Abstract
Due to the transformation of artificial intelligence (AI) tools and technologies, AI-driven drug discovery has come to the forefront. It reduces the time and expenditure. Due to these advantages, pharmaceutical industries are concentrating on AI-driven drug discovery. Several drug molecules have been discovered using AI-based techniques and tools, and several newly AI-discovered drug molecules have already entered clinical trials. In this review, we first present the data and their resources in the pharmaceutical sector for AI-driven drug discovery and illustrated some significant algorithms or techniques used for AI and ML which are used in this field. We gave an overview of the deep neural network (NN) models and compared them with artificial NNs. Then, we illustrate the recent advancement of the landscape of drug discovery using AI to deep learning, such as the identification of drug targets, prediction of their structure, estimation of drug-target interaction, estimation of drug-target binding affinity, design of de novo drug, prediction of drug toxicity, estimation of absorption, distribution, metabolism, excretion, toxicity; and estimation of drug-drug interaction. Moreover, we highlighted the success stories of AI-driven drug discovery and discussed several collaboration and the challenges in this area. The discussions in the article will enrich the pharmaceutical industry.
Keywords: MT: Bioinformatics, drug molecules, artificial intelligence, drug discovery, deep learning, pharmaceutics
Graphical abstract
Chakraborty and colleagues comprehensively illustrate the recent drug discovery and development advances using AI to DL. Furthermore, they highlight the expanded success stories of AI-driven drug discovery, several collaborations among pharmaceutical giants and big computer or tech companies, and the significant challenges in AI-driven drug discovery.
Introduction
Innovation is the backbone of the pharmaceutical industry. Innovation can create life-saving solutions, and it provides society with life-saving medicines. Therefore, it is essential to innovate new drug compounds and patent them. Presently, research is at the forefront of science, and it helps to invent and patent new drugs.1,2 Pharmaceutical industries are following this route to invent and patent new drugs. Through the drug discovery process, new drugs are discovered for diseases, even neglected diseases. Pharmaceutical companies are investing a considerable amount of money in this direction. However, the drug discovery and development process is very complex. A high level of expertise and various technologies have been used in drug discovery and development.3 Drug discovery requires a considerable amount of time. This process, known as from bench to bedside, encompasses the journey from the initial discovery of drug molecules to their market availability. It has been estimated that the from-bench-to-the-bedside process typically takes 10–15 years or more. Therefore, a new innovative technological landscape has emerged in the field of drug discovery. Innovative technologies help the pharmaceutical sector by providing faster methods of drug discovery. Recent advancements in innovative tools and technologies can make a massive difference in drug discovery and development. Today’s innovative tools and technologies have made drug discovery and development faster. Consequently, new technologies now provide a faster route to drug discovery. At the same time, the process is also more productive for the pharmaceutical industry. It is the utmost need for the pharmaceutical industry. At the same time, the pharmaceutical industry needs to be more productive in terms of drug discovery. The innovative technologies help to quickly foster a very productive way of drug discovery.1,4,5,6
The drug discovery and development procedure encompasses three major stages: drug discovery, preclinical development of drug molecules, and clinical development of the therapeutic molecule. Traditional approaches to drug discovery and development face considerable challenges. They are time consuming, expensive, and have low success rates. It have been noted that developing a new drug molecule costs approximately US$2.6 billion on average and takes more than 10–15 years to enter the market.7,8 A recent cost analysis of drug development showed that the capital cost of drug development is US$1.3. At the same time, the average out-of-pocket success cost is just US$200 million. Similarly, out-of-pocket failure costs US$1 billion. Therefore, a successful drug’s discovery cost can be reduced if we reduce the failure costs.9 The failure costs of drug discovery can be reduced by using new technology like AI. The capital cost of drug development can be used in new technology development to reduce the other costs and increase the chance of success. At the same time, the traditional method is still not optimal for finding a new drug for many diseases. Researchers worldwide have moved toward the new technological advancements to minimize these hurdles and challenges. At the same time, the pharmaceutical industry and its researchers are shifting from traditional approaches to new methods and using various technologies to foster innovation and improve outcome. They are using new computational technologies like artificial intelligence (AI), machine learning (ML), and deep learning (DL) to perform a new way drug discovery in a new way. These new technologies mitigate the cost and speed of the drug discovery procedure. At the same time, they are also more productive. Another disadvantage is that the traditional method of drug discovery is slow and labor intensive because it relies on identifying and improving existing compounds. In contrast, AI-based approaches can handle and solve the issue through the rapid and efficient design of new drug compounds.
Since the last decade, AI has been used in different areas of biological sciences, medical sciences, and general sciences. AI, also known as machine intelligence, directs computer systems' capability to learn from past data and input. AI is generally applied when a machine imitates cognitive behavior and behaves like a human. It is associated with the human brain’s learning and problem-solving capabilities.10 Due to massive multi-omics data and high-performance computer hardware availability, AI techniques were introduced as a fundamental application in various disciplines. Similarly, data digitalization has increased in the pharmaceutical sector, which has inspired the use of AI. At the same time, automation was enhanced, and AI was empowered to handle large volumes of data. Pharmaceutical industries have collaborated with the computational industries.11 In the last few years, progress has been made in drug discovery using AI-enabled drug discovery technologies. AI has also been used by drug discovery organizations, which has changed the drug discovery scenario in the last decade. Various AI techniques have been adopted in the different areas of drug discovery, such as virtual screening, target selection, and hit-to-lead generation. Other application areas are bioavailability prediction, retrosynthesis and reaction forecast, de novo drug design.3,7
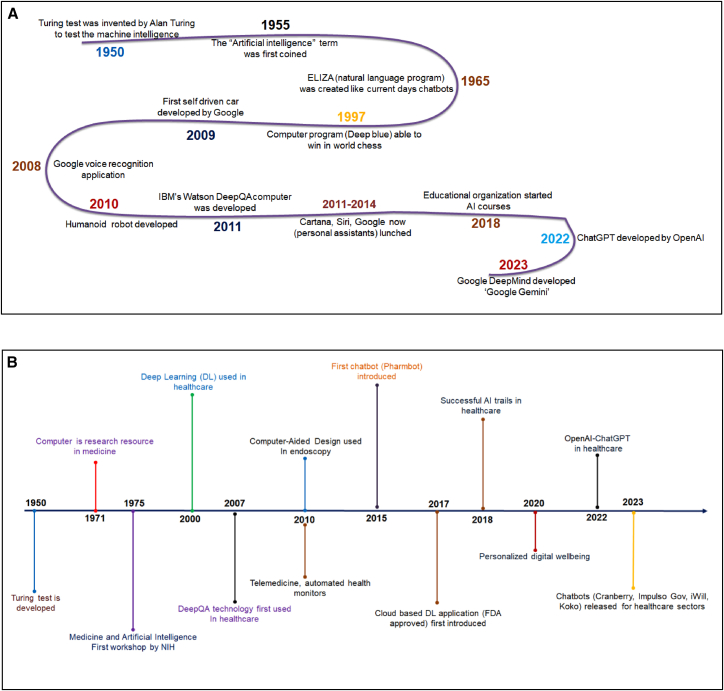
Many AI techniques, like traditional ML and DL, are associated with drug discovery and development analyses. After Alan Turing’s Turing test in 1950, several AI-related breakthrough discoveries were made, and different AI-related milestone achievements were occasionally created (Figure 1A). However, developing different models was a significant process in drug discovery and development. Therefore, model architectures also evolved to mitigate the healthcare sector. Model architectures such as convolutional neural networks (CNNs), graph NNs, recurrent NNs (RNNs), and transformers, were also evolved. At the same time, researchers noted the paradigm shift from supervised learning to self-supervised learning and reinforcement learning in drug discovery and development.
Figure 1.
Timelines illustrate the milestone achievements of AI and milestone of AI-related achievements in the healthcare sector, including the pharmaceutical sector
(A) The timeline depicts the achievements of AI. (B) The timeline depicts the milestone of AI-related achievements in the healthcare sector, including the pharmaceutical sector.
This article discusses the recent advancement of drug discovery using AI to DL with different examples. At the same time, we discussed several success stories and collaborations for AI-driven drug discovery. The article also noted several challenges in AI-related drug discovery.
AI: Our understanding
Overview of AI
AI algorithms are widely used in different sectors, from government to business. AI has been applied from time to time in different domains in the healthcare sector, including the pharmaceutical sector (Figure 1B). Data digitalization has increased in every sector, including the pharmaceutical sector, during the last few years. Digitalization has assisted in solving complex clinical problems through data acquisition, scrutinizing, and applying knowledge. It was the motivation for using AI.12 At the same time, automation was undertaken to manage the large volume of data. AI can mimic human intelligence using advanced technologies that involve several advanced tools and network systems. Therefore, a paradigm shift has been noted in every sector, including the pharmaceutical sector. Different researchers have stated that the rapid advancement of AI-guided automation will ultimately transform society’s work culture.
Data-driven AI
Data are required for any statistical inferences, including ML. Similarly, different models can be developed using data modalities. Data come in different forms, such as textual, image, and numerical. Extensive data analysis broadly transforms pharmaceutical and medicinal fields.13 Presently, data-driven digital transformation is noted in every sector, which is an emerging phenomenon.
In the pharmaceutical sector, digital transformation is swift and includes vast amounts of data. It has been noted that digital transformation through AI methods in the pharmaceutical sector is data driven. AI algorithms related to drug discovery depend on the pharmaceutical sector’s data. At the same time, AI model training data must be curated and accurate. Therefore, most pharmaceutical sector datasets are curated and accurate.14 Several data resources for AI model training in pharmacology sectors are the key components for drug discovery. The pharmaceutical sector’s data resources include high-quality datasets. These high-quality datasets are open resources. Therefore, these datasets are frequently used in drug discovery.
However, there is a long history of the use of different data-driven approaches in drug discovery. Recent advancement of ML and DL algorithms, data-driven approaches are used more in drug discovery and development.15 More advanced forms of DL take data-driven approaches to the next level, allowing them to handle more diverse and much larger datasets.15,16
These datasets are obtained from different data resources. Some data resources include ChEMBL, ChemDB, DGIdb, DrugBank, DTC, PubChem, and SIDER (Table 1).3 The drug discovery process can be performed using all the databases and their big data. The data quicken the drug discovery process. One example of a data resource is ChEMBL developed by EMBL-EBI. It currently contains more than 2 million compounds. These compounds show drug-like properties. It is a manually curated database. The database informs different properties such as, molecular properties, target interactions, and mechanisms of action.17,18 Similarly, another drug discovery-related database is ChemDB, which contains approximately 5 million small molecules. All these molecules are commercially available. It also includes their physicochemical properties such as solubility, molecular weight, and rotatable bond.19,20
Table 1.
Different data resources in the pharmaceutical sector for AI-driven drug discovery
| Sl. No. | Name of database | Web address | Remarks | Reference |
|---|---|---|---|---|
| 1. | ChemDB | http://cdb.ics.uci.edu/ | The chemical database encompasses approximately 5 million commercially accessible small molecules. It also holds the experimentally determined and predicted physicochemical properties |
Chen et al.19 |
| 2. | STITCH | http://stitch.embl.de/ | The database of identified and expected molecular interactions between proteins and chemicals, with 9,643,763 proteins derived from 2,031 organisms | Szklarczyk et al.21 |
| 3. | INPUT | http://cbcb.cdutcm.edu.cn/INPUT/ | The network pharmacology web database dedicated for traditional Chinese medicine it holds total 29,812 compounds, which was collected from 4,716 Chinese herbs |
Li et al.22 |
| 4. | DGIdb | http://www.dgidb.org/ | The specialized database offered information on drug testing index and druggable genomes over the 30 reliable sources | Freshour et al.23 |
| 5. | DTC | http://drugtargetcommons.fimm.fi/ | The crowd-sourcing platform delivers drug-target bioactivity data and cataloging of its targets | Tang et al.24 |
| 6. | SIDER | http://sideeffects.embl.de/ | The database affords information about the advertised medicines and its recorded adverse reactions | Campillos et al.25 |
| 7. | ChEMBL | https://www.ebi.ac.uk/chembl/ | The database consist of bioactive molecules having drug-like properties it collects bioactivity, chemical, and genomic data to support the translation of genomic evidence into operative new drugs. |
Mendez et al.26 |
| 8. | PubChem | https://pubchem.ncbi.nlm.nih.gov/ | The open access chemistry database platform that delivers significant information about the molecules, like as chemical structures, identifiers, chemical status, physical properties, and others associated biological activities | Kim et al.27 |
| 9. | COCONUT | https://coconut.naturalproducts.net/ | The database has 407,270 exclusive natural products, and it have the evidence about their molecular properties, molecular descriptors | Sorokina et al.28 |
| 10. | DrugBank | http://www.drugbank.ca/ | The database of drugs, its targets, 3D structures, and additional convenient information | Wishart et al.29 |
Frequent algorithms or techniques used for AI and ML
Several AI-related algorithms have been developed for drug discovery and development. Drug discovery and associated activities are performed very fast with the help of AI-enabled algorithms or techniques using different data resources. Two types of AI algorithms or techniques are commonly used in AI-enabled drug discovery and development: supervised and unsupervised learning algorithms or techniques.30,31 However, in ML, four algorithms are commonly used: supervised, semi-supervised, unsupervised, and reinforcement. In addition to these algorithms, a commonly used modeling regression analysis algorithm is called multiple linear regression.
