Abstract
Human herpes viruses (HHV) are ubiquitous and have been implicated in numerous long-term health conditions. Since the association between viral exposure and long-term health impacts is partially influenced by variation in human leukocyte antigen (HLA) genes, we evaluated in silico the binding affinities of 9 HHV envelope glycoproteins with 127 common HLA Class I and Class II molecules. The findings show substantial variability in HHV binding affinity across viruses, HLA Class, HLA genes, and HLA alleles. Specific findings were as follows: (1) the predicted binding affinities of HHVs were characterized by four distinct groupings—[HHV1, HHV2], [HHV3, HHV4, HHV5], [HHV6A], [HHV6B, HHV7, HHV8]—with relatively lower binding affinities for HHV1, HHV2, and HHV6a compared to other HHVs; (2) significantly higher binding affinity was found for HLA Class I relative to Class II; (3) analyses within each class demonstrated that alleles of the C gene (for Class I) and DRB1 gene (for Class II) had the highest binding affinities; and (4) for each virus, predicted binding affinity to specific alleles varied, with HHV6a having the lowest affinity for HHV-HLA complexes, and HHV3, HHV4, and HHV5 having the highest. Since HLA-antigen binding is the first step in initiating an immune response to foreign antigens, these relative differences in HHV binding affinities are likely to influence long-term health impacts such that the cells infected with viruses associated with higher binding affinities across common HLA alleles may be more reduced in numbers, thereby lowering the potential for long-term sequelae of their infections.
Keywords: Human herpes viruses, Binding affinity, Human leukocyte antigen (HLA)
Subject terms: Computational biology and bioinformatics, Genetics, Immunology, Microbiology, Diseases, Medical research
Introduction
Human herpes viruses (HHV) are practically ubiquitous viruses that can establish lifelong infection characterized by alternating periods of latency and reactivation1–3. HHVs include herpes simplex virus 1 (HSV1/HHV1) and HSV2/HV2, varicella zoster virus (VZV/HHV3), Epstein-Barr virus (EBV/ HHV4), cytomegalovirus (CMV/HHV5), HHV6A, HHV6B, HHV7, and Kaposi’s sarcoma virus (HHV8). As a family of viruses, herpesviridae tropism spans multiple systems4, and HHVs have been implicated in numerous long-term health conditions including autoimmune disorders, neoplasms, and neurodegenerative conditions5,6.
The association between viral exposure and long-term health impacts is partially influenced by individual variation in human leukocyte antigen (HLA) genes7–10. Located on chromosome 6, the HLA region is the most highly polymorphic of the human genome11. Small differences, even single amino acid changes in the binding groove, can alter HLA-antigen binding12, thereby influencing foreign antigen elimination and disease susceptibility8,11. HLA genes code for cell-surface glycoproteins that present bound viral epitopes to T cells, signaling immune system responses aimed at virus elimination. Each individual possesses 12 HLA alleles, inherited in a Mendelian fashion, including two of each of the HLA Class I genes (HLA-A, HLA-B, and HLA-C) and two of each of the HLA Class II genes (HLA-DR, HLA-DQ, HLA-DP). Glycoproteins of the two classes operate in concert albeit via different mechanisms and timeframes. HLA Class I, which are expressed on all nucleated cells, signal destruction of an infected cell by binding and transporting cytosolic virus epitopes to the cell surface for presentation to cytotoxic CD8 + T cells. HLA Class II molecules, which are expressed on lymphocytes and professional antigen presenting cells, bind and present endocytosed exogenous antigen epitopes to CD4 + T cells to stimulate antibody production and long-term adaptive immunity. Developing long-term immunity in the event of virus re-exposure occurs over weeks to months compared to the rapid elimination of infected cells via the HLA Class I system13.
It is reasonable to hypothesize that rapid elimination of viral antigens by HLA Class I molecules may reduce viral latency and, consequently, reduce long-term sequelae of viral infections. In light of the high degree of HLA polymorphism, however, the immune response to a HLA-virus antigen complex varies, meaning that some HLA-antigen complexes will be more efficient in mounting an immune response to a given virus than others. Indeed, this is exemplified by slowed progression and control of human immunodeficiency virus (HIV) in carriers of certain HLA alleles (for example, HLA-B*27:05 and B*57:01), as compared to rapid disease progression associated with other HLA alleles (e.g., B*35:01)14. A recent review synthesizing genetic associations of HLA with herpesvirus infection and disease found that most HLA genetic associations are virus- or disease- specific, although certain allotypes were broadly associated with susceptibility or control across herpes viruses15. We hypothesize that observed differences in HLA associations with HHVs are related to immune response of HHV-HLA pairs, the first step of which hinges on sufficient HLA-virus antigen binding affinity. Thus, in this study we used an in silico approach to evaluate the binding affinity of 9 HHV proteins with 127 common HLA Class I (n = 69) and Class II (n = 58) antigens.
Results
Predicted binding affinities (PBA) of HHV proteins with HLA class I and II molecules
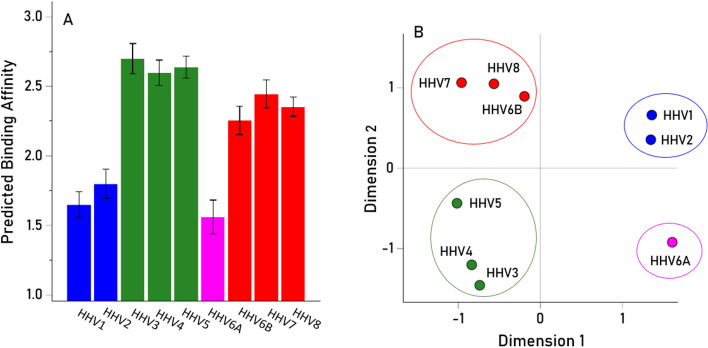
The HHV PBAs of the 9 HHV (Table 1) and 127 HLA Class I and II alleles (Tables 2 and 3) are given in Table S1 in Supplementary Material. The effects of HLA Class on these PBAs were evaluated using a repeated measures analysis of variance (ANOVA), where HHV PBA was the “Within-Subjects” factor and HLA Class was the “Between-Subjects” fixed factor. The results revealed a highly significant effect of HHV on PBA (Fig. 1A; P < 0.001, Greenhouse–Geisser test) and four distinct PBA groupings (color-coded in Fig. 1A): [HHV1, HHV2], [HHV3, HHV4, HHV5], [HHV6A], [HHV6B, HHV7, HHV8]. The same groupings were found using multidimensional scaling (MDS), occupying 4 distinct quadrants in the MDS map (Fig. 1B).
Table 1.
Viral proteins used.
| Index | Virus | Protein description | UniprotKB ID | AA |
|---|---|---|---|---|
| 1 | HHV1 | Envelope glycoprotein D | Q69091 | 394 |
| 2 | HHV2 | Envelope glycoprotein D | P03172 | 393 |
| 3 | HHV3 | Envelope glycoprotein E | Q9J3M8 | 623 |
| 4 | HHV4 | Envelope glycoprotein B | P03188 | 897 |
| 5 | HHV5 | Envelope glycoprotein B | P06473 | 906 |
| 6 | HHV6A | Envelope glycoprotein Q2 | P0DOE0 | 214 |
| 7 | HHV6B | Envelope glycoprotein Q1 | Q9QJ11 | 516 |
| 8 | HHV7 | Envelope glycoprotein H | P52353 | 690 |
| 9 | HHV8 | Envelope glycoprotein H | F5HAK9 | 730 |
Table 2.
