Abstract
Radiation therapy (RT) is a common treatment for lung cancer. Still, it can lead to irreversible loss of pulmonary function and a significant reduction in quality of life for one-third of patients. Preexisting comorbidities, such as chronic obstructive pulmonary disease (COPD), are frequent in patients with lung cancer and further increase the risk of complications. Because lung stem cells are crucial for the regeneration of lung tissue following injury, we hypothesized that airway stem cells from patients with COPD with lung cancer might contribute to increased radiation sensitivity. We used the air-liquid interface model, a three-dimensional (3D) culture system, to compare the radiation response of primary human airway stem cells from healthy and patients with COPD. We found that COPD-derived airway stem cells, compared to healthy airway stem cell cultures, exhibited disproportionate pathological mucociliary differentiation, aberrant cell cycle checkpoints, residual DNA damage, reduced survival of stem cells and self-renewal, and terminally differentiated cells post-irradiation, which could be reversed by blocking the Notch pathway using small-molecule γ-secretase inhibitors. Our findings shed light on the mechanisms underlying the increased radiation sensitivity of COPD and suggest that airway stem cells reflect part of the pathological remodeling seen in lung tissue from patients with lung cancer receiving thoracic RT.
Keywords: COPD, radiation therapy, lung, basal airway stem cells, DNA damage response, NOTCH pathway
Graphical Abstract
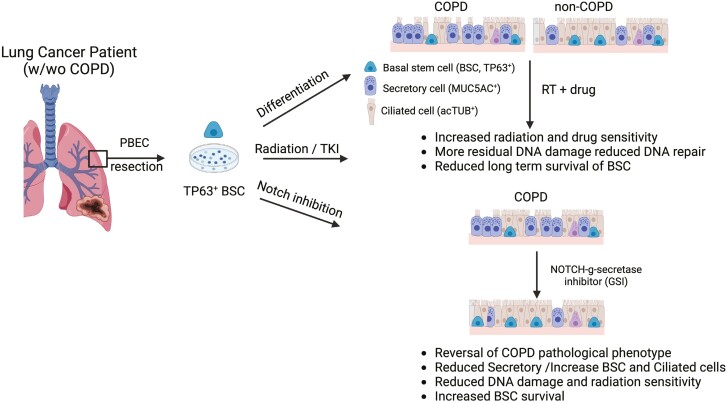
Graphical Abstract.
Significance statement.
Radiation pneumonitis and fibrosis are severe side effects of lung cancer treatment, exacerbated by common preexisting conditions such as chronic obstructive pulmonary disease (COPD). Existing research mostly focuses on healthy lung tissue or animal models. Using primary human patient tissue ex vivo, our study uncovers some underlying mechanisms of heightened radiation sensitivity of COPD-derived patient lung cells. Notably, the Notch stem cell pathway plays a crucial role. These findings offer potential for new interventions to alleviate cancer treatment side effects and improve the survival of patients with lung cancer.
Introduction
The main limitation of cancer radiotherapy is the irreversible damage it causes to normal tissues within the radiation field.1 In patients with lung cancer, radiation treatment can lead to radiation-induced lung injury, a dose-limiting complication resulting in life-threatening loss-of-lung function. Consequently, normal tissue tolerance often restricts tumor control. Additionally, radiation therapy (RT) induces DNA damage and increases the production of reactive oxygen species (ROS), leading to cell death, vascular permeability, and the recruitment of inflammatory mediators that trigger radiation pneumonitis (RP).2,3 Prolonged inflammation leads to irreversible remodeling of lung tissue, culminating in radiation-induced fibrosis and a progressive reduction in functional lung volume, which can be fatal.4 Furthermore, combined radiotherapy, chemotherapy, immunotherapy, and targeted agents exacerbate radiation-induced lung toxicity.5-7
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide, characterized by chronic airflow obstruction and impaired gas exchange, which progressively declines lung functioning.8 COPD is a comorbidity in 50%-70% of patients with lung cancer and a risk factor.9 Oxidative stress and chronic inflammation in the lung tissue are major factors linking COPD and lung cancer.9,10 Patients with COPD receiving RT are more susceptible to dose-limiting toxicities such as RP and experience worse outcomes.11,12
The upper airway is a pseudostratified epithelium with a basal layer containing the basal stem cells (BSC) and a luminal layer with the differentiated ciliated and secretory cells.13 The integrity of the lung epithelium is crucial for preventing infections by clearing inhaled pathogens. Airway BSC proliferates upon injury to repair the damage.14,15 A hallmark of COPD pathogenesis is a perturbed lung stem cell homeostasis resulting in goblet cell metaplasia, increased mucous secretion, and decreased mucous clearance.16,17 Detailed studies on COPD airway stem cells’ self-renewal, differentiation capacity, and injury response are limited.
We, and others, previously showed that irradiation induces a dose-dependent decrease of BSC with reduced long-term survival and differentiation.18-20
Modulating the stem cell signaling pathways may be important for preventing or repairing radiation-induced lung injury.21 For instance, the Notch signaling pathway is a crucial regulator of BSC self-renewal, inducing and maintaining secretory cell fates in the bronchial epithelium.22,23 Earlier work showed that blocking Notch signaling can protect against radiation-induced loss of airway stem cells and epithelial barrier integrity in the human cultured bronchial epithelium.24 Cigarette smoke and other agents strongly associated with COPD trigger an enhanced and abnormal inflammatory response.25 Cigarette smoke activates NOTCH, leading to an increase in mucous-producing secretory cells. Furthermore, it triggers airway epithelial cells to produce pro-inflammatory mediators and induces the epithelial-to-mesenchymal transition. By blocking NOTCH, these effects can be reversed.26-28 Conversely, ROS produced by inflammatory conditions may activate Notch.29 The exact role of the NOTCH pathway on COPD response to radiation-induced damage, however, remains unclear.
Currently, there are no predictive biomarkers for identifying patients at risk for developing lung toxicities due to RT. This study investigates whether aberrant stem cell function and response to radiation contribute to patients’ with COPD increased sensitivity to RP. Primary human bronchial epithelial 3D cell models were used to compare radiation sensitivity, DNA damage response, and differentiation capacity between healthy and patients with COPD. The results demonstrate that COPD airway stem cells have defects contributing to radiation sensitivity and histopathological consequences. Our results show that stem cell therapies could be a potential therapeutic approach to mitigate radiation-induced lung injury in patients with cancer.
Methods
Primary human bronchial epithelial cells
PBECs were kindly provided by the primary lung culture (PLUC) facility MUMC+, Maastricht, The Netherlands. Lung tissue used for the isolation of PBECs was obtained from the Maastricht Pathology Tissue Collection (MPTC) and originated from tissue resected during lobectomies or pneumonectomies (Patient#107, 108, 115, 123, 126, 127, 133, 139, 158, 159, and 161) of patients who underwent surgery for lung cancer. Collection, storage, and use of tissue and patient data were performed in agreement with the “Code for Proper Secondary Use of Human Tissue in the Netherlands” (http://www.fmwv.nl). The scientific board of the MPTC approved the use of materials for this study under MPTC2010-019. In addition, formal permission was obtained from the local Medical Ethical Committee (METC) code 2017-0087, and patients have provided written consent to use the material for research. PneumaCult-ex basal medium supplemented Pneumacult-ex 50x supplement (#05008 STEMCELL Technologies), 96 ng/mL hydrocortisone (#07925 STEMCELL Technologies), and 1% penicillin/streptomycin (L0022 VWR) were used as culture media before seeding the cells, the flasks were coated with 10 μg/mL fibronectin (ref 354008 corning), 10 μg/mL bovine serum albumin (8076.5 Carl Roth), and 30 μg/mL Collagen (11563550 Thermo Fisher Scientific). After trypsinization, the cells were centrifuged for 5 minutes at 150 g, collected, and finally counted using an automatic counter (Beckman Coulter).
Irradiation of cells
PBECs were irradiated with 2 and 4 Gy using the X-ray cabinet (Philips 225 kV, 10 mA or PXI, Multirad 225 kV, 17.8 mA). For the stem cell regeneration and differentiation capacity, the stem cells were irradiated in 2D cultures and, immediately after irradiation, were seeded in air-liquid interface (ALI). The samples were collected 24 hours, 7, 15, or 21 days after airlift and processed for immunofluorescence (IF). Additionally, for DNA damage, stem cell, and ciliated cell assays, fully differentiated 3D ALI cultures were irradiated on day 21 post-airlift, and the samples were collected 24 hours after irradiation.
