC binding groove
The flexibility of the loaded peptide within each pMHC complex was evaluated by analyzing the B-factor obtained during the generation phase of the MD simulation (last 50 ns, Fig. 3). In all structures, the coupling of β2M to pMHC exhibited moderate flexibility (data available upon request). Across all structures, the results revealed a consistent pattern of flexibility. KTFPPTEPK displayed average flexibility in the α-helix region of the F pocket at the end of the binding groove (Fig. 3A). Similarly, all four predicted peptides and the negative control (FVGFFTDV) loaded onto the MHC displayed the most flexibility in the α-helix region surrounding the F pocket. This observation indicates that VINTTDHLK, KLIMTQVSK, and FVGFFTDV exhibited tremendous flexibility within these particular regions of the MHC binding groove during the production phase of the MD simulation, as depicted by the blue-to-red color (Fig. 3). Intriguingly, all structures exhibited a highly rigid region at the β-sheet in the pMHC structure's binding groove. Notably, the flexibility of the loaded predicted peptides corresponded to the flexibility of their respective MHC molecules. The second amino acid position (T, I, L, and V) tightly occupied the A pocket of the binding groove, resulting in the structures' most rigid region.
Fig. 3.
The flexibility of the pMHC in each peptide was evaluated using the B-factor obtained from the last 50 ns of the MD simulation production phase. The degree of flexibility is displayed as the structure's color gradient and radiance. For the color gradient, deep blue indicates the most rigidity, and deep red indicates the most flexibility. The radiance of the structure ranged from zero to one to reflect the most rigidity to most flexibility, respectively.
3.5. Per-residue decomposition (DC) energy of the complex
A comprehensive investigation was conducted to determine how particular residues within the MHC binding grooves and the loaded peptides affect the binding free energy. This evaluation used the molecular mechanics/generalized Born surface area (MM/GBSA) method, utilizing 500 snapshots obtained from the equilibrium state of the MD simulation's production phase. Notably, the reference and predicted pMHC complexes displayed significant interactions at the anchor residues (precisely, the second and third amino acid positions), as indicated by the total energy values (Fig. 4 and Supplementary Fig. S1). Additionally, each peptide displayed a unique level of binding energy and residue pattern at other positions. Among the investigated structures, only the second position of the negative control peptide, FVGFFTDV, exhibited significant binding energy.
Fig. 4.
Averaged decomposition energy contributions in HLA-A*11:01 binding to predicted GPC3 peptides. The The loaded peptides are indicated using a rainbow color gradient from blue to white, reflecting the lowest to highest decomposition energy.
The energy profile of the binding groove for all seven pMHC complexes is depicted in Fig. 5 and Supplementary Fig. S2. To evaluate the residue binding of MHC to loaded peptides, we focused on values less than −2 kcal/mol, according to established procedures [32]. Each of the seven pMHC structures exhibited a distinct decomposition of the free energy pattern, with the interaction between the MHC molecule and the loaded peptide primarily centered on the α1 and α2 of MHC molecules. Nevertheless, specific residues from the β-sheet also contributed to capturing the loaded peptide (Fig. 5 and Supplementary Fig. S2). Among the amino acid residues found within the binding grooves of all six evaluated MHC systems, including Y7, Y9, E63, D66, Q70, Y99, K146, W147, Y159, and W167, there was a noticeable level of . However, in the negative control structure FVGFFTDV, despite exhibiting nearly the same positively charged amino acids responsible for binding the loaded peptide, the values for those positions within the binding groove were shallow compared to the other structures, particularly compared to the reference structures (Fig. 5 and Supplementary Fig. S2). Therefore, the binding affinity involves the free energy decomposition at each position on the loaded peptide and the position responsible for binding on the MHC molecule.
Fig. 5.
Energy contributions of HLA residue fingerprint plots. The The binding grooves are indicated using a rainbow color gradient from blue to white, representing the lowest to highest decomposition energy, respectively. The structure also mentions the potential residues that interacted with the peptide. The loaded peptide is presented in a purple stick-and-ball structure.
3.6. H-bond patterns between the groove of the MHC and the loaded predicted peptides
The intermolecular hydrogen bonding between the MHC groove and the loaded predicted peptides was thoroughly examined using snapshots captured during the production phase, specifically the last 50 ns of the MD simulation. To identify hydrogen bond occupation, we considered a distance of less than 3.5 Å and an angle greater than 120° between the donor (D) and acceptor (A) atoms. Fig. 6 and Supplementary Fig. S3 present only the hydrogen bonds that occupied over 40 % during the production phase. Interestingly, we observed that the preferred hydrogen bonds predominantly formed at the anchor positions of the loaded peptides. Furthermore, upon analyzing all predicted pMHC structures, we found that amino acid positions 1, 2, 3, and 9 displayed the majority of hydrogen bonding interactions with the surrounding amino acids of the MHC molecule (Fig. 6 and Supplementary Fig. S3).