The 69 HLA Class I alleles used.
| Index | Gene A | Index | Gene B | Index | Gene C |
|---|---|---|---|---|---|
| 1 | A*01:01 | 21 | B*07:02 | 57 | C*01:02 |
| 2 | A*02:01 | 22 | B*08:01 | 58 | C*03:03 |
| 3 | A*02:05 | 23 | B*13:02 | 59 | C*04:01 |
| 4 | A*03:01 | 24 | B*14:01 | 60 | C*05:01 |
| 5 | A*11:01 | 25 | B*14:02 | 61 | C*06:02 |
| 6 | A*23:01 | 26 | B*15:01 | 62 | C*07:01 |
| 7 | A*24:02 | 27 | B*15:17 | 63 | C*07:02 |
| 8 | A*25:01 | 28 | B*15:18 | 64 | C*07:04 |
| 9 | A*26:01 | 29 | B*18:01 | 65 | C*12:02 |
| 10 | A*29:01 | 30 | B*27:02 | 66 | C*12:03 |
| 11 | A*29:02 | 31 | B*27:05 | 67 | C*14:02 |
| 12 | A*30:01 | 32 | B*35:01 | 68 | C*15:02 |
| 13 | A*30:02 | 33 | B*35:02 | 69 | C*16:01 |
| 14 | A*31:01 | 34 | B*35:03 | ||
| 15 | A*32:01 | 35 | B*35:08 | ||
| 16 | A*33:01 | 36 | B*37:01 | ||
| 17 | A*33:03 | 37 | B*38:01 | ||
| 18 | A*36:01 | 38 | B*39:01 | ||
| 19 | A*68:01 | 39 | B*39:06 | ||
| 20 | A*68:02 | 40 | B*40:01 | ||
| 41 | B*40:02 | ||||
| 42 | B*41:01 | ||||
| 43 | B*41:02 | ||||
| 44 | B*44:02 | ||||
| 45 | B*44:03 | ||||
| 46 | B*44:05 | ||||
| 47 | B*45:01 | ||||
| 48 | B*47:01 | ||||
| 49 | B*49:01 | ||||
| 50 | B*50:01 | ||||
| 51 | B*51:01 | ||||
| 52 | B*52:01 | ||||
| 53 | B*55:01 | ||||
| 54 | B*56:01 | ||||
| 55 | B*57:01 | ||||
| 56 | B*58:01 | ||||
Table 3.
The 58 HLA Class II alleles used.
| Index | Gene | Index | Gene | Index | Gene |
|---|---|---|---|---|---|
| 1 | DPB1*01:01 | 16 | DQB1*02:01 | 30 | DRB1*01:01 |
| 2 | DPB1*02:01 | 17 | DQB1*02:02 | 31 | DRB1*01:02 |
| 3 | DPB1*02:02 | 18 | DQB1*03:01 | 32 | DRB1*01:03 |
| 4 | DPB1*03:01 | 19 | DQB1*03:02 | 33 | DRB1*03:01 |
| 5 | DPB1*04:01 | 20 | DQB1*03:03 | 34 | DRB1*04:01 |
| 6 | DPB1*04:02 | 21 | DQB1*04:02 | 35 | DRB1*04:02 |
| 7 | DPB1*05:01 | 22 | DQB1*05:01 | 36 | DRB1*04:03 |
| 8 | DPB1*06:01 | 23 | DQB1*05:02 | 37 | DRB1*04:04 |
| 9 | DPB1*09:01 | 24 | DQB1*05:03 | 38 | DRB1*04:05 |
| 10 | DPB1*10:01 | 25 | DQB1*06:01 | 39 | DRB1*04:07 |
| 11 | DPB1*11:01 | 26 | DQB1*06:02 | 40 | DRB1*04:08 |
| 12 | DPB1*13:01 | 27 | DQB1*06:03 | 41 | DRB1*07:01 |
| 13 | DPB1*14:01 | 28 | DQB1*06:04 | 42 | DRB1*08:01 |
| 14 | DPB1*17:01 | 29 | DQB1*06:09 | 43 | DRB1*08:03 |
| 15 | DPB1*19:01 | 44 | DRB1*09:01 | ||
| 45 | DRB1*10:01 | ||||
| 46 | DRB1*11:01 | ||||
| 47 | DRB1*11:02 | ||||
| 48 | DRB1*11:03 | ||||
| 49 | DRB1*11:04 | ||||
| 50 | DRB1*12:01 | ||||
| 51 | DRB1*13:01 | ||||
| 52 | DRB1*13:02 | ||||
| 53 | DRB1*13:03 | ||||
| 54 | DRB1*13:05 | ||||
| 55 | DRB1*14:01 | ||||
| 56 | DRB1*15:01 | ||||
| 57 | DRB1*15:02 | ||||
| 58 | DRB1*16:01 | ||||
Fig. 1.
(A) Mean predicted binding affinity (± SEM) of the 9 HHV proteins analyzed across the 127 HLA alleles. (B) Plot of the derived HHV protein configuration yielded by the multidimensional scaling analysis. Viral proteins are color-coded to highlight the 4 distinct PBA groups.
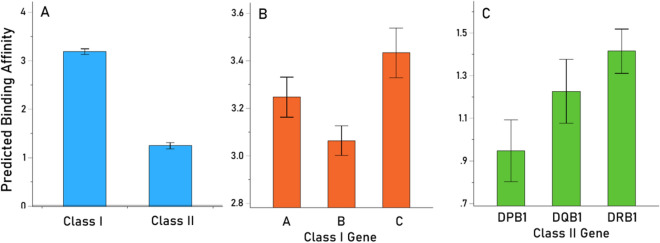
With respect to HLA Class, Class I PBA was significantly higher than Class II (Fig. 2A, P < 0.001, F-test). Finally, with respect to HLA genes, we used a repeated measures ANOVA to evaluate the effect of Gene (“Between-Subjects fixed factor) within each Class. We found a statistically significant effect of Gene for both Class I (P = 0.01, F-test) and Class II (P = 0.038, F-test), with genes C and DRB1 having the highest PBAs (Fig. 2B,C, respectively).
Fig. 2.
A Mean predicted binding affinity (± SEM) of the 9 HHV proteins in the 2 HLA Classes (A), Class I genes (B), and Class II genes (C).
Variation of PBA across HLA alleles
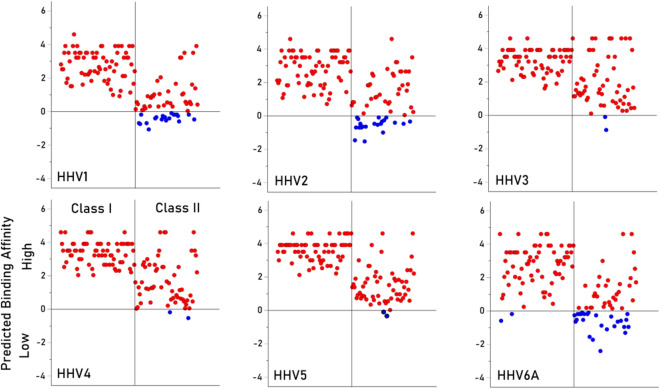
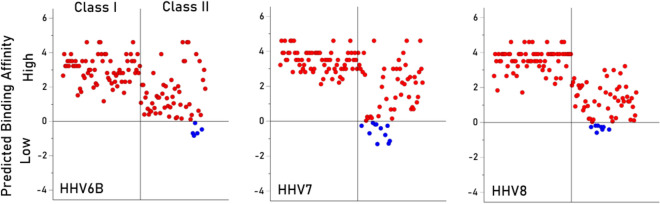
The analyses above evaluated the overall effects of Virus, Class and Gene on PBA. Here we show the individual PBA values for all 127 HLA alleles and the 9 HHV viruses in Figs. 3 and 4. The PBA value of zero corresponds to lowest percentile rank of 1 (PBA = ln(1) = 0; see Methods), a conservative threshold for high binding affinity. It can be seen that (a) most PBAs are of high affinity (> 0), (b) the number of low affinity PBA differ across viruses, being highest for HHV6A and lowest for HHV3, HHV4 and HHV5, and (c) in all viruses but HHV6A, low affinity PBAs are confined to Class II. This variation in PBA across alleles and viruses is captured in the heatmap of Fig. 5.
