Abstract
The 5′-cap structure of most spliceosomal small nuclear RNAs (snRNAs) and certain small nucleolar RNAs (snoRNAs) undergoes hypermethylation from a 7-methylguanosine to a 2,2,7-trimethylguanosine structure. 5′-Cap hypermethylation of snRNAs is dependent upon a conserved sequence element known as the Sm site common to most snRNAs. Here we have performed a mutational analysis of U3 and U14 to determine the cis-acting sequences required for 5′-cap hypermethylation of Box C/D snoRNAs. We have found that both the conserved sequence elements Box C (termed C′ in U3) and Box D are necessary for cap hypermethylation. Furthermore, the terminal stem structure that is formed by sequences that flank Box C (C′ in U3) and Box D is also required. However, mutation of other conserved sequences has no effect on hypermethylation of the cap. Finally, the analysis of fragments of U3 and U14 RNAs indicates that the Box C/D motif, including Box C (C′ in U3), Box D and the terminal stem, is capable of directing cap hypermethylation. Thus, the Box C/D motif, which is important for snoRNA processing, stability, nuclear retention, protein binding, nucleolar localization and function, is also necessary and sufficient for cap hypermethylation of these RNAs.
INTRODUCTION
The 5′-ends of primary RNA polymerase II transcripts, including mRNAs, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs), are capped with a monomethyl guanosine structure (m7GpppN or simply m7G). In some of these RNAs the 5′-m7G cap structure has been shown to be an important determinant in RNA transport, stability, splicing and initiation of protein translation (1–8). The m7G cap structures of most newly synthesized snRNAs and certain snoRNAs (but not mRNAs) are converted post-transcriptionally to 2,2,7-trimethylguanosine (m3G) cap structures via the addition of two extra methyl groups to the guanine base (9,10).
5′-Cap hypermethylation has been extensively characterized for spliceosomal snRNAs that function in pre-mRNA splicing (e.g. U1, U2, U4 and U5 snRNAs). m7G-capped precursor snRNAs become hypermethylated in the cytoplasm after being exported from the nucleus (11). Cap hypermethylation of snRNAs requires a sequence element known as the Sm site [consensus PuA(U)4–6GPu] that is present in each snRNA (12,13). The Sm site and flanking RNA sequences/structures mediate binding of the Sm proteins, a group of eight core snRNP proteins (B′, B, D1, D2, D3, E, F and G) (14–17). Sm protein binding appears to be essential for 5′-cap hypermethylation since mutation of the Sm site results in loss of both binding of Sm proteins and cap hypermethylation (18). The hypermethylated cap (which itself is a binding site for an importin α-like adaptor protein called snurportin; 19) together with the Sm proteins function as a bipartite nuclear localization signal that directs the snRNA back into the nucleus (4,20–22).
Less is known regarding the 5′-cap hypermethylation of snoRNAs. SnoRNAs are a large family of conserved RNAs (∼150 species in human cells) that function in the processing and modification (2′-O-methylation and pseudouridylation) of rRNA within the nucleolus (23–27). Depending on their mode of biogenesis and the cell type in which they are expressed, some members of each of the two major classes of snoRNAs (Box C/D and Box H/ACA) receive a trimethylguanosine cap. snoRNAs that are transcribed from independent genes by RNA polymerase II undergo 5′-cap hypermethylation (23,28). However, numerous snoRNAs are processed from the introns of pre-mRNAs or polycistronic pre-snoRNA transcripts and contain 5′-phosphate termini rather than cap structures (23,27,29). Interestingly, if artificially provided with an m7G cap structure, intronic Box C/D snoRNAs will become hypermethylated, indicating that sequences required for snoRNA cap hypermethylation are also present in intronic snoRNAs (30,32). Unlike most other cellular RNAs, snoRNAs are actively retained in the nucleus and snoRNA cap hypermethylation appears to normally occur in the nucleoplasm rather than in nucleoli (31,32). A conserved functional role of the trimethylated cap structure in snoRNP biogenesis or function has not been demonstrated but the m3G cap may be important for transport of certain snoRNAs to the nucleolus in some cell types (31) but not others (33–35).