Fig. 6.
Hydrogen bond interactions between the peptide and the HLA molecule. H-bonding between loaded peptides and their counterpart residues of the MHC binding groove. The peptide residues are shown in light blue, and residues of the MHC molecule that provided the H-bond are shown in light pink. Oxygen and nitrogen atoms are indicated in red and blue, respectively.
Moreover, each subsequent amino acid residue exhibited a robust formation of hydrogen bonds with the binding groove of its corresponding MHC molecule. For instance, L5 of TTDHLKFSK, T5, and L8 of VINTTDHLK, T5, and S8 of KLIMTQVSK, and S6 and Q8 of STIHDSIQY displayed such interactions (Fig. 6 and Supplementary Fig. S3). Interestingly, even in the negative control structure FVGFFTDV, hydrogen bond occupation was observed at amino acid positions 1, 2, 7, 8, and 9. However, the number of atoms involved in forming these bonds was significantly lower than the other structures, particularly when compared to the KTFPPTEPK.
The majority of H-bonds on the MHC molecule were mainly concentrated in the α1 (N63, N66, Q70, T73, and D77) and α2 (T143, W147, Y159, and Y171) of the binding groove (Fig. 6 and Supplementary Fig. S3). These specific positions correspond to the A pocket (N63, N66, Q70, Y99, Y159, and Y171) and F pocket (D77, D116, T143, and W147) of the MHC binding groove. Interestingly, Y99 and D116, situated on the β-sheet of the MHC binding groove, showed the potential to form hydrogen bonds with the loaded peptides. Particularly noteworthy, we observed a significantly high percentage of hydrogen bonding for Y159 and Y99 of the MHC molecule with the amino acid position 1 and 3, respectively, among all peptides included in this study. Moreover, it is essential to highlight that the abundance of hydrogen bond occupancies in the pMHC structures (Fig. 6 and Supplementary Fig. S3) correlated with the crucial amino acid positions responsible for binding to the loaded peptides, as evidenced by the low of decomposition-free energy (Fig. 5 and Supplementary Fig. S2).
3.7. In silico binding affinity evaluation of the predicted peptides on HLA*11:01
The in-silico binding affinity of predicted peptides within the HLA*11:01 molecule's groove was evaluated using two distinct approaches. Firstly, data from 500 snapshots during the production phase were used to calculate the binding free energy, employing solvated interaction energy (SIE) calculations (Supplementary Table S2) and molecular mechanics/generalized Born surface area (MM/GBSA) measures (Supplementary Table S3) [33]. The conformational entropy change in pMHC complexity during MD simulation was considered by incorporating the entropy term (TΔS), which was further calculated through normal-mode (N) analysis [34]. The binding energy (ΔGbind) was subsequently derived by combining the solute entropy term and ΔGtotal, as assessed by MM/GBSA (Fig. 7). The results showed that the binding free energy values from SIE and MM/GBSA for the KTFPPTEPK were −14.43 ± 0.77 and −26.40 ± 5.18 kcal/mol, respectively. Conversely, the negative control structure, FVGFFTDV, exhibited the highest ΔGbind values for both SIE (−12.34 ± 0.48 kcal/mol) and MM/GBSA (−19.63 ± 1.95 kcal/mol). The affinity of KTFPPTEPK is significantly higher than that of the negative control.
Fig. 7.
Evaluating predicted peptides' binding affinity on HLA-A*11:01: insights from in silico and in vitro analyses. The total free energy of the predicted peptides bound to HLA-A*11:01 was computed based on data obtained from 500 snapshots during the production phase. This comprehensive evaluation employed SIE (A) and MM/GBSA (B) methods. To validate the binding affinity of the peptides in an experimental in vitro setting, we utilized the TAP-deficient T2 cell line, known for expressing HLA-A*11:01. Quantitative assessment of the pMHC complex presentation on the T2 cell surface was performed using flow cytometry, with specific staining involving an antibody against the pan-HLA-ABC family, conjugated with FITC. The in vitro binding affinity of the peptides was determined by assessing the IC50 (C). All bar graphs depict the mean ± SEM, with data obtained from three independent experiments. The statistical analysis employed one-way ANOVA with Tukey's correction, effectively identifying significant differences among the samples (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001), while instances of non-significant differences were denoted as “ns” (not significantly different).
Regarding the predicted GPC3 peptides, the binding free energy values indicated that the pMHC complexed with VINTTDHLK demonstrated the most potent binding ability among the four peptides predicted. The ΔGbind for VINTTDHLK was estimated to be −14.46 ± 0.53 kcal/mol and −30.79 ± 0.49 kcal/mol for SIE and MM/GBSA, respectively. In contrast, TTDHLKFSK was found to be the least favorable peptide for loading onto HLA*11:01, with the lowest binding free values of −12.00 ± 0.24 kcal/mol and −19.63 ± 1.95 kcal/mol for SIE and MM/GBSA, respectively. The ΔGbind value indicated that VINTTDHLK was comparable to STIHSIQY but had a more remarkable ability to bind to the target MHC molecule than KLIMTQVSK. Consequently, the rank of GPC3 peptides predicted by the selected web-based algorithms, based on their binding abilities from strongest to weakest, was VITTDHLK ≈ STIHSIQY > KLIMTQVSK > TTDHLKFSK (Fig. 7).
