Abstract
In vitro selections for catalytic activity have been designed for the isolation of genes encoding enzymes from libraries of proteins displayed on filamentous phages. The proteins are generally expressed as C-terminal fusions with the N-terminus of the minor coat protein p3 for display on phages. As full-length cDNAs generally contain several stop codons near their 3′ end, this approach cannot be used for their expression on the surface of phages. Here we show that in vitro selection for catalytic activity is compatible with a system for expression of proteins as N-terminal fusions on the surface of bacteriophages. It is highlighted for the Stoffel fragment of Taq DNA polymerase I and makes use of (p3–Jun/Fos–Stoffel fragment) fusions. The efficiency of the selection is measured by an enrichment factor found to be about 55 for a phage polymerase versus a phage not expressing a polymerase. This approach could provide a method for the functional cloning of nucleotidyl transferases from cDNA libraries using filamentous phage display.
INTRODUCTION
Phage display has been extensively used for the isolation of binding proteins and their encoding genes from large libraries. Phage display may also provide a method for the isolation of enzymes from protein libraries: in vitro selection of phage proteins for catalytic activity has been recently designed as affinity selections of the reaction product coupled to the phage enzymes that catalysed the reaction from substrate to product (1–4).
Such in vitro selection for catalytic activity using phage display has been shown for several enzymes which are substrate cleaving catalysts such as nuclease (1) and peptidase (2), or enzymes catalysing synthetic reactions such as DNA polymerase (3) and peptide ligase (4). In these selections, enzymes were fused to the N-terminus of minor coat protein p3 for display on filamentous phage. This approach cannot be used for full-length cDNA expression products as their genes contain stop codons at their 3′ end which would prevent translation of the fusion protein.
Two main systems for expression of full-length cDNAs on filamentous phage have been designed (5,6) and used successfully in affinity selections (7–11). Full-lengths cDNAs have been fused at the 3′ end of the sequence encoding the leucine zipper of Fos: when used with g3-jun fusions, the (p3–Jun/Fos–cDNA expression product) complex is assembled in the periplasm of Escherichia coli and further displayed on filamentous phage (5). This system relies on the leucine zippers Jun and Fos which form heterodimers. Cysteine residues were introduced at the N- and C-termini of Jun and Fos peptides for formation of two disulfide bridges between the proteins (p3–Jun) and (Fos–cDNA expression product) (5). Full-length cDNAs have been also fused at the 3′ end of g6, the gene encoding minor coat protein p6 of filamentous phage (6,12).
In this report we show that proteins can be selected in vitro for nucleotidyl transferase activity when expressed on filamentous phage as p3–Jun/Fos fusions.
MATERIALS AND METHODS
Reagents
Synthesis, purification and characterisation of maleimidyl derivatised primer 1 (Fig. 1) was done in the laboratory as described earlier (3). Biotin-dUTP is biotin-ɛ-aminocaproyl-γ-aminobutyryl-[5-(3-aminoallyl)-2′-deoxy-uridine-5′-triphosphate] (Roche, Meylan, France). Standard oligonucleotides were from Genset (Paris, France), except template 2 which was synthesised and purified by HPLC in the laboratory.
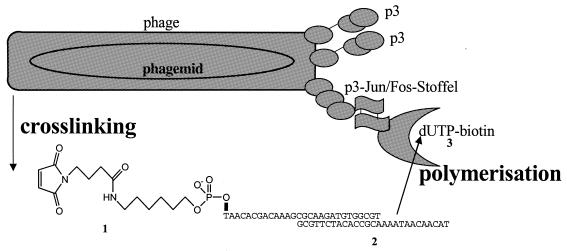
Figure 1.
Principle for selection according to the catalytic activity of nucleotidyltransferases displayed on filamentous phage. The Stoffel fragment polymerase is expressed on phage as a heterodimer of two protein fusions, phage coat protein p3 fused to the leucine zipper of Jun and the leucine zipper of Fos fused to the Stoffel fragment of Taq DNA polymerase I. Phages displaying an enzyme or not are cross-linked with a substrate, a maleimidyl derivatised primer 1 hybridised to oligodeoxynucleotide 2. If a phage particle displays an enzyme, the cross-linked substrate is converted to the product by addition of biotin-dUTP 3; the biotinylated phage enzyme can be isolated by affinity chromatography on streptavidin-coated beads. If a phage particle does not display an enzyme, a cross-linked substrate is not converted to product and is not recovered by affinity chromatography.
