Abstract
Real-time reverse transcription followed by polymerase chain reaction (RT–PCR) is the most suitable method for the detection and quantification of mRNA. It offers high sensitivity, good reproducibility and a wide quantification range. Today, relative expression is increasingly used, where the expression of a target gene is standardised by a non-regulated reference gene. Several mathematical algorithms have been developed to compute an expression ratio, based on real-time PCR efficiency and the crossing point deviation of an unknown sample versus a control. But all published equations and available models for the calculation of relative expression ratio allow only for the determination of a single transcription difference between one control and one sample. Therefore a new software tool was established, named REST© (relative expression software tool), which compares two groups, with up to 16 data points in a sample and 16 in a control group, for reference and up to four target genes. The mathematical model used is based on the PCR efficiencies and the mean crossing point deviation between the sample and control group. Subsequently, the expression ratio results of the four investigated transcripts are tested for significance by a randomisation test. Herein, development and application of REST© is explained and the usefulness of relative expression in real-time PCR using REST© is discussed. The latest software version of REST© and examples for the correct use can be downloaded at http://www.wzw.tum.de/gene-quantification/.
INTRODUCTION
Reverse transcription (RT) followed by polymerase chain reaction (PCR) is a powerful tool for the detection and quantification of mRNA. Nowadays real-time RT–PCR is widely and increasingly used, because of its high sensitivity, good reproducibility and wide quantification range (1,2). It is the most sensitive method for the detection and quantification of gene expression levels, in particular for low abundance mRNA (1,2), in tissues with low concentrations of mRNA (e.g. bone marrow, fatty tissues), from limited tissue samples (e.g. biopsies, single cells) (3,4) and to elucidate small changes in mRNA expression levels (1,2,5). However, it is a very complex technique with various substantial problems associated with its true sensitivity, reproducibility and specificity and, as a fully quantitative methodology, it suffers from the problems inherent in real-time RT–PCR. Generally, two quantification strategies can be performed: an absolute and a relative quantification. In absolute quantification the absolute mRNA copy number per vial or capillary is determined by comparison with appropriate external calibration curves (2). An absolute quantification makes it easier to compare expression data between different days and laboratories, because the calibration curve is a non-changing solid and reliable basis. The relative expression is based on the expression ratio of a target gene versus a reference gene and is adequate for most purposes to investigate physiological changes in gene expression levels. Trends can be better explained by relative quantification, but the results are strongly dependent on the reference gene and the normalisation procedure used. Some mathematical models have already been developed to calculate the relative expression ratios of single samples (6–8; http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf), with or without efficiency correction. Equation 1 shows the most convenient mathematical model, which includes an efficiency correction for real-time PCR efficiency of the individual transcripts (6).
Ratio = (Etarget)ΔCPtarget(control – sample)/(Eref)ΔCPref(control – sample) 1
The relative expression ratio of a target gene is computed, based on its real-time PCR efficiencies (E) and the crossing point (CP) difference (Δ) of an unknown sample versus a control (ΔCPcontrol – sample). In mathematical models the target gene expression is normalised by a non-regulated reference gene expression, e.g. derived from housekeeping genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), albumin, actins, tubulins, cyclophilin, 18S ribosomal RNA (rRNA) or 28S rRNA (9–11). But all published equations and available models for the calculation of relative expression ratios allow for the determination of only a single transcription difference between one control and one sample (n = 1), e.g. given in an DNA array experiment, and not for a group-wise comparison for more samples (n > 2), given in an experimental trial.
Therefore, a new software tool was established, named REST© (relative expression software tool), which compares two groups, with up to 16 data points in the sample group versus 16 data points in the control group, and tests the group differences for significance with a newly developed randomisation test. Nevertheless, the successful application of real-time RT–PCR and REST© depends on a clear understanding of the practical problems. Therefore, a clear experimental design, application and validation of the applied real-time RT–PCR remains essential for accurate and fully quantitative measurement of mRNA transcripts. This paper explains the development of REST© application, discusses the technical aspects involved in an experimental trial and illustrates the usefulness of relative expression in real-time RT–PCR using REST©.
MATERIALS AND METHODS
Animal experiment, total RNA extraction and reverse transcription
Total RNA extraction was performed from rat liver as described previously (12). Adult rats were either fed with physiological zinc concentrations (control group, 58 p.p.m. Zn, n = 7) or suffered 22–29 days under zinc depletion (sample group, 2 p.p.m. Zn, n = 6) (W.Windisch, manuscript submitted). Isolated total RNA integrity was electrophoretically verified by ethidium bromide staining and by an average optical density (OD) OD260/OD280 nm absorption ratio of 1.97 (range 1.78–2.09). Either 330, 1000 or 3000 ng total RNA was reverse transcribed with 100 U Superscript II Plus RNase H– reverse trancriptase (Gibco Life Technologies, Gaithersburg, MD) in a volume of 40 µl, using 100 µM random hexamer primers (Pharmacia Biotech, Uppsala, Sweden) according to the manufacturers’ instructions. Therefore, concentrations of 8.25, 25 or 75 ng cDNA (= reverse transcribed total RNA) per µl were achieved.
