Abstract
Purpose
The current study aims to investigate the significance of N6-methyladenosine (m6A) methylationrelated genes in the clinical prognosis of childhood relapsed B-cell acute lymphoblastic leukemia (B-ALLL) patient.
Methods
Transcriptome data and corresponding clinical data on m6A methylation-related genes (including 20 genes) were obtained from the Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database.
Results
The bone marrow (BM) samples of 134 newly diagnosed (naive) and 116 relapsed B-ALL from TARGET were enrolled in the current study. Three genes (FTO, HNRNPC, RBM15B) showed significant up-regulation in relapsed B-ALL compared with that in naive B-ALL.The three genes had a significantly worse survival (P < 0.05). The LASSO Cox regression model was used to select the most predictive genes as prognostic indicators, and YTHDC1 and FTO were identified as prognostic factors for relapsed B-ALL. Finally, the results of multivariate regression analysis showed that the risk score of m6A methylation-related genes was an independent prognostic factor in relapsed B-ALL (P < 0.05).
Conclusion
We found that the expression levels of m6A methylation-related genes were different in naive and relapsed patients with B-ALL and correlated with survival and prognosis.This implies that m6A methylation-related genes may be promising prognostic indicators or therapeutic targets for relapsed B-ALL.
Keywords: m6A methylation-related genes, Relapsed B-ALL, Children, Prognosis
Introduction
Although the prognosis of childhood B-cell acute lymphoblastic leukemia (B-ALL) has improved significantly in recent years, 10-15% will experience relapse [1–3]. This is partly because pediatric B-ALL tend to have more unfavorable cytogenetic profiles [4, 5]. N6-methyladenosine (m6A) is the most common epigenetic modification on eukaryotic messenger RNA (mRNA) and plays an important role in many basic biological processes [6, 7]. The dysregulation of m6Amodification plays a crucial role in the growth and advancement of several types of cancers, such as acute myeloid leukemia (AML) [8–10]. Nevertheless, research on the m6A modification in relapse B-ALL is confined. Although the extent of m6A modification in relapse B-ALL remains scantily investigated. Our objective in this study is to evaluate the activity of m6A machinery (writers, erasers, and readers) in bone marrow (BM) samples from primary B-ALL patients, both naive and relapsed. Additionally, we will examine the relationship between gene expression related to m6A methylation and the clinical prognosis of patients. Through this research, our goal is to identify new prognostic biomarkers and potential therapeutic targets for relapsed B-ALL.We present the following article in accordance with the MDAR reporting checklist.
Methods and materials
Data collection
We downloaded the transcriptome data and clinical data of naïve and relapsed B-ALL from The Therapeutically Applicable Research To Generate Effective Treatments (TARGET) database (https://ocg.cancer.gov/programs/target). MRNA expression data, as well as clinical information such as age, gender, early treatment response, and fusion gene status, were collected from 134 newly diagnosed (naive) and 116 relapsed B-ALL BM samples. The results published here are entirely or partially based on data generated from studies applicable to treatment, in order to generate effective treatment methods(https://ocg.cancer.gov/programs/target). The data used for this analysis can be found in the https://portal.gdc.cancer.gov/projects. The Ethics Committee of the Office of Cancer Genomics (OCG) approved this study, and the informed consent forms were signed by the guardians of the patients involved. This study followed the guidelines outlined in the Helsinki Declaration (clinical trial numbers: NCT 00070174, NCT00372593, NCT01371981, and NCT00002798).
Bioinformatic analysis/statistics
The differential expression of 20 m6A methylation-related genes, including writers (METTL3, METTL14, VIRMA, RBM15, RBM15B, RMBX, WTAP, and ZC3H13), readers (HNRNPC, HNRNPA2B1, YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, IGF2BP2, IGF2BP3), and erasers (ALKBH5, FTO), in samples of naive and relapse B-ALL BM were assessed using R sofeware.