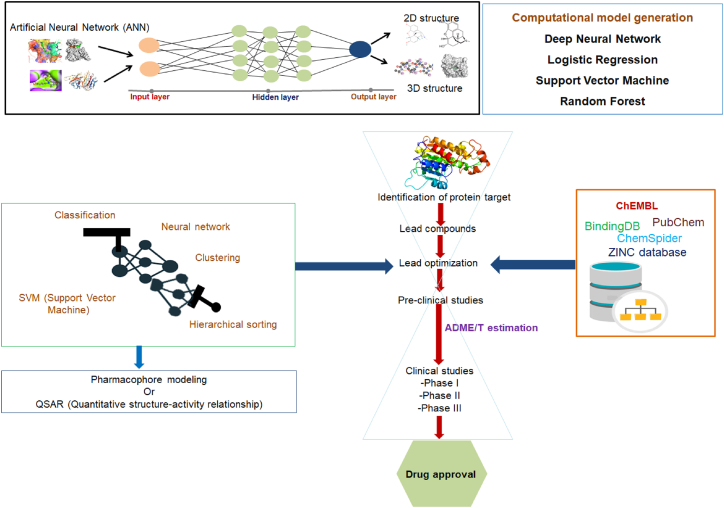
AI involves various areas, such as knowledge-based representation, reasoning-based domain, and solution search domain, and among them, a fundamental paradigm is ML. Therefore, ML is a subfield of Al. Similarly, a promising subfield of ML is DL. DL involves artificial NN (ANN) algorithms (Figure 3). There are various types of ANNs, which include CNNs, multilayer perceptron networks, and RNNs.
Figure 3.
A schematic diagram shows different steps of drug discovery and development where AI and DL models have been successfully implemented
Unsupervised learning algorithms or techniques
The unsupervised learning algorithm is very commonly used in AI-related algorithms. It can perform more complex processing tasks and efficiently group the unlabeled datasets into a set of classes. The algorithm can group the members in similar given classes or separate them into other classes.30,31,32 Therefore, it can identify recurring patterns during the grouping of the datasets. One example is the unsupervised learning algorithm Seq2seq fingerprint, which uses SMILE strings to generate the molecular fingerprint. Therefore, it can be trained with the unlabeled datasets of SMILE.33
Similarly, Lo et al.34,35 developed the program ShapeAlign for unsupervised three-dimensional (3D) chemical clustering to understand similarity. It unites both two-dimensional (2D) and 3D metrics based on the Obabel PF2 fingerprint. Finally, the program can shape pharmacophoric points.34,35 The commonly used unsupervised learning algorithms are K-means clustering, hierarchical clustering, and principal component analysis.36
Supervised learning algorithms or techniques
This group of algorithms is very commonly used in AI-related algorithms. It is used to train labeled datasets and estimate outcomes. Finally, it helps to identify patterns.37,38 Recently, using supervised learning algorithms, Lo et al. developed a self-organizing map (SOM) model for QSAR methods in drug discovery and development. The model is modified using the supervised method and is now entitled as supervised SOM. It can illustrate more precise predictions than standard QSAR.39 Supervised learning algorithms are multiple regression analysis, logistic regression, k-nearest neighbor, decision trees, random forest plots, support vector machines (SVMs), and so on.36
Semi-supervised learning algorithms or techniques
The semi-supervised learning group of algorithms is a division of ML. This model can be trained in both labeled and unlabeled data. Therefore, the algorithm combines supervised and unsupervised learning to train AI models for classification and regression tasks.40,41 It uses a small portion of the data (supervised and unsupervised). Semi-supervised algorithms or techniques include co-training, self-training, and graph-based labeling. The algorithm is used in text classification. Several semi-supervised learning algorithms have been developed for drug discovery and development. Recently, Sahoo et al.42 developed a MultiCon model to estimate the drug function from chemical structure analysis. The model was developed based on a semi-supervised learning algorithm. According to therapeutic applications, the MultiCon model classifies drugs into 12 categories. It attained an accuracy of 97.74% for class prediction of drugs.42 Similarly, researchers developed another semi-supervised model for synergistic drug combination prediction. The name of the model is NLLSS. The model can evaluate the potentiality of combinations of synergistic drugs by integrating various categories of information.43 Similarly, Wu et al.44 developed a model based on a semi-supervised learning algorithm to predict drug-disease interactions using a three-layer data-integrated model. This model was implemented for the case studies on four diseases. Using the Kyoto Encyclopedia of Genes and Genomes Comparative Toxicogenomics database (CTD), the researcher confirmed the top-ranked drug-disease associations.44 Likewise, Chen et al.45 developed a model to predict the chemical toxicology of drugs using the semi-supervised learning algorithm. The model can efficiently perform the prediction tasks using current chemical databases.
Reinforcement learning algorithms or techniques
This ML algorithm is trained to solve multi-level problems using the trial and error method for optimal output. It is a feedback-based ML algorithm. Here, the model learns from the real-life scenarios to perform the actions. Stahl et al. 46 developed the multiparameter optimization process in drug design. The model can develop the design of safe compounds. The model was developed using reinforcement learning algorithms or techniques.46 Pereira et al.47 developed a generator for de novo drug design. The model was developed to understand the drugs that can cross the blood-brain barrier. The developed drugs should have permeability and solubility when crossing the blood-brain barrier so that the molecule can reach the site of action in the brain. It can also help in neural drug development. It uses deep reinforcement learning.47 Liu et al.48 generated a model for the de novo design of a drug molecule called DrugEx. The model was developed based on multi-objective reinforcement learning. The model helps to improve the generated drug molecule. The drug molecule can have one specific or multiple drug targets, while the molecules avoid off-targets.48
Deep NN of DL models
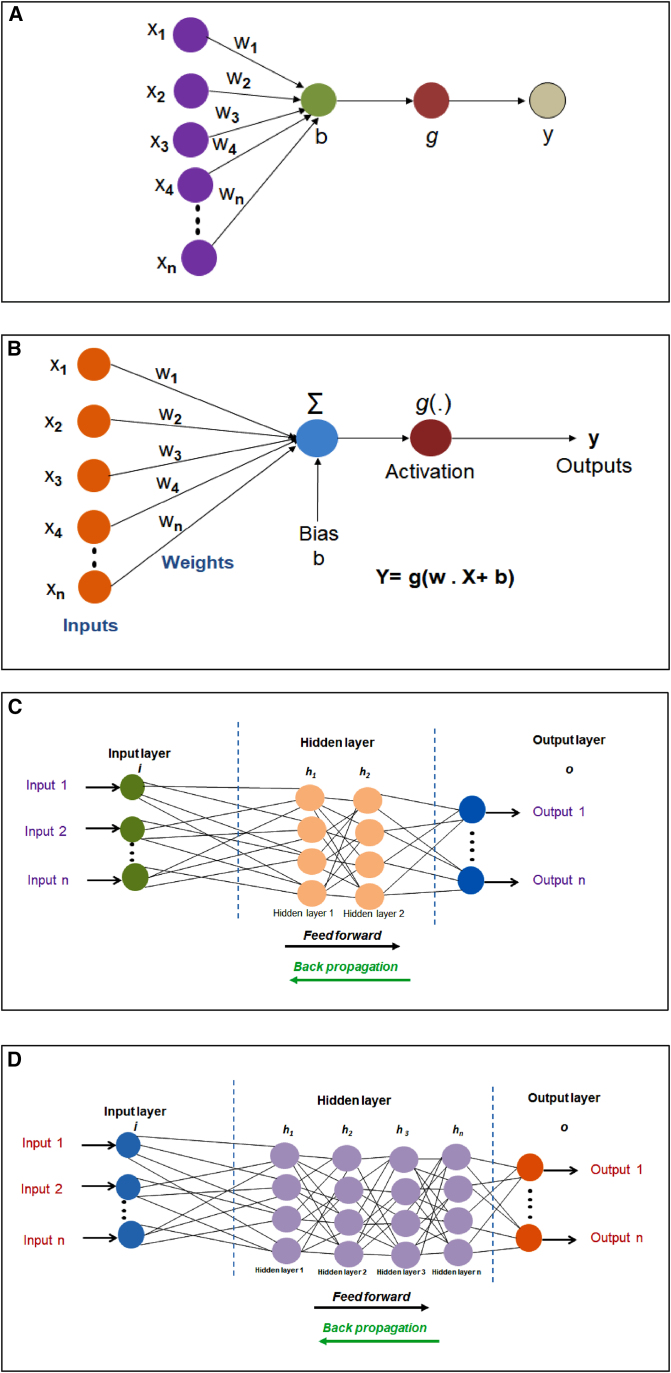
DL models are the modern form of ANNs. However, ANN models are the primary models of AI and were developed a long time ago. If we look back, in 1943, Warren McCulloch and Walter Pitts described a NN based on algorithms and mathematics, which they called threshold logic. It might be the earliest ANN (ANN model).49,50 The basic structure of the ANN was developed from the structure of the human brain, consisting of a network of interconnected neurons. Depending on the type of ANN, the neuron’s nodes are varied.50 The input and output nodes describe the ANN input and output values (Figures 2A and 2B). The output value of an ANN is calculated from its input values, and the equation is shown as follows:
| (Equation 1) |
Figure 2.
The basic architecture and framework of ANNs
It also illustrates basic architecture and framework differences of ANNs and DNNs. (A) The basic framework of ANN (B) The architecture of ANN which shows the input, weight, bias, and output. (C) The ANN with two hidden layers, h1 and h2. (D) An architecture of a DNN model. It has been noted that an ANN has one or two hidden layers, while DNN has several hidden layers.
In Equation 1, input values are denoted as ×1, ×2, ×3, ×4, … xn. The output value is denoted as y. Here, w1, w2, w3, w4, … wn are denoted as weights, and a bias term, b, has been used. In the above equation, g is denoted as the activation function. To train an ANN model effectively, researchers should follow certain steps. In the first step, the problem should be defined. In the second step, researchers should collect the datasets. At the same time, the data needs to be divided into training datasets, testing datasets, and validation sets. Then, the researchers need to optimize the parameters. The weight values and biases should be adjusted in the hidden layers. Then, it should be applied to the ANN model and monitor the preference of the model. The ANN algorithm was designed from the 1960s to the 1980s and has been applied since then. When researchers used the algorithm, they found that the method has various problems, such as diminishing gradients and over fitting. Due to this lacuna, ML algorithms such as SVM and RF (random forest) are used to replace the existing methods in due course.
The models of DL are very sophisticated forms of multi-level NNs called deep NNs (DNNs). Using large amounts of labeled or unlabeled training datasets, the DNNs perform the detection.14,51 The difference between traditional ANN and DNN is the complication of the networks and the scale used. In ANN, there is an input feature called the input layer. After that, there are some hidden layers: several nonlinear transformations. Finally, in an output layer, the predictions are made. Every output node refers to a task that will be predicted through the algorithm. Output node refers to a task that is a specific class. It has been noted that traditional ANN has used one or two hidden layers (Figure 2C). Here, powerful hardware is needed to operate the ANN. In contrast, DNN has several hidden layers (Figure 2D). DNN uses more powerful hardware in terms of graphics processing unit (GPU). A critical difference between ANNs and DNNs is the number of layers. DNNs use a more significant number of hidden layers, whereas traditional ANNs usually contains one or two. Therefore, DNNs can use more nodes in each layer due to the more powerful GPU and CPU hardware, which allows it to compute more complicated data. One example is the DropOut and DropConnect methods, which can address the overfitting problem and solve a more complicated problem.50,52,53,54,55 Using the DropOut and DropConnect methods, one can either drop the weights of synaptic connections or drop the states of neural units. These two are significant and effective strategies for enhancing ANN inference performance.54,55 Data science researchers use several DL packages, software and libraries, the most popular of which are Keras, PyTorch, Caffe, Caffe2, and TensorFlow. Most of them are open source and popular DL packages. These open-source packages and libraries have been developed due to the rapid development of the DL technique. DNNs have been applied in different areas of drug discovery. This algorithm is used in the pathological image classification and analysis,56,57 de novo drug discovery,58,59 prediction of protein-ligand interaction,60 and target-based drug design.61 Several researchers have tried to apply DNN in the area of drug discovery.61 Shi et al.61 have developed Pocket2Drug, which will help to design target-based drugs. This model is significant for the discovery of new biopharmaceuticals. It uses encoder-decoder DNNs.61 Similarly, Shi et al.62 recently developed a graph-based model called GraphSite, which can classify ligand binding sites using deep graph learning. Likewise, Wu et al.63 developed a model for anticancer drug discovery. It performs target-based and cell-based anticancer drug discovery. It is a multipurpose DL platform. In addition to these models, several others have been applied in drug discovery and development research, such as DTI-CNN, DeepDTA, WideDTA, PADME, DeepAffinity, and DeepChem (Table 2). The models are primarily developed for researchers to generate novel molecules and predict absorption, distribution, metabolism, excretion, and toxicity (ADMET) effects. These models are essential for performing translational research.64,65 The significant problems during the process of drug discovery and development comprise the unfavorable ADMET properties of the probable drug molecules. This issues are understood to be a primary reason for the failure of drug candidates during development. Additionally, estimating ADMET properties consumes substantial capital, time, and resources.66,67 Therefore, most DL models focus on the ADMET estimation process. In contrast, DL models are also successful in building QSAR models and applying these models in drug discovery and development research.65
Table 2.