Optimisation of RT–PCR
Highly purified salt-free primer for target gene metallothionein (MT) (forward primer, CTC CTG CAA GAA GAG CTG CT; reverse primer, TCA GGC GCA GCA GCT GCA CTT) and for reference gene GAPDH (forward primer, GTC TTC ACT ACC ATG GAG AAG G; reverse primer, TCA TGG ATG ACC TTG GCC AG) were generated commercially (MWG Biotech, Ebersberg, Germany). The MT primer set is able to amplify the transcripts of MT isoform 1 and MT isoform 2 mRNA. Conditions for real-time PCRs were optimised in a gradient cycler (Mastercycler Gradient, Eppendorf, Germany) with regard to Taq DNA polymerase (Roche Molecular Biochemicals, Basel, Switzerland), forward and reverse primers, MgCl2 concentrations (Roche Molecular Biochemicals) and various annealing temperatures (54–66°C). RT–PCR amplification products were separated on a 4% high resolution NuSieve agarose (FMC Bio Products, Rockland, ME) gel electrophoresis and analysed with the Image Master system (Pharmacia Biotech). Optimised conditions were transferred to the following LightCycler real-time PCR protocol.
LightCycler real-time PCR
For determination of test and software variations all applications of different total cDNA input were performed in triplets (MT 1–3 and GAPDH 1–3). Real-time PCR mastermix was prepared as follows (to the indicated end-concentration): 6.4 µl water, 1.2 µl MgCl2 (4 mM), 0.2 µl forward primer (0.4 µM), 0.2 µl reverse primer (0.4 µM) and 1 µl LightCycler–Fast Start DNA Master SYBR Green I (Roche Molecular Biochemicals). Nine microlitres of master-mix was filled in the glass capillaries and a 1 µl volume of cDNA (either 8.25, 25 or 75 ng) was added as PCR template. Capillaries were closed, centrifuged and placed into a cycling rotor. A four-step experimental run protocol was used: (i) denaturation program (10 min at 95°C); (ii) amplification and quantification program repeated 40 times (15 s at 95°C; 10 s at 60°C for MT or 10 s at 58°C for GAPDH; 20 s at 72°C; 5 s at 86°C for MT or 5 s at 84°C for GAPDH with a single fluorescence measurement); (iii) melting curve program (60–99°C with a heating rate of 0.1°C per s and a continuous fluorescence measurement); (iv) cooling program down to 40°C. To improve SYBR Green I quantification a high temperature fluorescence measurement point at the end of the fourth segment was performed (13). It melts the unspecific PCR products below the chosen temperature, e.g. primer dimers, eliminates the non-specific fluorescence signal and ensures accurate quantification of the desired GAPDH and MT real-time RT–PCR product, respectively. For the described mathematical model it is necessary to determine the CPs for each transcript. The CP is defined as the point at which the fluorescence rises appreciably above the background fluorescence. In this study the Second Derivate Maximum Method was performed for CP determination, using LightCycler Software 3.5 (Roche Molecular Biochemicals).
Statistics
For statistical evaluations of the determined CP variations and calculated relative expression variations (Tables 1–3), data were analysed for significant differences by ANOVA using approximate tests (Sigma Stat for Windows Software®, Version 2.0; Jandel Corporation).
Table 1. ANOVA of CV of inter-assay variation of MT and GAPDH CPs determined in rat liver by real-time RT–PCR started either with 8.25 (1–3), 25 (4–6) or 75 ng (7–9) cDNA per capillary.
| Control | Sample | |||
|---|---|---|---|---|
| Mean CP | CV (%) | Mean CP | CV (%) | |
| MT 1–3 (8.25 ng cDNA)a | 24.559 | 7.87 | 31.167 | 8.77 |
| 24.563 | 7.94 | 31.167 | 7.85 | |
| 22.669 | 3.82 | 28.842 | 6.43 | |
| MT 4–6 (25 ng cDNA)a | 25.884 | 10.03 | 32.612 | 5.29 |
| 21.520 | 5.85 | 27.862 | 6.31 | |
| 20.464 | 5.84 | 26.642 | 3.43 | |
| MT 7–9 (75 ng cDNA)a | 20.801 | 8.76 | 27.397 | 3.63 |
| 22.340 | 4.48 | 28.955 | 3.67 | |
| 20.794 | 8.74 | 27.397 | 2.43 | |
| GAPDH 1–3 (8.25 ng cDNA)b | 23.957 | 4.58 | 23.537 | 4.15 |
| 23.342 | 6.85 | 23.432 | 2.52 | |
| 23.350 | 1.59 | 22.235 | 4.33 | |
| GAPDH 4–6 (25 ng cDNA)b | 22.530 | 7.94 | 22.350 | 3.09 |
| 22.899 | 10.42 | 21.730 | 12.89 | |
| 21.801 | 6.35 | 20.758 | 5.27 | |
| GAPDH 7–9 (75 ng cDNA)b | 19.510 | 8.56 | 19.023 | 7.45 |
| 21.314 | 9.23 | 20.525 | 7.16 | |
| 20.279 | 11.19 | 19.307 | 7.86 | |
Given are the mean CP and the CV, each one based on n = 7 for control and n = 6 for sample, respectively.
aANOVA: testing for differences of CV values, neither control nor sample, nor concentration, nor interaction showed a significant effect.
bANOVA: for GAPDH there was a significant effect (P < 0.05) for concentration.