Statistical analyses
Univariate Cox regression was used to analyze the set of 20 identified genes. Candidate genes were chosen based on whether they met the screening requirement of P<0.05. Following that, we employed LASSO regression on high-dimensional data in order to select the most pertinent prognostic factors using package in R software. Using this method, we identified five genes and calculated their corresponding risk scores. Based on the median expression of m6A methylation-related genes, patients were categorized into high-risk and low-risk groups. We then utilized the K-M survival approach to examine the association between these m6A-related genes and the survival rates. The P-value of the K-M survival curves was determined using log-rank tests. To assess the accuracy of the model, we generated a receiver operating characteristic (ROC) curve. Univariate and multivariate Cox regression analyses were carried out to identify prognostic factors for relapsed B-ALL.
The regression coefficients of 5 optimal prognostic genes were derived from the multivariate Cox proportional hazards regression model. Subsequently, a linear combination method was adopted to assemble expression level and coefficient of each gene to get a risk score formula. The samples of BM were divided into two groups using “sample” function of R software. Heatmap of ALL was plotted using “pheatmap” R package with zero-mean normalization. PCA was used to estimate batch effect and clustering result using “ggfortify” R package. Two groups of boxplot were analyzed using Wilcoxon-test. For Kaplan-Meier curves, p-values and hazard ratio (HR) with 95% confidence interval (CI) were generated by log-rank tests and univariate Cox proportional hazards regression. All analytical methods above and R packages were performed using R software version 3.6.1 (The R Foundation for Statistical Computing, 2019). All statistical tests were two-sided. P < 0.05 was considered as statistically significant.
Results
TARGET dataset and patients’ characteristics
The current study included samples of 134 newly diagnosed (naive) B-ALL and 116 relapsed B-ALL extracted from the BM collection of TARGET. After excluding samples with incomplete clinical data, a group of 116 patients diagnosed with relapsed B-ALL (consisting of 64 males and 52 females) were included for analysis. The age range from 1.1 years to 18.7 years and the mean age of the individuals in the dataset was 8.4 years. The available clinical information encompassed demographic characteristics, initial response to treatment, and the presence of fusion genes.