ML and DL-based applications in different areas of drug discovery and development
| Sl. No. | Software/tools | Method | Web address | Remarks | Reference |
|---|---|---|---|---|---|
| 1. | AutoGrow4 | Genetic algorithm | http://durrantlab.com/autogrow4 | It is used for the de novo drug designing and lead optimization purposes | Spiegel and Durrant68 |
| 2. | TrixX | ML | – | The structure-based molecule catalog applied for extensive virtual screening in sublinear time | Schellhammer and Rarey69 |
| 3. | LS-align | ML | http://zhanglab.ccmb.med.umich.edu/LS-align/ | The atomic-level, flexible ligand structural alignment algorithm used for high-throughput virtual screening | Hu et al.70 |
| 4. | StackCBPred | ML | https://bmll.cs.uno.edu/ | The prediction of protein-carbohydrate binding sites from the available sequence using the stacking-based | Gattani et al.71 |
| 5. | DrugFinder | ML | https://drugfinder.ca/ | In silico virtual screening service for search a drug or medical condition | Lagarde et al.72 |
| 6. | LigGrep | ML | http://durrantlab.com/liggrep/ | The web tool applied for filtering docked complex to improve virtual-screening hit rates | Ha et al.73 |
| 7. | LSA | Conventional similarity algorithms | – | The local-weighted structural alignment web tool used for virtual pharmaceutical screening | Li et al.74 |
| 8. | DEEPScreen | CNNs | https://github.com/cansyl/DEEPscreen | The web tool used for high-performance DTI prediction | Rifaioglu et al.75 |
| 9. | DLIGAND2 | Distance-scaled | https://github.com/sysu-yanglab/DLIGAND2 | This web toll used for analysis of improved knowledge-based energy purpose for protein–ligand interactions | Chen et al.76 |
| 10. | Dr.VAE | ML | https://github.com/rampasek/DrVAE | It models both the drug response in relations of viability and the cellular transcriptomic perturbations | Rampášek et al.77 |
| 11. | SMDIP | ML | – | It shows the pharmacokinetic and pharmacodynamic profiles of the drug molecules | Ibrahim et al.78 |
| 12. | MoleculeNet | ML | https://moleculenet.org/ | Used for accurate predictions about molecular properties of drug –and its comparison | Wu et al.79 |
| 13. | DTI-CNN | DL | – | The DTI prediction tool performed to outperform the prevailing state-of-the-art methods by the intelligent interface | Nag et al.67 |
| 14. | DeepDTA | DL | https://github.com/hkmztrk/DeepDTA | The non-structure-based method and usages SMILES as input data for drugs. The amino acids sequences are likewise encoded in SMILES | Ozturk et al.80 |
| 15. | WideDTA | DL | – | The web tool holds text-based sources of information as input, where the proteins are signified by smaller lengths of residues are not identified in full-length sequence | Nag et al.67 |
| 16. | PADME | DL | – | This tool predict method, which usages drug molecules-target landscapes and fingerprints (as the input) | Feng et al.81 |
| 17. | DeepAffinity | DL | – | Structural property sequence representation that annotates the sequence with structural information, which are shorter than the other representations, provides structural details efficiently and gives higher resolution of the sequences | Karimi et al.82 |
| 18. | DeepChem | DL | https://github.com/deepchem/deepchem | The DNNs applied to analyze medicines and predict drug-related features, including as bioactivities and physicochemical qualities | Altae-Tran et al.83 |
| 19. | DeepConv-DTI | DL | https://github.com/GIST-CSBL/DeepConv-DTI. | This tool predict the model capturing local residue patterns of proteins in identification of DTIs | Lee et al.84 |
| 20. | DeepCPI | DL | https://github.com/FangpingWan/DeepCPI | It accurately predict and identification of compound-protein interactions at a large scale | Wan et al.85 |
| 21. | DeepDTnet | DL | https://github.com/ChengF-Lab/deepDTnet | This DL-based tool offers a potent network-based methodology for identification of target to expedite drug repurposing and reduce the translational gap in drug development | Zeng et al.86 |
| 22. | DeepGRMF | DL | https://github.com/renshuangxia/DeepGRMF | It offers anintegrated graph models, NNs, and matrix-factorization methods to operate diverse information from drug chemical structures and predict cell response to drugs | Ren et al.87 |
| 23. | DeepLIFT | DL | https://github.com/kundajelab/deeplift | This tool predicts drug response in cancer cell lines and their mechanism of action and evaluated its performance using three cross-validation schemes | Sada Del Real and Rubio88 |
| 24. | DeepSide | DL | http://github.com/OnurUner/DeepSide | It exploits data concerning the drug targets, structural fingerprints, and drug side effects | Arshed et al.89 |
ML- and DL-based applications in drug discovery and development
All ML procedures belong to AI methods that use vast amounts of data. Over the past decade, ample data availability has successfully transformed AI methods into improved ML methods to solve critical problems. ML is considered as one of the best choices for solving issues using various variables and big data.14,67,90,91 ML algorithms are classified into supervised and unsupervised techniques.90,92
Recently, ML methods have evolved into DL methods, which are more efficient and powerful tools for handling the vast amounts of data generated across various fields. DL is a subset of ML, and it has evolved to deal with high-complex data and decision-making from the analysis. DL methods have been applied in modern drug discovery to efficiently deal with the extensive data generated from the drug discovery and development field.90
Several ML- and DL-based applications have been used occasionally and applied in drug discovery and development. Some significant ML-based models are TrixX, LS-align, StackCBPred, DrugFinder, and LigGrep (Table 2).
DL methods have shown better performance than ML methods. Therefore, they have recently appeared as one of the most promising tools in drug discovery research. Some significant DL-based models are DTI-CNN, DeepDTA, WideDTA, PADME, and DeepAffinity. All these models are very significant for drug discovery and development (Table 2).
Recent landscape of drug discovery: The role of AI to DL
AI-integrated drug discovery has led to a revolution in the drug discovery landscape. Substantial progress has been made in AI-integrated drug discovery. Therefore, significant changes have been made in the pharmaceutical industry. The pharmaceutical industry, which focuses on drug discovery, is implementing the AI platform for drug discovery and development. At the same time, the industries are trying to collaborate with technology companies.11
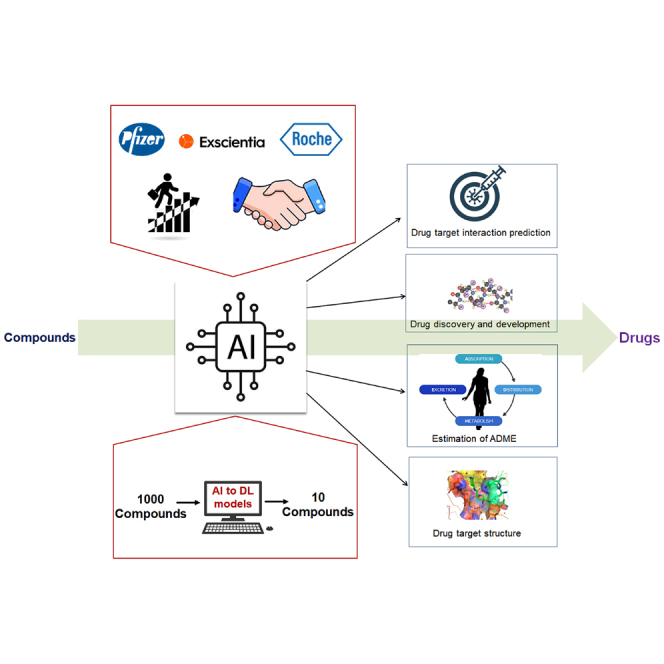
The first step of drug discovery is small molecule discovery. AI facilitates the development of a large number of drug molecules. AI also helps hits to lead generation speedily.11,93 Similarly, drug target identification is a significant step in drug discovery. AI and DL models have been implemented to identify drug targets faster and more accurately. AI and DL models have occasionally been generated from the initial to final steps of drug discovery and development (Figure 3).
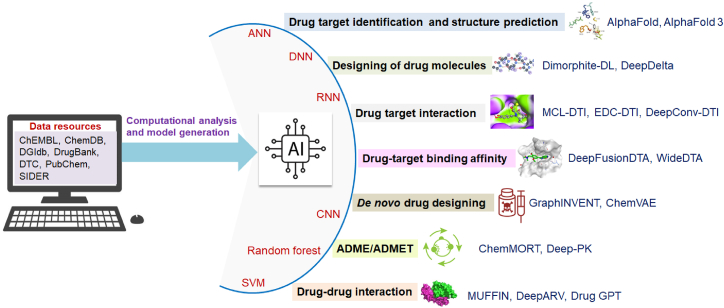
Similarly, several AI, ML, and DL models have been developed to understand the different properties of drug-target interactions (DTIs). At the same time, AI, ML, and DL models have been developed in different areas of drug discovery and development, such as lead molecule development, drug administration, distribution, metabolism, excretion, toxicity (ADMET), and drug-drug interaction (DDI), etc (Figure 4).
Figure 4.
A schematic diagram is depicted as an overall framework, visually demonstrating the application of AI technology in drug discovery
Identification of drug target and prediction of its structure
It has been noted that most of the drug targets are proteins. Other than the experimental and multi-omics approaches, ML and DL approaches have been applied to target identification. Here, researchers develop the disease network and perform a high-level analysis. During the network construction, a biological network was developed to capture the associations between genes, proteins, and molecular entities. Potential targets that involve a disease can be identified from these networks. AI-identified targets are validated using different kinds of experiments, and the target validation can be performed through cell culture experiments and animal models. Several researchers have successfully validated AI-identified targets.94 One example is Zhang et al.,95 who developed an ML-enabled technique to determine the association of the KANK1 gene to amyotrophic lateral sclerosis. Santos et al.66 analyzed 1,578 US Food and Drug Administration-approved drugs and found that proteins are the significant drug target (human and pathogen proteins). The most important classes of protein-based drug targets in humans are the G-protein couple receptors (approximately 12%), ion channels (approximately 19%), kinases (approximately 10%), and nuclear receptors (approximately 3%). Other than the proteins, other biomolecules (human and pathogen proteins) such as DNA, RNA, and peptides are also found as drug targets.96,97 However, protein plays a significant role in cell-cell transduction and cell signaling. AI modeling approaches may help to solve the structure of previously unsolved protein-based drug targets. Presently, AI approaches have been used for protein structure prediction. ANN-based AlphaFold is able to forecast the 3D structures of proteins. AlphaFold model was developed by DeepMind.98,99 The AlphaFold model predicted the 3D structure of human proteins, which is essential. For a given protein, using all heavy atoms, it molded the protein’s 3D structure directly from the primary amino acid sequence, aligned the homologous sequences, formed the 3D coordinates, and finally developed the model 3D structure of the protein. During the prediction of the protein’s 3D structure, AlphaFold uses the ANN and GPU.99 Recently, AlphaFold 3 has been introduced by Isomorphic Labs and Google DeepMind. Using a single unified deep-learning framework, the AlphaFold 3 has the capacity for high-accuracy modeling of the structure in the biomolecular landscape.100 Therefore, for the advancement of protein structure prediction, AlphaFold has significantly contributed in the field, and it will revolutionize drug discovery. However, the change in protein structures has been noted in different environments. Similarly, under the same conditions, proteins may show multiple coexisting structures. Recent ML and DL methods efficiently determine these structures, which will help in drug discovery and development.3,101
Design of drug molecules using ML and DL
Drug molecular design is an integral part of drug discovery, and ML, DL algorithms have helped to design a drug molecule’s structure. DL algorithms such as RNNs and autoencoders are used in molecular design during drug discovery and development.102 During the molecular design of drug molecules, molecular representation can be performed using two steps: representations of 3D geometry and molecular graphs. Representations of molecular graphs can be further developed using some steps: SMILES and string-based representations, image-based representations, tensor representations, and other graph-based representations. SMILES-enabled methods often struggle to achieve a high percentage.102,103,104 One example of a DL-based tool for small molecular design is the Gypsum-DL. To design compound libraries, it accepts flat SDF formats or SMILES. Different properties, such as cis/trans isomeric states, chiral, tautomeric, and ionization, are considered from the input. Then, it predicts the structural model by changing the structure from a 2D structural model into a 3D one.105 Another DL-based model, dimorphite DL, can estimate the ionization states of small molecules to understand the drug-like properties.106 Recently, Ivanenkov et al.107 developed Chemistry42, an AI-based platform to design novel small molecules with optimized properties. Finally, a designed drug-like molecule should be characterized to understand its properties. The different ML-to-DL methods of ADMET and DL-based QSAR methods help to characterize the properties of designed drug molecules.108 Several AI-to-DL-enabled tools have been developed to understand the ADMET properties of the developed molecules. One such ML-based ADME platform is ADME-AI. It can predict accurate and fast ADMET properties. It is an open-source web server.109 Another one is DeepDelta, a DL-enabled tool that predicts molecules' ADMET. It performs using algorithms (random forest plots andothers). Through direct training on molecular pairs, it predicts molecular properties accurately by directly training on molecular pairs.110
Estimation of DTI
DTI estimation is one of the critical areas in drug discovery and development. It illustrates the interaction between protein targets and chemical molecules.67,98,99 DTI has been determined using several experimental methods, such as phage display technology, yeast two-hybrid method, and co-immunoprecipitation techniques.3,111,112 These wet laboratory methods are time consuming. Using the increasing biological data and faster prediction, DTI has applied ML and DL to estimate DTI quickly. Several ML and DL models have been developed occasionally to predict the DTI estimation.67,113,114,115 Yang et al.116 have developed one ML-based model for DTI prediction. The model is entitled ML-DTI. Here, they applied four different methods, which yielded similar results. The model tested common targets and drug interaction prediction. Orphan-drug and orphan-target interaction is one of the critical areas of understanding. During the application of this model, it was found to increase the performance of orphan-drug and orphan-target intercations.116 Similarly, Rayhan et al.117 developed another deep CNN-based DTI prediction model called FRnet-DTI. It has auto-encoder-based feature manipulation of DTI.117 Likewise, Zhou et al.118 developed a model related to an augmented graph attention network (AGAT) to understand a binding site estimation of DTI. The model is called AGAT-PPIS. It can map the identity and initial residual.118
DTI identification is not limited to the drug’s or protein’s structural features. It can capture information about the drug-protein complex and the feature representation known as hybrid features. These hybrid features can be constructed using molecular docking, molecular dynamics simulations, or ML-to-DL models. Other than these methods, ligand-based, gene ontology-based, network-based, and text mining-based methods are also important. Bagherian et al.115 have illustrated these methods in a review article. Qian et al.119 developed a model for DTI using multimodal information of a drug molecule called MCL-DTI. It uses the bidirectional multi-head cross-attention method to understand the association between drugs and targets.119 Another model was developed using the DL model, called EDC-DTI, to predict DTIs, and it attains the low computational costs and highest predictive performance. As an example, the model was used to estimate the interaction of drugs and targets such as afatinib (DB08916), nifedipine (DB01115), and simvastatin (DB00641).120 Lee et al.84 have developed the DeepConv-DTI, where DTI can be predicted using DL. For DTIs, the model has some advantages, like identifying the binding sites of proteins.84
Estimation of drug-target binding affinity
The interactions between drug-target pairs have been studied in the context of binding affinity prediction. This illustrates the potency of the drug-target pair and is broadly enlightening for the field of drug discovery. Binding affinity can be predicted through computational methods.