Air-liquid interface culture
Non-COPD(N) and COPD(C) primary bronchial epithelial cells (PBEC; 6 × 104 to 9 × 104) cells were seeded in 500 µL of K-SFM plus complement medium on top of 0.4 µm pore polyester membrane inserts of an ALI transwell system (Corning). After reaching confluence, approximately 3-5 days post-seeding, the PBECs were subjected to airlift with medium supplementation only in the bottom well. Once in the ALI system, the cells were grown in Pneumocult ALI-basal medium (STEMCELL Technologies) complemented with Hydrocortisone (STEMCELL Technologies), Heparin (STEMCELL Technologies), and Pneumacult 100X supplement (STEMCELL Technologies) up to 28 days. The PBECs were cultured in Pneumocult ALI medium plus complement at 37 °C in a 5% CO2 humidified incubator and replaced every 2 days until sample collection, essentially as described.24
Immunostaining and confocal microscopy
PBECs were fixed in 4% paraformaldehyde for 10 minutes. Cells were permeabilized with PBS with 0.3% Triton-X-100 for 2.5 hours. Samples were blocked with a solution of PBS-0.1% Triton, 3% bovine serum albumin, and 10% normal goat serum for 2 hours. The samples were incubated overnight at 4 °C with the primary antibodies dissolved in blocking solution (MUC5 (1:1000; Cat #ab3649; Abcam), Ac-TUB (1:1000; Cat #T7451-200UL; Sigma-Aldrich), TP63 (1:1000; Cat #ab124762; Abcam), CK5 (1:1000; Lot# B241498; Biolegend), and 53BP1 (1:750; Lot #612522; BD Biosciences). The washes were performed with PBS-0.1% Triton, 10 minutes each, 3 times. The samples were incubated with secondary fluorescent antibodies goat anti-mouse Alexa 488 (1:500; Thermo Fisher Scientific) and goat anti-rabbit Alexa Fluor 555 (1:500; Thermo Fisher Scientific) for 1 hour at room temperature in the dark. The fluorescent protein 4ʹ,6-diamidino-2-fenylindool (DAPI; 1:5000; Thermo Fisher Scientific) was used for 15 minutes to detect the nuclei. The samples were then mounted in DAKO fluorescent mounting medium (Lot #11246755; Dako) and stored in the dark at 4 °C until usage. A confocal microscope (Leica TCS) was used to obtain stacks of 2 µm of each sample.
RAD51 staining and quantification
Just before fixation, cells were washed with cold PBS, incubated with ice-cold 0.5% Triton-X-100 extraction buffer (0.5% Triton-X-100, 20 mM HEPES-KOH (pH 7.9), 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 1 minute, washed with cold PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 15 minutes at room temperature. Cells were washed twice with 0.1% Triton and subsequently blocked with PBS + buffer (5 mg Bovine Serum Albumine and 1.5 mg glycine/mL PBS). Cells were incubated overnight at 4 °C with the primary antibodies dissolved in PBS + buffer (Rad51; in-house; rabbit; 1:10 000). Cells were permeabilized with 0.1% Triton and washed with PBS + buffer. Cells were incubated with secondary antibodies dissolved in PBS + buffer (anti-rabbit Alexa594; Life Technologies) for 1 hour at room temperature in the dark. Subsequently, the coverslips were mounted on microscope slides using an Antifade mounting medium with DAPI (Vectashield). The coverslips were sealed with nail polish to prevent the samples from drying out. A Leica STELLARIS 5 confocal microscope was used to visualize immunofluorescence in cells. The following laser lines were used: DAPI (405 laser), Alexa 594 (561 laser). For each sample, 8 Z-stack images were captured using a 40× objective. Using these images, Rad51 foci number was analyzed for each nucleus. This was accomplished using homemade FIJIImageJ scripts. In short, cell nuclei were segmented based on the DAPI signal employing a sequential process of thresholding, the watershed Plug-In, and the “analyze particles” function in FIJI. For the identification of foci within the segmented nuclei, individual segmentation masks were created for each nucleus. Segmentation masks of the foci were generated using thresholds based on the mean fluorescence of the RAD51 signal plus 2 times (for control samples) and 1.3 times (for 24-hour samples) the + factor*standard deviation of the Rad51 signal.30 The number of the segmented foci per nucleus were then measured using the ‘analyze particles ‘measurement function within FIJIImageJ.
ALI IF quantification
The cells in these 5 fields were then counted to obtain a total of 500-1000 cells per condition (100-200 cells per image). Stainings with Ac-TUB and MUC5AC were captured using a 63× objective. The Z-stack was used as the image in the paper. ImageJ was used to count the positive cells and the foci in the nucleus. Comparable results were obtained in at least 3 independent COPD and healthy donors. TP63 immunofluorescence staining quantification was achieved by determining the number of positive cell nuclei as a proportion of the total number of nuclei in 3 representative fields of each sample and analyzed using ImageJ win 6.4 (Fiji). Likewise, 53BP1 quantification was achieved by determining the number of positive 53BP1 foci in TP63-positive cells as a proportion of the total number of TP63 cells in 5 representative fields of each sample.
Proliferation of BSC
BSC proliferation was monitored using the automated live cell analysis with the IncuCyte ZOOM (Essen Bioscience). COPD and non-COPD PBECs were seeded at 2000 cells/well in a 96-well plate, 6 replicates per donor, and the proliferation was monitored for 10 days until cells reached confluence. Analysis was performed in GraphPad Prism 10.2.1 using nonlin-fit, exponential growth equation to calculate the doubling time.
Western blotting
Western blotting was performed according to standard protocol. To extract proteins, RIPA buffer (50 mM Tris-HCl, 0.5% DOC, 0.1% SDS, 1 mM EGTA, 2 mM EDTA, 10% Glycerol, 1% Triton X-100, 150 mM NaCl, and 1 mM PMSF) was used. The protein concentrations were determined with Bradford Protein Assay (Bio-Rad). Proteins (40 µg) were separated on a 7.5% or 12% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dry bovine milk and TBS and subsequently incubated (O/N, 4 °C) with primary antibodies (1:1000) and visualized using HRP-linked secondary antibodies (goat anti-mouse and goat anti-rabbit, 1:2500, Cell-Signaling) and Super ECL Luminescence.
RNA expression analysis
Total RNA was isolated from PBEC’s using NucleoSpin RNA (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. One microgram RNA was used as input material to generate cDNA by using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) followed by Reverse transcription quantitative PCR (qRT-PCR) using SensiMix SYBR high-ROX kit (GC Biotech, Waddinxveen, the Netherlands). mRNA expression was analyzed with CFX Connect Real-Time System (Bio-Rad) and HPRT and RPL13A were used as housekeeping genes. Primers used for gene expression for qPCR are in Table 1.
Table 1.
Primers sequences used in gene expression studies.
| Target | Forward primer | Reverse primer |
|---|---|---|
| RPL13A | CCGGGTTGGCTGGAAGTACC | CTTCTCGGCCTGTTTCCGTAG |
| HPRT | TATTGTAATGACCAGTCAACAG | GGTCCTTTTCACCAGCAAG |
| NOTCH1 | AGGACCTCATCAACTCACACGC | TCTTTGTTAGCCCCGTTCTTCAG |
| NOTCH2 | CCGTGTTGACTTCTGCTCTCTCAC | CCTACTACCCTTGGCATCCTTTG |
| NOTCH3 | TC TCAGACTGGTCCGAATCCAC | ACACTTGCCTCTTGGGGGTAAC |
| NOTCH4 | ATGCGAGGAAGATACGGAGTGG | TCGGAATGTTGGAGGCAGAAC |
| HES1 | AGGCGGACATTCTGGAAATG | CGGTACTTCCCCAGCACACTT |
| HES4 | CACCGCAAGTCCTCCAAG | TCACCTCCGCCAGACACT |
| HEY1 | GAAACTTGAGTTCGGCTCTAGG | GCTTAGCAGACTCTTGCTCCAT |
| HEY2 | GGCGTCGGGATCGGATAAATA | AAGTAGCCTTTACCCCCTGTT |
| JAGGED 1 | ATCGTGCTGCCTTTCAGTTT | ACTGTCAGGTTGAACGGTGTC |
| JAGGED 2 | GTCGTCATCCCCTTCCAGT | CTCCTCATTCGGGGTGGTAT |
| DLL1 | CTACTACGGAGAGGGCTGCT | CCAGGGTTGCACACTTTCTC |
Cell cycle analysis
Cell cycle analysis was performed using Click-iT Plus EdU Pacific Blue Flow Cytometry Assay Kit (Thermo Fisher Scientific) and propidium iodide (PI) staining. 5-Ethynyl-2-deoxyuridine (EdU) was incorporated for 4 hours at 37 °C in cell culture conditions. The Click-iT reaction was performed for 30 minutes with a Click-it buffer containing Alexa fluorazide and CuSO4 according to the manufacturer’s protocol (Thermo Fisher Scientific). For PI staining, cells were incubated with 1 µg/mL of PI, 100 µg/mL of RNaseA, and 0.1% Triton X-100 in PBS for 30 minutes at room temperature. Unstained, PI-only, EdU-only, and Click-iT-only samples were used for compensation, background correction, and autofluorescence. A FACS Canto II cytometer with BD FACSDiva 6.1.1 software was used for cell cycle analysis. FlowJo V10.1 was used to exclude doublets and cellular debris and determine cells’ distribution within G0-G1, S, and G2-M phases.