A systematic analysis of the cis-acting sequences required for 5′-cap hypermethylation of snoRNAs has not been reported. Previously, the Box D sequence element was shown to be critical for 5′-cap hypermethylation of Box C/D snoRNAs in vivo and in vitro (32). More recently, we have observed that four U3 snoRNA variants, each containing substitution mutations in two conserved Box elements (i.e. Boxes B + C′, B + D, C + C′ and C + D), failed to be hypermethylated in vivo (36). Two of these double mutants did not include Box D mutations, indicating that sequences in addition to Box D were required for cap hypermethylation of U3 snoRNA. In this work we have determined the cis-acting sequences of Box C/D snoRNAs required for efficient cap hypermethylation in Xenopus oocytes through a mutational analysis of U3 and U14 snoRNAs. Our findings indicate that the Box C/D motif, consisting of the conserved Box C (C′ in U3) and Box D elements and a stem structure that tethers the two elements in close proximity, is both necessary and sufficient for 5′-cap hypermethylation of U3 and U14 and likely all other Box C/D snoRNAs.
MATERIALS AND METHODS
Generation of snoRNA mutant constructs
All of the U3 mutants and subfragments utilized in this manuscript were produced by PCR methods as previously described (35,36). The U14 constructs were generated by PCR methods using the U14.5 gene (37) as previously described (35). Briefly, the following base substitutions were introduced into U14: ΔC, TGATGA→ACTACT; ΔTS, GCGAAT→CGCTTA; ΔTSR, ATTCGC→TAAGCG. The U14ΔAV construct (38) was amplified by PCR using the oligonucleotides ATTTAGGTGACACTATAGATTCGCTGTGATGATGGATTCCA and TTCGCTCAGACATCCAAGG to eliminate flanking intronic sequences and to add an SP6 promoter (underlined sequence).
In vitro transcription
PCR products or linearized plasmids were utilized as templates for transcription. SP6 or T7 RNA polymerase were used to generate m7G-capped, 32P-labeled RNAs using reaction conditions essentially as previously described (8). In vitro transcription was also utilized to produce control RNAs as previously described: Xenopus U8 (34), Xenopus U1, U1sm– and U6 (8). All RNAs received m7GpppG caps during in vitro transcription except U6 snRNA, which received its natural methyl-pppG cap structure (39).
Xenopus oocyte nuclear injections
The procedure by which oocytes were microinjected and micromanipulated has been described elsewhere in detail (35,40). In brief, 10 nl of a 20 mg/ml blue dextran solution containing ∼1 fmol of each 32P-labeled RNA was injected into the nucleus of a stage V or VI oocyte. The injected oocytes were maintained in MBSH buffer at 18°C until they were manually dissected in J buffer (41) into nuclear and cytoplasmic fractions. The RNA in the nuclear and cytoplasmic fractions was isolated from between three and five oocytes by proteinase K digestion, phenol extraction and ethanol precipitation. RNA stability and nucleocytoplasmic distribution were determined by PAGE analysis of 32P-labeled RNA from one nuclear or one cytoplasmic equivalent on a denaturing 8% urea–polyacrylamide gel. The labeled RNAs were detected by autoradiography.
Immunoprecipitations
Immunoprecipitations were performed using purified RNA from one oocyte nuclear equivalent as described (35). Polyclonal antibodies that specifically recognize either the m7G (42) or m2,2,7G (43) cap were utilized.
RESULTS
Nuclear hypermethylation of U3 depends on elements of the Box C/D motif
The aim of this study was to determine the RNA sequence requirements for hypermethylation of Box C/D snoRNAs. U3, the major focus of this study, contains six short conserved sequence elements called Boxes A′, A, C′, B, C and D (Fig. 1A). The primary sequence of the so-called ‘hinge’ region is not conserved, but like the conserved elements in the 5′-domain, Box A′ and Box A, the hinge is of functional importance in U3 base pairing with pre-rRNA (44–47). The relative order of the U3 Box elements and the hinge region in the primary sequence is invariant in U3 molecules from diverse organisms (28,46,47,49–53). While distant from one another in primary sequence, RNA folding brings the Box B and Box C as well as Box C′ and Box D elements adjacent to one another within loop structures (Fig. 1A). Both the resultant Box B/C and Box C′/D motifs of U3 are located in the 3′-domain of the molecule.