Understanding the binding affinity among a drug molecule and its target is one of the areas in drug discovery and development. However, researchers have not studied this area much. Experimentally, analysis of drug-target binding affinity is expensive and time consuming. Therefore, computational methods are essential to reduce the time and cost. First, Ozturk et al.80 developed the drug-target binding affinity model in 2018. The work is represented as compound 1D form and modeled protein sequences. The prediction method uses the CNNs.80 Similarly, Pu et al.59 developed a model for the affinity prediction of drug-target binding. The model is entitled DeepFusionDTA. The two-stage DNN model was developed based on the Hybrid Deep-Learning Ensemble Model. The model was tested on the two datasets i.e., Davis and KIBA. The model shows a 1.0% Concordance Index (CI) increase in the first dataset and a 1.5% increase in the second dataset.59 Using co-regularized variational autoencoders, Li et al.121 developed another model for the affinity prediction of drug-target binding. It consists of two VAEs for generating target sequences and drug SMILES strings.121 Other ML/DL-based models have been proposed in this area, such as DeepAffinity82 and WideDTA.122 All these ML/DL-based models are helpful for the estimation of drug-target binding affinity. Recently, Thafar et al.123 designed a model called Affinity2Vec to predict the binding affinity of drug targets. Researchers have tested the model using a weighted heterogeneous graph incorporating different data, such as drug-target binding affinities, target-target similarity, and drug-drug similarity-related data.123 Another model uses cross-scale graph contrastive learning to estimate the drug binding affinity to targets. The model is called CSCo-DTA, which was proposed by Wang et al.124 Using the erlotinib molecule, the model verified the analyzed targets with the docking.
Design of de novo drug
It refers to the design of drug-like molecules through computational methods. The method started the design of drug-like molecules without a starting template. The boom in AI techniques has opened new possibilities for de novo drug design and accelerated drug discovery. In this case, novel molecular structures are generated from atomic building blocks with no previous relations. However, there are some main differences between conventional drug design and de novo drug design. In the case of conventional design, a structure-based approach was considered, and it depends on the active site’s properties of a biological target.
AI, including ML or DL, is an emerging field that has influenced the drug discovery process. Therefore, the de novo drug design approach was also influenced by AI.125 It designs novel chemical entities based on information such as receptors and ligands. Here, the biological targets are called receptors or their active binders, known as ligands. In de novo drug design, the receptor active site or ligand pharmacophore modeling is the main element in constructing the molecule.125,126 Several ML/DL-based models have been proposed for de novo drug design. During this decade, several DL-based models have been developed occasionally for de novo drug design. Some exciting models are MolRNN, an RNN-based model,127 and GraphINVENT, a GAN-based model,128 ChemVAE, an encoder-decoder-based model,129 and ReLeaSE, a reinforcement learning-based model.130 Another de novo drug molecule design model, druGAN, was recently noted. The method applied a deep generative adversarial autoencoder (AAE) model for developing new molecules with anticancer effects.131 Similarly, combining reinforcement learning algorithms with hybrid VAE, another model was developed for de novo drug design, known as PaccMannRL. This model efficiently designs anticancer molecules using transcriptomic data.132 Recently, Macedo et al.133 developed a de novo drug design model using graph convolutional networks. The model efficiently develops novel quinoline scaffold molecules and analyzes drug-related properties such as synthetic accessibility, toxicity, and pharmacokinetics.133 Therefore, all of these examples reflect that ML/DL models offer new openings in the de novo drug design and, thus, accelerate and revolutionize the drug discovery and development process.
Several other recent de novo drug design models have been developed, exploiting evolutionary algorithms occasionally. Some of these significant models are Dock_GA,134 MoleGear,135 AutoGrow4,68 and SECSE.136
Prediction of drug toxicity
Drug toxicity estimates undesirable or adversative effects of drug-like molecules. It is one of the main contributors to the costly process of drug development.137 This attribute is related to drug safety. During drug development, prediction of side effects and drug safety measurement are significant components.138 However, laboratory estimation of drug toxicity studies during the drug development process is time consuming. Therefore, computational models reduce time and cost in this case. Recently, using three-layer DNN, a model was developed to predict the toxicity of drug-like molecules or compounds known as DeepTox. In this model, the input of DNN has been used as 0D to 3D molecular descriptors.139 Recently, a DL-based toxicity prediction model known as Deep-PK has been developed. This model estimates the toxicity and pharmacokinetics of small molecules.140 Therefore, ML/DL models are essential for predicting drug toxicity.
Estimation of ADME
Over the past decade, drug discovery and development have evaluated the ADME property, one of the most critical issues. Previously, experimental evaluation methods (in vivo and in vitro) were used, but these methods are time consuming, laborious, and costly.104 Therefore, estimation of the ADME properties of drug molecules is essential. AI models play a significant role in assessing the ADME, and several AI models have been developed from time to time. It is essential to understand the drug’s ADME properties, which are essential for the drug discovery and development process. Therefore, it is necessary to comprehend these four properties in detail. The four properties are absorption, distribution, metabolism, and excretion. However, some scientists have illustrated the ADME as ADMET, where toxicity has been included in the ADME model as a fifth property. To understand the ADME-toxicity feature of a drug, researchers must understand the inhibition of cytochrome CYP2D6 (P450 2D6), which commonly affects the molecules passing through the blood-brain barrier. Additionally, studying plasma protein binding is essential.141,142,143 Recently, Yi et al.144 developed a model for estimating ADME-Toxicity properties entitled ChemMORT. The model predicts the ADME-toxicity features of drug molecules using multi-objective particle swarm optimization and DL algorithms.144 Gu et al.145 developed another model for estimating ADME-toxicity properties entitled asadmetSAR3.0. The web-based model efficiently predicts the ADME-toxicity.145 Another important web-based model that efficiently predicts the ADME-toxicity model is OptADMET. This model improves lead compounds' ADMET properties through substructure alterations.146
Drug toxicity prediction is one of the crucial parameters for humans. Several types of toxicities have been predicted from time to time, such as liver toxicity (drug-induced liver injury [DILI]), carcinogenesis, and heart toxicity.147 Drug-induced cardiotoxicity and DILI are significant adverse effects triggered by many essential drugs.148 Using data-driven approaches to comprehend toxicity is an important area. In this direction, several data-driven databases or libraries have been introduced for predicting toxicity, such as ClinTox,149 ToxCast,150 and Tox21.151
Some chemical datasets with SMILE format data are available for the ADMET for molecule property prediction, such as Llipophilicity, FreeSolv, and ESOL.152 These datasets are essential to predict the ADMET. From the molecular structure, Delaney153 developed a method to directly evaluate the aqueous solubility of the molecule. Similarly, Mobley and Guthrie154 developed a FreeSolv database to evaluate the calculated and experimental hydration-free energies. It might help to compute hydration-free energies for small molecules in water and molecular structures.154 It is beneficial to understand the properties of a molecule’s structure. Therefore, it may help to understand the ADME of a drug molecule. Lipophilicity is one of the critical parameters of the drug that defines solubility, the ability to penetrate through cell barriers and transport to the molecular target. Therefore, it is essential for drug discovery and development.155 It affects the pharmacokinetics and, ultimately, the ADME. Waring156 suggested understanding lipophilicity is necessary for drug development. AI-enabled ADME might help in this direction.
Several AI-, ML-, or DL-enabled tools or models have recently been developed, focusing on ADME or ADMET. One such ML-based platform is ADMET-AI. It is one of the fastest web-based ADMET prediction platforms using the Python package. It reduces time by approximately 45% compared with other available platforms performing ADMET analysis.109 One recent tool, Deep-PK, has been designed using DL to comprehend the pharmacokinetics and toxicity of input drug molecules. In this model, researchers have used graph NN and graph-based signatures to extract the feature. Finally, it produces the soundest predictive performance.73
Similarly, Yi et al.144 developed a DL-based automatic platform, ChemMORT, to optimize ADMET. The analytical platform uses three modules during optimization: encoder, decoder, and optimizer. These three modules are an SMILES encoder, a descriptive decoder, and a molecular optimizer.144 Using AI-, ML-, or DL-enabled ADME or ADMET tools or models, the researchers can quickly comprehend a molecule’s pharmacokinetics and safety properties during drug discovery and development.
Estimation of DDI
When we consume two or more drugs, the combination may create unwanted side effects. Therefore, DDIs are described as the unavoidable side effects that result from the intake of two or more drugs.157 It is associated with treatment failure or clinical toxicity. DDIs can be studied through an experimental approach using in vitro and in vivo methods to assess DDI potential.157,158 Predicting DDIs is essential for human health. Conversely, a substantial amount of cost and time is required to predict DDIs using an experimental approach. Presently, using AI, one can analyze the DDIs very quickly. The advantage of AI is that enabling DDI prediction can take less time and lower the cost. Therefore, several AI-to-DL models have been developed in this direction.159
DDIs also need to be understood during drug discovery development, and this is an emerging area of research. It is an essential threat to public health. Therefore, researchers are trying to understand the properties of DDI.160 DDIs were studied through the different models of AI.159 Recent DNN models can analyze the DDI. Recently, Liu et al.161 developed a model DANN-DDI to estimate DDIs. Another model was developed to access DDI. It is a multi-scale feature model called MUFFIN.162 Another DL model can analyze the DDI, which is known as AttentionDDI. This multi-modal NN can benefit drug development through better DDI prediction.163 Pham et al.164 developed a model for predicting DDI using DL. The model, called DeepARV, aims to understand the DDI between ARVs and comedications.164 Similarly, Rohani and Eslahchi165 proposed an NDD model to study the DDI through the NN using a heuristic similarity selection process. The model can evaluate unknown DDIs.165 Similarly, another recent DL-enabled DDI model, SSF-DDI, was developed. The method uses substructure features using the drug molecule graph and drug sequence for DDI prediction. The model integrates drug sequence features and structural features from the drug molecule graph. The encoder captures sequence features pulled out from drug molecules using MixAttention and multilayer CNNs. It also uses a directed message-passing NN to feature the extraction of substructures.166 However, these DDI models solved the DDI problem in the future through proper prediction and understanding.
Large language models: From drug target identification to drug discovery and development
After the release of ChatGPT on November 30, 2022, large language models (LLMs) gained high interest, and millions of people have been using them.167,168 LLMs perform human-like conversations with cutting-edge technology. Other LLM-based chatbots include Google’s Gemini, Models of Meta’s LLaMA family, and Mistral AI’s models. LLMs can train vast amounts of text through the supervised training process. It uses several fields of medical science for drug discovery.169 LLMs can help to provide the necessary information during drug discovery and development, such as drug target discovery, pharmacokinetics, and pharmacodynamics. It can also help to understand the AMDE properties of a drug molecule. It also helps to comprehend DDI during drug discovery and development.8,170 Recently, several next-generation, domain-specific LLMs have been developed, such as the DrugChat model. The model uses LLM, an adapter, and a graph NN. It can produce drug-molecule graphs.169 Now, LLMs have been used in different areas of drug discovery and development.
However, LLMs have several limitations, such as accuracy and plagiarism.168 By addressing those limitations, LLMs can open up immense future possibilities in drug discovery and development.
AI-designed molecules entering clinical trial: Some success stories
The preliminary result for AI-designed molecules shows very promising. Some AI-designed molecules have crossed the preclinical barrier and entered clinical trials (Table 3). An AI-designed A2A receptor antagonist molecule EXS-21546 also entered into the clinical trial. The molecule is an immuno-oncology molecule that will be used to treat solid tumors carrying high adenosine signatures. It was developed from the collaborative effect between Evotec and Exscientia.171 Exscientia AI Ltd. is a leading pharma-technology company that performs AI-related drug discovery. Evotec is a biotechnology-based drug discovery company in Germany. Exscientia AI Ltd. developed another AI-designed molecule, DSP-1181. This 5-HT1a agonist for obsessive-compulsive disorder.172,173 Exscientia collaborated with Sumitomo Dainippon Pharma, Japan, to develop this AI-generated drug.
Table 3.