Apoptosis assay
To measure apoptosis, a Pacific Blue Annexin V/SYTOX AADvanced Apoptosis Kit for flow cytometry (Thermo Fisher Scientific) was used according to the manufacturer’s protocol. PBEC cells were treated with etoposide (10 µM) for 24 hours as a positive control (data not shown). The number of cells in early and late apoptosis was analyzed using a FACS Canto II cytometer with BD FACSDiva 6.1.1 software. Using FLowV10.1, doublets and cellular debris were excluded.
Cell viability
Cell viability was measured using trypan blue (cat. no.15250061; Thermo Fisher Scientific) in ratio 1:1 with trypsinized cells. Cell viability was determined as the percentage of living cells 24 hours after 10 μM KU55933 (ATM inhibitor) using an automatic Coulter counter (TC20; Bio-Rad). The percentage of living cells was plotted.
Human bronchial organoids
COPD-derived PBEC cells (5 × 104) were seeded in 100% Cultrex plug (Trevigen) and pipetted in a 24-well plate. Organoid culture medium containing 10% RSPO-conditioned medium, 10% Noggin-conditioned medium, 1× B27 supplement, 1.25 mM N-acetyl-cysteine, 10 mM nicotinamide, 500 nM A83-01, 1 µM SB202190, 25 ng/mL Human FGF-7, 100 ng/mL FGF-10, and 5 µM Y-27632 was then applied over the Cultrex mound and refreshed every other day.31 The organoids were kept in culture for 21 days.
Alkaline comet assay
DNA damage after RT exposure was assessed by using the alkaline comet assay. Briefly, COPD and non-COPD BSCs 24 hours after RT treatment were washed with PBS (pH 7.4 w/o Ca++/Mg++), detached by using trypsinization (150 µL/well for 5 minutes), and suspended in PBS (pH 7.4 w/o Ca++/Mg++). Cells were then centrifuged (300 × g for 5 minutes), the supernatant was discarded, and the pellets were resuspended in ice-cold PBS to reach 106 cells per condition. Next, the cells were mixed, embedded in a 3:7 ratio with a 1% low melting point agarose, and kept at 37 °C to avoid solidification. Next, 2 × 70 µL of each sample was placed on the slides, and a coverslip was put on top. After agarose polymerization (4 °C for 10 minutes), the coverslip was removed. Next, the slides were incubated at 4 °C for 1 hour, protected from light in lysis solution (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris-base pH 10, 10 M of NaOH at pH 10 and 1% Triton-X-100). Subsequently, the slides were immersed in electrophoresis buffer (1 mM of Na2EDTA and 300 mM of NaOH at pH of 12) for 20 minutes at 4 °C in the electrophoresis platform for DNA unwinding. Then, the electrophoresis ran for 20 minutes at a constant ~1 V/cm at 4 °C. At the end of the electrophoresis, the slides were washed with PBS (1 × 10 minutes) and deionized H2O (1 × 10 minutes). Then, the slides were dried overnight and protected from light at room temperature. Finally, all the slides were stained with GelRed (Biotium cat. no. 41003; Merck KGaA, cat. no. SCT123) and analyzed using the Comet III Perceptives image analysis software. The DNA percentages in the comet tails (% tail intensity) were obtained for 100 (2 × 50) cells per experimental condition.
Statistical analysis
GraphPad Prism Software (v7) was used to perform statistical analysis. For all experiments, means ± SE were reported. In all experiments, a minimum of 3 individual donors were used per group (non-COPD and COPD). One-way ANOVA test post hoc Tukey multiple comparisons were used to analyze statistical differences or Mann-Whitney. P < .05 was considered statistically significant.
Results
Lung BSCs from patients with COPD have perturbed mucociliary differentiation
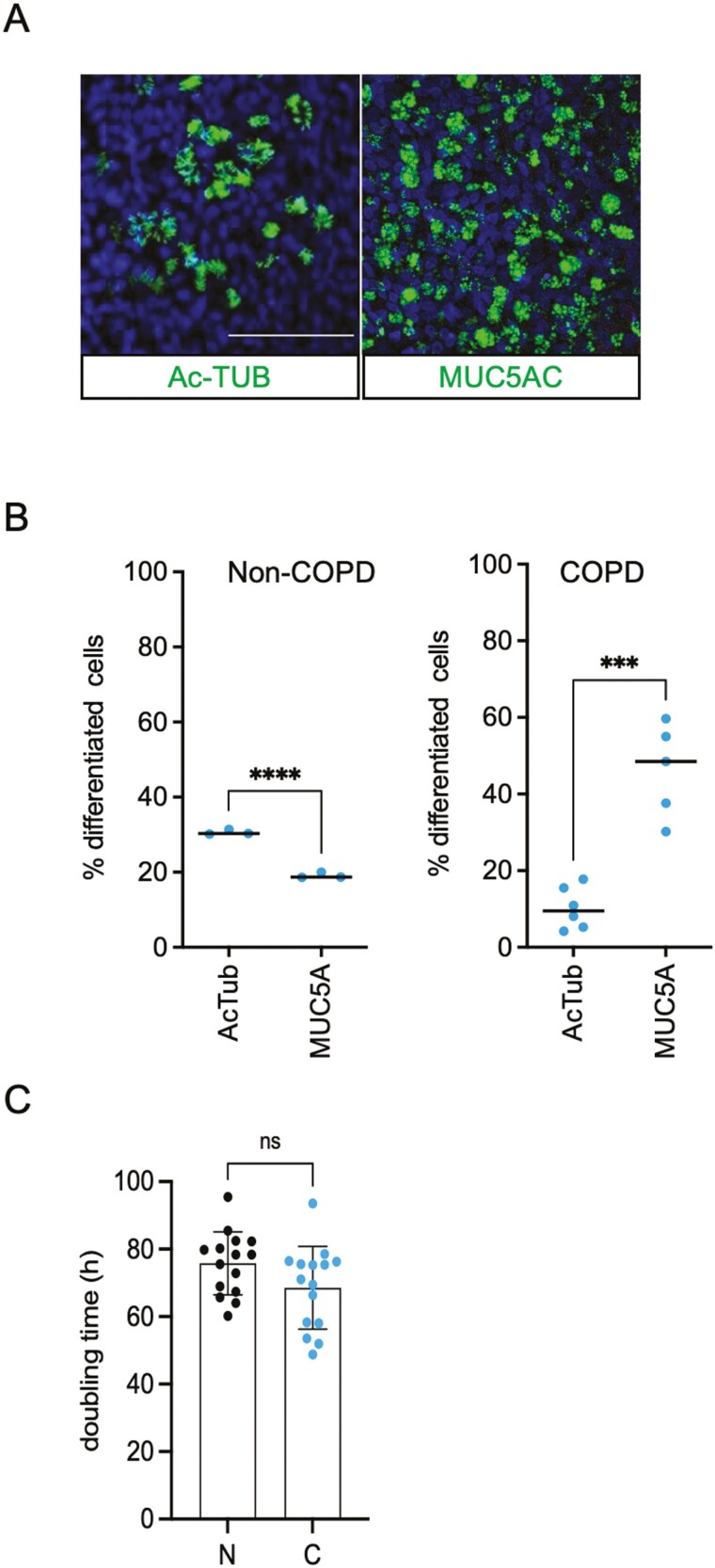
We first sought to investigate if BSCs derived from PBEC from patients with lung cancer with COPD differ in their differentiation capacity from non-affected patient with lung cancer donors. Using 3D ALI culture, we compared the composition and differentiation capacity of non-COPD (N) and COPD (C) derived BSC following earlier methodology.24 Using IF staining, we show that COPD-derived BSC cultures that are induced to differentiate in ALI are composed of 2.4-fold increased mucous cell numbers (MUC5A+) and 30% fewer ciliated cells (Ac-TUB+) compared to PBECs from non-COPD donors (Figure 1A, B). To investigate whether COPD BSCs have impaired proliferation compared to non-COPD BSC, we monitored their growth using live cell imaging. We found that the doubling time measured over 10 days in culture between COPD-derived airway stem cells (TP63+) did not differ significantly from non-COPD BSCs, albeit there were differences among individuals (Figure 1C, Supplementary Figure S1A). We then exposed non-COPD and COPD BSCs to irradiation (2-4 Gy) and characterized their differentiation capacity in ALI. We found an altered ratio of Ac-TUB and MUC5AC expressing cells in both non-COPD and COPD ALIs derived from irradiated BSCs. The impaired differentiation capacity of BSCs is more pronounced in COPD-derived BSCs (Supplementary Figure S1B).