Fig. 3.
Individual PBA values are plotted for viruses HHV1–HHV6A, as indicated. Red, high affinity PBA; blue, low affinity PBA. N = 127 for each plot.
Fig. 4.
Individual PBA values are plotted for viruses HHV6B, HHV7 and HHV8, as indicated. Red, high affinity PBA; blue, low affinity PBA. N = 127 for each plot.
Fig. 5.
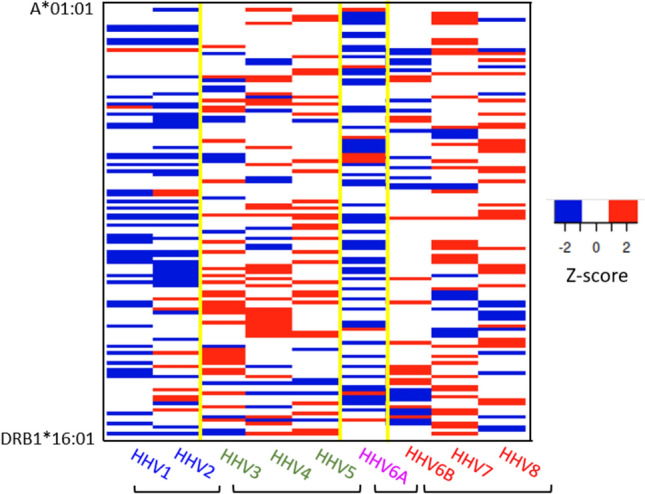
Heatmap of PBAs across the 127 HLA alleles (rows) and the 9 viral proteins (columns). Red, high PBA values (Z-score ≥ 2); blue, low PBA values (Z-score ≤ -2).
Association of PBA with protein length
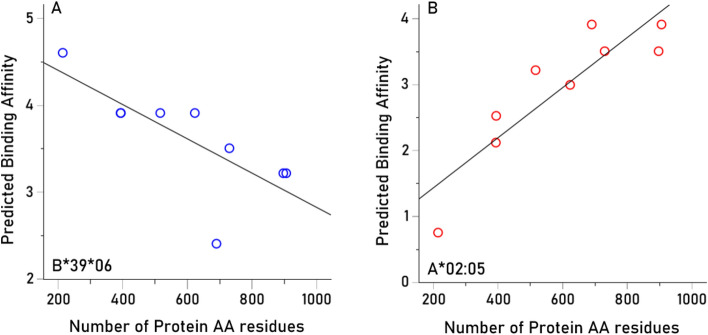
Since we tested all possible 9-AA (for Class I) and 15-AA (for Class II) epitopes, it is possible that PBA estimates could depend on protein length since longer proteins would afford more AA sequences to which HLA molecules could potentially bind. We evaluated this hypothesis by computing, for each allele, the Pearson correlation between PBA values and the number of amino acids in a protein (Table 1). We found the following (Table 4). (a) 18/127 (14.1%) of the correlations were negative (10 Class I, 8 Class II), speaking against the hypothesis above; an example is shown in Fig. 6A. (b) 95/127 (74.8%) correlations were not statistically significant, at a nominal threshold of P < 0.05, uncorrected for multiple comparisons; no correlation was significant at a threshold of P < 0.05/127, i.e. P < 0.000394, after the conservative Bonferroni correction or P < 0.000404, after the less conservative Šidák correction (see Methods); and (c) the percent of PBA variance explained by protein AA length (i.e. 100 x ), irrespective of statistical significance, ranged from 0.001 to 78.5%, but was heavily skewed towards small values (median = 16.7%, mean ± SEM 24.9 ± 2.1%). An example of high positive correlation is shown in Fig. 6B. Overall, these results indicate that there is only a small overall contribution of protein length to PBA values.
Table 4.
Correlation between HHV PBA and the number of amino acids in the HHV protein (N = 9). *, P < 0.05.
| Index | HLA allele | Class | Gene |
Pearson |
P-value | Percent of variance explained |
|---|---|---|---|---|---|---|
| 1 | A*01:01 | 1 | 1 | 0.1414 | 0.7167 | 1.999 |
| 2 | A*02:01 | 1 | 1 | 0.7334 | 0.0245* | 53.793 |
| 3 | A*02:05 | 1 | 1 | 0.8864 | 0.0015* | 78.568 |
| 4 | A*03:01 | 1 | 1 | 0.3691 | 0.3284 | 13.620 |
| 5 | A*11:01 | 1 | 1 | 0.7155 | 0.0302* | 51.197 |
| 6 | A*23:01 | 1 | 1 | 0.5215 | 0.1499 | 27.193 |
| 7 | A*24:02 | 1 | 1 | 0.4754 | 0.1958 | 22.605 |
| 8 | A*25:01 | 1 | 1 | 0.6672 | 0.0496* | 44.516 |
| 9 | A*26:01 | 1 | 1 | 0.8258 | 0.0061* | 68.197 |
| 10 | A*29:01 | 1 | 1 | 0.5064 | 0.1642 | 25.647 |
| 11 | A*29:02 | 1 | 1 | 0.5064 | 0.1642 | 25.647 |
| 12 | A*30:01 | 1 | 1 | 0.5019 | 0.1686 | 25.190 |
| 13 | A*30:02 | 1 | 1 | 0.1697 | 0.6625 | 2.879 |
| 14 | A*31:01 | 1 | 1 | − 0.0005 | 0.9989 | 0.000 |
| 15 | A*32:01 | 1 | 1 | 0.7196 | 0.0288* | 51.786 |
| 16 | A*33:01 | 1 | 1 | − 0.0346 | 0.9296 | 0.120 |
| 17 | A*33:03 | 1 | 1 | − 0.1377 | 0.7239 | 1.896 |
| 18 | A*36:01 | 1 | 1 | − 0.5571 | 0.1192 | 31.038 |
| 19 | A*68:01 | 1 | 1 | 0.6239 | 0.0726 | 38.920 |
| 20 | A*68:02 | 1 | 1 | 0.8610 | 0.0029* | 74.129 |
| 21 | B*07:02 | 1 | 2 | 0.3514 | 0.3538 | 12.347 |
| 22 | B*08:01 | 1 | 2 | 0.4557 | 0.2177 | 20.766 |
| 23 | B*13:02 | 1 | 2 | 0.7441 | 0.0215* | 55.363 |
| 24 | B*14:01 | 1 | 2 | 0.2255 | 0.5596 | 5.085 |
| 25 | B*14:02 | 1 | 2 | 0.2255 | 0.5596 | 5.085 |
| 26 | B*15:01 | 1 | 2 | 0.6041 | 0.0849 | 36.497 |
| 27 | B*15:17 | 1 | 2 | 0.3886 | 0.3013 | 15.102 |
| 28 | B*15:18 | 1 | 2 | 0.5747 | 0.1055 | 33.030 |
| 29 | B*18:01 | 1 | 2 | 0.8092 | 0.0082* | 65.481 |
| 30 | B*27:02 | 1 | 2 | 0.1412 | 0.7171 | 1.993 |
| 31 | B*27:05 | 1 | 2 | 0.1222 | 0.7541 | 1.494 |
| 32 | B*35:01 | 1 | 2 | 0.6533 | 0.0564 | 42.684 |
| 33 | B*35:02 | 1 | 2 | − 0.1272 | 0.7443 | 1.619 |
| 34 | B*35:03 | 1 | 2 | − 0.2384 | 0.5368 | 5.682 |
| 35 | B*35:08 | 1 | 2 | 0.6203 | 0.0747 | 38.483 |
| 36 | B*37:01 | 1 | 2 | 0.6997 | 0.0359* | 48.952 |
| 37 | B*38:01 | 1 | 2 | 0.3403 | 0.3701 | 11.584 |
| 38 | B*39:01 | 1 | 2 | − 0.1490 | 0.7019 | 2.221 |
| 39 | B*39:06 | 1 | 2 | − 0.7481 | 0.0204* | 55.959 |
| 40 | B*40:01 | 1 | 2 | 0.5616 | 0.1156 | 31.535 |
| 41 | B*40:02 | 1 | 2 | 0.7892 | 0.0114* | 62.286 |
| 42 | B*41:01 | 1 | 2 | 0.7848 | 0.0123* | 61.584 |
| 43 | B*41:02 | 1 | 2 | 0.4623 | 0.2102 | 21.375 |
| 44 | B*44:02 | 1 | 2 | 0.1335 | 0.7320 | 1.783 |
| 45 | B*44:03 | 1 | 2 | 0.2514 | 0.5140 | 6.322 |