Phagemid construction
In a first cloning step, the gene 3 domain 3, g3D3, of the plasmid pJuFo (5) was substituted by the full-length gene 3, g3, as described in the following (Fig. 2). The full-length gene 3 was amplified by PCR from pHEN1-Stoffel (3) with deoxyoligonucleotides 5′-ATTCATTCAGGATCCGGTGCCGCAGAAACTGTTGAAAGTTGTTTAGC-3′ and 5′-TCATACTCTGCTAGCTTATTAAGACTCCTTATTACGC-3′ using the temperature/time sequence (°C/min) (94/3) (94/1, 53/1, 72/2)25 (72/15). The purified PCR product was digested by NheI, partially digested by BamHI and ligated into the BamHI–NheI restricted pJuFo vector to yield the phagemid pJLE.
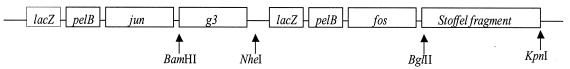
Figure 2.
Representation of the gene fusions of phagemid pJLE-Stoffel. This phagemid differs from pJuFo by a substitution of g3 domain 3 by a full copy of g3 and by the insertion of the gene encoding the Stoffel fragment of Taq DNA polymerase I at the 3′ end of fos. jun and fos are the sequences encoding the leucine zippers of proteins Jun and Fos, respectively. Restriction sites used in this work are indicated by arrows.
In a second cloning step, the Stoffel fragment of Taq polymerase was inserted as a BglII–KpnI fragment (Fig. 2). The Stoffel fragment was amplified from pHEN1-Stoffel (3) with deoxyoligonucleotides 5′-GCTCTAGAAGATCTGGCGGTGGCAGCC-3′ and 5′-TCTTATTGTGGTACCTTACTCCTTGGCGGAGAGC-3′ using the temperature/time sequence (94/3) (94/1, 59/1, 72/2)25 (72/15). The purified PCR product was digested by BglII and KpnI prior to ligation into the vector pJLE restricted at these sites to yield pJLE-Stoffel.
Phage preparation
The phage particles were prepared either from phagemid pHEN1 (13) or from phagemid pJLE-Stoffel using the helper phage KM13 (14) as described earlier (15), except that the cultures were grown at 30°C in 2× YT medium aqueous solution of 16 g bactotryptone, 10 g bacto-yeast extract and 5 g NaCl at pH 7.0 in 1 l) containing 0.1 g/l ampicillin, 0.025 g/l kanamycin and 0.1 mM isopropyl-β-d-thiogalactoside (IPTG). The phage particles were purified by two-step PEG precipitation and by size-exclusion chromatography using a S300 Sephacryl column (Pharmacia, Saclay) to remove potential free polymerases from the phage preparation.
In vitro selections for catalytic activity
A mixture of ∼3 × 107 infective phage particles in 15 µl, after trypsin catalysed p3 cleavage, expressing or not the Stoffel fragment of Taq polymerase, was incubated at 37°C for 2 h with 10 mM MgCl2, 10 µM maleimidyl-derivatised primer 1 and 50 µM template 2 in 0.1 M NaCl and 25 mM Na2HPO4 (PBS) to allow for hybridisation of primer and template, and for cross-linking of primer on phage (Fig. 1). Addition of deoxynucleotide triphosphates (dUTP-biotin, dCTP, dGTP and dATP at a final concentration of 10 µM each) to the mixture was required for the polymerisation. After 5 min at 37°C, addition of 15 µl 250 mM EDTA stopped the polymerisation reaction. Affinity chromatography was done using 200 µl streptavidin-coated superparamagnetic beads (Dynal, M280) to isolate biotinylated phage particles as described earlier (3) by washing seven times with 200 µl of 0.3 M Tris–HCl, 33 mM EDTA and 1.3 M NaCl at pH 9. N-tosyl-l-phenylalanine-chloromethylketone (TPCK) treated trypsin from bovine pancreas (Sigma, Saint Quentin) at a final concentration of 1 g/l was added to the beads resuspended in 100 µl PBS for cleavage of helper phage p3 copies. The mixture of phage polymerases and of phages not expressing a polymerase prior to selection was treated similarly with trypsin.
To allow for infection, a 10-fold excess in volume of a culture of E.coli TG1 at an optical density of 0.5 at 600 nm was added to the phage mixtures after trypsin cleavage and left for 15 min at 37°C without stirring. The cells were then plated on 0.1 g/l ampicillin-containing plates.