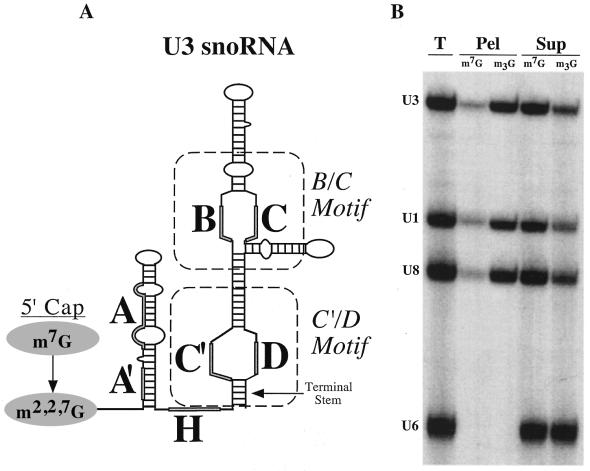
Figure 1.
(A) Secondary structural model of the Xenopus U3 snoRNA. The locations of the phylogenetically conserved Box sequences (A′, A, C′, B, C and D) as well as the hinge region (H) are shaded. The 5 bp terminal stem is denoted by an arrow. The general areas of the C′/D and B/C motifs are marked with dashed lines. The two cap forms (m7G and m2,2,7G) associated with precursor and mature snoRNAs, respectively, are indicated. (B) Identification of the 5′-cap structure (either m7G or m2,2,7G) by immunoprecipitation. 32P-labeled, m7G-capped U3, U1 and U8 and methyl-pppG capped U6 RNAs were injected into oocyte nuclei. RNAs present in the nucleus 8 h after nuclear injection were isolated and subsequently precipitated using anti-m7G (m7G) (42) or anti-m2,2,7G (m3G) (43) cap antibodies as indicated. RNAs from the input total (T), pellet (Pel) and supernatant (Sup) fractions were separated by denaturing polyacrylamide gel electrophoresis and assessed by autoradiography.
To determine precisely which conserved sequence elements of U3 RNA are necessary for cap hypermethylation, we systematically mutated U3 by replacing every nucleotide of each Box element and the hinge region with its Watson–Crick complement (Materials and Methods). 32P-labeled, m7G-capped U3 RNA variants were synthesized by in vitro transcription and subsequently injected into Xenopus laevis oocyte nuclei. Co-injected with the U3 RNAs were U1, U8 and U6 control RNAs. Eight hours after injection, the nuclear RNAs were recovered and assayed for cap hypermethylaton by immunoprecipitations using antibodies specific to the monomethyl- (m7G) and trimethylguanosine (m3G) cap structures. The RNAs were separated and assessed by polyacrylamide gel electrophoresis. As expected, wild-type m7G-capped U3 and U8 Box C/D snoRNAs were efficiently hypermethylated in oocyte nuclei (Fig. 1B). m7G-capped U1 snRNA was also hypermethylated and served as a positive control for cap hypermethylation in our experiments. U6 snRNA was used as a negative control since its unique methyl-pppG cap structure (39) is not recognized by either antibody.
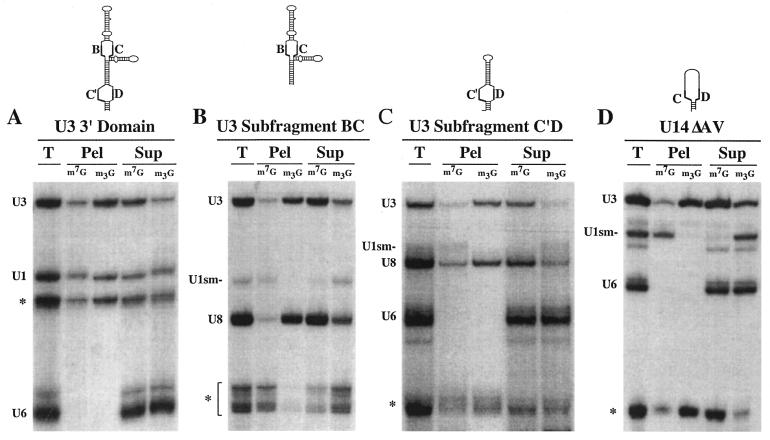
Through our analysis we discovered that 5′-cap hypermethylation was unaffected by substitution mutation (Δ) of U3 Box elements A′, A, B and C and the hinge region (Fig. 2 and data not shown). However, mutation of either Box C′ or Box D disrupted cap hypermethylation (U3ΔC′ and U3ΔD, Fig. 2). No detectable amount of the Box D variant and only a very small fraction of the Box C′ variant of U3 were immunoprecipitated by the trimethylguanosine (m3G) cap antibodies. These results, in conjunction with the previous finding that the terminal stem of U3 is required for cap hypermethylation (32,54), indicate that elements of the Box C/D motif (comprised of Boxes C′ and D and the terminal stem) but not other conserved elements of U3 are necessary for 5′-cap hypermethylation of U3.