| 46 | B*44:05 | 1 | 2 | 0.1068 | 0.7846 | 1.140 |
| 47 | B*45:01 | 1 | 2 | 0.8818 | 0.0017* | 77.754 |
| 48 | B*47:01 | 1 | 2 | 0.2151 | 0.5784 | 4.627 |
| 49 | B*49:01 | 1 | 2 | 0.7972 | 0.0101* | 63.560 |
| 50 | B*50:01 | 1 | 2 | 0.7929 | 0.0108* | 62.868 |
| 51 | B*51:01 | 1 | 2 | − 0.2813 | 0.4634 | 7.913 |
| 52 | B*52:01 | 1 | 2 | 0.2988 | 0.4348 | 8.926 |
| 53 | B*55:01 | 1 | 2 | 0.4628 | 0.2096 | 21.422 |
| 54 | B*56:01 | 1 | 2 | 0.1705 | 0.6610 | 2.907 |
| 55 | B*57:01 | 1 | 2 | − 0.0663 | 0.8654 | 0.440 |
| 56 | B*58:01 | 1 | 2 | 0.1480 | 0.7040 | 2.190 |
| 57 | C*01:02 | 1 | 3 | 0.0073 | 0.9850 | 0.005 |
| 58 | C*03:03 | 1 | 3 | 0.3188 | 0.4030 | 10.164 |
| 59 | C*04:01 | 1 | 3 | 0.6436 | 0.0614 | 41.419 |
| 60 | C*05:01 | 1 | 3 | 0.7428 | 0.0219* | 55.175 |
| 61 | C*06:02 | 1 | 3 | 0.1482 | 0.7036 | 2.196 |
| 62 | C*07:01 | 1 | 3 | 0.8779 | 0.0019* | 77.075 |
| 63 | C*07:02 | 1 | 3 | 0.6494 | 0.0584 | 42.169 |
| 64 | C*07:04 | 1 | 3 | 0.7295 | 0.0257* | 53.212 |
| 65 | C*12:02 | 1 | 3 | 0.3373 | 0.3747 | 11.377 |
| 66 | C*12:03 | 1 | 3 | 0.0723 | 0.8533 | 0.523 |
| 67 | C*14:02 | 1 | 3 | 0.2842 | 0.4585 | 8.079 |
| 68 | C*15:02 | 1 | 3 | 0.5929 | 0.0924 | 35.158 |
| 69 | C*16:01 | 1 | 3 | 0.2565 | 0.5052 | 6.581 |
| 70 | DPB1*01:01 | 2 | 4 | 0.4997 | 0.1708 | 24.969 |
| 71 | DPB1*02:01 | 2 | 4 | 0.4088 | 0.2746 | 16.714 |
| 72 | DPB1*02:02 | 2 | 4 | 0.4069 | 0.2771 | 16.554 |
| 73 | DPB1*03:01 | 2 | 4 | 0.7532 | 0.0191* | 56.736 |
| 74 | DPB1*04:01 | 2 | 4 | 0.5051 | 0.1655 | 25.509 |
| 75 | DPB1*04:02 | 2 | 4 | 0.4019 | 0.2836 | 16.153 |
| 76 | DPB1*05:01 | 2 | 4 | 0.8815 | 0.0017* | 77.703 |
| 77 | DPB1*06:01 | 2 | 4 | 0.7597 | 0.0175* | 57.714 |
| 78 | DPB1*09:01 | 2 | 4 | 0.7099 | 0.0322* | 50.392 |
| 79 | DPB1*10:01 | 2 | 4 | 0.7479 | 0.0205* | 55.933 |
| 80 | DPB1*11:01 | 2 | 4 | 0.2491 | 0.5181 | 6.205 |
| 81 | DPB1*13:01 | 2 | 4 | 0.4097 | 0.2735 | 16.784 |
| 82 | DPB1*14:01 | 2 | 4 | 0.7369 | 0.0235 | 54.305 |
| 83 | DPB1*17:01 | 2 | 4 | 0.6677 | 0.0494* | 44.579 |
| 84 | DPB1*19:01 | 2 | 4 | 0.4835 | 0.1873 | 23.380 |
| 85 | DQB1*02:01 | 2 | 5 | 0.2349 | 0.5430 | 5.517 |
| 86 | DQB1*02:02 | 2 | 5 | 0.2349 | 0.5430 | 5.517 |
| 87 | DQB1*03:01 | 2 | 5 | 0.1720 | 0.6581 | 2.959 |
| 88 | DQB1*03:02 | 2 | 5 | 0.3846 | 0.3068 | 14.790 |
| 89 | DQB1*03:03 | 2 | 5 | 0.0720 | 0.8539 | 0.519 |
| 90 | DQB1*04:02 | 2 | 5 | 0.0819 | 0.8341 | 0.670 |
| 91 | DQB1*05:01 | 2 | 5 | 0.4846 | 0.1862 | 23.483 |
| 92 | DQB1*05:02 | 2 | 5 | 0.3653 | 0.3337 | 13.343 |
| 93 | DQB1*05:03 | 2 | 5 | 0.2830 | 0.4607 | 8.006 |
| 94 | DQB1*06:01 | 2 | 5 | 0.7545 | 0.0188* | 56.927 |
| 95 | DQB1*06:02 | 2 | 5 | 0.7915 | 0.0110* | 62.651 |
| 96 | DQB1*06:03 | 2 | 5 | − 0.0974 | 0.8031 | 0.950 |
| 97 | DQB1*06:04 | 2 | 5 | 0.5744 | 0.1057 | 32.993 |
| 98 | DQB1*06:09 | 2 | 5 | 0.6394 | 0.0637 | 40.879 |
| 99 | DRB1*01:01 | 2 | 6 | 0.1778 | 0.6471 | 3.163 |
| 100 | DRB1*01:02 | 2 | 6 | 0.2987 | 0.4349 | 8.923 |
| 101 | DRB1*01:03 | 2 | 6 | 0.1621 | 0.6770 | 2.626 |
| 102 | DRB1*03:01 | 2 | 6 | − 0.1666 | 0.6684 | 2.775 |
| 103 | DRB1*04:01 | 2 | 6 | − 0.1313 | 0.7363 | 1.724 |
| 104 | DRB1*04:02 | 2 | 6 | 0.4284 | 0.2499 | 18.356 |
| 105 | DRB1*04:03 | 2 | 6 | 0.1163 | 0.7657 | 1.353 |
| 106 | DRB1*04:04 | 2 | 6 | 0.3001 | 0.4327 | 9.006 |
| 107 | DRB1*04:05 | 2 | 6 | 0.0633 | 0.8714 | 0.401 |
| 108 | DRB1*04:07 | 2 | 6 | 0.0113 | 0.9771 | 0.013 |
| 109 | DRB1*04:08 | 2 | 6 | 0.0170 | 0.9653 | 0.029 |
| 110 | DRB1*07:01 | 2 | 6 | 0.7908 | 0.0112* | 62.534 |
| 111 | DRB1*08:01 | 2 | 6 | − 0.4362 | 0.2405 | 19.025 |
| 112 | DRB1*08:03 | 2 | 6 | − 0.4704 | 0.2013 | 22.130 |
| 113 | DRB1*09:01 | 2 | 6 | 0.5371 | 0.1359 | 28.848 |
| 114 | DRB1*10:01 | 2 | 6 | 0.1782 | 0.6465 | 3.175 |
| 115 | DRB1*11:01 | 2 | 6 | − 0.6965 | 0.0371* | 48.514 |
| 116 | DRB1*11:02 | 2 | 6 | 0.2902 | 0.4488 | 8.421 |
| 117 | DRB1*11:03 | 2 | 6 | 0.3671 | 0.3311 | 13.475 |
| 118 | DRB1*11:04 | 2 | 6 | 0.7600 | 0.0175* | 57.761 |
| 119 | DRB1*12:01 | 2 | 6 | 0.0863 | 0.8253 | 0.744 |
| 120 | DRB1*13:01 | 2 | 6 | 0.2902 | 0.4488 | 8.421 |
| 121 | DRB1*13:02 | 2 | 6 | 0.2990 | 0.4345 | 8.939 |
| 122 | DRB1*13:03 | 2 | 6 | − 0.0736 | 0.8508 | 0.541 |
| 123 | DRB1*13:05 | 2 | 6 | − 0.6965 | 0.0371* | 48.514 |
| 124 | DRB1*14:01 | 2 | 6 | 0.7490 | 0.0202* | 56.100 |
| 125 | DRB1*15:01 | 2 | 6 | 0.2329 | 0.5465 | 5.424 |
| 126 | DRB1*15:02 | 2 | 6 | 0.0310 | 0.9369 | 0.096 |
| 127 | DRB1*16:01 | 2 | 6 | 0.5425 | 0.1313 | 29.432 |
Fig. 6.
