ABSTRACT
Helicobacter pylori infection has been linked to gastrointestinal diseases including gastric cancer. High rates of H. pylori infection and gastric cancer have been reported in indigenous populations within the United States. We report whole-genome sequencing of three H. pylori isolates originating from Native American patients presenting with gastric disease.
KEYWORDS: Helicobacter pylori, gastric cancer, gastrointestinal diseases, Navajo Nation, Native American
ANNOUNCEMENT
Helicobacter pylori, a Gram-negative bacterium, is the causative agent of most human gastric infections and is linked to gastric cancer (1–3). Indigenous communities in the United States have elevated rates of infection and gastric cancer. Members of the Navajo Nation have rates of infection ranging between 56% and 70% (4, 5) and gastric cancer rates three to four times higher than non-Hispanic white populations (6). Navajo patients with gastric symptoms had a gastric biopsy during endoscopy and H. pylori culturing or PCR was performed. H. pylori was present in ~23% of these biopsy samples (7). We describe whole-genome sequencing of three H. pylori isolates from these patients.
Patients were enrolled by informed consent (Navajo Nation IRB #NNR16.263). Gastric pinch biopsy samples were collected during a routine scheduled patient endoscopy. Samples were ground in sterile phosphate-buffered saline, inoculated onto Columbia Agar plates containing 5% defibrinated sheep blood and H. pylori selective supplement (Dent), and incubated for 72 h at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2). Minimum inhibitory concentrations (MICs) for clarithromycin and metronidazole were determined with ETESTs (bioMérieux) by inoculating 100 µL of a 3 McFarland equivalent isolate suspension onto Mueller-Hinton II plates with 5% defibrinated sheep blood and incubating at 37°C for 4 days under microaerophilic conditions.
Isolate genomic DNA was extracted using a Blood and Tissue Kit (Qiagen) following the manufacturer’s protocol with the additional pretreatment for Gram-negative bacteria. Whole-genome sequencing libraries were generated as previously described (8, 9) except quality was assessed with a Fragment Analyzer using the High Sensitivity NGS fragment kit. Samples were sequenced on the MiSeq platform. Contaminating sequencing reads were identified and removed with the BBsplit tool (BBMap v38.93—sourceforge.net/projects/bbmap/) using phiX (J02482.1) and human (GCF_000001405.39) genomes as references, followed by assignment of taxonomic classifications to reads with kraken2 v2.1.2 (10) and removal of contaminating reads. H. pylori genomes were assembled using SPAdes v3.15.3 (--careful, --cov-cutoff auto) (11). Depth of coverage was calculated from minimap2 v2.24 (-ax sr) (12) alignments using Samtools v1.16.1 (13). Contigs with anomalously low depth of coverage were removed. Assembly metrics were calculated with the statswrapper.sh tool (sourceforge.net/projects/bbmap/ v39.01). Assemblies were annotated with the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v6.6 (14). Core genome single nucleotide polymorphisms (SNPs) were called from nucmer v3.1 (15) alignments (reference = GCA_017821535.1) within NASP v1.2.1 (16), and a phylogeny was inferred with IQ-TREE v2.2.2.3 (17, 18) from proximity filtered SNPs (distance of 5). The vacA and cagA genotypes were determined with in silico PCR (usearch v11.0.667_i86linux32 – search_pcr, -maxdiffs 2) (19) using previously described primers (20–23). Genomes were screened for antibiotic-resistance markers listed in the Comprehensive Antimicrobial Resistance Database (24).
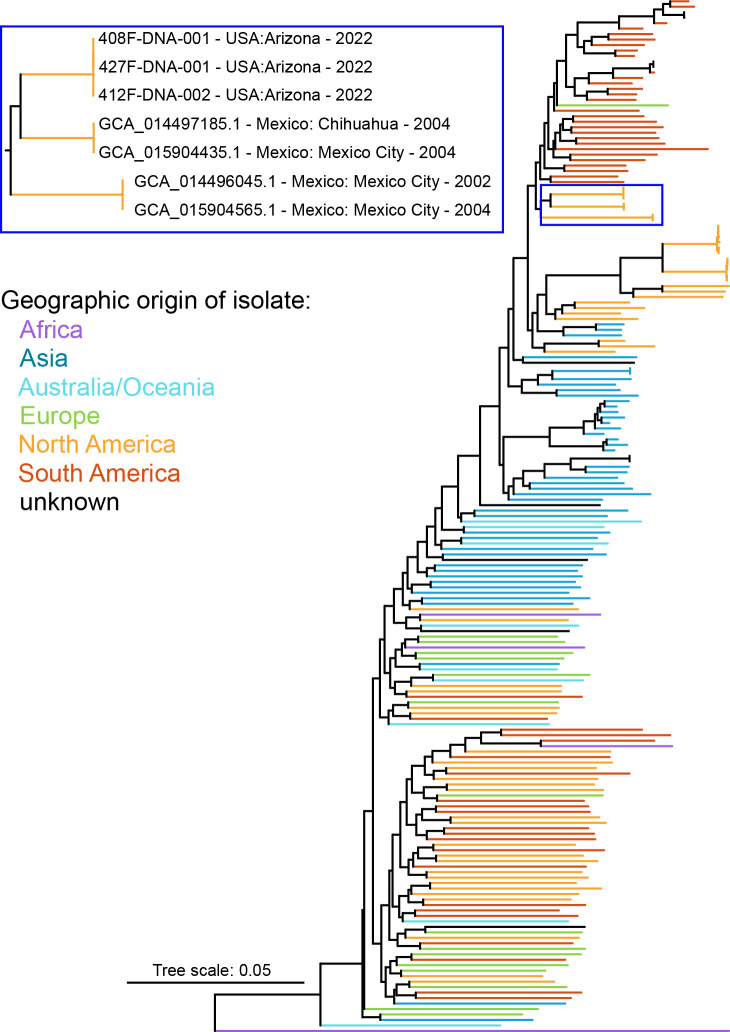
Genome assembly information is presented in Table 1. The three isolates are putatively genotyped as cagA− and vacA type s2i2m2. A SNP phylogeny (Fig. 1) indicates that the isolates are closely related to isolates originating from Indigenous or Mestizo individuals presenting with gastritis in Mexico (25, 26). ETESTS indicate some isolates are resistant to clarithromycin and metronidazole (Table 1).