Expression of m6A methylation-related genes in relapsed B-ALL
A gene expression heatmap was created using 20 m6A methylation-related genes to gain a comprehensive understanding of their expression in naive and relapsed B-ALL samples. Analysis of the TARGET data revealed that three genes (FTO, HNRNPC, RBM15B) were significantly up-regulated, while four genes (METTL14, YTHDC1, YTHDC2, RBMX) were significantly down-regulated in relapsed B-ALL compared to naive B-ALL (Fig. 1A-B). Based on Fig. 1C, statistical analysis using Pearson correlation confirmed positive correlations among four genes.The correlation coefficient of 0 indicates a strong positive correlation between METTL3 and YTHDC1.66.METTL4 and YTHDC2, as well as YTHDC1 and YTHDC2, exhibited robust positive correlations with correlation coefficients of 0.64 and 0.56.
Fig. 1.
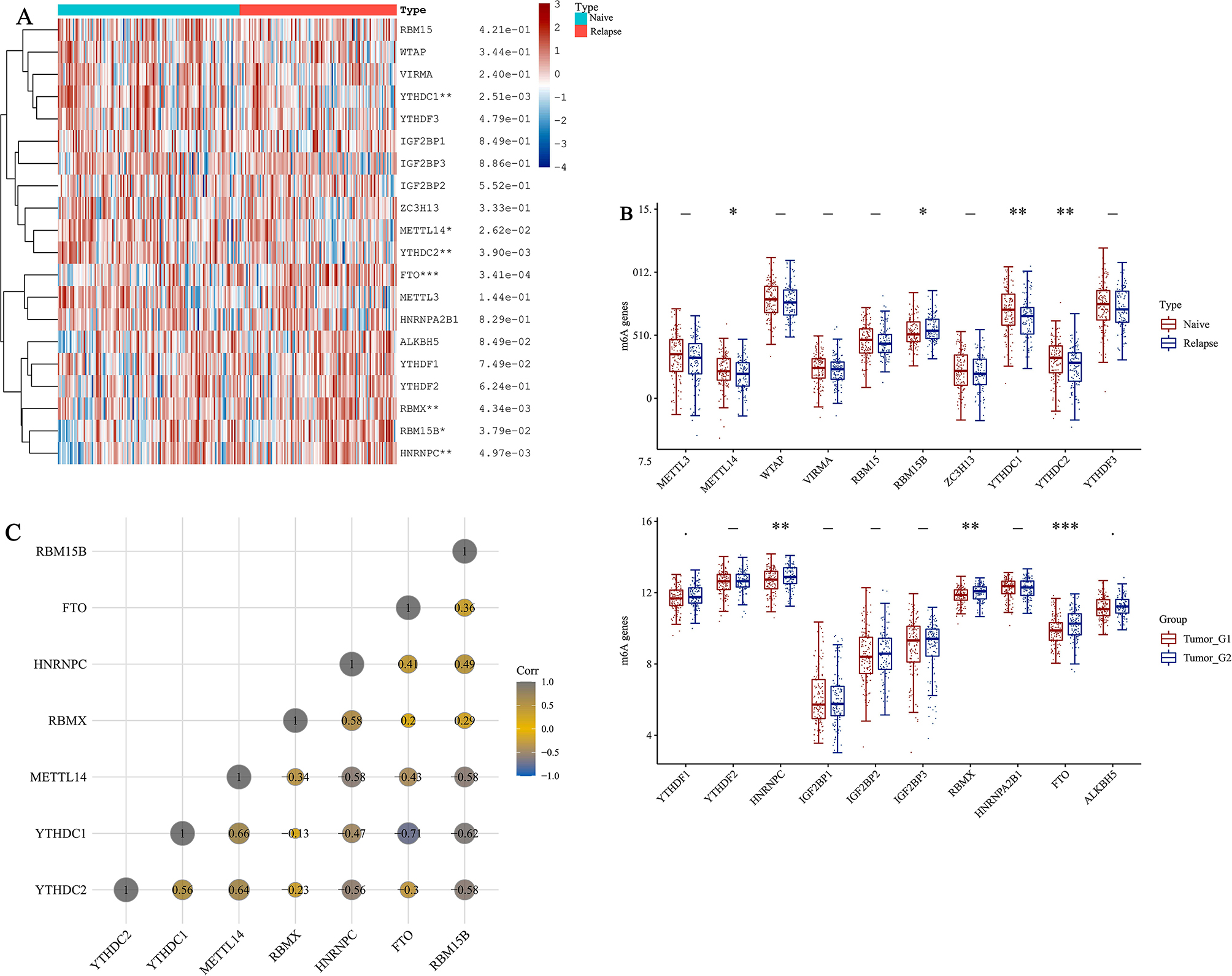
Expression, correlation, and prognostic information of m6A methylation-related genes. (A) Heatmaps of m6A methylationrelated genes expressed in naive and relapsed B-ALL. (***P < 0.001, **P < 0.01, *P < 0.05) (B) The expression distribution of m6A in naive and relapsed B-ALL.The abscissa represents different m6A, and the ordinate represents the expression distribution of gene, different colors represent different groups. *P < 0.05, **P < 0.01,***P < 0.001, asterisks (*) stand for significance levels. The statistical difference of two groups was compared through the Wilcox test, significance difference of two groups was tested with Kruskal-Wallis test. (C) Correlation matrix of interaction in m6A methylation-related genes. Correlation coefficients are plotted with negative correlation (gray) and positive correlation (yellow)
Survival analysis of m6A methylation-related genes
The results revealed that patients with elevated expression of three genes (FTO, HNRNPC, RBM15B) experienced considerably poorer survival rates P < 0.05). Conversely, individuals with decreased expression of two genes (YTHDC1, YTHDC2) exhibited significantly better survival outcomes (P < 0.05). The remaining two genes demonstrated no significant difference in terms of survival (P > 0.05) (Fig. 2A-E).
Fig. 2.