Figure 1.
Growth and differentiation of human COPD BSC. (A) Representative immunofluorescent staining of 3D ALI culture at day 28 from COPD BSC in mucous cells (MUC5AC), ciliated cells (Ac-TUB), and DAPI nuclear stain. (B) Quantification of Ac-TUB+, MUC5AC + cells from non-COPD and COPD ALI cultures as a percentage of total cells. (C) Doubling time of non-COPD and COPD BSC cell cultures with Incucyte cell imaging. N = 3 independent donors non-COPD and COPD per group *P < .05; **P < .01; ***P < .001; ****P < .0001. Scale bar = 50 μm.
COPD BSCs have impaired growth, differentiation, and self-renewal after radiation and TKI treatment
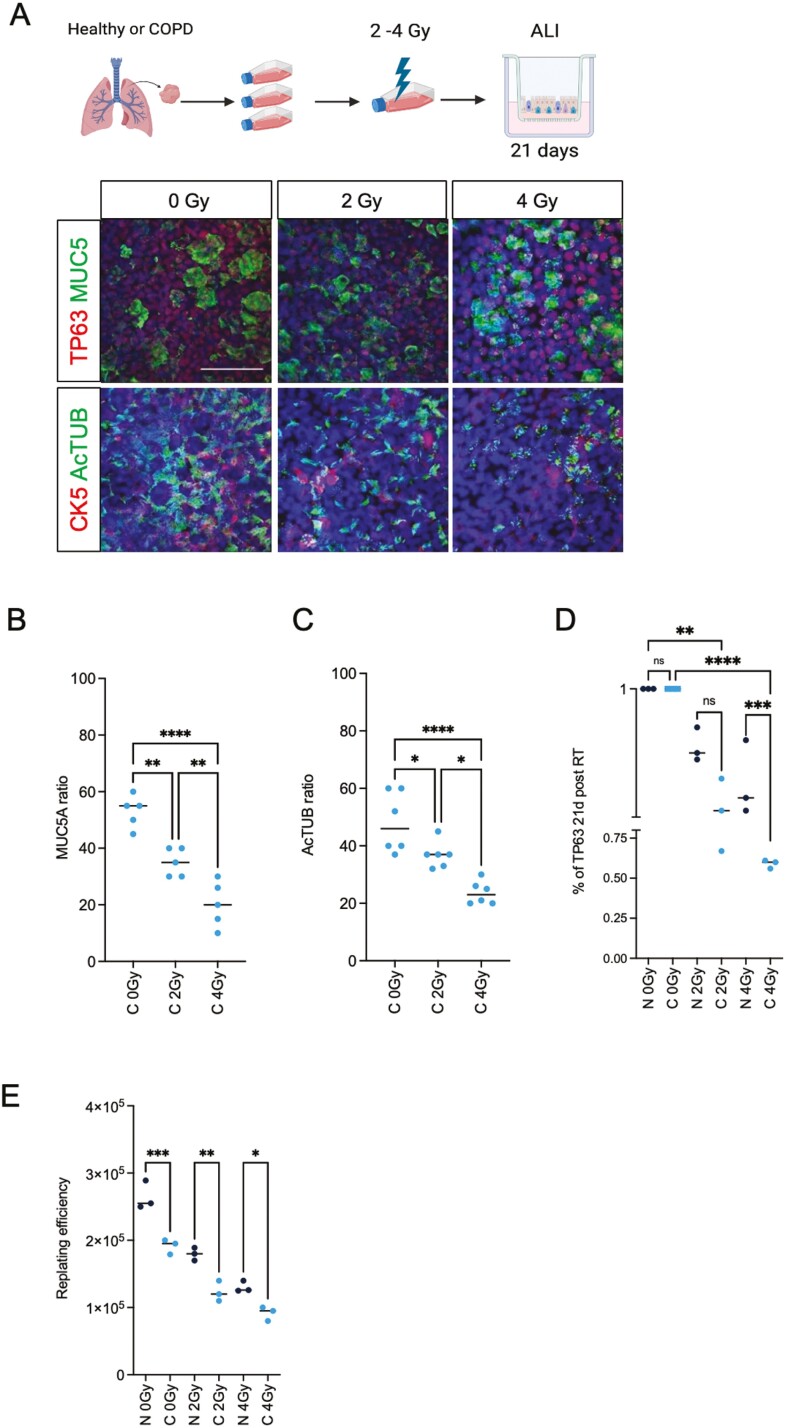
We previously reported that residual DNA damage 24 hours post-RT is a robust measure of human upper airway stem cells’ long-term survival and regenerative capacity.18 To investigate the radiation sensitivity of COPD-derived airway stem cells, we irradiated both healthy and COPD BSCs with 2 and 4 Gy, respectively; harvested the cells, seeded them in ALI, and monitored their ability to generate a stratified and polarized epithelium using IF for Cytokeratin-5 (CK5) and TP63, respectively, for BSCs, MUC5AC for mucous cells and acetylated tubulin (Ac-TUB) for ciliated cells (Figure 2A). The ALIs from irradiated BSCs show a defect in mucociliary differentiation compared to non-irradiated BSC. We observed 36% and 64% fewer mucous cells at 2 and 4 Gy, respectively, and 22% and 52% fewer ciliated cells, at 2 and 4 Gy, respectively (Figure 2B and C). Next, we measured the number and replating efficiency of CK5+ and TP63+ BSCs after irradiation. We observed a 15% reduction in survival at 2 Gy and 31% reduction at 4 Gy of COPD BSCs compared to non-affected BSCs (Figure 2D). We performed a replating assay and passaged BSCs 3 passages to assess the long-term survival and repopulation of COPD-derived BSCs. We found that COPD BSCs have a dose-dependent decrease in replating efficiency of 30% at 2 Gy and 32% at 4 Gy, respectively, compared to healthy BSCs (Figure 2E). The reduction in differentiation capacity for irradiated BSCs from COPD PBECs was consistently lower than that of non-COPD in a dose-dependent manner (Supplementary Figure S1B). Next, we asked if COPD BSCs are also more sensitive to first-line EGFR-targeted lung cancer treatment with small-molecule tyrosine kinase inhibitors (TKI’s). We derived bronchospheres from COPD- and non-affected donor BSCs and measured their viability after 48 hours of treatment with first-and third-generation tyrosine kinase inhibitors (TKI’s); Gefitinib, and Osimertinib. Both TKI’s acted on-target and reduced the phosphorylation of EGFR (Y1068) receptor at 1 μM (Supplementary Figure S2A) and caused a reduction in both viability (fold 1.3 for Gefitinib and 1.5 for Osimertinib), although this reduction was significantly stronger in COPD BSCs (Supplementary Figure S2B). A reduction in long-term survival was also more pronounced in the COPD-derived bronchospheres (40% reduction with 4 Gy; Supplementary Figure S2C).
Figure 2.
Differential sensitivity to COPD- and non-COPD PBEC irradiation in ALI culture. (A) Schematic representation of the treatment plan and representative examples of immunofluorescent staining of 3D ALI culture at day 21 ALI from COPD for TP63, CK5, MUC5AC, and Ac-TUB. (B) Quantification of MUC5AC and (C) AcTUB, and (D) TP63 in the non-COPD and COPD cultures upon irradiation (0-2-4 Gy) at 21 days in ALI expressed as ratio of total cells. (E) Replating efficiency of non-COPD and COPD from ALI shows a reduced capacity for replating before and after 2, 4 Gy irradiation. N = 3 independent donors non-COPD and COPD per group. *P < .05; **P < .01; ***P < .001.; ****P < .0001. Scale bar = 50 μm. Created with Biorender.