Table 3. Factor of down-regulation of MT and GAPDH expression levels in rat liver under zinc depletion (= NOT normalised by the GAPDH expression).
| 8.25 ng cDNA | MT 1 | MT 2 | MT 3 | Mean ratio, CV |
|---|---|---|---|---|
| 29.630 | 29.565 | 23.706 | 27.634, 12.3% | |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 3) | |
| GAPDH 1 | GAPDH 2 | GAPDH 3 | ||
| 0.767 | 1.059 | 0.495 | 0.774, 36.46% | |
| P < 0.370 | P < 0.876 | P < 0.031 | (n = 3) | |
| 25 ng cDNA | MT 4 | MT 5 | MT 6 | |
| 31.499 | 25.846 | 23.758 | 27.038, 14.83% | |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 3) | |
| GAPDH 4 | GAPDH 5 | GAPDH 6 | ||
| 0.893 | 0.478 | 0.517 | 0.629, 36.34% | |
| P < 0.771 | P < 0.341 | P < 0.121 | (n = 3) | |
| 75 ng cDNA | MT 7 | MT 8 | MT 9 | |
| 29.435 | 29.735 | 29.543 | 29.571, 0.51% | |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 3) | |
| GAPDH 7 | GAPDH 8 | GAPDH 9 | ||
| 0.735 | 0.607 | 0.541 | 0.628, 15.72% | |
| P < 0.477 | P < 0.198 | P < 0.124 | (n = 3) | |
| MT: overall mean and CV | 28.081, 10.22% (n = 9) | |||
| No significant difference between concentrations | ||||
| GAPDH: overall mean and CV | 0.677, 29.79% (n = 9) | |||
| No significant difference between concentrations | ||||
Raw CP data sets are identical to Table 1. ANOVA (n = 9) was performed for MT and GAPDH separately to test the influence of the cDNA starting concentration on expression ratio without normalisation.
DEVELOPMENT OF REST©
Our goal was the development of a software tool that allows for a relative quantification between groups, and a subsequent test for significance of the derived results with a suitable statistical model. Further, the software must be able to run on a widely available platform, which can be used worldwide on different computer systems. For that reason it was programmed to run in Microsoft Excel® (Microsoft Corporation). In what follows, the four pages of REST© and the statistical model, a Pair Wise Fixed Reallocation Randomisation Test© are described in detail.
Page 1—Introduction
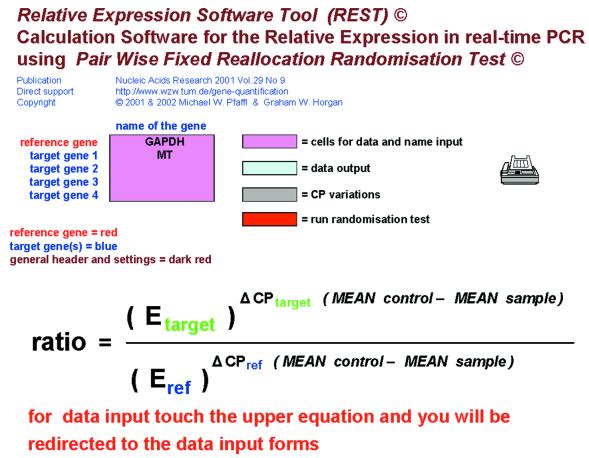
On the introduction page the basic settings are made for the REST© application (Fig. 1). Up to four genes and one reference gene can be labelled. Different background colours in the spreadsheets and the print command are shown and described. Pink cells indicate cells for data input, blue cells indicate data output, grey cells are used for calculation purposes and output of the CP variation, the red box will start the Randomisation Test itself and the printer icon indicates ‘print this page’. Further, the relative expression equation is given with direct links to the data input section on page CP input + randomisation test.
Figure 1.
Page 1—Introduction.
Page 2—PCR efficiency
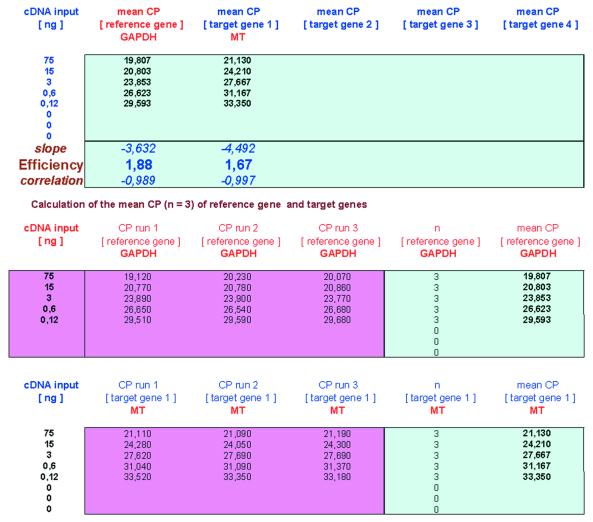
The PCR efficiency calculation is facultative and not obligatory for the user (Fig. 2). To generate the data basis for the determination of PCR efficiency of each transcript, it is recommended to use various dilutions in triplets of a pool of all available cDNAs. This ensures the best estimation of the PCR efficiency. If the user wants to determine the real-time PCR efficiencies, an import via copy and paste of cDNA starting concentrations in dilution row and the corresponding CP values measured by the real-time PCR machine is possible. Depending on the real-time PCR platform used, CP values can be determined either by the Threshold Cycles = Fit Point Method (all platforms) or Second Derivate Maximum Method (only LightCycler). Up to three CPs can be inserted in the table (run 1–3) per cDNA starting concentration and REST© determines the slope with a logarithmic algorithm, as published earlier (1,6,14), as well as an indication of the linearity of this logarithmic alignment using Pearson’s correlation coefficient. The real-time PCR efficiencies were calculated from the slope, according to the established equation E = 10[–1/slope] (1,14). E is in the range from 1 (minimum value) to 2 (theoretical maximum and optimum). If no real-time PCR efficiencies are calculated here, REST© assumes an optimal efficiency of E = 2.0 on the following pages and further procedures.