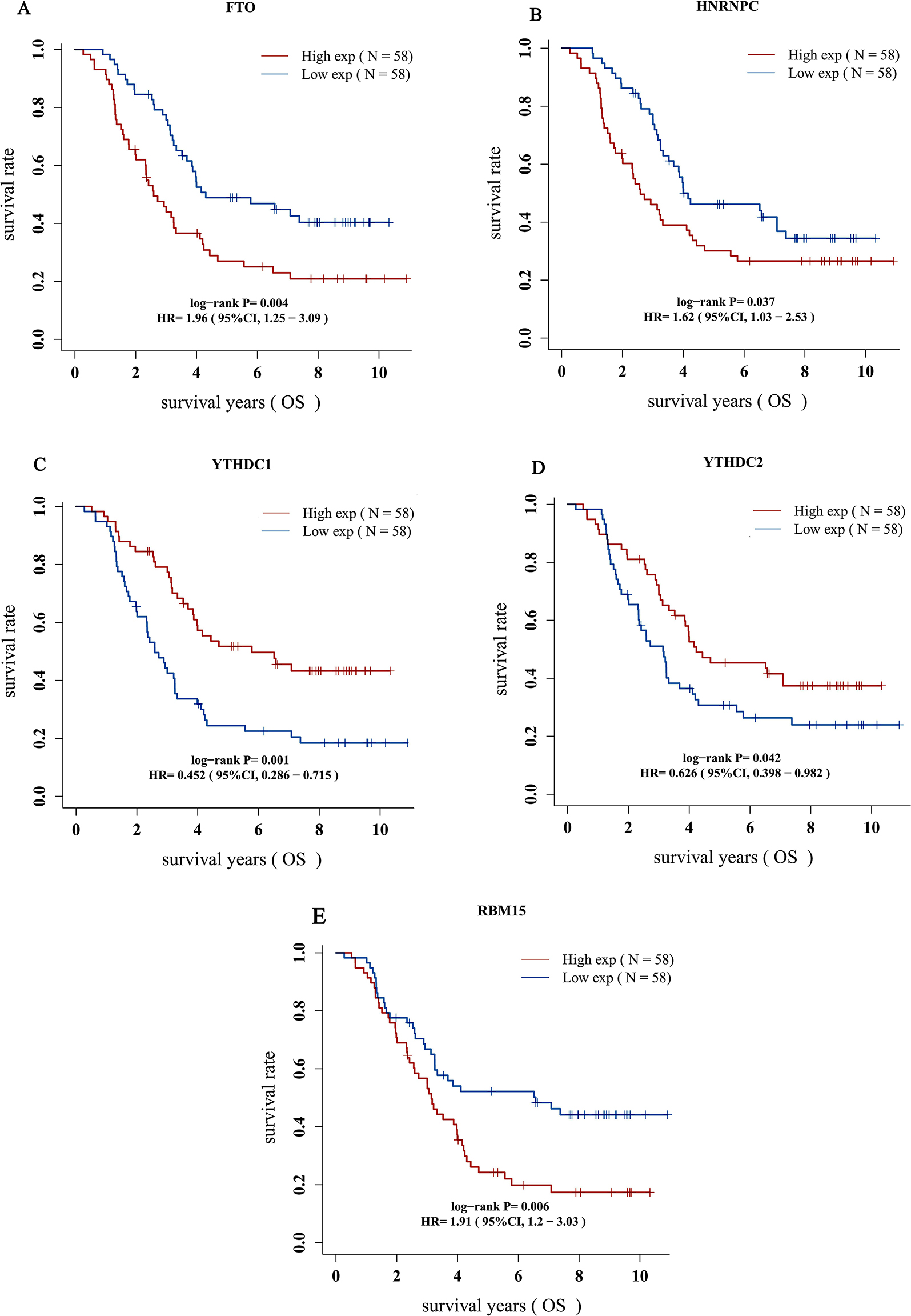
(A-E) Prognostic information for five genes, which had a significantly survival rate (P < 0.05)
Construction of LASSO model
The univariate Cox regression was used to analyze 20 genes and five candidate genes with P < 0.05 were as screening criteria (Fig. 3A). We used the LASSO Cox regression model to select the most predictive genes as prognostic indicators. When the median of the sum of squared residuals is the smallest, select λ (Fig. 3B–C).
Fig. 3.