The proportions of phage polymerases and of phages not expressing any polymerase in populations before or after selection were determined by PCR screening of about 100 colonies using the cycles (temperature in °C/time in min) (94/10) (94/1, 59/1, 72/2)35 (72/10) and four oligonucleotides: 5′-TTTAATCATCTGCAGTACCGGGAGCTC-3′, 5′-GGAAGCTCTGAAAGTAGC-3′ (yielding a 515 bp fragment of the gene encoding the Stoffel fragment) and 5′-CTATGCGGCCCCATTCA-3′, 5′-ACAGCTATGACCATGATTACGCC-3′ (yielding a 217 bp fragment deriving from pHEN1), and by further gel electrophoresis of the PCR products yielding band patterns which characterised each phagemid.
RESULTS
For in vitro selection of catalytic activity from cDNA expression products libraries, a phagemid vector was first constructed from a phagemid pJuFo (5) known to efficiently express cDNAs on phage (Fig. 2). In short, domain 3 of g3 was substituted by the entire g3 and the gene encoding the Stoffel fragment of Taq DNA polymerase I (16) was inserted at the 3′ end of fos, the sequence coding for the leucine zipper of Fos. Three phagemids showing expected restriction patterns after cleavage with BamHI and NheI restriction enzymes within g3 were chosen for phage preparation. Phagemids were rescued using an engineered helper phage KM13, whose p3 contains a protease cleavable site between domains 2 and 3 (14). Phages not expressing any p3–Jun fusion display only cleavable p3 copies and are thereby rendered non-infective by proteolysis. Phages expressing one p3–Jun fusion display one full p3 copy after proteolysis and remain infective (14). The protease-resistant fraction of phage is therefore a measure of the fraction of phage expressing the p3–Jun fusion.
Of the three phage supernatants prepared without addition of IPTG to the culture medium, two of them contained a fraction of phage displaying the p3–Jun fusion <10–4 and the other ∼10–3. This last phage preparation corresponded to the phagemid named pJLE. For this phagemid, the fraction of phage displaying the p3–Jun fusion was found to be 1.3 × 10–3, 1.5 × 10–2 and 9.6 × 10–3 when phage was prepared in a 2× TY medium containing 0, 0.1 and 1 mM IPTG, respectively.
The Stoffel fragment of Taq DNA polymerase I was then cloned into pJLE as a BglII–KpnI fragment to yield pJLE-Stoffel for expression of the (p3–Jun/Fos–Stoffel) fusion protein (Figs 1 and 2). Phage was further prepared in a 2× TY medium containing 0.1 mM IPTG from two phagemids with appropriate BglII and KpnI restriction patterns. The trypsin-resistant fraction of phage was evaluated for two clones 1 and 2 to be 7.9 × 10–3 and 1.9 × 10–2.
For the in vitro selection of polymerase activity, we prepared model libraries composed of two types of phage particles, the ones expressing a polymerase and the others not expressing any polymerase. The substrate is cross-linked to phage particles and converted to product if the phage displays an active enzyme in proximity to the substrate (Fig. 1). The selection is an affinity chromatography for the reaction product, which is coupled to the phage polymerase that catalysed the reaction.
To measure the efficiency of the selection, two parameters were defined: the yield, which is the proportion of phage polymerases recovered after selection, and the enrichment factor, which is the ratio of phage polymerases in the selected population divided by the ratio of phage polymerases in the population prior to selection. The yield was measured by titering infective phage particles before or after the selection. The enrichment factors were measured by counting phage polymerases and phages not expressing a polymerase (which are characterised by the size of phagemid fragments amplified by PCR) either in the initial phage mixture or in the selected phage mixture.
For the selection experiment using clone 2, the phage titer before selection and after trypsin cleavage was 1.1 × 109 phages/ml; the phage titer after the selection and after trypsin cleavage was 5.25 × 103 phages/ml. These titers correspond to a yield of 4.8 × 10–6.
For the selection experiment involving clone 1, the titers before and after selection were 1.1 × 109 and 9.0 × 105 phages/ml, respectively: the yield of recovered phage is 8.2 × 10–4. For this clone, the enrichments into active phage polymerases were further measured. In the phage mixture before selection, a 1.32-fold excess in phages displaying no polymerase was found by PCR screening and in the phage mixture after selection a 43-fold excess in phage polymerases was observed by PCR screening. The enrichment factor into phage polymerases was then 57. In an independent selection experiment carried out with distinct phage preparations of clone 1 phage polymerases and of phages not expressing a polymerase, an enrichment of 55 was measured. This corresponded to a 23-fold excess of phages displaying no polymerase before selection and a 2.4-fold excess of phage polymerases after the selection.