Different AI-designed drugs that enter clinical trials
| Sl. No. | Drug | Clinical trials number | Phase | Organization/sponsor | Remark |
|---|---|---|---|---|---|
| 1. | REC-4881 | NCT05552755 | 1 and 2 | Recursion Pharmaceuticals Inc., USA |
Inhibitor component of mitogen-activated protein kinase kinase 1 and 2 for individual with familial adenomatous polyposis |
| 2. | BEN-2293 | NCT04737304 | 1 and 2 | BenevolentAI, UK |
It is an inhibitor of pan-tyrosine kinase inhibitor applies to patients who have atopic dermatitis (mild to moderate) |
| 3. | RLY-4008 | NCT04526106 | 1 and 2 | Relay Therapeutics, USA | The inhibitor of fibroblast growth factor receptor 2 used against in the patients with unresectable or metastatic cholangiocarcinoma and other solid tumors |
| 4. | BEN-8744 | NCT06118385 | 1 | BenevolentAI, UK |
The phosphodiesterase 10 inhibitor for treating inflammatory bowel diseases such as ulcerative colitis |
| 5. | REC-2282 | NCT05130866 | 2 and 3 | Recursion Pharmaceuticals Inc., USA | The histone deacetylase inhibitor used for in patients with progressive NF2-mutated meningiomas |
| 6. | INS018_055 | NCT05975983 | 2 | In Silico Medicine Hong Kong Limited, China | Consider as small molecules inhibitor of adults with idiopathic pulmonary fibrosis |
| 7. | REC-994 | NCT05085561 | 2 | Recursion Pharmaceuticals Inc., USA |
The superoxide scavenger molecule used against symptomatic cerebral cavernous malformation |
| 8. | EXS-21546 | NCT05920408 | 1 and 2 | Exscientia AI Limited, Scotland |
Adenosine A2A receptor antagonist molecule used against advanced solid tumors carrying high adenosine signatures |
| 9. | GTAEXS617 | NCT05985655 | 1 and 2 | Exscientia AI Limited, Scotland |
The CD4/CDK6 inhibitor, used for the treatment of advanced solid tumors |
| 10. | ISM3312 | CTR20230768 | 2 | Insilico Medicine, China |
Used for the treatment of coronavirus disease 2019 patients as 3CL protease inhibitor compound |
| 11. | NDI-010976/GS-0976 |
NCT02876796, NCT02856555, NCT02891408, NCT03987074 |
1 and 2 | Gilead Sciences, USA |
The oral dose administration in against of nonalcoholic fatty liver disease |
| 12. | OPL-0401 | NCT05393284 | 2 | Valo Health, Inc, USA |
The drug used for patients with diabetes mellitus with non-proliferative diabetic retinopathy or mild proliferative diabetes retinopathy with or without diabetic macular edema |
| 13. | ISM3091 | NCT05932862 | 1 | Exelixis, USA |
The combination therapy with olaparib in patients with advanced solid tumors |
| 14. | RLY-1971/RG-6433 | NCT04252339 | 1 | Hoffmann-La Roche, Switzerland |
Used as highly potent and selective SHP2 inhibitor, for advanced or metastatic solid tumors |
| 15. | RLY-2608 | NCT05216432 | 1 | Relay Therapeutics, Inc. USA |
The single agent in advanced solid tumor patients and in combination with fulvestrant in patients with advanced breast cancer |
| 16. | ANPA-0073 | ACTRN12621000644864 | 1 | Structure Therapeutics, USA |
The testing single incremental doses of oral capsules used for patient having pulmonary arterial hypertension, idiopathic pulmonary fibrosis |
| 17. | PHI-101 |
NCT04842370 NCT04678102 |
1 | Seoul National University Hospital, South Korea |
The PHI-101, and novel FLT3 inhibitor in the treatment of relapsed or refractory acute myeloid leukemia for patients who have received standard therapy |
| 18. | SGR-1505 | NCT05544019 | 1 | Schrödinger, Inc. USA |
The oral inhibitor of MALT1 used against for the non-Hodgkin’s lymphoma |
| 19. | OPL-0301 | NCT05327855 | 2 | Valo Health, Inc, USA |
The supportive impromevnt drug used for the patients having post-myocardial infarction left ventricular dysfunction |
| 20. | GSBR-1290 | NCT05762471 | 1 and 2 | Gasherbrum bio inc. USA |
This drug used for the patients having overweight/obesity and type 2 diabetes mellitus on metformin |
Exscientia developed the next drug candidate, an AI-designed molecular candidate. The molecule is known as EXS4318. The drug candidate is a selective protein kinase C-theta inhibitor.171 Another AI-designed molecule, INS018_055, is developed by InSilico Medicine. It is a biotechnology-based drug design company in Hong Kong. The molecule is a TRAF2- and NCK-interacting kinase inhibitor. The molecule is developed for idiopathic pulmonary fibrosis.94,174
Another drug candidate is RLY-4008, developed by BenevolentAI and a small-molecule phosphodiesterase 10 inhibitor. BenevolentAI performs an AI-based innovation and is a global leader in AI located in the UK. The molecules are developed for ulcerative colitis.
Collaboration for AI-enabled drug discovery and development
Several collaborations were made occasionally in the pharmaceutical sector to foster drug discovery and development (Figure 5).175,176 The collaboration between academia-industry is frequently noted in pharmaceutical companies. It occurs because academia can shine in the discovery of drugs with fundamental concepts. In contrast, the industry excels in the translational process of product development.177 However, different types of collaborations were found in the pharmaceutical sector for drug discovery and development, such as academia and pharmaceutical collaboration, a collaboration between two pharmaceutical companies, and a collaboration between one pharmaceutical company and one venture capitalist.177,178 However, all these collaborations were made for faster drug discovery and development methods with less economic input. Some specific collaboration has been noted from time to time. We noted some collaborative efforts for the neglected or rare disease drug development.179,180 During the pandemic, we found several collaborations for coronavirus disease 2019 vaccine development and deployment.181,182,183,184 However, several molecules have been discovered and entered the market due to collaborative efforts, and several examples are noted in this direction.
Figure 5.
Collaborations between pharmaceutical and technology companies for AI-enabled drug discovery and development
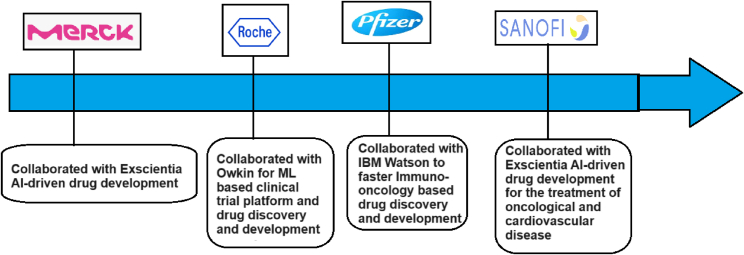
Big pharma companies have collaborated with computer giants for AI-enabled drug discovery and development. These collaborations are fostering the discovery of new drug molecules. Several new drug molecules have resulted from these collaborations. One example is the collaboration between Evotec and Exscientia, which resulted in the molecule EXS-21546. The molecule has entered the clinical trial and will be used to treat solid tumors. Several other collaborative efforts have been noted for AI-enabled drug discovery. Marck has collaborated with Exscientia for AI-related drug development. Roche has collaborated with Owkin to develop an ML-based clinical trial platform for drug discovery and development.
Similarly, the pharmaceutical giant Pfizer has collaborated with technology company IBM Watson to speed up drug molecule discovery and development in immuno-oncology. Pfizer, IBM, and Microsoft have collaborated to solve healthcare-related problems comprehensively. Sanofi collaborates with Exscientia for AI-related drug development in oncology and cardiovascular disease.11 These collaboration efforts have made AI-enabled drug discovery and development more potent and successful. We will see that these collaborative efforts will result in several new drugs shortly.
Challenges
Researchers have documented several challenges in AI-driven drug discovery and development, which are as follows.
Issues related to the availability of quality data
Several challenges have been noted in this area of drug development. AI models need to be trained to high-quality data. The main challenge is the availability of high-quality and suitable datasets for training the model. Although chemical and biological data are increasing, the data quality is not so good. Therefore, data curation can be done. At the same time, a cost is involved in accessing data from the database. It is an additional cost to a company and might increase drug development costs. However, more high-quality datasets in pharmacological science and pharmaceutical chemistry must be developed immediately. These datasets can help to train and test AI models, which might solve the data availability problem.
Interpretability of AI or DL models
Developed AI or DL-based drug discovery and development models should be adequately understood and explained. Understandability and explainability of AI model predictions remain challenging. DL models deal with a large number of parameters and multiple layers. It is challenging for nonexpert users to explain the model in the case of drug discovery and development. Therefore, understanding and explaining the DL models is vital for drug discovery and development, but sometimes it is challenging.185,186,187 High-end skills and trained workforces with knowledge of both areas, such as computer engineering with AI specialization and pharmaceutical science knowledge, are immediately required. These skills help to modify algorithms in this direction and understand and predict the outcomes of algorithms in pharmaceutical science and drug development. They might solve the problem of interpretability of AI models.
Similarly, the models developed through the AI technique are not explainable. Likewise, we cannot explain the AI-derived result due to its black box nature.188,189
Computational constraints of high-end AI models
These high-end AI models are not feasible for smaller research entities because they need to train for a prolonged time with a large amount of data. To run these models requires enormous storage resources and vast computational infrastructure. Parallel computation might be helpful for computing high-end AI models.187,190
In this direction, significant research entities require enormous storage resources, and vast computational infrastructure should be established by every country. As the infrastructure cost is too high, every company might not establish the infrastructure. Therefore, the country should support it for the companies. However, the high-end infrastructure must be accessible to companies dealing with AI-enabled drug discovery. It might solve the infrastructure-related issues associated with the computational constraints of high-end AI models.
The need for a more skilled and trained workforce
A skilled and trained workforce is another problem. We need more software engineers with the knowledge of AI technology and more skilled data scientists. With an explicit knowledge of these fields and pharmaceutical expertise, the workforce can efficiently perform AI-driven drug discovery. However, we also need a more skilled and trained workforce.
Conclusion
AI has made significant progress in disease diagnostics, helping healthcare professionals such as radiologists and clinical pathologists, and revolutionizing these fields. AI has also made significant progress in precision medicine.13,191 Different models developed using AI have been developed to solve radiological and pathological diagnostics and precision medicine problems. Such experiences and success stories might support AI-enabled drug discovery.
Drug discovery is a critical field that deals with multi-step search, multi-dimensional, and optimization problems. With powerful problem-solving capacity, AI has been applied in different fields of drug discovery to solve complicated problems. Over the last decade, AI-enabled techniques have been vastly used in various drug discovery and development steps, and we have witnessed it. The revolution of AI techniques has substantially impacted drug discovery, accelerating the process. Conversely, recent LLM applications like ChatGPT have enriched the drug discovery and development field. Researchers are applying this NPL-based DL model in drug target discovery and advance it to clinical trials more quickly.8,170,192 Our discussion indicates that an AI-enabled, fast-approaching wave has been created in the drug discovery field among pharmaceutical companies with the potential to change drug discovery in the future. Although several challenges exist, AI techniques and technologies will address and overcome them. AI techniques will bring drastic changes in AI-driven drug discovery. DL and ML technology-driven research have successfully generated several models and platforms for drug discovery research. Many researchers have developed AI-driven models, technology, and drug discovery and development tools. In contrast, venture capitalists invest vast amounts of money in different start-up companies focusing on AI-based drug development. Therefore, all factors support the growth of AI-assisted drug discovery and development. Therefore, all assume that the sector will likely grow very fast. At the same time, it is very promising that major pharmaceutical companies will collaborate with technology giants to enter the market by leveraging faster drug discovery and development methods through AI-assisted techniques. Therefore, AI-assisted drug discovery is likely to grow very fast. With faster discovery timelines, AI-enabled drug discovery might be a game changer for research and development in the pharmaceutical sector, from small molecules to therapeutic antibody drug discovery. It will also revolutionize pharmaceutical research and development. We are very hopeful that several AI-driven drugs will enter the market very soon.
Acknowledgments
This study was supported by research grants from the Zuoying Armed Forces General Hospital (KAFGH-ZY-A-110011, KAFGH-ZY-A-10901).
Author contributions
C.C. conceptualized the manuscript, performed investigation, writing original manuscript draft, writing – review & editing and supervised the whole project. M.B. performed validation, figures and tables development. S.S.L. and Z.H.W. did the validation and formal analysis. Y.H.L. performed formal analysis and fund acquisition.
Declaration of interests
No potential conflict of interest was declared by authors.
Contributor Information
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
Yi-Hao Lo, Email: a0948060004@mail.ngh.com.tw.