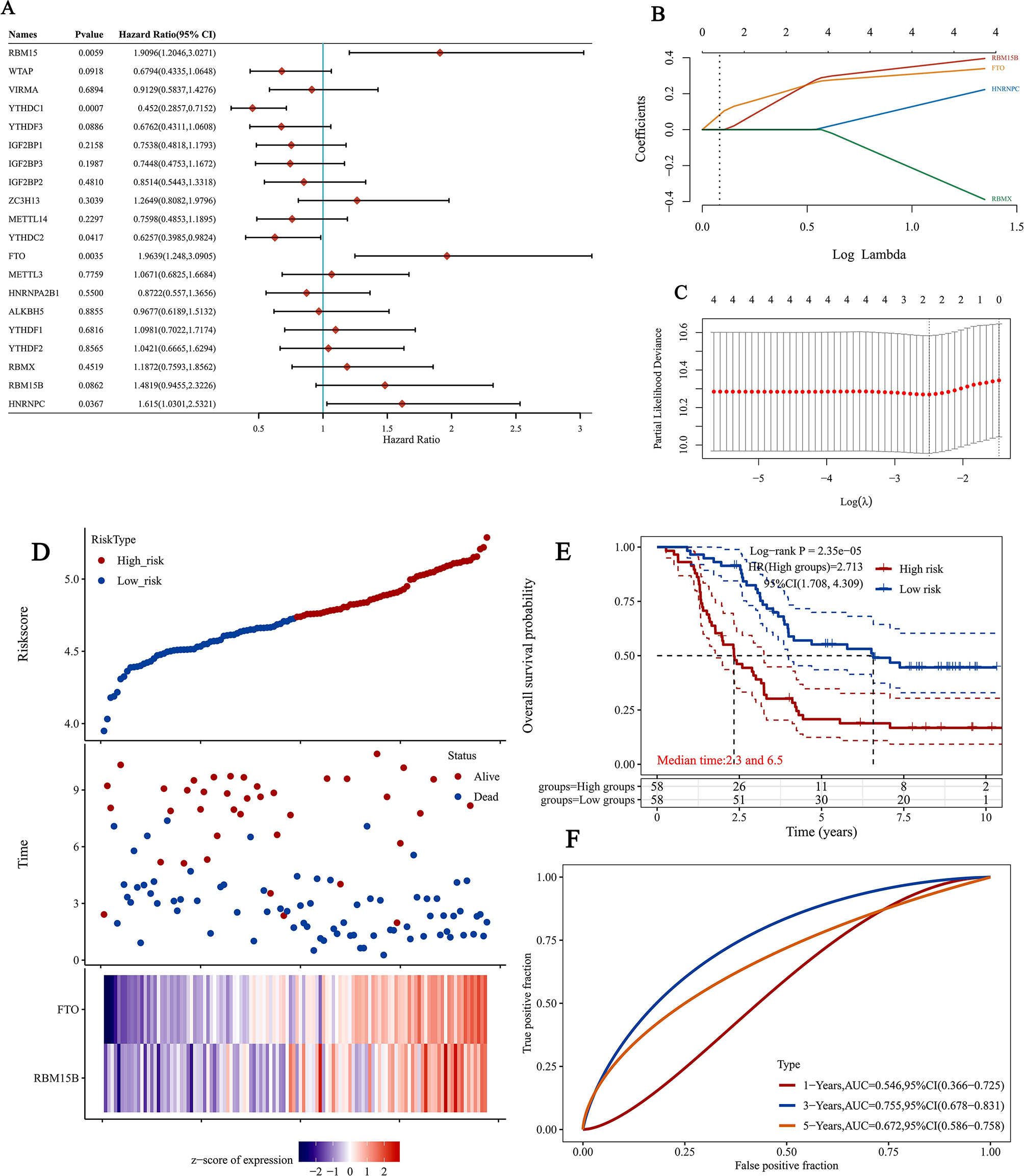
Gene selection and survival analysis in relapsed B-ALL prognosis prediction. (A) Forest plots for hazard ratios (HRs) of survival-associated mA methylation-related genes in relapsed B-ALL. (B) Coefficients of selected features are shown by lambda parameter. (C) Partial likelihood deviance versus log (λ) was drawn using LASSO Cox regression model. (D) The Riskscore, survival time and survival status of selected dataset. The top scatterplot represents the Riskscore from low to high. Different colors represent different groups. The scatter plot distribution represents the Riskscore of different samples correspond to the survival time and survival status. The buttom heatmap is the gene expression from the signature. (E) Kaplan-Meier survival analysis of the risk model from dataset, comparison among different groups was made by log-rank test. HR(High exp) represents the hazard ratio of the low-expression samples relatives to the high-expression samples. HR > 1 indicates the gene is a risk factor, and HR < 1 indicates the gene is a protective factor. HR(95%Cl), the median survival time (LT50) for different groups, in years. (F) The ROC curve and AUC of the gene. The higher values of AUC corresponding to higher predictive power
YTHDC1 and FTO have been identified as prognostic factors for relapsed ALL. We also calculated the risk scores for these two genes. According to the combination model of the cutoff values of the median expression of two candidate genes, patients are divided into high-risk and low-risk groups. The prognosis of the low-risk group is always better than that of the high-risk group. Draw the survival curve using the K-M method (Fig. 3D-E). We also compared the prognostic efficiency of risk factors using receiver operating characteristic (ROC) curves. The results showed that the area under the curve (5-year area under the curve (AUC)) was 67.2% (Fig. 3F), indicating that m6A methylation related genes can serve as biomarkers for the prognosis of relapsed B-ALL.