Figure 2.
Page 2—PCR efficiency.
Page 3—CP input + randomisation test
On the top the calculated PCR efficiencies or alternatively E = 2.0 are shown and will be the basis for the calculation and randomisation test (Fig. 3). Up to 16 CP data per group (control or sample group) can be inserted for the reference gene and up to four target genes (input section of page 3 is not shown). On clicking the red box, the Randomisation Test application window will appear. Here the range of the data set must be defined, for the control group and sample group, by touching the last cell containing the last CP data point (on the bottom right of the pink input window). Further, the number of randomisations can be chosen and the randomisation test will be started on clicking OK. It is recommended that at least 2000 randomisations be performed (see next section statistical model).
Figure 3.
Page 3—CP input + randomisation test.
The numeric results of the randomisation test are given in the Randomisation Data Output box: the concerned Genes, the CP mean of control group (Control Means), the CP mean of sample group (Sample Means), the Expression Ratios normalised by the reference gene, the corresponding p-Values, the Expression Ratios-nn not normalised by the reference gene, the corresponding p-Values-nn and the number of Randomisations performed. To simplify matters for the user, additional answer sentences were created according to the calculated results. They are divided into the Randomisation Test Results (normalised by reference gene expression) and Randomisation Test Results (not normalised by reference gene expression). The sentences tell the user if the sample group in comparison with the control group is up- or down-regulated and illustrates the factor of regulation and if this up- or down-regulation is significantly different or not. For up-regulation, the factor of regulation is equal to the given value in the Randomisation Data Output box. In the case of down-regulation, the regulation factor is illustrated as a reciprocal value (1/expression ratio or 1/expression ratio-nn, respectively).
Page 4—Ratio + variation output
The mean CP of the genes, the CP variations and the coefficient of variation (CV) are calculated and shown to illustrate the reproducibility and variation of the investigated group data subsets (Fig. 4).
Figure 4.
Page 4—Ratio + variation output.
Statistical model: Pair Wise Fixed Reallocation Randomisation Test©
Differences in expression between control and treated samples were assessed in group means (Fig. 1) for statistical significance by randomisation tests (15,16; http://www.bioss.ac.uk/smart/unix/mrandt/slides/frames.htm). Permutation or randomisation tests are a useful alternative to more standard parametric tests for analysing experimental data. They have the advantage of making no distributional assumptions about the data, while remaining as powerful as more standard tests (16).
The rationale for the randomisation test is that standard parametric tests (such as analysis of variance or t-tests) depend on assumptions, such as normality of distributions, whose validity is doubtful. In our case, where the quantities of interest are derived from ratios and variances can be high, normal distributions would not be expected, and it is unclear how a parametric test could best be constructed. A randomisation test avoids making any assumptions about distributions, and is instead based on one we know to be true: that treatments were randomly allocated. The test is conducted as follows.
A statistical test is based on the probability of an effect as large as that observed occurring under the null hypothesis of no treatment effect. If this hypothesis is true, the values in one treatment group were just as likely to have occurred in the other group. The randomisation test repeatedly and randomly reallocates the observed values to the two groups, and notes the apparent effect (expression ratio in our case) each time. The proportion of these effects which are as great as that actually observed in the experiment gives us the P-value of the test.
They calculate P-values by obtaining the proportion of random allocations of the mean observed data to the control and treated sample groups that would give greater indications of a treatment effect than that observed. If this is small, then there is evidence that the observed treatment effect is not simply the result of random allocation. Thus, the test makes no assumptions concerning the distribution of measured gene expression in any hypothesised population—it assumes only the random allocation of treatment. In practice, it is impractical to examine all possible allocations of data to treatment groups, and a random sample is drawn. If 2000 or more samples are taken, a good estimate of the P-value (SE < 0.005 at P = 0.05) is obtained. In the applied Pair Wise Fixed Reallocation Randomisation Test© for each sample, the CP values for reference and target genes are jointly reallocated to control and sample groups (= pair wise fixed reallocation), and the expression ratios are calculated on the basis of the mean values as described above. They are deemed to give greater indications of a treatment effect than that actually observed if │log R│ > │log R0│ where R0 is the true expression ratio and R the result of reallocation. In the Pair Wise Fixed Reallocation Randomisation Test© a two-sided test was performed. The randomisation tests were carried out using a Microsoft Excel® macro (Microsoft Corporation) attached to a purpose-built spreadsheet and running in the background of REST©.