References
- 1.Khanna I. Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discov. Today. 2012;17:1088–1102. doi: 10.1016/j.drudis.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Strohbehn G.W., Kacew A.J., Goldstein D.A., Feldman R.C., Ratain M.J. Combination therapy patents: a new front in evergreening. Nat. Biotechnol. 2021;39:1504–1510. doi: 10.1038/s41587-021-01137-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Liu X., Zhang S., Chen S. Artificial intelligence for drug discovery: Resources, methods, and applications. Mol. Ther. Nucleic Acids. 2023;31:691–702. doi: 10.1016/j.omtn.2023.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kneller R. The importance of new companies for drug discovery: origins of a decade of new drugs. Nat. Rev. Drug Discov. 2010;9:867–882. doi: 10.1038/nrd3251. [DOI] [PubMed] [Google Scholar]
- 5.Brown D.G., Wobst H.J., Kapoor A., Kenna L.A., Southall N. Clinical development times for innovative drugs. Nat. Rev. Drug Discov. 2022;21:793–794. doi: 10.1038/d41573-021-00190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecker D.J., Aiello C.D., Arron J.R., Bennett C.F., Bernard A., Breakefield X.O., Broderick T.J., Callier S.L., Canton B., Chen J.S., et al. Opportunities and challenges for innovative and equitable healthcare. Nat. Rev. Drug Discov. 2024;23:321–322. doi: 10.1038/d41573-024-00032-4. [DOI] [PubMed] [Google Scholar]
- 7.Deng J., Yang Z., Ojima I., Samaras D., Wang F. Artificial intelligence in drug discovery: applications and techniques. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbab430. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborty C., Bhattacharya M., Lee S.S. Artificial intelligence enabled ChatGPT and large language models in drug target discovery, drug discovery, and development. Mol. Ther. Nucleic Acids. 2023;33:866–868. doi: 10.1016/j.omtn.2023.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentwich I. Pharma's Bio-AI revolution. Drug Discov. Today. 2023;28 doi: 10.1016/j.drudis.2023.103515. [DOI] [PubMed] [Google Scholar]
- 10.Frankish K., Ramsey W.M. Cambridge University Press; 2014. The Cambridge Handbook of Artificial Intelligence. [DOI] [Google Scholar]
- 11.Paul D., Sanap G., Shenoy S., Kalyane D., Kalia K., Tekade R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today. 2021;26:80–93. doi: 10.1016/j.drudis.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramesh A.N., Kambhampati C., Monson J.R.T., Drew P.J. Artificial intelligence in medicine. Ann. R. Coll. Surg. Engl. 2004;86:334–338. doi: 10.1308/147870804290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty C., Bhattacharya M., Pal S., Lee S.-S. From machine learning to deep learning: An advances of the recent data-driven paradigm shift in medicine and healthcare. Curr. Res. Biotechnol. 2023;7 doi: 10.1016/j.crbiot.2023.100164. [DOI] [Google Scholar]
- 14.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., Li B., Madabhushi A., Shah P., Spitzer M., Zhao S. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019;18:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadybekov A.V., Katritch V. Computational approaches streamlining drug discovery. Nature. 2023;616:673–685. doi: 10.1038/s41586-023-05905-z. [DOI] [PubMed] [Google Scholar]
- 16.Askr H., Elgeldawi E., Aboul Ella H., Elshaier Y.A.M.M., Gomaa M.M., Hassanien A.E. Deep learning in drug discovery: an integrative review and future challenges. Artif. Intell. Rev. 2023;56:5975–6037. doi: 10.1007/s10462-022-10306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zdrazil B., Felix E., Hunter F., Manners E.J., Blackshaw J., Corbett S., de Veij M., Ioannidis H., Lopez D.M., Mosquera J.F., et al. The ChEMBL Database in 2023: a drug discovery platform spanning multiple bioactivity data types and time periods. Nucleic Acids Res. 2024;52:D1180–D1192. doi: 10.1093/nar/gkad1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padalino G., Coghlan A., Pagliuca G., Forde-Thomas J.E., Berriman M., Hoffmann K.F. Using ChEMBL to Complement Schistosome Drug Discovery. Pharmaceutics. 2023;15:1359. doi: 10.3390/pharmaceutics15051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Swamidass S.J., Dou Y., Bruand J., Baldi P. ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics. 2005;21:4133–4139. doi: 10.1093/bioinformatics/bti683. [DOI] [PubMed] [Google Scholar]
- 20.Chen J.H., Linstead E., Swamidass S.J., Wang D., Baldi P. ChemDB update--full-text search and virtual chemical space. Bioinformatics. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Tang Q., Meng F., Du P., Chen W. INPUT: An intelligent network pharmacology platform unique for traditional Chinese medicine. Comput. Struct. Biotechnol. J. 2022;20:1345–1351. doi: 10.1016/j.csbj.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freshour S.L., Kiwala S., Cotto K.C., Coffman A.C., McMichael J.F., Song J.J., Griffith M., Griffith O.L., Wagner A.H. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J., Tanoli Z.U.R., Ravikumar B., Alam Z., Rebane A., Vähä-Koskela M., Peddinti G., van Adrichem A.J., Wakkinen J., Jaiswal A., et al. Drug Target Commons: A Community Effort to Build a Consensus Knowledge Base for Drug-Target Interactions. Cell Chem. Biol. 2018;25:224–229.e2. doi: 10.1016/j.chembiol.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campillos M., Kuhn M., Gavin A.C., Jensen L.J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 26.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorokina M., Merseburger P., Rajan K., Yirik M.A., Steinbeck C. COCONUT online: Collection of Open Natural Products database. J. Cheminf. 2021;13:2. doi: 10.1186/s13321-020-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatansever S., Schlessinger A., Wacker D., Kaniskan H.Ü., Jin J., Zhou M.M., Zhang B. Artificial intelligence and machine learning-aided drug discovery in central nervous system diseases: State-of-the-arts and future directions. Med. Res. Rev. 2021;41:1427–1473. doi: 10.1002/med.21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talevi A., Morales J.F., Hather G., Podichetty J.T., Kim S., Bloomingdale P.C., Kim S., Burton J., Brown J.D., Winterstein A.G., et al. Machine Learning in Drug Discovery and Development Part 1: A Primer. CPT Pharmacometrics Syst. Pharmacol. 2020;9:129–142. doi: 10.1002/psp4.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Celebi M.E., Aydin K. Springer International Publishing. Part of: Springer Professional "Wirtschaft+Technik" , Springer Professional "Technik" , Springer Professional "Wirtschaft"; 2016. Unsupervised Learning Algorithms. [DOI] [Google Scholar]
- 33.Cord M., Cunningham P. Springer Berlin Heidelberg; 2008. Machine Learning Techniques for Multimedia: Case Studies on Organization and Retrieval. [DOI] [Google Scholar]
- 34.Lo Y.C., Senese S., Damoiseaux R., Torres J.Z. 3D Chemical Similarity Networks for Structure-Based Target Prediction and Scaffold Hopping. ACS Chem. Biol. 2016;11:2244–2253. doi: 10.1021/acschembio.6b00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo Y.C., Senese S., France B., Gholkar A.A., Damoiseaux R., Torres J.Z. Computational Cell Cycle Profiling of Cancer Cells for Prioritizing FDA-Approved Drugs with Repurposing Potential. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lo Y.C., Rensi S.E., Torng W., Altman R.B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today. 2018;23:1538–1546. doi: 10.1016/j.drudis.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alloghani M., Al-Jumeily D., Mustafina J., Hussain A., Aljaaf A.J. In: Supervised and Unsupervised Learning for Data Science. Berry M.W., Mohamed A., Yap B.W., editors. Springer International Publishing; 2020. A Systematic Review on Supervised and Unsupervised Machine Learning Algorithms for Data Science; pp. 3–21. [DOI] [Google Scholar]
- 38.Xu Z., Wang S., Zhu F., Huang J. Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics. 2017. Seq2seq fingerprint: An unsupervised deep molecular embedding for drug discovery; pp. 285–294. [DOI] [Google Scholar]
- 39.Xiao Y.D., Clauset A., Harris R., Bayram E., Santago P., 2nd, Schmitt J.D. Supervised self-organizing maps in drug discovery. 1. Robust behavior with overdetermined data sets. J. Chem. Inf. Model. 2005;45:1749–1758. doi: 10.1021/ci0500839. [DOI] [PubMed] [Google Scholar]
- 40.Van Engelen J.E., Hoos H.H. A survey on semi-supervised learning. Mach. Learn. 2020;109:373–440. [Google Scholar]
- 41.Zhou X., Belkin M. Vol. 1. Elsevier; 2014. Semi-supervised learning; pp. 1239–1269. (Academic press library in signal processing). [Google Scholar]
- 42.Sahoo P., Roy I., Wang Z., Mi F., Yu L., Balasubramani P., Khan L., Stoddart J.F. MultiCon: A Semi-Supervised Approach for Predicting Drug Function from Chemical Structure Analysis. J. Chem. Inf. Model. 2020;60:5995–6006. doi: 10.1021/acs.jcim.0c00801. [DOI] [PubMed] [Google Scholar]
- 43.Chen X., Ren B., Chen M., Wang Q., Zhang L., Yan G. NLLSS: Predicting Synergistic Drug Combinations Based on Semi-supervised Learning. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G., Liu J., Wang C. Predicting drug-disease interactions by semi-supervised graph cut algorithm and three-layer data integration. BMC Med. Genom. 2017;10:79. doi: 10.1186/s12920-017-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J., Si Y.W., Un C.W., Siu S.W.I. Chemical toxicity prediction based on semi-supervised learning and graph convolutional neural network. J. Cheminf. 2021;13:93. doi: 10.1186/s13321-021-00570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahl N., Falkman G., Karlsson A., Mathiason G., Bostrom J. Deep Reinforcement Learning for Multiparameter Optimization in de novo Drug Design. J. Chem. Inf. Model. 2019;59:3166–3176. doi: 10.1021/acs.jcim.9b00325. [DOI] [PubMed] [Google Scholar]
- 47.Pereira T., Abbasi M., Oliveira J.L., Ribeiro B., Arrais J. Optimizing blood-brain barrier permeation through deep reinforcement learning for de novo drug design. Bioinformatics. 2021;37:i84–i92. doi: 10.1093/bioinformatics/btab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X., Ye K., van Vlijmen H.W.T., Emmerich M.T.M., IJzerman A.P., van Westen G.J.P. DrugEx v2: de novo design of drug molecules by Pareto-based multi-objective reinforcement learning in polypharmacology. J. Cheminf. 2021;13:85. doi: 10.1186/s13321-021-00561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCulloch W.S., Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biol. 1990;52:99–115. [PubMed] [Google Scholar]
- 50.Chen H., Engkvist O., Wang Y., Olivecrona M., Blaschke T. The rise of deep learning in drug discovery. Drug Discov. Today. 2018;23:1241–1250. doi: 10.1016/j.drudis.2018.01.039. [DOI] [PubMed] [Google Scholar]
- 51.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- 53.Wan L., Zeiler M., Zhang S., Le Cun Y., Fergus R. International Conference on Machine Learning. 2013. Regularization of neural networks using dropconnect; pp. 1058–1066. [Google Scholar]
- 54.Katuwal R., Suganthan P.N. 2018 IEEE Symposium Series on Computational Intelligence (SSCI) 2018. Dropout and dropconnect based ensemble of random vector functional link neural network; pp. 1772–2177. [Google Scholar]
- 55.Iosifidis A., Tefas A., Pitas I. DropELM: Fast neural network regularization with Dropout and DropConnect. Neurocomputing. 2015;162:57–66. [Google Scholar]
- 56.Pei L., Jones K.A., Shboul Z.A., Chen J.Y., Iftekharuddin K.M. Deep Neural Network Analysis of Pathology Images With Integrated Molecular Data for Enhanced Glioma Classification and Grading. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.668694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amerikanos P., Maglogiannis I. Image Analysis in Digital Pathology Utilizing Machine Learning and Deep Neural Networks. J. Personalized Med. 2022;12:1444. doi: 10.3390/jpm12091444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atz K., Cotos L., Isert C., Håkansson M., Focht D., Hilleke M., Nippa D.F., Iff M., Ledergerber J., Schiebroek C.C.G., et al. Prospective de novo drug design with deep interactome learning. Nat. Commun. 2024;15:3408. doi: 10.1038/s41467-024-47613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pu Y., Li J., Tang J., Guo F. DeepFusionDTA: Drug-Target Binding Affinity Prediction With Information Fusion and Hybrid Deep-Learning Ensemble Model. IEEE ACM Trans. Comput. Biol. Bioinf. 2022;19:2760–2769. doi: 10.1109/TCBB.2021.3103966. [DOI] [PubMed] [Google Scholar]
- 60.Verma N., Qu X., Trozzi F., Elsaied M., Karki N., Tao Y., Zoltowski B., Larson E.C., Kraka E. SSnet: A Deep Learning Approach for Protein-Ligand Interaction Prediction. Int. J. Mol. Sci. 2021;22:1392. doi: 10.3390/ijms22031392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi W., Singha M., Srivastava G., Pu L., Ramanujam J., Brylinski M. Pocket2Drug: An Encoder-Decoder Deep Neural Network for the Target-Based Drug Design. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.837715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi W., Singha M., Pu L., Srivastava G., Ramanujam J., Brylinski M. GraphSite: Ligand Binding Site Classification with Deep Graph Learning. Biomolecules. 2022;12:1053. doi: 10.3390/biom12081053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J., Xiao Y., Lin M., Cai H., Zhao D., Li Y., Luo H., Tang C., Wang L. DeepCancerMap: A versatile deep learning platform for target- and cell-based anticancer drug discovery. Eur. J. Med. Chem. 2023;255 doi: 10.1016/j.ejmech.2023.115401. [DOI] [PubMed] [Google Scholar]
- 64.Rubio D.M., Schoenbaum E.E., Lee L.S., Schteingart D.E., Marantz P.R., Anderson K.E., Platt L.D., Baez A., Esposito K. Defining translational research: implications for training. Acad. Med. 2010;85:470–475. doi: 10.1097/ACM.0b013e3181ccd618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jing Y., Bian Y., Hu Z., Wang L., Xie X.Q. Deep Learning for Drug Design: an Artificial Intelligence Paradigm for Drug Discovery in the Big Data Era. AAPS J. 2018;20:58. doi: 10.1208/s12248-018-0210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin J., Sahakian D.C., de Morais S.M.F., Xu J.J., Polzer R.J., Winter S.M. The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr. Top. Med. Chem. 2003;3:1125–1154. doi: 10.2174/1568026033452096. [DOI] [PubMed] [Google Scholar]