Furthermore, MM/GBSA proved to be more suitable for evaluating the binding energy of the pMHC than SIE, as it could statistically differentiate potential binder peptides for the desired MHC molecule. While SIE indicated the trend of the predicted peptides' binding range to the MHC molecule, it lacked the statistical confidence to rank their binding effectiveness definitively. Therefore, MM/GBSA is the preferred method for assessing pMHC binding energy.
3.8. The binding ability of predicted peptides with HLA-A*11:01 on TAP-deficient T2 cell line
The ability of predicted peptides to bind to HLA-A*11:01 presented on the TAP-deficient T2 cell line was also determined in this study. Different concentrations of peptides were incubated with TAP-deficient T2 cell line overexpressing HLA-A*11:01 in the presence of 3 μg/ml of β2M. Flow cytometry was used to evaluate the stability of each pMHC after incubating the peptides with the T2 cell line for 24 h to determine the binding ability of the predicted peptides. To quantify the binding ability of the peptides to the MHC molecules, we utilized the half-maximal inhibitory concentration (IC50). For the reference peptide (KTFPPTEPK), the IC50 value was determined to be 0.22 ± 0.02 mM, whereas the IC50 was not able to be evaluated in the negative control peptide (FVGFFTDV) (Fig. 7, Fig. 8). Among the predicted peptides, VITTDHLK displayed a low IC50 value (0.45 ± 0.01 nM), indicating its strong potential for binding to HLA-A*11:01 on T2 cells. Similarly, STIHSIQY exhibited favorable binding to the MHC molecule, with no significant difference in IC50 value (0.35 ± 0.02 nM) compared to VITTDHLK (Fig. 7, Fig. 8). Next in line for favorable binding was KLIMTQVSK, with an IC50 value of 1.15 ± 0.02 nM. Conversely, TTDHLKFSK demonstrated the highest IC50 value (2.80 ± 0.33 nM), suggesting its lower binding ability than the other predicted peptides in this study (Fig. 7, Fig. 8). Hence, the binding capabilities of the predicted peptides to the T2 cell line, as determined by their IC50 values, ranked as follows: VITTDHLK ≈ STIHSIQY > KLIMTQVSK > TTDHLKFSK, aligning with the results from the MM/GBSA binding energy evaluation.
Fig. 8.
The binding ability of the peptide to the MHC molecule in the T2 over expressing HLA-A*11:01 cell line. The antibody against the pan-HLA-ABC family conjugated with FITC was used to evaluate the binding affinity of the peptide to the MHC molecule. The stained cells were then subjected to flow cytometry for signal detection. No peptide condition was used for subtraction, which influenced positive binding activity between the peptide and the MHC molecule, as shown in the left image under each of the six peptides. The IC50, to reflect the degree to which the peptide binds to the MHC, is shown as the standard error of the mean in the right image under each of the five peptides.
4. Discussion
The growing understanding of how antigens are presented on the MHC molecule and their crucial role in triggering human immunity has led to promising advancements in peptide vaccines for cancer treatment, especially in early-phase clinical trials [35]. Various computational algorithms have been developed to identify short peptides derived from antigens of interest that can specifically bind to MHC molecules [8,9]. However, not all predicted peptides effectively bind to the desired MHC molecules and activate T-cells. Therefore, to validate the functionality of these predicted peptides, extensive and time-consuming in vitro evaluations, including binding assays and other functional studies, are essential. Our primary objective in this study was to employ MD simulation to comprehensively analyze the dynamic behavior and atomic-level binding affinity of predicted peptides loaded onto the MHC before conducting in vitro functional testing. By establishing a robust correlation between in silico and in vitro binding affinity, we can significantly enhance our confidence in peptide prediction through MD simulation, thus reducing the number of peptides that need expensive and unnecessary in vitro functional testing. Our study focused on GPC3, a tumor-associated antigen in hepatocellular carcinoma, loaded onto HLA-A*11:01.