Prognostic value of the m6A methylation-related genes
Univariate analysis indicated that several factors, including gender, age, white blood cell counts (WBC), and the risk score of m6A methylation-related genes, had an impact on patient prognosis (P<0.05). Despite the lack of correlation between the prognosis of relapsed B-ALL and the early treatment response and fusion gene status, the P-value was greater than 0.05 (Table 1). The findings from the analysis of multiple variables revealed that the risk score associated with m6A methylation-related genes played a significant role in predicting prognosis for relapsed B-ALL patients, showing its independence (P < 0.05) (Table 2).
Table 1.
Univariate analysis for OS among pediatric patients with relapsed B-ALL
| OS | |||
|---|---|---|---|
| Variables | Total | HR(95%CI) | P value |
| Gender | 0.035 | ||
| Male | 64 (55.2%) | 1.0 | |
| Female | 52 (44.8%) | 0.6 (0.4, 1.0) | |
| Age (mean ± SD) | 8.4 ± 5.1 | 1.1 (1.0, 1.1) | <0.001 |
| WBC (mean ± SD) | 60.3 ± 134.3 | 1.0 (1.0, 1.0) | 0.002 |
| D29 MRD | 0.719 | ||
| Negative | 85 (73.9%) | 1.0 | |
| Positive | 30 (26.1%) | 1.1 (0.7, 1.8) | |
| D43 MRD | 0.795 | ||
| Negative | 112 (96.6%) | 1.0 | |
| Positive | 4 (3.4%) | 0.8 (0.2, 3.4) | |
| ETV6/RUNX1 Status | 0.682 | ||
| Negative | 93(85.3%) | 1.0 | |
| Positive | 16(14.7%) | 0.8 (0.3, 2.1) | |
| KMT2A Status | 0.542 | ||
| Negative | 86 (95.6%) | 1.0 | |
| Positive | 4 (4.4%) | 1.4 (0.4, 4.6) | |
| TCF3 PBX1 Status | 0.306 | ||
| Negative | 65 (86.7%) | 1.0 | |
| Positive | 10 (13.3%) | 1.5 (0.7, 3.2) | |
| BCR/ABL Status | 0.178 | ||
| Negative | 115 (99.1%) | 1.0 | |
| Positive | 1 (0.9%) | 3.9 (0.5, 29.1) | |
| RISKSCORE (mean ± SD) | -3.0 ± 0.2 | 5.9 (2.2, 15.4) | < 0.001 |
Table 2.