These results indicate that phage polymerases can be isolated in vitro from mixtures of phages expressing or not expressing a polymerase.
DISCUSSION
In vitro selection of proteins for catalytic activity using phage display has the potential to become a method for the isolation of enzymes endowed with new catalytic activities from large protein libraries. Several selections have been described in the literature which all make use of enzymes expressed as fusions with the N-terminus of the minor coat protein 3 of filamentous phage (1–4). On the contrary, the novel system described in this work has been developed for the isolation of a given catalytic activity from libraries of cDNA expression products on phage. We have demonstrated the feasibility of such a system for DNA polymerase activity.
The phagemid/helper phage strategy is especially useful to create large libraries of proteins on phage. In this system, many phages do not contain any p3 fusion, but only wild-type p3 copies provided by the helper phage; these infective phage particles represent a potential background of the selection. Helper phage engineering by introduction of a protease cleavable helper p3 provides a method to render non-infective by trypsin digestion all phage particles not expressing any fusion, and therefore to reduce the background in selections (4,14).
cDNAs have been expressed on filamentous phage as fusions with the C-terminus minor coat protein p6 (6,12). The background reduction method described for protein 3, which is an essential protein for host infection, may not, however, be adaptable to minor coat protein p6, as it has not been shown to be essential for infection. Therefore, for expression of cDNAs on phage we used the phagemid pJuFo developed by Crameri and Suter (5), which codes for p3D3–Jun and Fos–protein fusions, with one modification. This modification is the substitution of the domain 3 of protein p3 by a complete p3, so that a phage expressing only one copy of non-cleavable p3 fusion remains infective.
The extremely low yield of recovery of phage polymerases for clone 2 may be ascribed to a mutation arising during PCR amplification within the gene encoding the Stoffel fragment of Taq polymerase which may render the enzyme inactive or which may prevent the formation of the Fos–polymerase fusion. The recovery of ∼0.1% of phage displaying a fusion observed for clone 1 can be compared with the yield of ∼1% for phage displaying a polymerase–p3 fusion, which was measured in experiments described earlier (3). This difference may be due to several factors. For example, the two fusion proteins Jun–p3 and Fos–Stoffel fragment may not be produced in stoechiometric amounts. One fusion may also be more efficiently folded or exported in the bacterial periplasm than the other.
The substrate cross-linked on phage may also be more easily accessible to a (p3–polymerase) fusion than to a (p3–Jun/Fos–polymerase) fusion. Substrate cross-linking sites on phage were indeed found on the major coat protein p8 primary amines (3). But other less frequent cross-linking sites may exist although they are not known yet. The insertion of Jun and Fos leucine zippers in the construct might have increased the distance between the polymerase and the substrate cross-linking site. The overall efficiency of the in vitro selection for polymerase activity was therefore not a trivial problem. Furthermore, the structure of filamentous phages’ tip including proteins p3 and p6 are not known. Accordingly, the accessibility of a phage enzyme’s active site by substrates cross-linked to phage may be best evaluated by measuring significant enrichment factors in in vitro selections for catalytic activity.
Using the system described here, enrichment factors of more than 50 have been measured for in vitro selections of polymerase activity from model libraries containing phage polymerases and phages not expressing any polymerase. Such enrichments should be sufficient for the isolation of enzymes and their genes from libraries of more than 107 proteins displayed on phage. It has, for example, been shown that with an enrichment factor around 50, optimal signal sequences can be selected in vitro from a library of more than 107 signal sequences after a sufficiently large number of cycles of amplification and selection (17).
The in vitro selection is described here for the Stoffel fragment of Taq DNA polymerase I. When applied to cDNA expression libraries on filamentous phage, this selection shall yield to the isolation of DNA polymerases in particular, but more generally to the isolation of nucleotidyltransferases which catalyse the addition of at least one biotin-labelled nucleotide (Fig. 1).
The in vitro selection for polymerase activity described here is an affinity selection for the reaction product cross-linked to the phage enzyme that catalysed the reaction from substrate to product. This is the first report of such selections for proteins expressed as N-terminal fusions on phage. We expect that this system will allow the isolation of nucleotidyl transferases from libraries of full-length cDNA expression products. It may be more generally used as a tool for the functional cloning of enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. M. Suter for allowing the use of pJuFo and Dr P. England for discussions and for critical reading of the manuscript. We are grateful to C. Gouyette and to O. Helynck for technical assistance. E.B. acknowledges a CNRS grant (bourse de docteur-ingénieur). Financial support was provided by the MENRT (ACI 1999), by the CNRS (PCV) and by Institut Pasteur (PTR).