MHC-binding peptide prediction systems employ several methodologies for predicting the binding of peptides to MHC molecules. None of the MHC class-I binding predictors showed superior performance compared to others in a previous investigation of 13 predictors. The performance of each approach varied depending on the HLA type and peptide length. Prior research has shown that methods based on ANN have performed better than those based on matrices. Despite being considered the top predictor, NetMHCpan 4.0 did not surpass other methods in accuracy for all HLA types and peptide lengths. As the study did not assess prediction models, it is impossible to determine a single model that is the best for all situations [36]. Given the context-dependent nature of prediction tools, the majority vote technique cannot determine the most effective strategy. Instead, we propose implementing a consensus approach that utilizes many highly ranked predictors in each prediction tool to enhance the accuracy of MHC class-I binding predictions. Therefore, this study employed five prediction algorithms, namely SYFPEITHI, IEDB-MHCI, Rankpep, NetMHC 4.0, and NetCTL, to predict peptides that have the potential to bind to HLA-A*11:01 [[10], [11], [12], [13],26]. As shown in Table 1, the algorithms generated intriguingly diverse peptide predictions. All five prediction algorithms assigned the peptide TTDHLKFSK a high score and rank. However, at least three algorithms gave high levels and scores to other peptides, including VINTTDHLK, KLIMTQVSK, and STIHDSIQY. Due to their high binding potential with the HLA-A*11:01 molecule, these four predicted peptides were selected for inclusion in the subsequent MD simulation.
For the MD simulation, we used the positive control of the pMHC complex of a nonamer peptide derived from the SARS protein, loaded onto HLA-A*11:01. The crystal structure of this specific pMHC complex (KTFPPTEPK, PDB ID: 1X7Q) served as the reference structure. Prior research has validated its reliability regarding binding capability and atomic properties after the MD simulation and additional analyses [32]. Additionally, the application of APE-Gen in our study has been demonstrated to be highly effective in generating initial structures for MD simulations of peptide-loaded MHC complexes. According to the validation results, APE-Gen can accurately reproduce native-like conformations with minimal RMSD, which suggests that the initial structure generation process is highly precise. The robustness of APE-Gen is further bolstered by the successful reproduction of crystal structures, accurate generation from sequence information, and effective modeling of non-canonical peptides [18].
To compare the reliability of the structure generated by APE-Gen and the structure resolved by X-ray crystallography, both structures underwent a comparable RMSD analysis of the loaded peptide and the MHC molecule during 200 ns of MD simulation. This assessment was also confirmed for two other structures retrieved from the PDB database (ATIGTAMYK for 6JOZ and SALEWIKNK for 7S8R). Furthermore, the subsequent evaluation of atomic interactions and binding free energy during the productive phase indicated that the stability shown by RMSD (Fig. 1, Fig. 2) exhibited no significant differences. These findings suggest that APE-Gen is well-suited for preparing initial structures for MD simulations in studies such as ours, where the precise modeling of peptide-MHC interactions and the subsequent evaluation is essential.
In the present study, we utilized APE-GEN to generate an initial MD simulation model of four anticipated peptides bound to HLA-A*11:01. We acquired the MHC molecule from the reference structure (KTFPPTEPK). We employed APE-GEN to dock the four expected peptides. This allowed us to generate the four pMHC structures that were utilized in this investigation. For the negative control in assessing HLA-A*11:01 binding, we employed the GPC3 peptide (FVGFFTDV), which demonstrated positive binding to HLA-A*02:01 [20]. APE-GEN subsequently formed the pMHC complex. Following the addition of the solvent, we conducted a 200-ns MD simulation after the minimization process. The peptides remained firmly and securely within their pMHC binding grooves during the MD simulations. During the last 50 ns of the manufacturing phase (as shown in Fig. 2), a slight fluctuation in RMSD of less than 1.0 Å was seen in the binding groove of all pMHCs. Prior studies have demonstrated a correlation between reduced variability in peptide binding within the MHC groove and the binding of peptide-MHC molecules [37].
We comprehensively evaluated the in silico binding properties of the predicted peptides to the HLA-A*11:01 molecule, considering total binding energy and H-bonding. The results demonstrated that all peptides' amino and carboxyl termini were firmly located in the A and F pockets of the HLA-A*11:01 binding groove, respectively (Fig. 4, Fig. 5, Fig. 6). Notably, throughout the MD simulation, the second amino acid position exhibited strong binding to the A pocket of the MHC molecule, resulting in a significant reduction in the total per-residue decomposition of the free energy (Fig. 4) and a substantial occurrence of H-bonding around this specific position across all peptides (Fig. 6). Even the additional third amino acid position of the predicted peptides KLIMTQVSK, STIHDSIQY, and the reference peptide KTFPPTEPK showed potential binding to the A pocket of the MHC (Fig. 4, Fig. 6). In contrast, the final amino acid position at the carboxyl terminus of every peptide interacted with the F pocket of the MHC binding groove. These specific amino acid positions in the peptides have previously been identified as anchor sites, which are known to facilitate binding to the grooves of diverse MHC class-I molecules [3,9,21,38,39]. Additionally, it is notable that only the second position of the negative control peptide FVGFFTDV exhibited significant binding energy (Fig. 4). Nonetheless, additional research is required to evaluate the functional testing, specifically T-cell activation in HLA-A*11:01 molecules presenting the four predicted peptides analyzed in this study.