Irradiated COPD BSCs show cell cycle defects, reduced DNA repair, and increased cell death
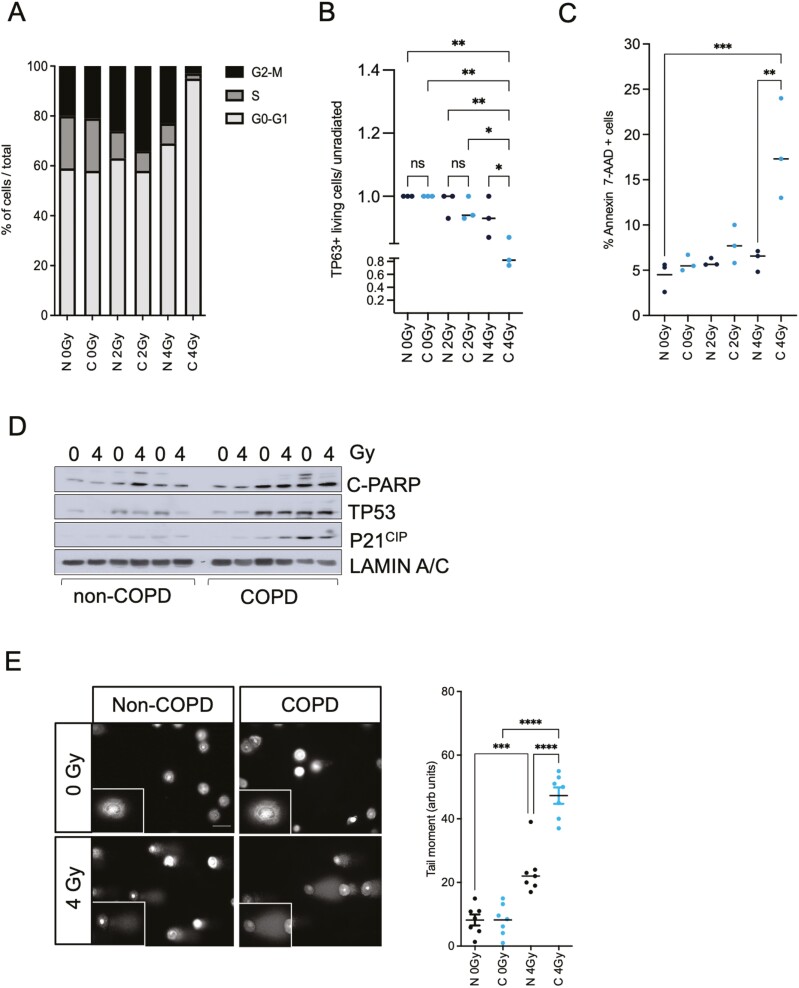
To uncover whether the increased COPD stem cell sensitivity to irradiation is due to an impaired cell cycle arrest, we performed cell cycle analysis (Edu/PI staining) 24 hours after irradiation, using FACS (Supplementary Figure S1C). We observed that irradiated COPD stem cells have a higher accumulation in the G2/M phase when compared to healthy stem cells at 2 Gy, but at 4 Gy show a strong increase of G0-G1 and a concomitant decrease of G2/M compared to healthy BSC (Figure 3A). These results correlated with a decreased viability of TP63-positive BSC in COPD cultures upon 4 Gy irradiation (Figure 3B). Using Annexin V-7AAD staining, we observed a 2- to 3-fold increase in apoptosis 24 hours after 4 Gy irradiation in COPD BSCs compared to non-COPD BSCs (Figure 3C). These results suggest that the increased sensitivity to irradiation is due to an impaired cell cycle distribution and a higher susceptibility to cell death. We analyzed c-PARP, TP53, and P21CIP (CDKN2A1) protein expression 24 hours post-irradiation to investigate why COPD stem cells are more susceptible to cell death upon irradiation. COPD BSCs showed increased basal levels of C-PARP, TP53, and P21 compared to healthy BSC, although irradiation did not change this (Figure 3D). Finally, we performed an alkaline comet assay to assess whether the increased cell death in the patient with COPD cells was due to increased DNA damage and repair upon irradiation. We found that irradiated COPD BSCs have a 2-fold higher percentage of unrepaired DNA breaks remaining 24 hours after irradiation than healthy BSCs (Figure 3E), indicating that COPD BSCs exhibit increased persistence of unrepaired DNA damage post-irradiation compared to BSCs from non-COPD donors.
Figure 3.
Irradiated chronic obstructive pulmonary disease (COPD) BSCs have an impaired cell cycle distribution with increased TP53, P21 levels compared to non-COPD BSCs. (A) COPD BSCs have an increased accumulation in the G2-M phase upon 2 Gy irradiation and decreased S phase distribution compared to non-COPD. At 4 Gy, a strong and significant increase in G0/G1 and a reduction in G2/M are observed. (B) Reduction in percentage of TP63 + cells upon 2-4 Gy irradiation (C) Annexin-V-PI FACS staining in COPD and non-COPD BSC’s upon irradiation. Decreased percentage of living cells and increased apoptosis in COPD stem cells upon 4 Gy irradiation. (D) Cleaved PARP, TP53, P21CIP Western blot in non-COPD and COPD BSC upon 4 Gy irradiation (E) Representative images and quantification of the alkaline comet assay of BSC’s upon 4 Gy irradiation. Right panel. Tail-moment quantitation shows irradiated COPD BSCs have increased tail moment compared. N = 3 independent donors non-COPD and COPD per group *P < .05; **P < .01; ***P < .001; ****P < .0001.
Reduced DNA damage repair and enhanced DNA damage response in COPD BSCs
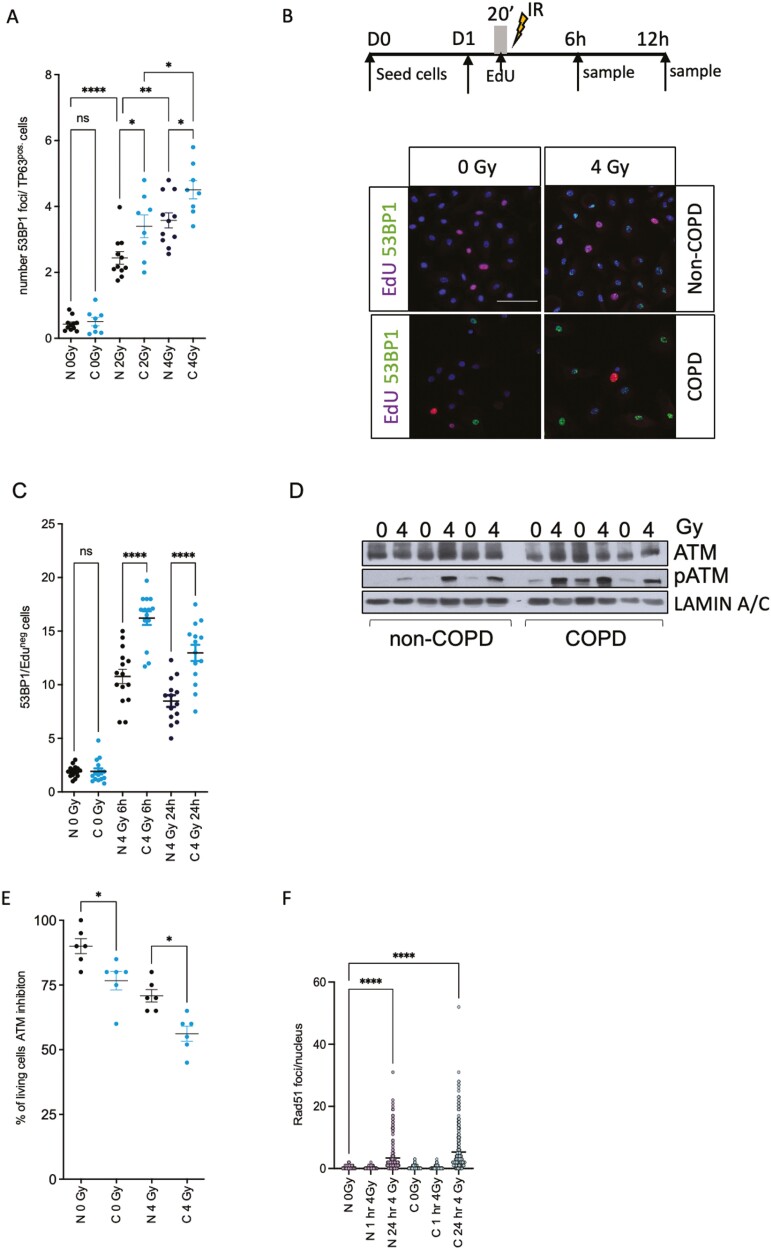
To directly address why COPD BSCs retain more DNA damage post-irradiation, we quantified the DNA damage response by measuring 53BP1 foci—a marker for dsDNA break repair (DSB) after irradiation in TP63 + COPD and non-COPD BSCs 24 hours post-irradiation. COPD stem cells showed a 1.6-fold increased number of 53BP1 foci upon 4 Gy irradiation compared to non-irradiated and irradiated non-COPD BSCs (Figure 4A). We next asked if the accumulation of the cells in G2-M was also associated with a delayed repair capacity upon irradiation. To do so, we incubated BSCs with EdU for 20 minutes before 4 Gy irradiation and performed a co-staining with 53BP1 and EdU, 6 and 24 hours post-RT, respectively (Figure 4B). We found that COPD BSCs have increased 53BP1 foci in the EdU negative (non-replicating) cells 6 and 24 hours after irradiation (Figure 4C). No significant differences were observed in the number of 53BP1 foci in the EdU-positive populations (not shown). We observed that COPD BSCs have a higher basal and radiation-induced phosphorylation of ATM kinase, indicating an intact but preactivated and more robust DNA damage response (Figure 4D) and the downstream activation of pCHK2 in 2 out of 3 patients (Supplementary Figure S3A). To address whether replication stress can explain these differences, we analyzed the expression of RPA32 24 hours post-irradiation, but no significant differences were observed (Supplementary Figure S3B). We next asked if blocking the DNA damage response with the ATM inhibitor KU55933 could affect their viability upon irradiation. KU55933 efficiently blocked pATM (not shown) in COPD BSCs. It significantly reduced the percentage of living cells compared to non-COPD BSCs upon 4 Gy irradiation, suggesting that ATM signaling protects against radiation-induced cell death (Figure 4E). We noted that irradiated COPD BSCs have a higher G2M fraction. Therefore, we addressed whether the reduced capacity for homologous recombination (HR) that occurs in G2/M and S-phase might explain the increased radiation sensitivity of COPD-derived BSC. We stained non-COPD (n = 3) and COPD (n = 3) BSCs for RAD51; a marker for HR-dependent dsDNA break repair 1 and 24 hours post-irradiation. We observed no differences between non-COPD and COPD donors, although radiation (4 Gy), as expected, increased RAD51 foci 24 hours post-RT in both (Figure 4F, Supplementary Figure S4A, B).