Multivariate analysis for OS among pediatric patients with relapsed B-ALL
| Outcome | Variable | HR (95% CI) | P value |
|---|---|---|---|
| OS | Gender | 0.6 (0.4, 1.1) | 0.130 |
| Age | 1.3 (0.7, 2.3) | 0.342 | |
| WBC | 1.0 (1.0, 1.0) | 0.467 | |
| D29 MRD | 1.0 (0.6, 1.6) | 0.937 | |
| D43 MRD | 1.8 (1.0, 3.3) | 0.071 | |
| ETV6/RUNX1 Status | 0.9 (0.4, 2.2) | 0.790 | |
| KMT2A Status | 0.9 (0.2, 3.6) | 0.859 | |
| TCF3 PBX1 Status | 1.1 (1.0, 1.2) | 0.153 | |
| BCR/ABL Status | 4.4 (0.5, 38.7) | 0.187 | |
| Risk score | 1.7 (1.1, 2.6) | 0.014 |
Discussion
Relapsed childhood patients with B-ALL have become a major global public health issue. Based on bioinformatics analysis conducted on the TARGET database, three m6A methylation-related genes showing up-regulation in relapsed B-ALL were identified, and their association with markedly poorer survival was established.The widespread distribution of m6A methylation-related genes in leukemic BM samples was revealed by our research, suggesting their significant implications in predicting the prognosis of relapsed B-ALL.Furthermore, there was a notable interconnection observed among m6A methylation-related genes within regulatory networks, indicating their collaborative involvement in the progression of leukemia. Additionally, the deleterious impact on patients with relapsed B-ALL may be attributed to the association of FTO, HNRNPC, and RBM15B with poorer survival outcomes.The indicated results suggest that m6A modulators could serve as potential targets for treating relapsed B-ALL.
The LASSO algorithm examines multiple independent variables at once and identifies the most impactful ones. With the LASSO algorithm, it is possible to concurrently evaluate numerous independent variables and pinpoint the variables with the greatest influence.According to the traditional regression methods are considerably less precise.Based on LASSO Cox analysis, prognostic factors for relapsed B-ALL were identified, including RBM15B, YTHDC1, YTHDC2, FTO, and HNRNPC among the 20 genes analyzed [11]. The ROC curve was used to evaluate the impact of m6A methylation related genes on the prognostic outcome of relapsed B-ALL.The findings indicated that genes associated with m6A methylation played a role in the survival of relapsed B-ALL.The risk score of m6A methylation related genes (YTHDC1 and FTO) may be a powerful biomarker for the survival rate of relapsed B-ALL. The high expression level of FTO indicates poor prognosis, while YTHDC1 can be considered a protective gene. We conducted a comprehensive biological analysis of the 20 most important m6A methylation related genes, which is more comprehensive than previous studies on the impact of individual genes on diseases. Due to the interactions between m6A methylation related genes, our study more accurately reflects their impact on relapsed B-ALL.
Totally, the combination of m6A methylation related genes and clinical parameters may have better predictable power than a single biomarker. Recently, m6A methylation related genes have shown bigger potential in predicting cancer prognosis [12–15]. Our research mainly showed that the expression levels of m6A methylation related genes had a vital part in relapsed B-ALL and may serve as a prognostic factor for the type of leukemia. The expression of m6A methylation related genes is highly related with the clinical characteristics of relapsed B-ALL, which can predict its prognosis and guide precise treatment. This present study provides vital rules for the future detection of the role of m6A methylation among relapsed B-ALL.
Acknowledgements
The study was supported by grant from Guangdong Science and Technology Department (2020B1212060018).
Author contributions
KY.Q wrote the manuscript and XY.L perform the study. JP.F and DH.Z reviewed the manuscript.All authors reviewed the manuscript.
Funding
This work was supported by Guangdong Basic and Applied Basic Research Foundation (NO.2024A1515012445), Special topic of science and technology for agriculture and social development of Guangzhou key R & D plan (2024B03J1247), Bethune Medical Scientific Research Fund Project (No.SCE111DS), Guangdong Medical Scientific Research Foundation (A2024057), Basic Research Project (Dengfeng Hospital) jointly funded by Universities (Institutes) in Guangzhou (No.202201020310), Guangzhou Basic and Applied Basic Research Foundation (2024A04J4686), Yat-sen Excellent Young Scientists Fund (2024A03J1185), Young Teacher Training Project of Sun Yat-sen University (24pnpy314) and Sun Yat-sen Pilot Scientific Research Fund (YXQH202205).
Data availability
The data in the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The Ethics Committee of the Office of Cancer Genomics (OCG) approved this study, and the informed consent forms were signed by the guardians of the patients involved.