This study assessed how well the predicted peptides can bind to the HLA-A*11:01 molecule. We utilized the Gibbs free energy (ΔG°) formula and employed SIE and MM/GBSA to analyze the binding energy. While the ΔGbind values from SIE showed slight variations among the peptides, the summation with the solute entropy term of MM/GBSA revealed more significant differences (Fig. 7). Nevertheless, all the energy evaluation methods consistently identified VINTTDHLK and STIHDSIQY as having a strong binding affinity to the desired MHC molecule among the predicted peptides (Fig. 7). Interestingly, despite receiving high scores and ranks from all prediction algorithms, the peptide TTDHLKFSK exhibited the lowest binding affinity. The difference in in silico predictions from MD simulation and the scores generated by the prediction tools can be attributed to factors like simulation time and the interactions of the binding groove in solvent environments. The foundation of the prediction algorithms employed by the prediction tools used in this study is the structural base of the binding pocket and the potential amino acid anchor positions on the loaded peptides [[10], [11], [12], [13],26]. The peptides are scored or ranked based on the snapshot of the relevant structures deposited in the database [9]. However, the MD simulation in the present study evaluated the interaction between peptides and the binding groove of the MHC in the solvent box at an atomic level. Simulations are also conducted for a specified time, which was 200 ns in our study. Differences in these factors among studies may explain why our study's prediction scores by MD simulation for some peptides were lower than the ratings generated by the five prediction tools that we used in this study.
To confirm the actual binding affinity of the predicted peptides in vitro, we conducted experiments using the TAP-deficient T2 cell line, which overexpresses HLA-A*11:01. While most prediction tools use a default IC50 cutoff value of <500 nM to select peptides that bind to a specific MHC molecule, a previous study reported no significant correlation between IC50 values from prediction tools and in vitro experimental data [40]. Our study similarly found higher IC50 values in vitro. Notably, the T2 cell line evaluation consistently demonstrated an excellent binding affinity for VINTTDHLK and STIHDSIQY, resulting in a low IC50 value, which agreed with the low ΔGbind value obtained from the MD simulation (Fig. 7, Fig. 8). The peptides KLIMTQVSK showed moderate binding affinity in the T2 cell line (Fig. 7, Fig. 8). On the other hand, the TTDHLKFSK peptide displayed the lowest binding affinity, consistent with the MD simulation results (Fig. 7, Fig. 8). The lower binding affinity for TTDHLKFSK can be attributed to its low total pre-residue decomposition energy and fewer H-bond occupations (Fig. 4, Fig. 6). Nevertheless, further investigation is necessary to comprehensively understand the concordance in binding affinity between in silico and in vitro results for this specific peptide.
In contrast, the counterpart interaction with the T-cell receptor, crucial for T-cell activation, was not included. Some previous studies reported that peptides did not constantly activate T-cells, which are thought to have a high affinity for binding to the MHC molecule [41,42]. Therefore, further investigation is warranted to explore T-cell activation in response to these predicted peptides.
5. Limitations
The findings of our MD simulation significantly contribute to the confidence in predicting peptides that can effectively bind to the desired MHC molecule. Although extending the MD simulation time from 500 ns to 1 μs (μs) may yield more precise results, it requires considerable computing resources and time. As an alternative, SIE may offer a faster evaluation of binding energy, but statistical evaluation is crucial for reliable support. MM/GBSA, commonly used to assess relative binding affinities, has consistently aligned with in vitro T2 binding results and is recommended as a criterion for selecting peptides suitable for loading onto the MHC molecule. MM/GBSA has been widely used to evaluate the relative binding affinities between proteins and peptides and between proteins and small molecules, particularly in the study of peptide and MHC molecule binding [[43], [44], [45]]. Together, these methods effectively differentiate binding energy among the predicted peptides and largely correlate with in vitro binding affinity results. However, it's essential to acknowledge that this study mainly focused on the peptide-MHC interaction. The differences between the IC50 values and binding energy obtained from our experiments and the earlier screening results can be attributed to the inherent changes in methodology. Prediction techniques rely on static images of peptide-MHC interactions, while our MD simulations provide a dynamic evaluation over a specified period. The application of this dynamic approach can reveal discrepancies that are not considered by static forecasting systems. The significance of our research lies in using a combination of predictive algorithms and dynamic simulations to improve the accuracy of peptide binding predictions. The information obtained from this approach emphasizes the importance of considering both static and dynamic assessments to understand peptide-MHC interactions thoroughly.
Our study provides important insights into the binding affinity and structural dynamics of GPC3 peptides with the HLA-A*11:01 MHC molecule using MD simulations. However, the study does not include an Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) analysis, which is essential for assessing the safety and effectiveness of these peptides in clinical settings. ADMET properties such as toxicity, allergenicity, half-life, and bioavailability are critical for determining whether these peptides can be safely used in patients. Addressing the comprehensive ADMET evaluations in future studies is crucial to advance these peptides toward potential clinical application. This will ensure that the peptides are effective and safe for patient treatment, thus supporting their potential use in cancer immunotherapy.