Figure 4.
Irradiated COPD BSCs have impaired non-homologous end joining and increased DNA damage. (A) 53BP1 nuclear Foci in TP63 + cells in COPD and non-COPD BSCs. (B) Schematic treatment plan and immunofluorescent staining of healthy and COPD BSCs for 53BP1 and EdUR 24 hours after irradiation (0-4 Gy) (C) Quantification of the 53BP1 foci in the EdU-positive cells 6 and 24 hours after irradiation. (D) Total ATM, phospho-ATM Western blot in non-COPD and COPD stem cells 24 hours post-irradiation (0-4 Gy). Lamin A/C is loading control. (E) Viability of non-COPD and COPD BSC upon phospho-ATM inhibition (10 µM KU55933) added 24 hours before irradiation and analysis 24 hours post-irradiation (0-4 Gy). (F) RAD51 nuclear foci in non-COPD vs COPD BSC 1 and 24 hours post-irradiation with 4 Gy. N = 3 independent donors non-COPD and COPD per group *P < .05; **P < .01; ***P < .001; ****P < .0001. Scale bar = 50 μm.
NOTCH inhibition reverts radiation-induced DNA damage in COPD stem cells and pathological phenotype
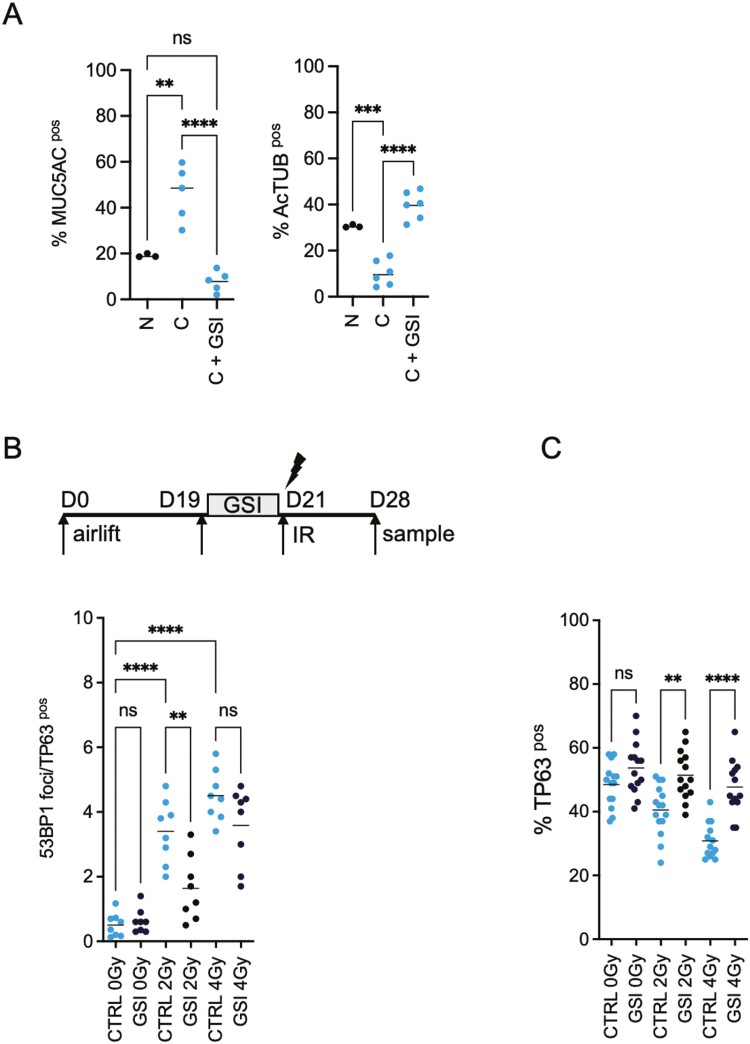
Our previous results showed that NOTCH inhibition using small-molecule γ-secretase inhibitors (GSI) reduces the radiation-induced DNA damage in healthy PBEC and promotes post-IR survival in 3D stratified and polarized cultured primary lung epithelia.18,24 We first assessed the expression of Notch Receptors N1-N4, Notch ligands (DLL1 and JAGGED 1, 2), and NOTCH targets (HES1, HES4, HEY1, and HEY2) between non-COPD and COPD-derived BSCs from PBECs. We observed significantly increased expression for NOTCH3 receptors and HES1 and HES4 targets in COPD BSC. While the absolute expression levels of other NOTCH receptors, ligands, and targets differed per donor these changes were not significant as a group (Supplementary Figure S1D). To assess if NOTCH signaling also affects radiation-induced DNA damage in COPD stem cells, ALI cultures were treated with NOTCH/γ-secretase inhibitor DBZ (1 μM) for 48 hours (19 days post-airlift) to block the NOTCH pathway and irradiated with 2 or 4 Gy, 21 days post-airlift. We demonstrated that endogenous NOTCH1 cleavage and signaling were active in COPD stem cells, and NICD1 cleavage was blocked with the Notch/γ-secretase DBZ (Supplementary Figure S3C). First, NOTCH/γ-secretase inhibition increased the percentage of ciliated cells and reduced the percentage of mucous (MUC5AC+)/secretory cells, reverting a typical characteristic of COPD pathology (Figure 5A). We observed a radiation dose-dependent increase in 53BP1 foci in COPD TP63 + BSCs. Furthermore, NOTCH inhibition reduced the number of 53BP1 foci in TP63 + (Figure 5B) and inhibited the radiation-induced reduction in TP63 + cells in BSCs (Figure 5C) whereas Notch inhibition with GSI, 48 hours before irradiation did not alter the number or timing of RAD51 foci in either group upon irradiation (Figure 4F).
Figure 5.
NOTCH inhibition reverts the pathological secretory phenotype and reduces radiation-induced DNA damage in COPD BSCs. A) Quantification of Ac-TUB+, MUC5A + cells in the ALI system shows that COPD cultures have a perturbed differentiation phenotype compared to non-COPD ALIs with an excess of MUC5A + cells at the expense of ciliated cells, which can be reverted with NOTCH inhibition. (B) Schematic representation of the treatment plan and quantification of the 53BP1 staining in TP63 + cells, 24 hours after RT in the presence or absence of NOTCH inhibition at day 19 to 21, 48 hours before RT (2-4 Gy). (C) Quantification of TP63, 24 hours after RT in the presence or absence of NOTCH inhibition at day 19 to 21, 48 hours prior to RT (2-4 Gy). N = 3 independent donors non-COPD and COPD per group *P < .05, **P < .01; ***P < .001; ****P < .0001.
Discussion
In the present study, we used patient-derived primary lung epithelial cultures to model the mechanism behind the enhanced radiation sensitivity of human COPD patients with lung cancer who receive thoracic RT. It has been shown that ex vivo ALI cultures from healthy donors represent the transcriptome from in vivo human bronchial epithelial, although the relative proportions of cell types differ.32 We find that ex vivo human COPD upper airway stem cells recapitulate important histopathological defects of COPD patients, most notably the aberrant mucociliary differentiation pattern and sensitivity to irradiation and tyrosine kinase inhibitors used in first-line treatment of EGFR-mutated lung cancer. We observed a higher increase in radiation-induced DNA damage and a reduced cell survival 24 hours post-irradiation in COPD upper airway BSC. Furthermore, COPD BSCs have reduced DNA repair capacity, possibly contributing to their G2/M cell cycle arrest and apoptosis.