RESULTS
Confirmation of primer specificity
Specificity of RT–PCR products was documented with high resolution gel electrophoresis and resulted in a single product with the desired length (MT, 106 bp; GAPDH, 197 bp). In addition, a LightCycler melting curve analysis was performed which resulted in single product-specific melting temperatures: 87.4°C (GAPDH) and 89.7°C (MT). No primer primer–dimer formations were generated during the applied 40 real-time PCR amplification cycles.
Real-time PCR amplification efficiencies and variation
Real-time PCR efficiencies were calculated from the slopes given in LightCycler software (Roche Molecular Biochemicals LightCycler Software®, Version 3.5). The corresponding real-time PCR efficiency (E) of one cycle in the exponential phase was calculated according to the equation: E = 10[–1/slope], as described earlier (1,6,14). Investigated transcripts showed real-time PCR efficiency rates for MT (EMT = 1.67) and GAPDH (EGAPDH = 1.88) in the investigated range from 120 pg to 75 ng cDNA input, repeated six times, with high linearity [Pearson correlation coefficient (r) > 0.989].
To mimic different reverse transcription efficiencies and to confirm precision and reproducibility of real-time PCR, as well as for REST©, three replicates of real-time RT–PCR at each of various cDNA input concentrations (three times more and three times less concentrated) were performed and real-time RT–PCR and REST© variations (CV) were determined. As shown in Table 1, variations of investigated transcripts are based on the CP variation and remained stable between 2.43 and 10.03% for MT and 1.59 and 12.89% for GAPDH; the latter showing a dependence on the cDNA input in real-time PCR. CP itself decreased with increasing cDNA input in both factors and groups.
Variation and reproducibility of REST©
On the basis of the previously published mathematical model (6), REST© calculates the relative expression ratios on the basis of group means for target gene MT versus reference gene GAPDH and tests the group ratio results for significance. Normalised and not-normalised expression results were compared.
Normalised by GAPDH expression. As presented in Table 2, the down-regulation factor (reciprocal value of ratio) of MT mRNA in the case of zinc deficiency was calculated by REST© starting from different cDNA concentrations. Further different runs (MT 1–3 and GAPDH 1–3, n = 3 × 9) were compared to calculate all possible combinations between individual real-time runs. Derived variations and the influence of deviating cDNA starting amounts on the REST© calculated relative expression ratio, and the significance of the performed randomisation test are presented in Table 2. Over all investigated combinations (n = 27) a mean factor of down-regulation of 44.505 (CV = 26.83%) was observed. No significant differences between cDNA starting concentration on expression ratio could be found.
Table 2. Factor of down-regulation of MT versus GAPDH expression levels in rat liver under zinc depletion (= normalised by the GAPDH expression).
| 8.25 ng cDNA | MT 1 | MT 2 | MT 3 | Mean ratio, CV |
|---|---|---|---|---|
| GAPDH 1 | 38.637 | 38.553 | 30.912 | 39.333, 35.18% |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 9) | |
| GAPDH 2 | 27.989 | 27.928 | 22.393 | |
| P < 0.001 | P < 0.001 | P < 0.001 | ||
| GAPDH 3 | 59.899 | 59.767 | 47.922 | |
| P < 0.001 | P < 0.001 | P < 0.001 | ||
| 25 ng cDNA | MT 4 | MT 5 | MT 6 | |
| GAPDH 4 | 35.290 | 28.956 | 26.617 | 46.358, 29.43% |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 9) | |
| GAPDH 5 | 65.868 | 54.046 | 49.680 | |
| P < 0.001 | P < 0.004 | P < 0.003 | ||
| GAPDH 6 | 60.852 | 49.931 | 45.896 | |
| P < 0.001 | P < 0.001 | P < 0.001 | ||
| 75 ng cDNA | MT 7 | MT 8 | MT 9 | |
| GAPDH 7 | 40.021 | 40.429 | 40.168 | 47.831, 13.12% |
| P < 0.001 | P < 0.001 | P < 0.001 | (n = 9) | |
| GAPDH 8 | 48.446 | 48.939 | 48.624 | |
| P < 0.001 | P < 0.001 | P < 0.001 | ||
| GAPDH 9 | 54.366 | 54.919 | 54.565 | |
| P < 0.001 | P < 0.001 | P < 0.001 | ||
| Overall mean (n = 27) |
|
|
44.504 |
| Overall CV (n = 27) | 26.83% |
Expressed factor of down-regulation (1/ratio) and P-values of control group (n = 7) versus zinc depletion (sample group; n = 6) were calculated by REST©. Data were determined in triplets at each of different stages of cDNA input (8.25, 25 and 75 ng), according to the CP values given in Table 1. ANOVA (n = 3 × 9) was performed to test the influence of cDNA starting concentration on expression ratio including normalisation by GAPDH. There was no significant difference between concentration, but a highly significant effect between MT and especially between GAPDH replicates.
No normalisation by GAPDH. In Table 3 the factor of down-regulation of MT mRNA in the case of zinc deficiency was calculated by REST© without normalisation by the reference gene. For MT a mean down-regulation factor of 28.081 (CV = 10.22%) and for GAPDH of 0.677 (CV = 29.79%) were observed. No significant differences between cDNA starting concentration on expression ratio could be found either for MT or GAPDH.