TABLE 1.
Genome assembly metrics and accession numbers
| Isolate | 408F-DNA-001 | 412F-DNA-002 | 427F-DNA-001 |
|---|---|---|---|
| Assembly accession | JAYXIY000000000 | JAYXIX000000000 | JAYXIW000000000 |
| SRA accession | SRR27606653 | SRR27606652 | SRR27606651 |
| Sequencing kit | 500-cycle Nano v2 | 500-cycle Nano v2 | 600-cycle v3 |
| Sequencing format | 2 × 251 bp | 2 × 251 bpa | 2 × 301 bp |
| Total number of paired reads | 373,565 | 441,603 | 1,201,328 |
| Average depth of coverage | 113× | 124× | 436× |
| Number of contigs | 28 | 19 | 18 |
| Genome size (bp) | 1,570,731 | 1,570,272 | 1,570,218 |
| L50 | 4 | 3 | 3 |
| N50 (bp) | 176,074 | 236,910 | 236,904 |
| Length of longest contig (bp) | 266,682 | 411,760 | 411,746 |
| Average GC content | 0.39 | 0.39 | 0.39 |
| Total CDSs (PGAP) | 1,502 | 1,487 | 1,490 |
| Minimum inhibitory concentration for clarithromycin (R > 0.25 µg/mL)b | 0.125 | 1.5 | 0.5 |
| Minimum inhibitory concentration for metronidazole (R > 8 µg/mL)c |
8 | 16 | 1 |
| Mutations potentially associated with clarithromycin resistanced |
Mutations within 23S rRNA (ARO:3004134) - T510C, G722A, G760del, T896C, T976G, T1024C, C1516del, T1568C, C1648T, T2199C | ||
| Genes/mutations potentially associated with metronidazole resistancee | Presence of major facilitator superfamily antibiotic efflux pump (ARO:3003964); mutations within frxA (ARO:3007059) - V7I, A16T, Q27E, I44V, L71I, F72S, G73S, T110A, N111D, N124S, M126I, A154V, E176K, C193S; mutations within rdxA (ARO:3007055) - T31E, D59N, L62V, S88P, G98S, A118S, V123T, R131K, E175Q | ||
Reverse reads trimmed to 228 nucleotides due to sequencing quality.
Resistance breakpoint for clarithromycin—EUCAST v13.1.
Resistance breakbpoint for metronidazole—EUCAST v13.1.
Genomic data queried against features associated with clarithromycin resistance in H. pylori in the Comprehensive Antimicrobial Resistance Database. The mutations in Table 1 were identified within the 23S rRNA gene for all three isolates, but specific mutations listed within the CARD were not identified.
Genomic data queried against features associated with metronidazole resistance in H. pylori in the Comprehensive Antimicrobial Resistance Database. An antibiotic efflux pump gene was identified in all three genomes. Mutations were identified within frxA and rdxA in all three genomes; specific mutations listed within the CARD are in bold text.
Fig 1.
Core genome SNP phylogeny (midpoint rooted) of 186 publicly available H. pylori genomes and three newly sequenced H. pylori genomes. Colors indicate the continent of origin for the H. pylori isolates included in the tree. The blue box highlights the three newly sequenced isolates (408F, 412F, and 427F) and closely related isolates. The three newly sequenced isolates are closely related to isolates collected from Indigenous or Mestizo individuals presenting with gastritis in Mexico.
ACKNOWLEDGMENTS
The authors would like to acknowledge staff and WIHCC (Winslow Indian Health Care Clinic) who recruited and consented these patients to the study. The authors thank Amber Jones for performing whole-genome sequencing.