Previous studies have indicated that expansion progenitor/stem cell pools in tissues can protect against radiation-induced salivary gland damage.33 Cell therapies with salivary gland stem cells can restore functionality.34 Despite being a relatively quiescent organ, the lung possesses remarkable proliferative and repair capabilities following exposure to DNA-damaging agents, highlighting the presence of a highly active lung stem cell population.35 Previous work from us and others showed that radiation exposure can damage airway stem cells, impairing lung tissue repair, differentiation, and regeneration ex vivo.18,36 Here we now show that airway cells cultured from patients with lung cancer with COPD are more sensitive to RT than airway stem cells from non-affected patients and that blocking Notch signaling can mitigate part of this defect. Importantly, in mice, Tp63/K5 + distal airway BSCs can repopulate damaged lungs, which is also a Notch-dependent process.37,38 Similar findings have been reported in upper airway club cells.39 These findings reinforce that our ex vivo cultured cell models may reflect the cell-autonomous roles of airway stem cells in complex diseases such as COPD. Our findings show that COPD-derived stem cells repair DNA damage like normal airway stem cells,40 but less efficiently. Blocking Notch with small-molecule inhibitors can reduce DNA damage 24 hours post-irradiation, reduce cell death, and increase the survival of airway stem cells. Similarly, Notch blocking in unirradiated COPD cultures reverses the goblet metaplasia by blocking secretory differentiation and restoring ciliated cell numbers. Interestingly, COPD-derived BSC showed higher expression of NOTCH3, HES1, and HES4 targets, suggesting that the Notch signaling pathway is more active than in non-COPD BSC. These findings support the relative increase in goblet cells, the reduction of ciliated cells in primary cultures, and the sensitivity to NOTCH inhibition by GSI. Our findings are in line with others, where high NOTCH3 expression is observed in Asthma patients and NOTCH3-specific silencing in airway epithelial cultures from Asthma donors reduced MUC5A expression.41
The exact mechanism behind the differential sensitivity of COPD vs healthy stem cells to radiation is unknown. Previously, we have shown that Notch inhibition caused enhanced basal and radiation-induced activation of the DNA damage response.24 Here, we show a higher basal and radiation-induced pATM level in COPD stem cells compared to non-affected individuals, which can be further enhanced by Notch inhibition. Previous studies have demonstrated that NOTCH gain-of-function (oncogenic) mutations in C-elegans and human Notch-driven T-ALL42 utilize HR to protect against radiation-induced cell death because silencing of HR components (eg, Rad51) radiosensitizes. In the bronchial epithelium, we and others have demonstrated important roles for Notch signaling in epithelial lung development homeostasis and response to radiation injury.18,24 Basal lung stem cells repair radiation-induced DNA damage using the error-prone non-homologous end-joining pathway.40 Here, we used 53BP1 as a marker for non-homologous end-joining repair.43 We did not find evidence that NOTCH engages HR but rather NHEJ in response to radiation. Possibly, this reflects differences between healthy and tumor tissue, the cell type, or the radiation dose and quality used. A more thorough analysis is needed to clarify further the interaction between Notch signaling and the DNA damage response. It is, however, noteworthy that Notch inhibition already engages pATM without any induced DNA damage and that targeting ATM and RAD51 enhances Notch-dependent tumors’ sensitivity to radiation.42,44
Radiation-induced lung injury is a complex multifactorial disease.45 More than 90% of COPD and lung cancer cases are smoking-related. Cigarette smoke increases ROS and DNA damage in lung epithelial cells. Consistently, many reports show that COPD cells have more oxidative (DNA) damage than non-COPD cells but reduced expression of the DNA repair protein KU86 in upper airway cells.46 These results may explain the cell-autonomous role of lung epithelial cells in the increased radiation sensitivity of patients with COPD to RP and their predisposition to lung cancer in the absence of immune cells. It is important to emphasize that COPD is a complex progressive disease in which lung epithelial cells, lung parenchyma, and immune cells at different locations within the airways and alveoli interact and contribute to the disease phenotype in a complex manner. The increased radiation sensitivity we observe in COPD BSC is only one part of this complex phenotype. Of note, the hypermucosecretion observed in patients with COPD may be mitigated with approaches that temporarily block Notch signaling and may be of therapeutic interest in improving COPD defects by contributing to increased regeneration and repair of damaged lungs before RT and reducing the adverse normal tissue effects post RT24,41
Recent studies show that Notch blockade can restore multi-ciliated cells in human nasal epithelium and in vivo in mice.47 We have not addressed the long-term quality of airway stem cells protected from radiation damage by Notch inhibition since ALI cultures cannot be maintained for longer than 1-2 months, and few functional studies are limited. This is important because non-homologous end-joining is the primary DSB repair mechanism in BSCs40 but an error-prone DNA repair process that may result in genetically unstable stem cells, which has been proposed to contribute to smoking-related mutagenesis in basal lung stem cells.40 We have previously shown that Notch inhibition can protect from radiation-induced cell death of TP63 + cells and improve the integrity of the luminal surface in 3D ALI.24 More advanced in vivo preclinical studies are needed to model COPD and further test such hypotheses and the therapeutic role of NOTCH targeting.
The data show that COPD-derived primary upper airway cultures may be developed as a patient-specific avatar. This will enable the study of radiation and drug-induced consequences that may lead to developing actionable targets to reduce lung damage during cancer treatment. Furthermore, it can be exploited to promote post-treatment tissue regeneration, protect lung function, and improve cancer patients’ long-term quality of life and survival.
Supplementary material
Supplementary material is available at Stem Cells Translational Medicine online.
Acknowledgments
We thank Maastro lab members and Maastro Clinic, for helpful discussions and support and Sabine Langie (Department of Pharmacology and Toxicology, Maastricht University) for technical help with the comet assay. Felix Meyer and Kerstin Borgmann (University of Hamburg) for valuable discussions.
Contributor Information
Lorena Giuranno, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Jolanda A F Piepers, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Evelien Korsten, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Reitske Borman, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Gerarda van de Kamp, Department of Molecular Genetics, Erasmus University Medical Center, Rotterdam, 3015 GD, The Netherlands; Oncode Institute, Erasmus University Medical Center, Rotterdam, 3015 GD, The Netherlands.
Dirk De Ruysscher, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Jeroen Essers, Department of Molecular Genetics, Erasmus University Medical Center, Rotterdam, 3015 GD, The Netherlands; Department of Radiotherapy, Erasmus University Medical Center, Rotterdam, 3015 GD, The Netherlands; Department of Vascular Surgery, Erasmus University Medical Center, Rotterdam, 3015 GD, The Netherlands.
Marc A Vooijs, Department of Radiation Oncology (MAASTRO)/GROW Research Institute for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, 6200 MD, The Netherlands.
Author contributions
Lorena Giuranno: design, collection, methods and assembly of data into figures, manuscript draft writing. Jolanda A.F. Piepers: design, collection, methods and assembly of data into figures, manuscript review. Evelien Korsten and Reitske Borman: collection of data. Gerarda van de Kamp: design, collection, and assembly of data into figures. Jeroen Essers: design. Dirk De Ruysscher: manuscript reviewing. Marc A. Vooijs: conception and design, financial support, data analysis and interpretation and assembly of figures, manuscript writing, and final approval. Lorena Giuranno and Jolanda A.F. Piepers contributed equally to this work.
Funding
L.G. was funded by a grant from the Maastro Cancer Foundation, R.B. was funded by EUROSTARS Ab-SENS (E155411).
Conflicts of interest
The authors declared no potential conflicts of interest.
Data availability
The data underlying this article are available in the article and in its online supplementary material and available upon request.