DISCUSSION
Today, real-time RT–PCR using fluorescence dyes significantly simplifies and accelerates the process of producing reproducible and reliable quantification of mRNA (1). This has led to the development of new kinetic RT–PCR methodologies that are revolutionising the possibilities of mRNA quantification (17). Absolute quantification is very common, where an appropriate external calibration curve is used to determine the absolute mRNA copy number (2). On the other hand, relative expression will be increasingly performed by researchers according to several established mathematical models (6–8). But until now no reliable application was available for a group-wise calculation of the relative expression ratio and a subsequent statistical comparison of the results by a statistical test. Herein, a new software tool is presented and described, which allows for such a group comparison and statistical analysis. REST© is based on an efficiency corrected mathematical model for data analysis. It calculates the relative expression ratio on the basis of the PCR efficiency (E) and crossing point deviation (ΔCP) of the investigated transcripts (6) and on a newly developed randomisation test macro.
Crossing-point determination
For the determination of CP in general two methods can be chosen: the Fit Point Method or adequate methodologies like Threshold Cycle (18,19) where CP will be measured at constant fluorescence level and the Second Derivative Maximum Method where CP will be measured at the maximum increase or acceleration of fluorescence, even if the fluorescence levels between curves are different (14). Besides the LightCycler, the Fit Point Method or Threshold Cycle are used in TaqMan® (PE Applied Biosystems, Foster City, CA), RotoGene® (Corbett Research, Sydney, Australia), iCycler® Thermal Cycler (Bio-Rad, Hercules, CA) and Multiplex Quantitative PCR System® (Stratagene, La Jolla, CA). The Second Derivate Maximum Method is an algorithm exclusively used in LightCycler software (Roche Molecular Biochemicals LightCycler Software®, Version 3.5).
Normalisation
The normalisation of the target gene with an endogenous standard is recommended. REST© allows for a normalisation of the target genes with a reference gene. On both mathematical models the Pair Wise Fixed Reallocation Randomisation Test© is performed and the results are presented in the appropriate output windows. Researchers can decide if they want to correct the data or not. The basis of data normalisation is the expression result of an endogenous desirable unregulated reference gene transcript to compensate inter-PCR variations (sample to sample variations) between the runs. If the CP deviation of the chosen reference gene has the same mean in the control as in sample group mean [ΔCPref (mean control – mean sample) = 0] then a stable and constant reference gene mRNA level is given. Real-time RT–PCR-specific errors in the quantification of mRNA transcripts are easily compounded with any variation in the amount of starting material between the samples. This is especially relevant when the samples have been obtained from different individuals, and will result in the misinterpretation of the derived expression profile of the target genes (1). Here some questions arise: what is the appropriate reference gene for an experimental treatment and investigated tissue (11,20)? Commonly used housekeeping genes (9) are suitable for reference genes, since they are present in all nucleated cell types and necessary for basic cell survival. The mRNA synthesis of housekeeping genes is considered to be stable in various tissues, even under experimental treatments (9–11). But numerous treatments and studies have already shown that housekeeping genes are regulated and vary under specific experimental conditions (21–24). This is a fundamental problem for each relative quantification and correction or normalisation in nucleic acid based models, given in array experiments as well as in mRNA expression analysis. If one desired reference is regulated in a specific experimental trial it remains to the investigator to decide which gene can fit the hypothesis of a non-regulated reference for a reliable normalisation. Therefore, he has to test for more housekeeping genes, and calculate a Housekeeping Gene Index© (publication in preparation). According to this Housekeeping Gene Index©, which is based on the expression of at least three housekeeping genes, a more reliable basis of normalisation in relative quantification using REST© can be postulated. The endogenous control or the calculated Housekeeping Gene Index© should be expressed at roughly the same CP range as the target gene (1). In same CP range, reference and target underwent already the same cycle condition, real-time RT–PCR kinetics, with respect to polymerase activation (heat activation of polymerase) or inactivation and reaction end product inhibition by the generated RT–PCR product (25). REST© can give you the first essential hints if a normalisation via the chosen reference gene is useful (by the factor of regulation and p-value-nn of the randomisation test of the reference), or if the reference is not suitable, because it is significantly regulated.
Efficiency correction
Beside the normalisation by a reference, the PCR efficiency in real-time PCR has a major impact on the accuracy of the calculated expression result (Roche Molecular Biochemicals LightCycler Relative Quantification Software, Version 1.0). A correction for efficiency, as performed in equations 1 and 2, is recommended and results in a more reliable estimation of the ‘real’ expression ratio compared with the no efficiency correction. Small efficiency differences between target and reference genes generate false expression ratio, and the researcher over or underestimates the ‘real’ initial RNA amount. When the difference (Δ) in PCR efficiency (E) is ΔE = 0.03 between target and reference gene, the falsely calculated difference in expression ratio is 46% in the case of Etarget < Eref and 209% in the case of Etarget > Eref after 25 performed cycles. This difference will increase dramatically by higher efficiency differences ΔE = 0.05 (27 and 338%) and ΔE = 0.10 (7.2 and 1083%) and higher cycles performed. Therefore, efficiency corrected quantification is calculated automatically by REST©, based on the method described on page 2 (Fig. 2). It is recommended to perform the determination of real-time PCR efficiency in triplets for every tissue separately in a pool of all starting RNAs to accumulate all possible impacts on PCR efficiency. As is known, each tissue exhibits an individual PCR efficiency, caused by RT and PCR inhibitors (purified in RNA extraction) and by variations in the total RNA pattern extracted.