Consent for publication
Not applicable.
Footnote
Reporting checklist: The authors have completed the MDAR reporting checklist.
Competing interests
The authors declare no competing interests.
Footnotes
†Dun-hua Zhou contributed equally to this work as corresponding author.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kun-yin Qiu, Xiong-yu Liao and Jian-pei Fang contributed equally to this work as first authors.
References
- 1.Brown PA, Ji L, Xu X, et al. Effect of postreinduction therapy consolidation with Blinatumomab vs Chemotherapy on Disease-Free Survival in Children, adolescents, and young adults with First Relapse of B-Cell Acute Lymphoblastic Leukemia: a Randomized Clinical Trial. JAMA. 2021;325(9):833–42. 10.1001/jama.2021.0669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell M, Kiss C, Zimmermann M, et al. Childhood Acute Lymphoblastic Leukemia: results of the Randomized Acute Lymphoblastic Leukemia Intercontinental-Berlin-Frankfurt-Münster 2009 Trial. J Clin Oncol. 2023;41(19):3499–511. 10.1200/JCO.22.01760 [DOI] [PubMed] [Google Scholar]
- 3.Shukla N, Sulis ML. Blinatumomab for treatment of children with high-risk relapsed B-Cell Acute Lymphoblastic Leukemia. JAMA. 2021;325(9):830–2. 10.1001/jama.2021.1395 [DOI] [PubMed] [Google Scholar]
- 4.Moorman AV, Antony G, Wade R, et al. Time to cure for Childhood and Young Adult Acute Lymphoblastic Leukemia is Independent of early risk factors: long-term Follow-Up of the UKALL2003 trial. J Clin Oncol. 2022;40(36):4228–39. 10.1200/JCO.22.00245 [DOI] [PubMed] [Google Scholar]
- 5.Attarbaschi A, Möricke A, Harrison CJ, et al. Outcomes of Childhood Noninfant Acute Lymphoblastic Leukemia with 11q23/KMT2A rearrangements in a modern therapy era: a Retrospective International Study. J Clin Oncol. 2023;41(7):1404–22. 10.1200/JCO.22.01297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun H, Li K, Liu C, Yi C. Regulation and functions of non-m6A mRNA modifications. Nat Rev Mol Cell Biol. 2023. 10.1038/s41580-023-00622-x. published online ahead of print, 2023 Jun 27. 10.1038/s41580-023-00622-x [DOI] [PubMed] [Google Scholar]
- 7.Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of RNA m6A modification in cancer. Nat Rev Clin Oncol. 2023;20(8):507–26. 10.1038/s41571-023-00774-x [DOI] [PubMed] [Google Scholar]
- 8.Weng H, Huang F, Yu Z, et al. The m6A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40(12):1566–e158210. 10.1016/j.ccell.2022.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, Xie W, Pickering BF, et al. N6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39(7):958–e9728. 10.1016/j.ccell.2021.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Y, Su R, Sheng Y, et al. Small-molecule targeting of oncogenic FTO demethylase in Acute myeloid leukemia. Cancer Cell. 2019;35(4):677–e69110. 10.1016/j.ccell.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S, Cong Z, Ji J, et al. Analysis of m6A-related signatures associated with the tumor immune microenvironment and predict survival in acute myeloid leukemia. Ann Transl Med. 2022;10(16):902. 10.21037/atm-22-3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng Y, Guan R, Hong W, et al. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann Transl Med. 2020;8(6):387. 10.21037/atm.2020.03.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Sun G, Pan S, et al. The Cancer Genome Atlas (TCGA) based m6A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered. 2020;11(1):759–68. 10.1080/21655979.2020.1787764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J, Zhao Z, Wan J, et al. N-6 methylation-related lncRNA is potential signature in lung adenocarcinoma and influences tumor microenvironment. J Clin Lab Anal. 2021;35(11):e23951. 10.1002/jcla.23951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzam SK, Alsafar H, Sajini AA. FTO m6A demethylase in obesity and Cancer: implications and underlying molecular mechanisms. Int J Mol Sci. 2022;23(7):3800. 10.3390/ijms23073800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data in the current study are available from the corresponding author on reasonable request.