- 67.Nag S., Baidya A.T.K., Mandal A., Mathew A.T., Das B., Devi B., Kumar R. Deep learning tools for advancing drug discovery and development. 3 Biotech. 2022;12:110. doi: 10.1007/s13205-022-03165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiegel J.O., Durrant J.D. AutoGrow4: an open-source genetic algorithm for de novo drug design and lead optimization. J. Cheminf. 2020;12:25. doi: 10.1186/s13321-020-00429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schellhammer I., Rarey M. TrixX: structure-based molecule indexing for large-scale virtual screening in sublinear time. J. Comput. Aided Mol. Des. 2007;21:223–238. doi: 10.1007/s10822-007-9103-5. [DOI] [PubMed] [Google Scholar]
- 70.Hu J., Liu Z., Yu D.J., Zhang Y. LS-align: an atom-level, flexible ligand structural alignment algorithm for high-throughput virtual screening. Bioinformatics. 2018;34:2209–2218. doi: 10.1093/bioinformatics/bty081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gattani S., Mishra A., Hoque M.T. StackCBPred: A stacking based prediction of protein-carbohydrate binding sites from sequence. Carbohydr. Res. 2019;486 doi: 10.1016/j.carres.2019.107857. [DOI] [PubMed] [Google Scholar]
- 72.Lagarde N., Goldwaser E., Pencheva T., Jereva D., Pajeva I., Rey J., Tuffery P., Villoutreix B.O., Miteva M.A. A Free Web-Based Protocol to Assist Structure-Based Virtual Screening Experiments. Int. J. Mol. Sci. 2019;20:4648. doi: 10.3390/ijms20184648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ha E.J., Lwin C.T., Durrant J.D. LigGrep: a tool for filtering docked poses to improve virtual-screening hit rates. J. Cheminf. 2020;12:69. doi: 10.1186/s13321-020-00471-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X., Yan X., Yang Y., Gu Q., Zhou H., Du Y., Lu Y., Liao J., Xu J. LSA: a local-weighted structural alignment tool for pharmaceutical virtual screening. RSC Adv. 2019;9:3912–3917. doi: 10.1039/c8ra08915a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rifaioglu A.S., Nalbat E., Atalay V., Martin M.J., Cetin-Atalay R., Doğan T. DEEPScreen: high performance drug-target interaction prediction with convolutional neural networks using 2-D structural compound representations. Chem. Sci. 2020;11:2531–2557. doi: 10.1039/c9sc03414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen P., Ke Y., Lu Y., Du Y., Li J., Yan H., Zhao H., Zhou Y., Yang Y. DLIGAND2: an improved knowledge-based energy function for protein-ligand interactions using the distance-scaled, finite, ideal-gas reference state. J. Cheminf. 2019;11:52. doi: 10.1186/s13321-019-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rampášek L., Hidru D., Smirnov P., Haibe-Kains B., Goldenberg A. Dr.VAE: improving drug response prediction via modeling of drug perturbation effects. Bioinformatics. 2019;35:3743–3751. doi: 10.1093/bioinformatics/btz158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ibrahim H., El Kerdawy A.M., Abdo A., Sharaf Eldin A. Similarity-based machine learning framework for predicting safety signals of adverse drug–drug interactions. Inform. Med. Unlocked. 2021;26 [Google Scholar]
- 79.Wu Z., Ramsundar B., Feinberg E.N., Gomes J., Geniesse C., Pappu A.S., Leswing K., Pande V. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 2018;9:513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ozturk H., Ozgur A., Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng Q., Dueva E., Cherkasov A., Ester M. Padme: A deep learning-based framework for drug-target interaction prediction. arXiv. 2018 doi: 10.48550/arXiv.1807.09741. Preprint at. [DOI] [Google Scholar]
- 82.Karimi M., Wu D., Wang Z., Shen Y. DeepAffinity: interpretable deep learning of compound-protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35:3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altae-Tran H., Ramsundar B., Pappu A.S., Pande V. Low Data Drug Discovery with One-Shot Learning. ACS Cent. Sci. 2017;3:283–293. doi: 10.1021/acscentsci.6b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee I., Keum J., Nam H. DeepConv-DTI: Prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput. Biol. 2019;15 doi: 10.1371/journal.pcbi.1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wan F., Zhu Y., Hu H., Dai A., Cai X., Chen L., Gong H., Xia T., Yang D., Wang M.W., Zeng J. DeepCPI: A Deep Learning-based Framework for Large-scale in silico Drug Screening. Dev. Reprod. Biol. 2019;17:478–495. doi: 10.1016/j.gpb.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y., Fang J., Huang Y., Guo H., Li L., et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020;11:1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren S., Tao Y., Yu K., Xue Y., Schwartz R., Lu X. De novo Prediction of Cell-Drug Sensitivities Using Deep Learning-based Graph Regularized Matrix Factorization. Pacific Symposium on Biocomputing. Pac. Symp. Biocomput. 2022;27:278–289. [PMC free article] [PubMed] [Google Scholar]
- 88.Sada Del Real K., Rubio A. Discovering the mechanism of action of drugs with a sparse explainable network. EBioMedicine. 2023;95 doi: 10.1016/j.ebiom.2023.104767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arshed M.A., Mumtaz S., Riaz O., Sharif W., Abdullah S. A deep learning framework for multi drug side effects prediction with drug chemical substructure. Int. J. Innov. Sci. Technol. 2022;4:19–31. [Google Scholar]
- 90.Dara S., Dhamercherla S., Jadav S.S., Babu C.M., Ahsan M.J. Machine Learning in Drug Discovery: A Review. Artif. Intell. Rev. 2022;55:1947–1999. doi: 10.1007/s10462-021-10058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Musella S., Verna G., Fasano A., Di Micco S. New Perspectives on Machine Learning in Drug Discovery. Curr. Med. Chem. 2021;28:6704–6728. doi: 10.2174/0929867327666201111144048. [DOI] [PubMed] [Google Scholar]
- 92.Wang Z., Liu K., Li J., Zhu Y., Zhang Y. Archives of computational methods in engineering; 2019. Various Frameworks and Libraries of Machine Learning and Deep Learning: A Survey; pp. 1–24. [Google Scholar]
- 93.Yadav S., Singh A., Singhal R., Yadav J.P. Revolutionizing drug discovery: The impact of artificial intelligence on advancements in pharmacology and the pharmaceutical industry. Intelligent Pharmacy. 2024;2:367–380. [Google Scholar]
- 94.Pun F.W., Ozerov I.V., Zhavoronkov A. AI-powered therapeutic target discovery. Trends Pharmacol. Sci. 2023;44:561–572. doi: 10.1016/j.tips.2023.06.010. [DOI] [PubMed] [Google Scholar]
- 95.Zhang S., Cooper-Knock J., Weimer A.K., Shi M., Moll T., Marshall J.N.G., Harvey C., Nezhad H.G., Franklin J., Souza C.D.S., et al. Genome-wide identification of the genetic basis of amyotrophic lateral sclerosis. Neuron. 2022;110:992–1008.e11. doi: 10.1016/j.neuron.2021.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I., Overington J.P. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 98.Editorial A step along the path towards AlphaFold - 50 years ago. Nature. 2024;628:509. doi: 10.1038/d41586-024-01094-5. [DOI] [PubMed] [Google Scholar]
- 99.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., Ronneberger O., Willmore L., Ballard A.J., Bambrick J., et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500. doi: 10.1038/s41586-024-07487-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qi X., Zhao Y., Qi Z., Hou S., Chen J. Machine Learning Empowering Drug Discovery: Applications, Opportunities and Challenges. Molecules. 2024;29 doi: 10.3390/molecules29040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Joshi R.P., Kumar N. Artificial Intelligence for Autonomous Molecular Design: A Perspective. Molecules. 2021;26:6761. doi: 10.3390/molecules26226761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen B., Pan Z., Mou M., Zhou Y., Fu W. Is fragment-based graph a better graph-based molecular representation for drug design? A comparison study of graph-based models. Comput. Biol. Med. 2024;169 doi: 10.1016/j.compbiomed.2023.107811. [DOI] [PubMed] [Google Scholar]
- 104.Guzman-Pando A., Ramirez-Alonso G., Arzate-Quintana C., Camarillo-Cisneros J. Deep learning algorithms applied to computational chemistry. Mol. Divers. 2023 doi: 10.1007/s11030-023-10771-y. [DOI] [PubMed] [Google Scholar]
- 105.Ropp P.J., Spiegel J.O., Walker J.L., Green H., Morales G.A., Milliken K.A., Ringe J.J., Durrant J.D. Gypsum-DL: an open-source program for preparing small-molecule libraries for structure-based virtual screening. J. Cheminf. 2019;11:34. doi: 10.1186/s13321-019-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ropp P.J., Kaminsky J.C., Yablonski S., Durrant J.D. Dimorphite-DL: an open-source program for enumerating the ionization states of drug-like small molecules. J. Cheminf. 2019;11:14. doi: 10.1186/s13321-019-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ivanenkov Y.A., Polykovskiy D., Bezrukov D., Zagribelnyy B., Aladinskiy V., Kamya P., Aliper A., Ren F., Zhavoronkov A. Chemistry42: An AI-Driven Platform for Molecular Design and Optimization. J. Chem. Inf. Model. 2023;63:695–701. doi: 10.1021/acs.jcim.2c01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta R., Srivastava D., Sahu M., Tiwari S., Ambasta R.K., Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol. Divers. 2021;25:1315–1360. doi: 10.1007/s11030-021-10217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Swanson K., Walther P., Leitz J., Mukherjee S., Wu J.C., Shivnaraine R.V., Zou J. ADMET-AI: a machine learning ADMET platform for evaluation of large-scale chemical libraries. Bioinformatics. 2024;40 doi: 10.1093/bioinformatics/btae416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fralish Z., Chen A., Skaluba P., Reker D. DeepDelta: predicting ADMET improvements of molecular derivatives with deep learning. J. Cheminf. 2023;15:101. doi: 10.1186/s13321-023-00769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nixon A.E., Sexton D.J., Ladner R.C. Drugs derived from phage display: from candidate identification to clinical practice. mAbs. 2014;6:73–85. doi: 10.4161/mabs.27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hamdi A., Colas P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol. Sci. 2012;33:109–118. doi: 10.1016/j.tips.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 113.Chen R., Liu X., Jin S., Lin J., Liu J. Machine Learning for Drug-Target Interaction Prediction. Molecules. 2018;23:2208. doi: 10.3390/molecules23092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu L., Ru X., Song R. Application of Machine Learning for Drug-Target Interaction Prediction. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bagherian M., Sabeti E., Wang K., Sartor M.A., Nikolovska-Coleska Z., Najarian K. Machine learning approaches and databases for prediction of drug-target interaction: a survey paper. Briefings Bioinf. 2021;22:247–269. doi: 10.1093/bib/bbz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Z., Zhong W., Zhao L., Chen C.Y.C. ML-DTI: Mutual Learning Mechanism for Interpretable Drug-Target Interaction Prediction. J. Phys. Chem. Lett. 2021;12:4247–4261. doi: 10.1021/acs.jpclett.1c00867. [DOI] [PubMed] [Google Scholar]
- 117.Rayhan F., Ahmed S., Mousavian Z., Farid D.M., Shatabda S. FRnet-DTI: Deep convolutional neural network for drug-target interaction prediction. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou Y., Jiang Y., Yang Y. AGAT-PPIS: a novel protein-protein interaction site predictor based on augmented graph attention network with initial residual and identity mapping. Briefings Bioinf. 2023;24 doi: 10.1093/bib/bbad122. [DOI] [PubMed] [Google Scholar]
- 119.Qian Y., Li X., Wu J., Zhang Q. MCL-DTI: using drug multimodal information and bi-directional cross-attention learning method for predicting drug-target interaction. BMC Bioinf. 2023;24:323. doi: 10.1186/s12859-023-05447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yuan Y., Zhang Y., Meng X., Liu Z., Wang B., Miao R., Zhang R., Su W., Liu L. EDC-DTI: An end-to-end deep collaborative learning model based on multiple information for drug-target interactions prediction. J. Mol. Graph. Model. 2023;122 doi: 10.1016/j.jmgm.2023.108498. [DOI] [PubMed] [Google Scholar]
- 121.Li T., Zhao X.M., Li L. Co-VAE: Drug-Target Binding Affinity Prediction by Co-Regularized Variational Autoencoders. IEEE Trans. Pattern Anal. Mach. Intell. 2022;44:8861–8873. doi: 10.1109/TPAMI.2021.3120428. [DOI] [PubMed] [Google Scholar]
- 122.Öztürk H., Ozkirimli E., Özgür A. WideDTA: prediction of drug-target binding affinity. arXiv. 2019 doi: 10.48550/arXiv.1902.04166. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thafar M.A., Alshahrani M., Albaradei S., Gojobori T., Essack M., Gao X. Affinity2Vec: drug-target binding affinity prediction through representation learning, graph mining, and machine learning. Sci. Rep. 2022;12:4751. doi: 10.1038/s41598-022-08787-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang J., Xiao Y., Shang X., Peng J. Predicting drug-target binding affinity with cross-scale graph contrastive learning. Briefings Bioinf. 2023;25 doi: 10.1093/bib/bbad516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mouchlis V.D., Afantitis A., Serra A., Fratello M., Papadiamantis A.G., Aidinis V., Lynch I., Greco D., Melagraki G. Advances in de Novo Drug Design: From Conventional to Machine Learning Methods. Int. J. Mol. Sci. 2021;22:1676. doi: 10.3390/ijms22041676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tang Y., Moretti R., Meiler J. Recent Advances in Automated Structure-Based De Novo Drug Design. J. Chem. Inf. Model. 2024;64:1794–1805. doi: 10.1021/acs.jcim.4c00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li Y., Zhang L., Liu Z. Multi-objective de novo drug design with conditional graph generative model. J. Cheminf. 2018;10:33. doi: 10.1186/s13321-018-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mercado R., Rastemo T., Lindelöf E., Klambauer G., Engkvist O., Chen H., Jannik Bjerrum E. Graph networks for molecular design. Mach. Learn, Sci. Technol. 2021;2 [Google Scholar]
- 129.Gomez-Bombarelli R., Wei J.N., Duvenaud D., Hernandez-Lobato J.M., Sanchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A., et al. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kadurin A., Nikolenko S., Khrabrov K., Aliper A., Zhavoronkov A. druGAN: An Advanced Generative Adversarial Autoencoder Model for de Novo Generation of New Molecules with Desired Molecular Properties in Silico. Mol. Pharm. 2017;14:3098–3104. doi: 10.1021/acs.molpharmaceut.7b00346. [DOI] [PubMed] [Google Scholar]