Relative quantification software
Up to now only one relative quantification software program for real-time PCR has been available and is distributed by Roche Molecular Biochemicals: the LightCycler Relative Quantification software (Version 1.0; Roche Molecular Biochemicals). The mathematical algorithm on which the Roche Molecular Biochemicals software is based is unpublished, and might be the one discussed earlier (6,8).
Ratio = [(Eref)CPsample/(Etarget)CPsample]/[(Eref)CPcalibrator/ (Etarget)CPcalibrator] 2
The LightCycler Relative Quantification software allows only for a comparison of maximal triplets (n = 3), of a target versus a calibrator (cal) gene (which is identical to the control), both corrected via a reference (ref). The relative and normalised expression ratio is calculated on the basis of the median of the performed triplets and computed according to the given equation 3 (Roche Molecular Biochemicals LightCycle Relative Quantifiation Software, Version 1.0). This equation contains a correction factor (CF) as well as a multiplication factor (MF) which are provided in the product-specific applications by Roche Molecular Biochemicals. Ratio concentration (conc) are derived from relative standard curves using the CP median values. Target to reference ratios of all samples are referenced to the target to reference ratio of the calibrator. Thus, it is important to correct for lot-to-lot differences of the calibrator for comparability of data (Roche Molecular Biochemicals LightCycle Relative Quantifiation Software, Version 1.0).
Ratio = [conc(target sample)/conc(reference sample) * MF]/ [conc(target calibrator/conc(reference calibrator) * CF] 3
Advantages of REST©
REST© allows a comparison of four target genes with a reference gene in two experimental groups with up to 16 data points per group. Relative quantification of a target transcript is based on the mean CP deviation of control and sample group, normalised by a reference transcript. Real-time PCR efficiency correction can be performed and is highly recommended. Normalisation via an endogenous standard can be performed according to the users demand, but it is recommended to compensate inter-RT–PCR (or sample to sample) variations (Roche Molecular Biochemicals LightCycle Relative Quantifiation Software, Version 1.0), variations in RNA integrity, RT efficiency differences and cDNA sample loading variations (26). Therefore, a high reproducibility of RT and RT efficiency which greatly varies between tissues, the applied RNA isolation methodology and the RT enzymes used (27,28) are not important any more. Herein, different cDNA input concentrations were tested (±300%) to mimic these huge RT variations and resulted in no significant changes of relative expression ratio evaluated by REST©. Also, the reproducibility of the developed mathematical model used in REST© was given, based on the exact determination of real-time amplification efficiencies and low LightCycler CP variability documented in REST©.
Pair Wise Fixed Reallocation Randomisation Test©
Randomisation tests with a pair-wise reallocation were seen as the most appropriate approach for this application. They make no assumptions about the distribution of observations in populations, which would always be questionable for gene expression measurements. Instead, they assume that animals were randomly allocated to control and treatment groups, which is known to be true if the experimental protocol was adhered. They are more flexible than non-parametric tests based on ranks (Mann–Whitney, Kruskal–Wallis, etc.) and do not suffer a reduction in power relative to parametric tests (t-tests, ANOVA, etc.) They can be slightly conservative (i.e. type I error rates lower than the stated significance level) due to acceptance of randomisations with group differences identical to that observed, but this mainly occurs when used with discrete data (which gene expression data are not) and small sample sizes.
CONCLUSIONS
REST© using the Pair Wise Fixed Reallocation Randomisation Test© is presented for a better understanding of relative quantification analysis in real-time RT–PCR. In rat liver the MT down-regulation in the zinc deficiency group versus the control group lead to similar results using either a normalisation or no normalisation via GAPDH. Real-time RT–PCR in combination with REST© is the method of choice for any experiments requiring sensitive, specific and reproducible quantification of mRNA. The software developed, based on the described mathematical model, exhibits suitable reliability as well as reproducibility in individual runs, confirmed by high accuracy and low variation independent of huge template concentration variations. The latest version of REST© and examples for the correct use can be downloaded at http://www.wzw.tum.de/gene-quantification/.
Acknowledgments
ACKNOWLEDGEMENTS
The author thanks D. Schmidt for technical assistance. The experimental trial was performed in collaboration with the Animal Nutrition and Production Physiology, Center of Life and Food Sciences, Technical University of Munich, under the supervision of Dr W. Windisch.
REFERENCES
- 1.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 2.Pfaffl M.W. and Hageleit,M. (2001) Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT–PCR. Biotechnol. Lett., 23, 275–282. [Google Scholar]
- 3.Lockey C., Otto,E. and Long,Z. (1998) Real-time fluorescence detection of a single DNA molecule. Biotechniques, 24, 744–746. [DOI] [PubMed] [Google Scholar]
- 4.Steuerwald N., Cohen,J., Herrera,R.J. and Brenner,C.A. (1999) Analysis of gene expression in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT–PCR. Mol. Hum. Reprod., 5, 1034–1039. [DOI] [PubMed] [Google Scholar]
- 5.Wittwer C.T., Ririe,K.M., Andrew,R.V., David,D.A., Gundry,R.A. and Balis,U.J. (1997) The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 6.Pfaffl M.W. (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res., 29, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ABI Prism (2001) Relative quantification of gene expression. 7700 Sequence Detection System User Bulletin 2.