This work was funded by 1R21CA248804-01, U54CA143925, and U54CA143924.
Contributor Information
Erik W. Settles, Email: Erik.Settles@nau.edu.
Catherine Putonti, Loyola University Chicago, Chicago, Illinois, USA.
DATA AVAILABILITY
The whole-genome sequencing project has been deposited under NCBI BioProject PRJNA1066305. Assembly accession numbers and Sequence Read Archive accession numbers are included in Table 1.
REFERENCES
- 1. Makola D, Peura DA, Crowe SE. 2007. Helicobacter pylori infection and related gastrointestinal diseases. J Clin Gastroenterol 41:548–558. doi: 10.1097/MCG.0b013e318030e3c3 [DOI] [PubMed] [Google Scholar]
- 2. de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. 2012. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 13:607–615. doi: 10.1016/S1470-2045(12)70137-7 [DOI] [PubMed] [Google Scholar]
- 3. Eusebi LH, Zagari RM, Bazzoli F. 2014. Epidemiology of Helicobacter pylori infection. Helicobacter 19 Suppl 1:1–5. doi: 10.1111/hel.12165 [DOI] [PubMed] [Google Scholar]
- 4. Stancioiu F. 2005. Helicobacter pylori: findings in a native American population. IHS Prim Care Provid 30:60–63. [Google Scholar]
- 5. Harris RB, Brown HE, Begay RL, Sanderson PR, Chief C, Monroy FP, Oren E. 2022. Helicobacter pylori prevalence and risk factors in three rural indigenous communities of northern Arizona. Int J Environ Res Public Health 19:797. doi: 10.3390/ijerph19020797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anonymous . 2013. Cancer among the Navajo 2005-2013
- 7. Monroy FP, Brown HE, Sanderson PR, Jarrin G, Mbegbu M, Kyman S, Harris RB. 2022. Helicobacter pylori in native Americans in northern Arizona. Diseases 10:19. doi: 10.3390/diseases10020019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stone NE, Hall CM, Ortiz M, Hutton SM, Santana-Propper E, Celona KR, Williamson CHD, Bratsch N, Fernandes LGV, Busch JD, Pearson T, Rivera-Garcia S, Soltero F, Galloway R, Sahl JW, Nally JE, Wagner DM. 2022. Diverse lineages of pathogenic leptospira species are widespread in the environment in Puerto Rico. PLoS Negl Trop Dis 16:e0009959. doi: 10.1371/journal.pntd.0009959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kozarewa I, Turner DJ. 2011. 96-Plex molecular Barcoding for the Illumina genome Analyzer. high-throughput next generation sequencing: Methods and applications 279-298 [DOI] [PubMed]
- 10. Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with kraken 2. Genome Biol. 20:257. doi: 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Heng, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44:6614–6624. doi: 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:1–9. doi: 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, Driebe EM, Drees KP, Hicks ND, Williamson CHD, Hepp CM, Smith DE, Roe C, Engelthaler DM, Wagner DM, Keim P. 2016. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb Genom 2:e000074. doi: 10.1099/mgen.0.000074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 20. Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vaca types with cytotoxin production and peptic ulceration (∗). J Biol Chem 270:17771–17777. doi: 10.1074/jbc.270.30.17771 [DOI] [PubMed] [Google Scholar]
- 21. Rhead JL, Letley DP, Mohammadi M, Hussein N, Mohagheghi MA, Eshagh Hosseini M, Atherton JC. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926–936. doi: 10.1053/j.gastro.2007.06.056 [DOI] [PubMed] [Google Scholar]
- 22. Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol 37:2274–2279. doi: 10.1128/JCM.37.7.2274-2279.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. 1998. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter 3:241–253. doi: 10.1046/j.1523-5378.1998.08056.x [DOI] [PubMed] [Google Scholar]
- 24. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, Edalatmand A, Petkau A, Syed SA, Tsang KK, et al. 2023. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res 51:D690–D699. doi: 10.1093/nar/gkac920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Camorlinga-Ponce M, Gómez-Delgado A, Aguilar-Zamora E, Torres RC, Giono-Cerezo S, Escobar-Ogaz A, Torres J. 2020. Phenotypic and genotypic antibiotic resistance patterns in Helicobacter pylori strains from ethnically diverse population in Mexico. Front Cell Infect Microbiol 10:539115. doi: 10.3389/fcimb.2020.539115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muñoz-Ramirez ZY, Pascoe B, Mendez-Tenorio A, Mourkas E, Sandoval-Motta S, Perez-Perez G, Morgan DR, Dominguez RL, Ortiz-Princz D, Cavazza ME, et al. 2021. A 500-year tale of co-evolution, adaptation, and virulence: Helicobacter pylori in the Americas. ISME J 15:78–92. doi: 10.1038/s41396-020-00758-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequencing project has been deposited under NCBI BioProject PRJNA1066305. Assembly accession numbers and Sequence Read Archive accession numbers are included in Table 1.