(A) negative association between PBA and number of amino acid (AA) residues of the HHV proteins (Table 1) for allele B*39:06. (B) positive association, for allele A*02:05.
Discussion
Here we evaluated binding affinities of 127 common HLA Class I and Class II alleles with envelope glycoproteins of 9 HHVs and documented substantial variability in the predicted binding affinities of HHVs with regard to HLA Class, gene, and allele. Since HLA–HHV antigen binding is a critical initial step in mounting an adaptive immune response to a viral infection, these findings highlight relative differences in binding affinities of specific HHVs with common HLA Class I and Class II alleles and point to enhanced ability of certain HLA alleles to facilitate a more effective adaptive immune system response to HHVs that bind with higher affinity to common HLA alleles. In the absence of high affinity HLA-HHV complex binding, the virus may persist16, establish latency17, and contribute to subsequent long-term health impacts18,19.
The HLA-HHV binding affinities here fell into four distinct groups, two of which were characterized by distinctly lower binding affinities. Specifically, HHV1/HHV2 and HHV6a, neurotropic viruses that have been implicated (to varying degrees) with neurological conditions20–23, had low binding affinities overall (Fig. 1) and particularly for HLA Class II (Figs. 3, 4, 5). HHV1 and HHV2, commonly referred to as herpes simplex virus -1 (HSV-1) and -2 (HSV-2), cause lifelong infections characterized by periods of latency and reactivation in the form of orolabial (HSV-1) or genital (HSV-2) lesions. The seroprevalence of HSV-1, which is typically acquired by oral contact, in individuals between the ages of 14 and 49 is estimated at 54% whereas the seroprevalence of HSV-2, which is typically sexually transmitted, is 16% in the same age range24. Extensive research supports a prominent role of HSV-1 in dementia20; HSV-2 has been shown to increase HIV acquisition25 and is associated with neurological complications26. Part of the roseola family, HHV6A infection is very common early in life, and, as a neurotropic virus, is commonly detected in the brains of healthy individuals as well as those with neurological diseases27–29. Notably, HHV6A has been shown to integrate into the host germline, allowing for generational transmission of the HHV6A viral genome30. The relatively low binding affinity of HHV1, HHV2, and HHV6A with common HLA alleles may hinder efficient elimination of those viruses, potentially contributing to the development of subsequent neurological effects. Despite their relatively higher overall binding affinities, several of the HHVs comprising the other two groupings (HHV3, HHV4, HHV5; HHV6b, HHV7, HHV8) have also been implicated in long-term health conditions including various cancers, neurological, and autoimmune disorders19,31–35. It is worth noting that even for the 6 viruses associated with higher overall predicted binding affinities to common HLA alleles, there was still substantial variability in predicted binding affinity across alleles (Figs. 3, 4, 5), indicating that some are preferentially able to bind with high affinity compared to others. Thus, the findings suggest that some HHVs are overall more readily handled by HLA-mediated adaptive immune system mechanisms than others, due to their superior binding affinity with common HLA alleles. Nonetheless, for all of the HHVs, the effectiveness of the adaptive immune system response is predicated on high-affinity HLA-HHV binding. To that end, we hypothesize that HHV latency and reactivation as well as long-term disease associations are HLA-dependent.
It is noteworthy that the predicted binding affinities of HHVs with Class I HLA molecules were significantly higher than those of Class II. HLA Class I and Class II play different albeit complementary roles in the adaptive immune response to viruses. HLA Class I promotes rapid elimination of infected cells via cytotoxic CD8 + T cells, whereas Class II contributes to long-term protection via antibody production and immunological memory, a process that can take months13. We propose that rapid elimination of HHVs by binding with HLA Class I to form high affinity HHV-HLA complexes may reduce the potential of the virus to establish latency, thereby reducing the likelihood of viral reactivation and associated diseases, as has been established in the case of early efficient elimination of HIV by certain Class I alleles36. This rapid elimination via Class I mediated cytotoxic T cells does not preclude development of Class II mediated antibody production; indeed, much of the population is seropositive for one or more HHVs (1,2,3). In some cases, seropositivity is associated with disease19,37, suggesting that antibodies reflective of seropositivity do not necessarily confer protection. For both HLA Class I and II, however, adaptive immune protection against HHVs are HLA-dependent in that absence of sufficient HHV-HLA binding affinity inhibits presentation to CD8 + or CD4 + T cells necessary for signaling destruction of infected cells (Class I) or antibody production (Class II), permitting the viral antigen to persist.
Class I HLA-C and Class II HLA-DRB1 genes were associated with higher binding affinity to HHVs than other genes within each respective HLA class. Compared to other classical HLA Class I genes (A and B), HLA-C is unique in that it is less frequently expressed on the cell surface but is the only HLA Class I gene for which virtually all allotypes serve as a natural ligand for multiple types of killer-cell immunoglobulin-like receptors (KIR) which are expressed on natural killer cells that are known to control infected cells efficiently38. As reviewed elsewhere38, mounting research has documented that HLA-C, in combination with KIR, influences control and/or progression of various viral infections including HIV, hepatitis C, and CMV, a member of the herpesviridae family (HHV5). Alleles of the DRB1 gene have been associated with both protection and susceptibility to various conditions including numerous autoimmune disorders7,39,40, many of which are associated with virus exposure41. The current study shines the spotlight more prominently on Class I HLA-C and Class II HLA-DRB1 in influencing the outcome and progression of various HHVs and points to superior binding affinity of HLA-C and HLA-DRB1 as an important underlying mechanism.