- 8.Soong R., Ruschoff,J. and Tabiti,K. (2000) Detection of colorectal micrometastasis by quantitative RT–PCR of cytokeratin 20 mRNA. Roche Molecular Biochemicals Internal Publication.
- 9.Marten N.W., Burke,E.J., Hayden,J.M. and Straus,D.S. (1994) Effect of amino acid limitation on the expression of 19 genes in rat hepatoma cells. FASEB J., 8, 538–544. [DOI] [PubMed] [Google Scholar]
- 10.Foss D.L., Baarsch,M.J. and Murtaugh,M.P. (1998) Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and beta-actin mRNA expression in porcine immune cells and tissues. Anim. Biotechnol., 9, 67–78. [DOI] [PubMed] [Google Scholar]
- 11.Thellin O., Zorzi,W., Lakaye,B., De Borman,B., Coumans,B., Hennen,G., Grisar,T., Igout,A. and Heinen,E. (1999) Housekeeping genes as internal standards: use and limits. J. Biotechnol., 75, 291–295. [DOI] [PubMed] [Google Scholar]
- 12.Pfaffl M.W., Meyer,H.H.D. and Sauerwein,H. (1998) Quantification of the insulin like growth factor-1 mRNA: development and validation of an internally standardised competitive reverse transcription-polymerase chain reaction. Exp. Clin. Endocrinol. Diabetes, 106, 502–512. [DOI] [PubMed] [Google Scholar]
- 13.Pfaffl M.W. (2001) Development and validation of an externally standardised quantitative Insulin like growth factor-1 (IGF-1) RT–PCR using LightCycler SYBR® Green I technology. In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR, Methods and Applications. Springer Press, Heidelberg, pp. 281–191.
- 14.Rasmussen R. (2001) Quantification on the LightCycler. In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR, Methods and Applications. Springer Press, Heidelberg, pp. 21–34.
- 15.Manly B. (1997) Randomization, Bootstrap and Monte Carlo Methods in Biology. Chapman & Hall.
- 16.Horgan G.W. and Rouault,J. (2000) Introduction to Randomisation Tests. Biomathematics and Statistics Scotland.
- 17.Orlando C., Pinzani,P. and Pazzagli,M. (1998) Developments in quantitative PCR. Clin. Chem. Lab. Med., 36, 255–269. [DOI] [PubMed] [Google Scholar]
- 18.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology, 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 19.Gibson U.E., Heid,C.A. and Williams,P.M. (1996) A novel method for real time quantitative RT–PCR. Genome Res., 6, 1095–1001. [DOI] [PubMed] [Google Scholar]
- 20.Haberhausen G., Pinsl,J., Kuhn,C.C. and Markert-Hahn,C. (1998) Comparative study of different standardization concepts in quantitative competitive reverse transcription–PCR assays. J. Clin. Microbiol., 36, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhatia P., Taylor,W.R., Greenberg,A.H. and Wright,J.A. (1994) Comparison of glyceraldehyde-3-phosphate dehydrogenase and 28S-ribosomal RNA gene expression as RNA loading controls for northern blot analysis of cell lines of varying malignant potential. Anal. Biochem., 216, 223–226. [DOI] [PubMed] [Google Scholar]
- 22.Bereta J. and Bereta,M. (1995) Stimulation of glyceraldehyde-3-phosphate dehydrogenase mRNA levels by endogenous nitric oxide in cytokine-activated endothelium. Biochem. Biophys. Res. Commun., 217, 363–369. [DOI] [PubMed] [Google Scholar]
- 23.Chang T.J., Juan,C.C., Yin,P.H., Chi,C.W. and Tsay,H.J. (1998) Up-regulation of beta-actin, cyclophilin and GAPDH in N1S1 rat hepatoma. Oncol. Rep., 5, 469–471. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J. and Snyder,S.H. (1992) Nitric oxide stimulates auto-ADP-ribosylation of glyceraldehydes 3 phosphate dehydrogenase. Proc. Natl Acad. Sci. USA, 89, 9382–9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kainz P. (2000) The PCR plateau phase—towards an understanding of its limitations. Biochim. Biophys. Acta, 1494, 23–27. [DOI] [PubMed] [Google Scholar]
- 26.Karge W.H., Schaefer,E.J. and Ordovas,J.M. (1998) Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol. Biol., 110, 43–61. [DOI] [PubMed] [Google Scholar]
- 27.Mannhalter C., Koizar,D. and Mitterbauer,G. (2000) Evaluation of RNA isolation methods and reference genes for RT–PCR analyses of rare target RNA. Clin. Chem. Lab. Med., 38, 171–177. [DOI] [PubMed] [Google Scholar]
- 28.Wong L., Pearson,H., Fletcher,A., Marquis,C.P. and Mahler,S. (1998) Comparison of the efficiency of M-MuLV reverse transcriptase, Rnase H-M-MuLV reverse transcriptase and AMV reverse transcriptase for the amplification of human immunglobulin genes. Biotechnol. Tech., 12, 485–489. [Google Scholar]