References
- 1. Kasmann L, Dietrich A, Staab-Weijnitz CA, et al. Radiation-induced lung toxicity - cellular and molecular mechanisms of pathogenesis, management, and literature review. Radiat Oncol. 2020;15(1):214. 10.1186/s13014-020-01654-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takeda A, Kunieda E, Ohashi T, et al. Severe COPD is correlated with mild radiation pneumonitis following stereotactic body radiotherapy. Chest. 2012;141(4):858-866. 10.1378/chest.11-1193 [DOI] [PubMed] [Google Scholar]
- 3. Wirsdorfer F, Jendrossek V.. Modeling DNA damage-induced pneumopathy in mice: insight from danger signaling cascades. Radiat Oncol. 2017;12(1):142. 10.1186/s13014-017-0865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Verma V, Simone CB 2nd, Werner-Wasik M.. Acute and late toxicities of concurrent chemoradiotherapy for locally-advanced non-small cell lung cancer. Cancers (Basel). 2017;9(9):120. 10.3390/cancers9090120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang K, Tepper JE.. Radiation therapy-associated toxicity: etiology, management, and prevention. CA Cancer J Clin. 2021;71(5):437-454. 10.3322/caac.21689 [DOI] [PubMed] [Google Scholar]
- 6. De Ruysscher D, Niedermann G, Burnet NG, et al. Radiotherapy toxicity. Nat Rev Dis Primers. 2019;5(1):13. 10.1038/s41572-019-0064-5 [DOI] [PubMed] [Google Scholar]
- 7. Xu K, Liang J, Zhang T, et al. Clinical outcomes and radiation pneumonitis after concurrent EGFR-tyrosine kinase inhibitors and radiotherapy for unresectable stage III non-small cell lung cancer. Thorac Cancer. 2021;12(6):814-823. 10.1111/1759-7714.13816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacNee W. Pathogenesis of chronic obstructive pulmonary disease. Proc Am Thorac Soc. 2005;2(4):258-66; discussion 290. 10.1513/pats.200504-045SR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Durham AL, Adcock IM.. The relationship between COPD and lung cancer. Lung Cancer. 2015;90(2):121-127. 10.1016/j.lungcan.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caramori G, Ruggeri P, Mumby S, et al. Molecular links between COPD and lung cancer: new targets for drug discovery? Expert Opin Ther Targets. 2019;23(6):539-553. [DOI] [PubMed] [Google Scholar]
- 11. Rancati T, Ceresoli GL, Gagliardi G, Schipani S, Cattaneo GM.. Factors predicting radiation pneumonitis in lung cancer patients: a retrospective study. Radiother Oncol. 2003;67(3):275-283. 10.1016/s0167-8140(03)00119-1 [DOI] [PubMed] [Google Scholar]
- 12. Kimura T, Togami T, Takashima H, et al. Radiation pneumonitis in patients with lung and mediastinal tumours: a retrospective study of risk factors focused on pulmonary emphysema. Br J Radiol. 2012;85(1010):135-141. 10.1259/bjr/32629867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rock JR, Onaitis MW, Rawlins EL, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA. 2009;106(31):12771-12775. 10.1073/pnas.0906850106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kotton DN, Morrisey EE.. Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat Med. 2014;20(8):822-832. 10.1038/nm.3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu M, Zhang X, Lin Y, Zeng Y.. Roles of airway basal stem cells in lung homeostasis and regenerative medicine. Respir Res. 2022;23(1):122. 10.1186/s12931-022-02042-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fahy JV, Dickey BF.. Airway mucus function and dysfunction. N Engl J Med. 2010;363(23):2233-2247. 10.1056/NEJMra0910061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O’Donnell R, Breen D, Wilson S, Djukanovic R.. Inflammatory cells in the airways in COPD. Thorax. 2006;61(5):448-454. 10.1136/thx.2004.024463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giuranno L, Wansleeben C, Iannone R, et al. NOTCH signaling promotes the survival of irradiated basal airway stem cells. Am J Physiol Lung Cell Mol Physiol. 2019;317(3):L414-L423. 10.1152/ajplung.00197.2019 [DOI] [PubMed] [Google Scholar]
- 19. Farin AM, Manzo ND, Kirsch DG, Stripp BR.. Low- and high-LET radiation drives clonal expansion of lung progenitor cells in vivo. Radiat Res. 2015;183(1):124-132. 10.1667/RR13878.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McConnell AM, Konda B, Kirsch DG, Stripp BR.. Distal airway epithelial progenitor cells are radiosensitive to High-LET radiation. Sci Rep. 2016;6:33455. 10.1038/srep33455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Serrano Martinez P, Giuranno L, Vooijs M, Coppes RP.. The radiation-induced regenerative response of adult tissue-specific stem cells: models and signaling pathways. Cancers. 2021;13(4):855-818. 10.3390/cancers13040855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rock JR, Gao X, Xue Y, et al. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell. 2011;8(6):639-648. 10.1016/j.stem.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lafkas D, Shelton A, Chiu C, et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature. 2015;528(7580):127-131. [DOI] [PubMed] [Google Scholar]
- 24. Giuranno L, Roig EM, Wansleeben C, et al. NOTCH inhibition promotes bronchial stem cell renewal and epithelial barrier integrity after irradiation. Stem Cells Transl Med. 2020;9(7):799-812. 10.1002/sctm.19-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hikichi M, Mizumura K, Maruoka S, Gon Y.. Pathogenesis of chronic obstructive pulmonary disease (COPD) induced by cigarette smoke. J Thorac Dis. 2019;11(suppl 17):S2129-S2140. 10.21037/jtd.2019.10.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Danahay H, Pessotti AD, Coote J, et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep. 2015;10(2):239-252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- 27. Bodas M, Moore AR, Subramaniyan B, et al. Cigarette smoke activates NOTCH3 to promote goblet cell differentiation in human airway epithelial cells. Am J Respir Cell Mol Biol. 2021;64(4):426-440. 10.1165/rcmb.2020-0302OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Vincenzo S, Ninaber DK, Cipollina C, et al. Cigarette smoke impairs airway epithelial wound repair: role of modulation of epithelial-mesenchymal transition processes and Notch-1 signaling. Antioxidants (Basel). 2022;11(10):2018. 10.3390/antiox11102018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paul MK, Bisht B, Darmawan DO, et al. Dynamic changes in intracellular ROS levels regulate airway basal stem cell homeostasis through Nrf2-dependent Notch signaling. Cell Stem Cell. 2014;15(2):199-214. 10.1016/j.stem.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Royen ME, Cunha SM, Brink MC, et al. Compartmentalization of androgen receptor protein-protein interactions in living cells. J Cell Biol. 2007;177(1):63-72. 10.1083/jcb.200609178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38(4):e100300. 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG.. Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol. 2011;44(4):465-473. 10.1165/rcmb.2009-0453OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lombaert IM, Brunsting JF, Wierenga PK, et al. Keratinocyte growth factor prevents radiation damage to salivary glands by expansion of the stem/progenitor pool. Stem Cells. 2008;26(10):2595-2601. [DOI] [PubMed] [Google Scholar]
- 34. Pringle S, Maimets M, van der Zwaag M, et al. Human salivary gland stem cells functionally restore radiation damaged salivary glands. Stem Cells. 2016;34(3):640-652. 10.1002/stem.2278 [DOI] [PubMed] [Google Scholar]
- 35. Weeden CE, Asselin-Labat ML.. Mechanisms of DNA damage repair in adult stem cells and implications for cancer formation. Biochim Biophys Acta Mol Basis Dis. 2018;1864(1):89-101. 10.1016/j.bbadis.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 36. Reynolds SD, Giangreco A, Hong KU, et al. Airway injury in lung disease pathophysiology: selective depletion of airway stem and progenitor cell pools potentiates lung inflammation and alveolar dysfunction. Am J Physiol Lung Cell Mol Physiol. 2004;287(6):L1256-L1265. 10.1152/ajplung.00203.2004 [DOI] [PubMed] [Google Scholar]
- 37. Vaughan AE, Brumwell AN, Xi Y, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517(7536):621-625. 10.1038/nature14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zuo W, Zhang T, Wu DZ, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517(7536):616-620. 10.1038/nature13903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xing Y, Li A, Borok Z, Li C, Minoo P.. NOTCH1 is required for regeneration of Clara cells during repair of airway injury. Stem Cells. 2012;30(5):946-955. 10.1002/stem.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weeden CE, Chen Y, Ma SB, et al. Lung basal stem cells rapidly repair DNA damage using the error-prone nonhomologous end-joining pathway. PLoS Biol. 2017;15(1):e2000731. 10.1371/journal.pbio.2000731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reid AT, Nichol KS, Chander Veerati P, et al. Blocking Notch3 signaling abolishes MUC5AC production in airway epithelial cells from individuals with asthma. Am J Respir Cell Mol Biol. 2020;62(4):513-523. 10.1165/rcmb.2019-0069OC [DOI] [PubMed] [Google Scholar]
- 42. Deng X, Michaelson D, Tchieu J, et al. Targeting homologous recombination in notch-driven C. elegans stem cell and human tumours. PLoS One. 2015;10(6):e0127862. 10.1371/journal.pone.0127862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shibata A, Jeggo PA.. Roles for 53BP1 in the repair of radiation-induced DNA double strand breaks. DNA Repair (Amst). 2020;93:102915. 10.1016/j.dnarep.2020.102915 [DOI] [PubMed] [Google Scholar]
- 44. Vermezovic J, Adamowicz M, Santarpia L, et al. Notch is a direct negative regulator of the DNA-damage response. Nat Struct Mol Biol. 2015;22(5):417-424. 10.1038/nsmb.3013 [DOI] [PubMed] [Google Scholar]
- 45. Giuranno L, Ient J, De Ruysscher D, Vooijs MA.. Radiation-Induced Lung Injury (RILI). Front Oncol. 2019;9:877. 10.3389/fonc.2019.00877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Caramori G, Adcock IM, Casolari P, et al. Unbalanced oxidant-induced DNA damage and repair in COPD: a link towards lung cancer. Thorax. 2011;66(6):521-527. 10.1136/thx.2010.156448 [DOI] [PubMed] [Google Scholar]
- 47. Vladar EK, Kunimoto K, Rojas-Hernandez LS, et al. Notch signaling inactivation by small molecule gamma-secretase inhibitors restores the multiciliated cell population in the airway epithelium. Am J Physiol Lung Cell Mol Physiol. 2023;324(6):L771-L782. 10.1152/ajplung.00382.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material and available upon request.