Figure 2.
Box C′ and Box D of U3 snoRNA are required for efficient 5′-cap hypermethylation. The 5′-cap structures (m7G or m2,2,7G) of variant U3 snoRNAs with substitution mutations were identified 8 h after nuclear injection by immunoprecipitation using anti-m7G (m7G) and anti-m2,2,7G (m3G) antibodies. RNAs from the total (T), pellet (Pel) and supernatant (Sup) fractions were analyzed by PAGE and autoradiography. Results of U3 variants with substitution mutations in the conserved Box A (ΔA), Box C′ (ΔC′), Box B (ΔB), Box C (ΔC) and Box D (ΔD) sequences as well as substitution mutation of the hinge region (ΔH) are shown. The mutant labeled ΔH contains a mutation in both the hinge region and Box A. Each U3 variant experiment included the use of co-injected control RNAs U1 and U6 and a typical immunoprecipitation distribution pattern for each is shown.
The Box C/D motif also mediates 5′-cap hypermethylation of U14
To determine if the Box C/D motif is of general importance in 5′-cap hypermethylation of Box C/D snoRNAs, we assayed wild-type and sequence variants of another Box C/D snoRNA, U14. In metazoans U14 is encoded within introns of pre-mRNA and lacks a 5′-cap structure (55). However, U14 is an efficient substrate for cap hypermethylation in vivo when it is synthesized with an m7G cap structure in vitro (32). Indeed, it was previously shown that cap hypermethylation of in vitro transcribed, capped U14 is dependent upon an intact Box D sequence element (32).
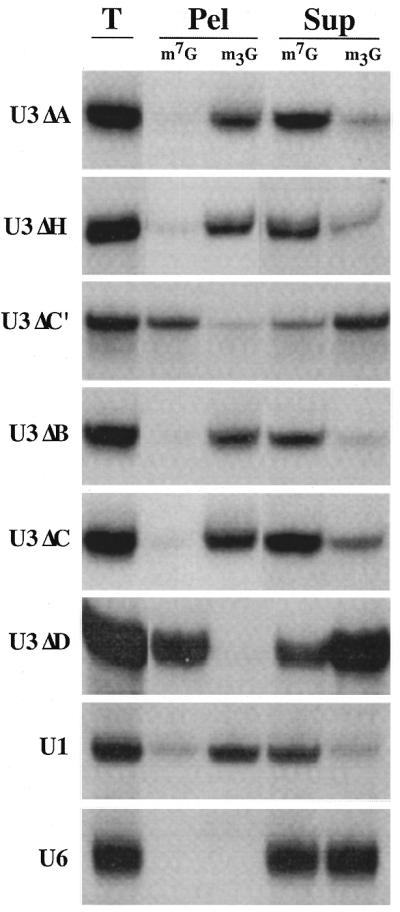
We have compared the hypermethylation of wild-type U14 and sequence variants of U14 (Fig. 3). Wild-type U14 with an m7G cap is efficiently hypermethylated in Xenopus oocyte nuclei (Fig. 3A; 32). However, the cap of the U14 variant containing a Box C substitution mutation did not undergo significant cap hypermethylation (U14ΔC, Fig. 3B). We next tested if an intact terminal stem was required for the 5′-cap hypermethylation of U14. Short stem structures are present at the termini of many Box C/D snoRNAs and bring Box C (C′ in U3) and Box D sequences adjacent to one another in the predicted secondary structures of the RNAs (35,36,38,56,57). The terminal stem of U14 was disrupted by substituting the 3′-strand of the stem with complementary sequences to abolish Watson–Crick base pairing. This variant remained m7G capped after injection into the oocyte, indicating the essential nature of the 3′-terminal stem in U14 cap hypermethylation (U14ΔTS, Fig. 3C). However, restoration of the stem by compensatory changes in the sequences of the 5′-strand of the U14 terminal stem (U14ΔTSR) restored the ability of the RNA to become hypermethylated at its 5′-cap (Fig. 3D). Thus, the helical structure and not the sequence identity of the terminal stem is critical for cap hypermethylation. Taken together, our results with U14 provide additional evidence that the Box C/D motif (including Box C and Box D and the terminal stem structure) is required for 5′-cap hypermethylation of Box C/D snoRNAs.
Figure 3.