The present findings, which document that binding affinity of a given HHV varies across HLA Class I and Class II alleles, must be considered with several qualifications. First, to ensure their survival, HHVs are notorious for utilizing immune evasion mechanisms, several of which involve downregulation of HLA or interference with transport or loading of antigenic peptides which may impair viral elimination even in the case of a strong antiviral immune response42. Second, the focus here is on virus antigen–HLA binding since that is a necessary first step in adaptive immunity; the extent of the human immune response is also partially dependent on immunogenicity of the antigen-HLA complex. Thus, it is possible that some of the high affinity virus-HLA associations documented here may not produce a sufficient immunogenic response against viral antigens. Third, the analyses focused on 127 common HLA Class I and Class II alleles; it is possible that other less common alleles that were not investigated here are capable of forming highly immunogenic complexes. Nonetheless, focusing on globally common alleles permits greater generalization of the findings. Finally, for each virus, we analyzed binding affinity of a single protein of a single strain—specifically, an envelope glycoprotein that is involved in viral entry into the cell. It is unclear to what extent the present findings extend to other proteins and other strains of each of the viruses investigated; although such analyses are beyond the scope of the present paper, they are currently underway.
Materials and methods
HHV proteins
We estimated the binding affinity (for each one of the 69 Class I alleles and 58 Class II alleles) of envelope glycoproteins of 9 HHV viruses (HHV1, HHV2, HHV3, HHV4, HHV5, HHV6A, HHV6B, HHV7, HHV8). Details of the proteins analyzed are given in Table 1 and their amino acid (AA) sequences are given in Table 2S in Supplementary Material. These proteins are involved in virus entry into the cell43 have been widely used in HHV-immunology research, including vaccine development44–53.
HLA alleles
We used 69 common HLA Class I alleles (Table 2) and 58 common HLA Class II alleles (Table 3) that we have employed in previous studies54. Briefly, we obtained the population frequency in 2019 of 127 common HLA Class I and Class II alleles from 14 Continental Western European Countries (Austria, Belgium, Denmark, Finland, France, Germany, Greece, Italy, Netherlands, Portugal, Norway, Spain, Sweden, and Switzerland). There was a total of 2746 entries of alleles from these countries, comprising 844 distinct alleles. Of those, 69 Class I alleles and 58 Class II alleles occurred in 9 or more countries, with a minimum frequency (in any country) of 0.01. Although those alleles were selected based on their frequency in Europe, they have been found to the common overall across 6 world populations55, namely African/African American (AFA), Asian/Pacific Islands (API), European/European descent (EURO), Middle East/North Coast of Africa (MENA), South or Central America/Hispanic/Latino (HIS), Native American (NAM), Unknown/not asked/multiple ancestries/other (UKN), and total (TOTAL). All but allele A*36:01 were Common in each one of the 6 populations above; allele A*36:01 was Intermediate in API and EURO populations, and Common in the remainder populations, and was Common across the 6 populations.
In silico determination of predicted binding affinity of HLA Class I and Class II molecules
Predicted binding affinities were obtained for viral protein epitopes using the Immune Epitope Database (IEDB) NetMHCpan (ver. 4.1) tool56,57. More specifically, we used the sliding window approach58–60 to test exhaustively all possible linear 9-mer (for HLA-I predictions) and 15-mer (for HLA-II predictions) AA residue epitopes of the 9 viral proteins analyzed (Table 1). The method is illustrated in Figs. 7 and 8 for the HHV4 virus protein. For each epitope-HLA molecule tested, this tool gives, as an output, the percentile rank of binding affinity of the HLA molecule and the epitope among predicted binding affinities of the same HLA molecule to a large number of different peptides of the same AA length; the smaller the percentile rank, the better the binding affinity. Now, given a protein of N amino acid length and an epitope length of k AA, there are N-k binding affinity predictions, i.e. N-k percentile ranks. Of these predictions, for each viral protein and HLA molecule tested, we retained the lowest percentile rank (LPR) as the best possible binding affinity of the protein-HLA molecule pair. We then applied two transformations on LPR. First, we took its inverse, so that higher values mean better binding affinities for more intuitive interpretation:
| 1 |
Fig. 7.

Sliding window method for HLA Class I analyses (window length = 9 AA), illustrated for HHV4.
Fig. 8.
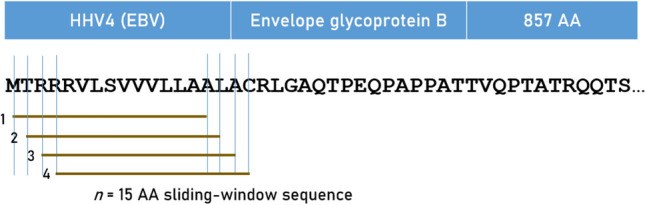
Sliding window method for HLA Class II analyses (window length = 15 AA), illustrated for HHV4.
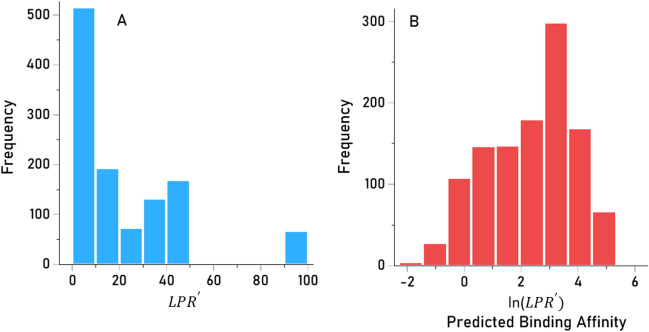
The distribution was heavily skewed to the left (Fig. 9A), resembling an exponential distribution. Therefore, values were (natural) log transformed to normalize its distribution for quantitative analyses (Fig. 9B):
| 2 |
Fig. 9.
Frequency distributions of (Eq. 1) (A, skewed to the left) and its log-transformed PBA values (Eq. 2) (B, unimodal).
Give the logarithmic transformation above, PBA > 0 indicate , whereas PBA < 0 indicate .
Statistical analyses
The IBM-SPSS statistical package (version 29.0.1.1 244) was used for implementing statistical analyses. Standard statistical methods were used, including descriptive statistics, ANOVA, Pearson correlation, etc. All P-values reported are 2-sided, .
Statistical significance uncorrected for multiple comparisons
For this condition, with , P < 0.05 indicated a statistically significant effect for each one of 127 correlations computed between viral PBA and number of amino acids in a viral protein.
Statistical significance corrected for multiple comparisons
Here we computed P values adjusted for the 127 correlations above in two ways, as follows. The first adjustment was the Bonferroni correction, where
| 3 |
The second was the Šidák correction, where
| 4 |
Multidimensional scaling
The potential groupings of the 9 HHV PBAs were evaluated by MDS using the ALSCAL procedure with the following parameters: Model: Euclidean distance; Level of measurement: ratio; Conditionality: matrix; Dimensions: minimum 2, maximum 2; S-stress convergence: 0.0001, minimum s-stress value: 0.005, maximum number of iterations: 30).
Visualization
For PBA visualization, the Heatmapper tool61 was used (http://www.heatmapper.ca/; accessed on July 13, 2024).
Supplementary Information
Author contributions
A.P.G. and L.M.J. retrieved the data. A.P.G. performed data analysis. A.P.G. made figures. L.M.J. and A.P.G. wrote, edited and approved the paper.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Partial funding for this study was provided by the University of Minnesota (the Anita Kunin Chair in Women’s Healthy Brain Aging, the Brain and Genomics Fund, the McKnight Presidential Chair of Cognitive Neuroscience, and the American Legion Brain Sciences Chair) and the U.S. Department of Veterans Affairs. The sponsors had no role in the current study design, analysis or interpretation, or in the writing of this paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Data availability
All data used were retrieved from freely accessible websites and, as such, are publicly and freely available at http://www.biostatistics.online/ineo-epp/neoantigen.php].
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-71558-1.