6. Conclusion
This study demonstrates the successful use of MD simulation and related calculations, such as analyzing atomic interactions and binding free energy, to accurately identify predicted peptides suitable for loading onto specific HLA molecules of interest. The strong concordance observed between the in silico and in vitro binding affinity outcomes enhances our confidence in the efficacy of in silico peptide prediction using MD simulation. Consequently, this approach can potentially diminish the necessity for resource-intensive and time-consuming in vitro functional testing, specifically by reducing the number of peptides that require such testing. Additionally, the application of MD simulation shows promising potential in advancing the development of peptide-based vaccines for cancer treatment. This progress holds significant importance in pursuing more efficient and targeted cancer therapies.
Funding disclosure
This study was supported by Mahidol University [R016010006, R016010005, R016034008, and Basic Research Fund: fiscal year 2022: BRF1-027/2565], Siriraj Research Fund, Faculty of Medicine Siriraj Hospital, Mahidol University [R015832018], and Program Management Unit-Competitiveness (PMUC) [C10F630063/1], and Chulalongkorn University [CE66_036_2300_008].
CRediT authorship contribution statement
Thaweesak Chieochansin: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Kamonpan Sanachai: Data curation, Methodology, Validation, Visualization, Writing – review & editing. Nitchakan Darai: Data curation, Methodology, Validation, Visualization, Writing – review & editing. Wannasiri Chiraphapphaiboon: Data curation, Writing – review & editing. Kornkan Choomee: Data curation, Writing – review & editing. Pa-thai Yenchitsomanus: Conceptualization, Supervision, Writing – review & editing. Chanitra Thuwajit: Conceptualization, Supervision, Writing – review & editing. Thanyada Rungrotmongkol: Conceptualization, Methodology, Supervision, Validation, Writing – review & editing.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used Grammarly and Quillbot as tools for grammatical checking. After using this tool, the authors reviewed and edited the content as needed and took full responsibility for the publication's content.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge The National Science and Technology Development Agency (NSTDA) Supercomputer Center (ThaiSC), Pathum Thani, Thailand, for providing computing resources for this study. The authors would also like to thank Prof. Dr. Maria Grazia Masucci of the Department of Cell and Molecular Biology, Karolinska Institutet, Stockholm, Sweden, for generously providing transporter associated with antigen processing TAP-deficient T2 cell line overexpressing HLA-A*11:01.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e36654.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F., Freund C. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak T.W., Saunders M.E., Jett B.D. In: Primer to the Immune Response. second ed. Mak T.W., Saunders M.E., Jett B.D., editors. Academic Cell; Boston: 2014. Chapter 6 - the major histocompatibility complex; pp. 143–159. [Google Scholar]
- 3.Abualrous E.T., Sticht J., Freund C. Major histocompatibility complex (MHC) class I and class II proteins: impact of polymorphism on antigen presentation. Curr. Opin. Immunol. 2021;70:95–104. doi: 10.1016/j.coi.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Geng J., Raghavan M. Conformational sensing of major histocompatibility complex (MHC) class I molecules by immune receptors and intracellular assembly factors. Curr. Opin. Immunol. 2021;70:67–74. doi: 10.1016/j.coi.2021.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saper M.A., Bjorkman P.J., Wiley D.C. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J. Mol. Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 7.Matsumura M., Fremont D.H., Peterson P.A., Wilson I.A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 8.Flower D.R. Towards in silico prediction of immunogenic epitopes. Trends Immunol. 2003;24:667–674. doi: 10.1016/j.it.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Desai D.V., Kulkarni-Kale U. T-cell epitope prediction methods: an overview. Methods Mol. Biol. 2014;1184:333–364. doi: 10.1007/978-1-4939-1115-8_19. [DOI] [PubMed] [Google Scholar]