REFERENCES
- 1.Pedersen H., Hölder,S., Sutherlin,D.P., Schwitter,U., King,D.S. and Schultz,P.G. (1998) A method for directed evolution and functional cloning of enzymes. Proc. Natl Acad. Sci. USA, 95, 10523–10528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Demartis S., Huber,A., Viti,F., Lozzi,L., Giovannoni,L., Neri,P., Winter,G. and Neri,D. (1999) A strategy for the isolation of catalytic activities from repertoires of enzymes displayed on phage. J. Mol. Biol., 286, 617–633. [DOI] [PubMed] [Google Scholar]
- 3.Jestin J.L., Kristensen,P. and Winter,G. (1999) A method for the selection of catalytic activity using phage display and proximity coupling. Angew. Chem. Int. Ed., 38, 1124–1127. [DOI] [PubMed] [Google Scholar]
- 4.Atwell S. and Wells,J.A. (1999) Selection for improved subtiligases by phage display. Proc. Natl Acad. Sci. USA, 96, 9497–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crameri R. and Suter,M. (1993) Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene, 137, 69–75. [DOI] [PubMed] [Google Scholar]
- 6.Jespers L.S., Messens,J.H., De Keyser,A., Eeckhout,D., Van den Brande,I., Gansemans,Y.G., Lauwereys,M.J., Vlasuk,G.P. and Stanssens,P.E. (1995) Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology, 13, 378–382. [DOI] [PubMed] [Google Scholar]
- 7.Crameri R., Jaussi,R., Menz,G. and Blaser,K. (1994) Display of expression products of cDNA libraries on phage surfaces. A versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur. J. Biochem., 226, 53–58. [DOI] [PubMed] [Google Scholar]
- 8.Kleber-Janke T., Crameri,R., Appenzeller,U., Schlaak,M. and Becker,W.M. (1999) Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int. Arch. Allergy Immunol., 119, 265–274. [DOI] [PubMed] [Google Scholar]
- 9.Crameri R., Hemmann,S. and Blaser,K. (1996) pJuFo: a phagemid for display of cDNA libraries on phage surface suitable for selective isolation of clones expressing allergens. Adv. Exp. Med. Biol., 409, 103–110. [DOI] [PubMed] [Google Scholar]
- 10.Shanmugavelu M., Baytan,A.R., Chesnut,J.D. and Bonning,B.C. (2000) A novel protein that binds juvenile hormone esterase in fat body tissue and pericardial cells of the tobacco hornworm Manduca sexta L. J. Biol. Chem., 275, 1802–1806. [DOI] [PubMed] [Google Scholar]
- 11.Eriksson T.L., Rasool,O., Huecas,S., Whitley,P., Crameri,R., Appenzeller,U., Gafvelin,G. and van Hage-Hamsten,M. (2001) Cloning of three new allergens from the dust mite Lepidoglyphus destructor using phage surface display technology. Eur. J. Biochem., 268, 287–294. [DOI] [PubMed] [Google Scholar]
- 12.Hufton S.E., Moerkerk,P.T., Meulemans,E.V., de Bruine,A., Arends,J.W. and Hoogenboom,H.R. (1999) Phage display of cDNA repertoires: the pVI display system and its applications for the selection of immunogenic ligands. J. Immunol. Methods, 231, 39–51. [DOI] [PubMed] [Google Scholar]
- 13.Hoogenboom H.R., Griffiths,A.D., Johnson,K.S., Chiswell,D.J., Hudson,P. and Winter,G. (1991) Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody Fab heavy and light chains. Nucleic Acids Res., 19, 4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristensen P. and Winter,G. (1998) Proteolytic selection for protein folding using filamentous bacteriophages. Fold Des., 3, 321–328. [DOI] [PubMed] [Google Scholar]
- 15.McCafferty J., Griffiths,A.D., Winter,G. and Chiswell,D.J. (1990) Phage antibodies: filamentous phage displaying antibody variable domains. Nature, 348, 552–554. [DOI] [PubMed] [Google Scholar]
- 16.Lawyer F.C., Stoffel,S., Saiki,R.K., Myambo,K., Drummond,R. and Gelfand,D.H. (1989) Isolation, characterisation and expression in E. coli of the DNA polymerase gene from Thermus aquaticus. J. Biol. Chem., 264, 6427–6437. [PubMed] [Google Scholar]
- 17.Jestin J.L., Volioti,G. and Winter,G. (2001) Improving the display of proteins on filamentous phage. Res. Microbiol., 152, 187–191. [DOI] [PubMed] [Google Scholar]