References
- 1.Staras, A. et al. Seroprevalence of cytomegalovirus infection in the United States, 1988–1994. Clin. Infect. Dis.43, 1143–1151. 10.1086/508173 (2006). 10.1086/508173 [DOI] [PubMed] [Google Scholar]
- 2.Wald, A. & Corey, L. Persistence in the population: epidemiology, transmission. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, Chapter 36 (eds Arvin, A. et al.) (Cambridge University Press, 2007). [PubMed] [Google Scholar]
- 3.McQuillan, G., Kruszon-Moran, D., Flagg, E. W. & Paulose-Ram, R. Prevalence of herpes simplex virus type 1 and type 2 in persons aged 14–49: United States, 2015–2016. NCHS Data Brief, no 304. Hyattsville, MD: National Center for Health Statistics; 2018. [PubMed]
- 4.Ye, S. et al. An atlas of human viruses provides new insights into diversity and tissue tropism of human viruses. Bioinformatics.38, 3087–3093. 10.1093/bioinformatics/btac275 (2022). 10.1093/bioinformatics/btac275 [DOI] [PubMed] [Google Scholar]
- 5.De Francesco, M. A. Herpesviridae, neurodegenerative disorders and autoimmune diseases: What is the relationship between them?. Viruses.16, 133. 10.3390/v16010133 (2024). 10.3390/v16010133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales-Sánchez, A. & Fuentes-Pananá, E. M. Human viruses and cancer. Viruses6, 4047–4079. 10.3390/v6104047 (2014). 10.3390/v6104047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dendrou, C. A., Petersen, J., Rossjohn, J. & Fugger, L. HLA variation and disease. Nat. Rev. Immunol.18, 325–339. 10.1038/nri.2017.143 (2018). 10.1038/nri.2017.143 [DOI] [PubMed] [Google Scholar]
- 8.Blackwell, J. M., Jamieson, S. E. & Burgner, D. HLA and infectious diseases. Clin. Microbiol. Rev.10.1128/cmr.00048-08 (2009). 10.1128/cmr.00048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James, L. M. & Georgopoulos, A. P. Breast cancer, viruses, and human leukocyte antigen (HLA). Sci. Rep.14, 16179. 10.1038/s41598-024-65707-9 (2024). 10.1038/s41598-024-65707-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madeleine, M. M. et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res.68, 3532–3539. 10.1158/0008-5472.CAN-07-6471 (2008). 10.1158/0008-5472.CAN-07-6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trowsdale, J. & Knight, J. C. Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet.14, 301–323. 10.1146/annurev-genom-091212-153455 (2013). 10.1146/annurev-genom-091212-153455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hov, J. R. et al. Electrostatic modifications of the human leukocyte antigen-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology.53, 1967–1976. 10.1002/hep.24299 (2011). 10.1002/hep.24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sego, T. J. et al. A modular framework for multiscale, multicellular, spatiotemporal modeling of acute primary viral infection and immune response in epithelial tissues and its application to drug therapy timing and effectiveness. PLoS Comput. Biol.16, e1008451. 10.1371/journal.pcbi.1008451 (2020). 10.1371/journal.pcbi.1008451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crux, N. B. & Elahi, S. Human leukocyte antigen (HLA) and immune regulation: How do classical and non-classical HLA alleles modulate immune response to human immunodeficiency virus and hepatitis c virus infections?. Front. Immunol.8, 832. 10.3389/fimmu.2017.00832 (2017). 10.3389/fimmu.2017.00832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer, W. H. & Norman, P. J. The impact of HLA polymorphism on herpesvirus infection and disease. Immunogenetics.75, 231–247. 10.1007/s00251-022-01288-z (2023). 10.1007/s00251-022-01288-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James, L. M. & Georgopoulos, A. P. Persistent antigens hypothesis: The human leukocyte antigen (HLA) connection. J. Neurol. Neuromed.3, 27–31. 10.29245/2572.942X/2018/6.1235 (2018). 10.29245/2572.942X/2018/6.1235 [DOI] [Google Scholar]
- 17.Grinde, B. Herpesviruses: Latency and reactivation–viral strategies and host response. J. Oral Microbiol.5, 22766. 10.3402/jom.v5i0.22766 (2013). 10.3402/jom.v5i0.22766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watson, A. M. M. et al. Persistent infection with neurotropic herpes viruses and cognitive impairment. Psychol. Med.43, 1023–1031. 10.1017/S003329171200195X (2013). 10.1017/S003329171200195X [DOI] [PubMed] [Google Scholar]
- 19.Chakravorty, S., Afzali, B. & Kazemian, M. EBV-associated diseases: Current therapeutics and emerging technologies. Front. Immunol.13, 1059133. 10.3389/fimmu.2022.1059133 (2022). 10.3389/fimmu.2022.1059133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itzhaki, R. F. Overwhelming evidence for a major role for Herpes Simplex Virus Type 1 (HSV1) in Alzheimer’s Disease (AD); Underwhelming evidence against. Vaccines9, 679. 10.3390/vaccines9060679 (2021). 10.3390/vaccines9060679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komaroff, A. L., Pellett, P. E. & Jacobson, S. Human Herpesviruses 6A and 6B in brain diseases: Association versus causation. Clin. Microbiol. Rev.34, e00143-e220. 10.1128/CMR.00143-20 (2020). 10.1128/CMR.00143-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren-Gash, C. et al. Human herpesvirus infections and dementia or mild cognitive impairment: A systematic review and meta-analysis. Sci. Rep.9, 4743. 10.1038/s41598-019-41218-w (2019). 10.1038/s41598-019-41218-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzeng, N. S. et al. Anti-herpetic medications and reduced risk of dementia in patients with herpes simplex virus infections—A nationwide, population-based cohort study in Taiwan. Neurotherapeutics.15, 417–429. 10.1007/s13311-018-0611-x (2018). 10.1007/s13311-018-0611-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley, H., Markowitz, L. E., Gibson, T. & McQuillan, G. M. Seroprevalence of herpes simplex virus types 1 and 2—United States, 1999–2010. J. Infect. Dis.209, 325–333. 10.1093/infdis/jit458 (2014). 10.1093/infdis/jit458 [DOI] [PubMed] [Google Scholar]
- 25.Looker, K. J. et al. Effect of HSV-2 infection on subsequent HIV acquisition: An updated systematic review and meta-analysis. Lancet Infect. Dis.17, 1303–1316. 10.1016/S1473-3099(17)30405-X (2017). 10.1016/S1473-3099(17)30405-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger, J. R. & Houff, S. Neurological complications of herpes simplex virus type 2 infection. Arch. Neurol.65, 596–600. 10.1001/archneur.65.5.596 (2008). 10.1001/archneur.65.5.596 [DOI] [PubMed] [Google Scholar]
- 27.Santpere, G., Telford, M., Andres-Benito, P., Navarro, A. & Ferrer, I. The presence of human herpesvirus 6 in the brain in health and disease. Biomolecules10, 1520. 10.3390/biom10111520 (2020). 10.3390/biom10111520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Readhead, B. et al. Multiscale analysis of independent Alzheimer’s cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron.99, 64–82. 10.1016/j.neuron.2018.05.023 (2018). 10.1016/j.neuron.2018.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eimer, W. A. et al. Alzheimer’s disease-associated β-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron.99, 56–63. 10.1016/j.neuron.2018.06.030 (2018). 10.1016/j.neuron.2018.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantry, S. N. & Medveczky, P. G. Latency, integration, and reactivation of human herpesvirus-6. Viruses9, 194. 10.3390/v9070194 (2017). 10.3390/v9070194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science375, 296–301. 10.1126/science.abj8222 (2022). 10.1126/science.abj8222 [DOI] [PubMed] [Google Scholar]