- 10.Rammensee H., Bachmann J., Emmerich N.P., Bachor O.A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 11.Reche P.A., Glutting J.P., Zhang H., Reinherz E.L. Enhancement to the RANKPEP resource for the prediction of peptide binding to MHC molecules using profiles. Immunogenetics. 2004;56:405–419. doi: 10.1007/s00251-004-0709-7. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S., Røder G., Peters B., Sette A., Lund O., Buus S. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gfeller D., Bassani-Sternberg M., Schmidt J., Luescher I.F. Current tools for predicting cancer-specific T cell immunity. OncoImmunology. 2016;5 doi: 10.1080/2162402X.2016.1177691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp B., Demharter S., Esmaielbeiki R., Deane C.M. Current status and future challenges in T-cell receptor/peptide/MHC molecular dynamics simulations. Brief Bioinform. 2015;16:1035–1044. doi: 10.1093/bib/bbv005. [DOI] [PubMed] [Google Scholar]
- 16.Satapornpong P., Jinda P., Jantararoungtong T., Koomdee N., Chaichan C., Pratoomwun J., Na Nakorn C., Aekplakorn W., Wilantho A., Ngamphiw C., Tongsima S., Sukasem C. Genetic diversity of HLA class I and class II alleles in Thai populations: contribution to genotype-guided therapeutics. Front. Pharmacol. 2020;11:78. doi: 10.3389/fphar.2020.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Wang P., Kim Y., Haste-Andersen P., Beaver J., Bourne P.E., Bui H.H., Buus S., Frankild S., Greenbaum J., Lund O., Lundegaard C., Nielsen M., Ponomarenko J., Sette A., Zhu Z., Peters B. Immune epitope database analysis resource (IEDB-AR) Nucleic Acids Res. 2008;36:W513–W518. doi: 10.1093/nar/gkn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abella J.R., Antunes D.A., Clementi C., Kavraki L.E. APE-gen: a fast method for generating ensembles of bound peptide-MHC conformations. Molecules. 2019;24 doi: 10.3390/molecules24050881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antunes D.A., Abella J.R., Hall-Swan S., Devaurs D., Conev A., Moll M., Lizée G., Kavraki L.E. HLA-arena: a customizable environment for the structural modeling and analysis of peptide-HLA complexes for cancer immunotherapy. JCO Clin Cancer Inform. 2020;4:623–636. doi: 10.1200/CCI.19.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshikawa T., Nakatsugawa M., Suzuki S., Shirakawa H., Nobuoka D., Sakemura N., Motomura Y., Tanaka Y., Hayashi S., Nakatsura T. HLA-A2-restricted glypican-3 peptide-specific CTL clones induced by peptide vaccine show high avidity and antigen-specific killing activity against tumor cells. Cancer Sci. 2011;102:918–925. doi: 10.1111/j.1349-7006.2011.01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blicher T., Kastrup J.S., Buus S., Gajhede M. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr D Biol Crystallogr. 2005;61:1031–1040. doi: 10.1107/S0907444905013090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case H.M.A.D.A., Belfon K., Ben-Shalom I.Y., Berryman J.T., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cisneros G.A., Cruzeiro V.W.D., Darden T.A., Forouzesh N., Giambaşu G., Giese T., Gilson M.K., Gohlke H., Goetz A.W., Harris J., Izadi S., Izmailov S.A., Kasavajhala K., Kaymak M.C., King E., Kovalenko A., Kurtzman T., Lee T.S., Li P., Lin C., Liu J., Luchko T., Luo R., Machado M., Man V., Manathunga M., Merz K.M., Miao Y., Mikhailovskii O., Monard G., Nguyen H., O'Hearn K.A., Onufriev A., Pan F., Pantano S., Qi R., Rahnamoun A., Roe D.R., Roitberg A., Sagui C., Schott-Verdugo S., Shajan A., Shen J., Simmerling C.L., Skrynnikov N.R., Smith J., Swails J., Walker R.C., Wang J., Wang J., Wei H., Wu X., Wu Y., Xiong Y., Xue Y., York D.M., Zhao S., Zhu Q., Kollman P.A. University of California; San Francisco: 2023. Amber 2023. [Google Scholar]
- 23.Tian C., Kasavajhala K., Belfon K.A.A., Raguette L., Huang H., Migues A.N., Bickel J., Wang Y., Pincay J., Wu Q., Simmerling C. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theor. Comput. 2020;16:528–552. doi: 10.1021/acs.jctc.9b00591. [DOI] [PubMed] [Google Scholar]
- 24.Duan Y., Wu C., Chowdhury S., Lee M.C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 25.Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J., Liles K., Chun M., Li P., Gohara D.W., Dolinsky T., Konecny R., Koes D.R., Nielsen J.E., Head-Gordon T., Geng W., Krasny R., Wei G.W., Holst M.J., McCammon J.A., Baker N.A. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018;27:112–128. doi: 10.1002/pro.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiewe A.J., Haworth I.S. Structure-based prediction of MHC-peptide association: algorithm comparison and application to cancer vaccine design. J. Mol. Graph. Model. 2007;26:667–675. doi: 10.1016/j.jmgm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 27.H.J.C.P, Berendsen J.P.M., van Gunsteren W.F., Di Nola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 28.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 29.Zajonc D.M. Unconventional peptide presentation by classical MHC class I and implications for T and NK cell activation. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21207561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huan X., Zhuo Z., Xiao Z., Ren E.C. Crystal structure of suboptimal viral fragments of Epstein Barr Virus Rta peptide-HLA complex that stimulate CD8 T cell response. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Habel J.R., Nguyen A.T., Rowntree L.C., Szeto C., Mifsud N.A., Clemens E.B., Loh L., Chen W., Rockman S., Nelson J., Davies J., Miller A., Tong S.Y.C., Rossjohn J., Gras S., Purcell A.W., Hensen L., Kedzierska K., Illing P.T. HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kongkaew S., Yotmanee P., Rungrotmongkol T., Kaiyawet N., Meeprasert A., Kaburaki T., Noguchi H., Takeuchi F., Kungwan N., Hannongbua S. Molecular dynamics simulation reveals the selective binding of human leukocyte antigen alleles associated with behçet's disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rungrotmongkol T., Nunthaboot N., Malaisree M., Kaiyawet N., Yotmanee P., Meeprasert A., Hannongbua S. Molecular insight into the specific binding of ADP-ribose to the nsP3 macro domains of chikungunya and Venezuelan equine encephalitis viruses: molecular dynamics simulations and free energy calculations. J. Mol. Graph. Model. 2010;29:347–353. doi: 10.1016/j.jmgm.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Wang Q., Pang Y.P. Normal-mode-analysis-monitored energy minimization procedure for generating small-molecule bound conformations. PLoS One. 2007;2 doi: 10.1371/journal.pone.0001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shemesh C.S., Hsu J.C., Hosseini I., Shen B.Q., Rotte A., Twomey P., Girish S., Wu B. Personalized cancer vaccines: clinical landscape, challenges, and opportunities. Mol. Ther. : the journal of the American Society of Gene Therapy. 2021;29:555–570. doi: 10.1016/j.ymthe.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonsack M., Hoppe S., Winter J., Tichy D., Zeller C., Küpper M.D., Schitter E.C., Blatnik R., Riemer A.B. Performance evaluation of MHC class-I binding prediction tools based on an experimentally validated MHC-peptide binding data set. Cancer Immunol. Res. 2019;7:719–736. doi: 10.1158/2326-6066.CIR-18-0584. [DOI] [PubMed] [Google Scholar]
- 37.Alba J., Rienzo L.D., Milanetti E., Acuto O., D'Abramo M. Molecular dynamics simulations reveal canonical conformations in different pMHC/TCR interactions. Cells. 2020;9 doi: 10.3390/cells9040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez-Trincado J.L., Gomez-Perosanz M., Reche P.A. Fundamentals and methods for T- and B-cell epitope prediction. J Immunol Res. 2017;2017 doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li L., Bouvier M. Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue. J. Immunol. 2004;172:6175–6184. doi: 10.4049/jimmunol.172.10.6175. [DOI] [PubMed] [Google Scholar]
- 40.Peters B., Bui H.H., Frankild S., Nielson M., Lundegaard C., Kostem E., Basch D., Lamberth K., Harndahl M., Fleri W., Wilson S.S., Sidney J., Lund O., Buus S., Sette A. A community resource benchmarking predictions of peptide binding to MHC-I molecules. PLoS Comput. Biol. 2006;2:e65. doi: 10.1371/journal.pcbi.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X., Liu X., Zhou Z., Ding Y., Wu Y., Qiu J., Shen C. Identification of HLA-A2 restricted epitopes of glypican-3 and induction of CTL responses in HLA-A2 transgenic mice. Cancer Immunol. Immunother. : CII. 2022;71:1569–1582. doi: 10.1007/s00262-021-03096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bethune M.T., Li X.H., Yu J., McLaughlin J., Cheng D., Mathis C., Moreno B.H., Woods K., Knights A.J., Garcia-Diaz A., Wong S., Hu-Lieskovan S., Puig-Saus C., Cebon J., Ribas A., Yang L., Witte O.N., Baltimore D. Isolation and characterization of NY-ESO-1-specific T cell receptors restricted on various MHC molecules. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E10702–e10711. doi: 10.1073/pnas.1810653115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian W., Liu X., Wang L., Zheng B., Jiang K., Fu G., Feng W. Deciphering the selective binding mechanisms of anaplastic lymphoma kinase-derived neuroblastoma tumor neoepitopes to human leukocyte antigen. J. Mol. Model. 2021;27:134. doi: 10.1007/s00894-021-04754-9. [DOI] [PubMed] [Google Scholar]
- 44.Ghobadi Z., Mahnam K., Shakhsi-Niaei M. In-silico design of peptides for inhibition of HLA-A*03-KLIETYFSK complex as a new drug design for treatment of multiples sclerosis disease. J. Mol. Graph. Model. 2022;111 doi: 10.1016/j.jmgm.2021.108079. [DOI] [PubMed] [Google Scholar]
- 45.Zoete V., Irving M., Ferber M., Cuendet M.A., Michielin O. Structure-based, rational design of T cell receptors. Front. Immunol. 2013;4:268. doi: 10.3389/fimmu.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The trajectory and topology files required for the MD simulation, the files necessary for evaluating the free energy changes (ΔGSIE and ΔGMM/GBSA), and the additional datasets utilized and examined in the present investigation has been included in the Supplement Material.