- 132.Born J., Manica M., Oskooei A., Cadow J., Markert G., Rodríguez Martínez M. PaccMann(RL): De novo generation of hit-like anticancer molecules from transcriptomic data via reinforcement learning. iScience. 2021;24 doi: 10.1016/j.isci.2021.102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Macedo B., Ribeiro Vaz I., Taveira Gomes T. MedGAN: optimized generative adversarial network with graph convolutional networks for novel molecule design. Sci. Rep. 2024;14:1212. doi: 10.1038/s41598-023-50834-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Prentis L.E., Singleton C.D., Bickel J.D., Allen W.J., Rizzo R.C. A molecular evolution algorithm for ligand design in DOCK. J. Comput. Chem. 2022;43:1942–1963. doi: 10.1002/jcc.26993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chu Y., He X. MoleGear: A Java-Based Platform for Evolutionary De Novo Molecular Design. Molecules. 2019;24:1444. doi: 10.3390/molecules24071444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu C., Liu S., Shi W., Yu J., Zhou Z., Zhang X., Lu X., Cai F., Xia N., Wang Y. Systemic evolutionary chemical space exploration for drug discovery. J. Cheminf. 2022;14:19. doi: 10.1186/s13321-022-00598-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guengerich F.P. Mechanisms of drug toxicity and relevance to pharmaceutical development. Drug Metabol. Pharmacokinet. 2011;26:3–14. doi: 10.2133/dmpk.dmpk-10-rv-062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pognan F., Beilmann M., Boonen H.C.M., Czich A., Dear G., Hewitt P., Mow T., Oinonen T., Roth A., Steger-Hartmann T., et al. The evolving role of investigative toxicology in the pharmaceutical industry. Nat. Rev. Drug Discov. 2023;22:317–335. doi: 10.1038/s41573-022-00633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mayr A., Klambauer G., Unterthiner T., Hochreiter S. DeepTox: toxicity prediction using deep learning. Front. Environ. Sci. 2016;3:80. [Google Scholar]
- 140.Myung Y., de Sá A.G.C., Ascher D.B. Deep-PK: deep learning for small molecule pharmacokinetic and toxicity prediction. Nucleic Acids Res. 2024;52:W469–W475. doi: 10.1093/nar/gkae254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y., Xing J., Xu Y., Zhou N., Peng J., Xiong Z., Liu X., Luo X., Luo C., Chen K., et al. In silico ADME/T modelling for rational drug design. Q. Rev. Biophys. 2015;48:488–515. doi: 10.1017/S0033583515000190. [DOI] [PubMed] [Google Scholar]
- 142.Das P., Gumma S.R., Nayak A., Menghani S., Mandhadi J.R., Prabhu P.P. A Rational Approach To Antitubercular Drug Design: Molecular Docking, Prediction of ADME Properties and Evaluation of Antitubercular Activity of Novel Isonicotinamide Scaffold. Recent Adv. Antiinfect. Drug Discov. 2024;19:148–158. doi: 10.2174/2772434418666230710142852. [DOI] [PubMed] [Google Scholar]
- 143.van de Waterbeemd H., Gifford E. ADMET in silico modelling: towards prediction paradise? Nat. Rev. Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 144.Yi J.C., Yang Z.Y., Zhao W.T., Yang Z.J., Zhang X.C., Wu C.K., Lu A.P., Cao D.S. ChemMORT: an automatic ADMET optimization platform using deep learning and multi-objective particle swarm optimization. Briefings Bioinf. 2024;25 doi: 10.1093/bib/bbae008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gu Y., Yu Z., Wang Y., Chen L., Lou C., Yang C., Li W., Liu G., Tang Y. admetSAR3.0: a comprehensive platform for exploration, prediction and optimization of chemical ADMET properties. Nucleic Acids Res. 2024;52:W432–W438. doi: 10.1093/nar/gkae298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yi J., Shi S., Fu L., Yang Z., Nie P., Lu A., Wu C., Deng Y., Hsieh C., Zeng X., et al. OptADMET: a web-based tool for substructure modifications to improve ADMET properties of lead compounds. Nat. Protoc. 2024;19:1105–1121. doi: 10.1038/s41596-023-00942-4. [DOI] [PubMed] [Google Scholar]
- 147.Lim S., Lee S., Piao Y., Choi M., Bang D., Gu J., Kim S. On modeling and utilizing chemical compound information with deep learning technologies: A task-oriented approach. Comput. Struct. Biotechnol. J. 2022;20:4288–4304. doi: 10.1016/j.csbj.2022.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ye L., Ngan D.K., Xu T., Liu Z., Zhao J., Sakamuru S., Zhang L., Zhao T., Xia M., Simeonov A., Huang R. Prediction of drug-induced liver injury and cardiotoxicity using chemical structure and in vitro assay data. Toxicol. Appl. Pharmacol. 2022;454 doi: 10.1016/j.taap.2022.116250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gayvert K.M., Madhukar N.S., Elemento O. A Data-Driven Approach to Predicting Successes and Failures of Clinical Trials. Cell Chem. Biol. 2016;23:1294–1301. doi: 10.1016/j.chembiol.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Richard A.M., Judson R.S., Houck K.A., Grulke C.M., Volarath P., Thillainadarajah I., Yang C., Rathman J., Martin M.T., Wambaugh J.F., et al. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 2016;29:1225–1251. doi: 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- 151.Huang R., Xia M., Nguyen D.T., Zhao T., Sakamuru S., Zhao J., Shahane S.A., Rossoshek A., Simeonov A. Tox21Challenge to build predictive models of nuclear receptor and stress response pathways as mediated by exposure to environmental chemicals and drugs. Front. Environ. Sci. 2016;3:85. [Google Scholar]
- 152.Wang X., Li Z., Jiang M., Wang S., Zhang S., Wei Z. Molecule Property Prediction Based on Spatial Graph Embedding. J. Chem. Inf. Model. 2019;59:3817–3828. doi: 10.1021/acs.jcim.9b00410. [DOI] [PubMed] [Google Scholar]
- 153.Delaney J.S. ESOL: estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Comput. Sci. 2004;44:1000–1005. doi: 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- 154.Mobley D.L., Guthrie J.P. FreeSolv: a database of experimental and calculated hydration free energies, with input files. J. Comput. Aided Mol. Des. 2014;28:711–720. doi: 10.1007/s10822-014-9747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arnott J.A., Planey S.L. The influence of lipophilicity in drug discovery and design. Expet Opin. Drug Discov. 2012;7:863–875. doi: 10.1517/17460441.2012.714363. [DOI] [PubMed] [Google Scholar]
- 156.Waring M.J. Lipophilicity in drug discovery. Expet Opin. Drug Discov. 2010;5:235–248. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- 157.Prueksaritanont T., Chu X., Gibson C., Cui D., Yee K.L., Ballard J., Cabalu T., Hochman J. Drug-drug interaction studies: regulatory guidance and an industry perspective. AAPS J. 2013;15:629–645. doi: 10.1208/s12248-013-9470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kusuhara H. How far should we go? Perspective of drug-drug interaction studies in drug development. Drug Metabol. Pharmacokinet. 2014;29:227–228. doi: 10.2133/dmpk.dmpk-14-pf-903. [DOI] [PubMed] [Google Scholar]
- 159.Chakraborty S., Chopra H., Akash S., Chakraborty C., Dhama K. Artificial intelligence (AI) is paving the way for a critical role in drug discovery, drug design, and studying drug-drug interactions - correspondence. Int. J. Surg. 2023;109:3242–3244. doi: 10.1097/JS9.0000000000000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Percha B., Altman R.B. Informatics confronts drug-drug interactions. Trends Pharmacol. Sci. 2013;34:178–184. doi: 10.1016/j.tips.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu S., Zhang Y., Cui Y., Qiu Y., Deng Y., Zhang Z., Zhang W. Enhancing Drug-Drug Interaction Prediction Using Deep Attention Neural Networks. IEEE ACM Trans. Comput. Biol. Bioinf. 2023;20:976–985. doi: 10.1109/TCBB.2022.3172421. [DOI] [PubMed] [Google Scholar]
- 162.Chen Y., Ma T., Yang X., Wang J., Song B., Zeng X. MUFFIN: multi-scale feature fusion for drug-drug interaction prediction. Bioinformatics. 2021;37:2651–2658. doi: 10.1093/bioinformatics/btab169. [DOI] [PubMed] [Google Scholar]
- 163.Schwarz K., Allam A., Perez Gonzalez N.A., Krauthammer M. AttentionDDI: Siamese attention-based deep learning method for drug-drug interaction predictions. BMC Bioinf. 2021;22:412. doi: 10.1186/s12859-021-04325-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pham T., Ghafoor M., Grañana-Castillo S., Marzolini C., Gibbons S., Khoo S., Chiong J., Wang D., Siccardi M. DeepARV: ensemble deep learning to predict drug-drug interaction of clinical relevance with antiretroviral therapy. NPJ Syst. Biol. Appl. 2024;10:48. doi: 10.1038/s41540-024-00374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rohani N., Eslahchi C. Drug-Drug Interaction Predicting by Neural Network Using Integrated Similarity. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu J., Che C., Jiang H., Xu J., Yin J., Zhong Z. SSF-DDI: a deep learning method utilizing drug sequence and substructure features for drug-drug interaction prediction. BMC Bioinf. 2024;25:39. doi: 10.1186/s12859-024-05654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chakraborty C., Pal S., Bhattacharya M., Dash S., Lee S.S. Overview of Chatbots with special emphasis on artificial intelligence-enabled ChatGPT in medical science. Front. Artif. Intell. 2023;6 doi: 10.3389/frai.2023.1237704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bhattacharya M., Pal S., Chatterjee S., Alshammari A., Albekairi T.H., Jagga S., Ige Ohimain E., Zayed H., Byrareddy S.N., Lee S.S., et al. ChatGPT’s scorecard after the performance in a series of tests conducted at the multi-country level: A pattern of responses of generative artificial intelligence or large language models. Current Research in Biotechnology. 2024;7 [Google Scholar]
- 169.Liang Y., Zhang R., Zhang L., Xie P. Drugchat: towards enabling chatgpt-like capabilities on drug molecule graphs. arXiv. 2023 doi: 10.48550/arXiv.2309.03907. Preprint at. [DOI] [Google Scholar]
- 170.Pal S., Bhattacharya M., Islam M.A., Chakraborty C. ChatGPT or LLM in next-generation drug discovery and development: pharmaceutical and biotechnology companies can make use of the artificial intelligence-based device for a faster way of drug discovery and development. Int. J. Surg. 2023;109:4382–4384. doi: 10.1097/JS9.0000000000000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Arnold C. Inside the nascent industry of AI-designed drugs. Nat. Med. 2023;29:1292–1295. doi: 10.1038/s41591-023-02361-0. [DOI] [PubMed] [Google Scholar]
- 172.Burki T. A new paradigm for drug development. Lancet. Digit. Health. 2020;2:e226–e227. doi: 10.1016/S2589-7500(20)30088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lowe D. AI-Generated Clinical Candidates, So Far. the Pipeline blog. 2021 www.science.org/content/blog-post/ai-generated-clinical-candidates-so-far [Google Scholar]
- 174.Ren F., Aliper A., Chen J., Zhao H., Rao S., Kuppe C., Ozerov I.V., Zhang M., Witte K., Kruse C., et al. A small-molecule TNIK inhibitor targets fibrosis in preclinical and clinical models. Nat. Biotechnol. 2024 doi: 10.1038/s41587-024-02143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cheng F., Ma Y., Uzzi B., Loscalzo J. Importance of scientific collaboration in contemporary drug discovery and development: a detailed network analysis. BMC Biol. 2020;18:138. doi: 10.1186/s12915-020-00868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takebe T., Imai R., Ono S. The Current Status of Drug Discovery and Development as Originated in United States Academia: The Influence of Industrial and Academic Collaboration on Drug Discovery and Development. Clin. Transl. Sci. 2018;11:597–606. doi: 10.1111/cts.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singh S.B., Martin G.E., McKittrick B., Crowther J., Fraenkel H., Lunn C., Bayne M., Perkins J.B., Gullo V. History and Prospects of Drug Discovery and Development Collaboration between Industry and Academia. J. Nat. Prod. 2024;87:1235–1245. doi: 10.1021/acs.jnatprod.4c00081. [DOI] [PubMed] [Google Scholar]
- 178.Ferrins L., Pollastri M.P. The Importance of Collaboration between Industry, Academics, and Nonprofits in Tropical Disease Drug Discovery. ACS Infect. Dis. 2018;4:445–448. doi: 10.1021/acsinfecdis.7b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pollastri M.P. Finding new collaboration models for enabling neglected tropical disease drug discovery. PLoS Neglected Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Litterman N.K., Rhee M., Swinney D.C., Ekins S. Collaboration for rare disease drug discovery research. F1000Res. 2014;3:261. doi: 10.12688/f1000research.5564.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Druedahl L.C., Minssen T., Price W.N. Collaboration in times of crisis: A study on COVID-19 vaccine R&D partnerships. Vaccine. 2021;39:6291–6295. doi: 10.1016/j.vaccine.2021.08.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chakraborty C., Sharma A.R., Bhattacharya M., Agoramoorthy G., Lee S.S. Asian-Origin Approved COVID-19 Vaccines and Current Status of COVID-19 Vaccination Program in Asia: A Critical Analysis. Vaccines. 2021;9:600. doi: 10.3390/vaccines9060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chakraborty C., Sharma A.R., Sharma G., Bhattacharya M., Saha R.P., Lee S.S. Extensive Partnership, Collaboration, and Teamwork is Required to Stop the COVID-19 Outbreak. Arch. Med. Res. 2020;51:728–730. doi: 10.1016/j.arcmed.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhou Q. International collaboration for global accessibility of COVID-19 vaccines. Natl. Sci. Rev. 2020;7:1269. doi: 10.1093/nsr/nwaa147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jiménez-Luna J., Grisoni F., Schneider G. Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2020;2:573–584. [Google Scholar]
- 186.Linardatos P., Papastefanopoulos V., Kotsiantis S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy. 2020;23:18. doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang Y., Liu C., Liu M., Liu T., Lin H., Huang C.B., Ning L. Attention is all you need: utilizing attention in AI-enabled drug discovery. Briefings Bioinf. 2023;25 doi: 10.1093/bib/bbad467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Savage N. Breaking into the black box of artificial intelligence. Nature. 2022 doi: 10.1038/d41586-022-00858-1. [DOI] [PubMed] [Google Scholar]
- 189.Chakraborty C., Bhattacharya M., Islam M.A., Agoramoorthy G. ChatGPT indicates the path and initiates the research to open up the black box of artificial intelligence. Int. J. Surg. 2023;109:4367–4368. doi: 10.1097/JS9.0000000000000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Suzuki J., Kazawa H., Zen H. Extracting representative subset from extensive text data for training pre-trained language models. Inf. Process. Manag. 2023;60 [Google Scholar]
- 191.Gupta N.S., Kumar P. Perspective of artificial intelligence in healthcare data management: A journey towards precision medicine. Comput. Biol. Med. 2023;162 doi: 10.1016/j.compbiomed.2023.107051. [DOI] [PubMed] [Google Scholar]
- 192.Ghim J.L., Ahn S. Transforming clinical trials: the emerging roles of large language models. Transl. Clin. Pharmacol. 2023;31:131–138. doi: 10.12793/tcp.2023.31.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]