- 32.Chang, Y., Moore, P. S. & Weiss, R. A. Human oncogenic viruses: nature and discovery. Philos. Trans. R. Soc. Lond. B. Biol. Sci.372, 20160264. 10.1098/rstb.2016.0264 (2017). 10.1098/rstb.2016.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söderberg-Nauclér, C., Geisler, J. & Vetcik, K. The emerging role of human cytomegalovirus infection in human carcinogenesis: A review of current evidence and potential therapeutic implications. Oncotarget10, 4333–4337. 10.18632/oncotarget.27016 (2019). 10.18632/oncotarget.27016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, K. H., Kwon, D. E., Do Han, K., La, Y. & Han, S. H. Association between cytomegalovirus end-organ diseases and moderate-to-severe dementia: A population-based cohort study. BMC Neurol.20, 1–9. 10.1186/s12883-020-01776-3 (2020). 10.1186/s12883-020-01776-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mody, P. H., Marvin, K. N., Hynds, D. L. & Hanson, L. K. Cytomegalovirus infection induces Alzheimer’s disease-associated alterations in tau. J. Neurovirol.29, 400–415. 10.1007/s13365-022-01109-9 (2023). 10.1007/s13365-022-01109-9 [DOI] [PubMed] [Google Scholar]
- 36.Payne, R. P. et al. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B 2705-restricted CD8+ T cells. J. Virol.84, 10543–10557. 10.1128/JVI.00793-10 (2010). 10.1128/JVI.00793-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hjalgrim, H., Friborg, J. & Melbye, M. The epidemiology of EBV and its association with malignant disease. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, Chapter 53 (eds Arvin, A. et al.) (Cambridge University Press, 2007). [PubMed] [Google Scholar]
- 38.Vollmers, S., Lobermeyer, A. & Körner, C. The new kid on the block: HLA-C, a key regulator of natural killer cells in viral immunity. Cells10, 3108. 10.3390/cells10113108 (2021). 10.3390/cells10113108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gough, S. C. & Simmonds, M. J. The HLA region and autoimmune disease: Associations and mechanisms of action. Curr. Genomics8, 453–465. 10.2174/138920207783591690 (2007). 10.2174/138920207783591690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furukawa, H. et al. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun.18, 1–7. 10.1038/gene.2016.40 (2017). 10.1038/gene.2016.40 [DOI] [PubMed] [Google Scholar]
- 41.Smatti, M. K. et al. Viruses and autoimmunity: A review on the potential interaction and molecular mechanisms. Viruses.11, 762. 10.3390/v11080762 (2019). 10.3390/v11080762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horst, D., Ressing, M. E. & Wiertz, E. J. Exploiting human herpesvirus immune evasion for therapeutic gain: Potential and pitfalls. Immunol. Cell Biol.89, 359–366. 10.1038/icb.2010.129 (2011). 10.1038/icb.2010.129 [DOI] [PubMed] [Google Scholar]
- 43.Heldwein, E. E. & Krummenacher, C. Entry of herpesviruses into mammalian cells. Cell Mol. Life Sci.65, 1653–1668. 10.1007/s00018-008-7570-z (2008). 10.1007/s00018-008-7570-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frye, T. D., Chiou, H. C., Hull, B. E. & Bigley, N. J. The efficacy of a DNA vaccine encoding herpes simplex virus type 1 (HSV-1) glycoprotein D in decreasing ocular disease severity following corneal HSV-1 challenge. Arch. Virol.147, 1747–1759. 10.1007/s00705-002-0830-6 (2002). 10.1007/s00705-002-0830-6 [DOI] [PubMed] [Google Scholar]
- 45.Leroux-Roels, G. et al. Immunogenicity and safety of different formulations of an adjuvanted glycoprotein D genital herpes vaccine in healthy adults: a double-blind randomized trial. Hum. Vaccin. Immunother.9, 1254–1262. 10.4161/hv.24043 (2013). 10.4161/hv.24043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heineman, T. C., Cunningham, A. & Levin, M. Understanding the immunology of Shingrix, a recombinant glycoprotein E adjuvanted herpes zoster vaccine. Curr. Opin. Immunol.59, 42–48. 10.1016/j.coi.2019.02.009 (2019). 10.1016/j.coi.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 47.Hong, J. et al. Glycoprotein B antibodies completely neutralize EBV infection of B Cells. Front. Immunol.13, 920467. 10.3389/fimmu.2022.920467 (2022). 10.3389/fimmu.2022.920467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirchmeier, M. et al. Enveloped virus-like particle expression of human cytomegalovirus glycoprotein B antigen induces antibodies with potent and broad neutralizing activity. Clin. Vaccine Immunol.21, 174–180. 10.1128/CVI.00662-13 (2014). 10.1128/CVI.00662-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jasirwan, C., Furusawa, Y., Tang, H., Maeki, T. & Mori, Y. Human herpesvirus-6A gQ1 and gQ2 are critical for human CD46 usage. Microbiol. Immunol.58, 22–30. 10.1111/1348-0421.12110 (2014). 10.1111/1348-0421.12110 [DOI] [PubMed] [Google Scholar]
- 50.Kawabata, A. et al. Analysis of a neutralizing antibody for human herpesvirus 6B reveals a role for glycoprotein Q1 in viral entry. J. Virol.85, 12962–12971. 10.1128/JVI.05622-11 (2011). 10.1128/JVI.05622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukai, T., Hata, A., Isegawa, Y. & Yamanishi, K. Characterization of glycoprotein H and L of human herpesvirus 7. Microbiol. Immunol.41, 43–50. 10.1111/j.1348-0421.1997.tb01171.x (1997). 10.1111/j.1348-0421.1997.tb01171.x [DOI] [PubMed] [Google Scholar]
- 52.Pertel, P. E. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol.76, 4390–4400. 10.1128/jvi.76.9.4390-4400.2002 (2002). 10.1128/jvi.76.9.4390-4400.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, H. Y. et al. Common polymorphisms in the glycoproteins of human cytomegalovirus and associated strain-specific immunity. Viruses13, 1106. 10.3390/v13061106 (2021). 10.3390/v13061106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.James, L. M. & Georgopoulos, A. P. Immunogenetic epidemiology of dementia and Parkinson’s Disease in 14 continental European countries: Shared human leukocyte antigen (HLA) profiles. J. Immunological Sci.5, 16–26. 10.29245/2578-3009/2021/12.1209 (2021). 10.29245/2578-3009/2021/12.1209 [DOI] [Google Scholar]
- 55.Hurley, C. K. et al. Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA.95, 516–531. 10.1111/tan.13811 (2020). 10.1111/tan.13811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reynisson, B., Alvarez, B., Paul, S., Peters, B. & Nielsen, M. NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res.48, 0449–0454. 10.1093/nar/gkaa379 (2020). 10.1093/nar/gkaa379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.IEDB Analysis Resource. Accessed on April 11, 2024. http://tools.iedb.org/mhci/
- 58.Charonis, S., James, L. M. & Georgopoulos, A. P. In silico assessment of binding affinities of three dementia-protective Human Leukocyte Antigen (HLA) alleles to nine human herpes virus antigens. Curr. Res. Transl. Med.68, 211–216. 10.1016/j.retram.2020.06.002 (2020). 10.1016/j.retram.2020.06.002 [DOI] [PubMed] [Google Scholar]
- 59.Charonis, S., Tsilibary, E. P. & Georgopoulos, A. SARS-CoV-2 virus and Human Leukocyte Antigen (HLA) Class II: Investigation in silico of binding affinities for COVID-19 protection and vaccine development. J. Immunol. Sci.4, 12–23. 10.29245/2578-3009/2020/4.1198 (2020). 10.29245/2578-3009/2020/4.1198 [DOI] [Google Scholar]
- 60.Charonis, S. A., Tsilibary, E. P. & Georgopoulos, A. P. In silico investigation of binding affinities between human leukocyte antigen class I molecules and SARS-CoV-2 virus spike and ORF1ab proteins. Explor. Immunol.1, 16–26. 10.37349/ei.2021.00003 (2021). 10.37349/ei.2021.00003 [DOI] [Google Scholar]
- 61.Babicki, S. et al. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res.44, W147–W153. 10.1093/nar/gkw419 (2016). 10.1093/nar/gkw419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used were retrieved from freely accessible websites and, as such, are publicly and freely available at http://www.biostatistics.online/ineo-epp/neoantigen.php].