The Box C/D motif of U14 is necessary for cap hypermethylation. Nuclear injections of in vitro transcribed RNAs were performed as in Figure 1. Four hours after injection the nuclear RNAs were isolated and analyzed using anti-m7G (m7G) or anti-m2,2,7G (m3G) antibodies. Diagrams representing the injected U14 and U14 mutant RNAs (adapted from Shanab and Maxwell; 79) are shown on the left (for precise sequences see Materials and Methods). The results of the cap hypermethylation experiments for wild-type U14 (A), the U14 Box C mutant (U14ΔC) (B), the U14 terminal stem mutant (U14ΔTS) (C) and the U14 stem restoration mutant (U14ΔTSR) (D) are shown on the right. Due to the relative instability and nuclear export of U14ΔC and U14ΔTS, a longer autoradiographic exposure (3- and 2-fold, respectively) is shown for these RNAs relative to the control U1 and U6 RNAs.
Minimal sequence sufficient for cap hypermethylation
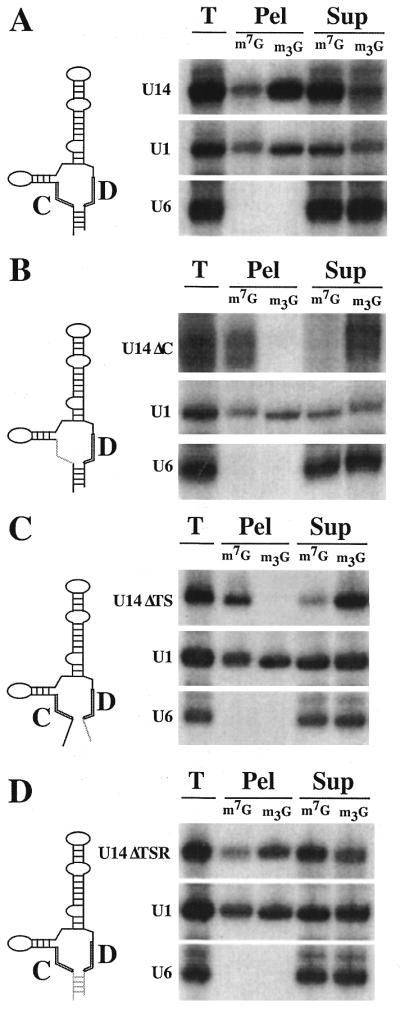
In an effort to identify the minimal nucleotide sequences required for cap hypermethylation, we tested a series of Box C/D snoRNA subfragments. A U3 snoRNA fragment in which the entire 5′-domain and hinge region (nt 1–75) was deleted underwent 5′-cap hypermethylation to a similar extent as co-injected wild-type U3 (Fig. 4A). This finding supports our mutational analysis which showed that the hinge region and conserved Boxes A′ and A were not required for cap hypermethylation (Fig. 2 and data not shown). Furthermore, this result indicates that the information required for efficient cap hypermethylation resides within the 3′-domain of U3, which includes the Box B/C and Box C′/D motifs (see Fig. 1A). The U3 3′-domain fragment was then further subdivided such that the B/C motif and the C′/D motif could be independently assayed for cap maturation. The lack of hypermethylation observed with the subfragment that includes the B/C motif (Box B, Box C and flanking stem regions) (Fig. 4B) is consistent with our findings that the conserved Box B and Box C elements were unnecessary for cap hypermethylation (Fig. 2). In contrast, a subfragment containing the C′/D motif, including Box C′, Box D and flanking stem regions, underwent significant 5′-cap hypermethylation (Fig. 4C). Furthermore, a U14 snoRNA subfragment (U14ΔAV) in which the majority of internal sequence between Box C and Box D is deleted (38) was hypermethylated to an extent comparable with the co-injected wild-type U3 (Fig. 4D). These results indicate that the Box C/D motif is sufficient for 5′-cap maturation.
Figure 4.
Analysis of the ability of stable subfragments of U3 and U14 to undergo 5′-cap hypermethylation. Nuclear RNAs were recovered from oocytes 4 h (U3 3′ Domain, U3 Subfragment BC and U14ΔAV) or 8 h (U3 Subfragment C′D) after nuclear injection and analyzed by immunoprecipitation. Diagrams of the U3 3′-domain fragment (A), U3 subfragment BC (B), U3 subfragment C′D (C) and U14ΔAV subfragment (D) are shown above the corresponding cap hypermethylation results (see Materials and Methds for descriptions of fragments). In each panel the subfragment is marked with an asterisk (*). Wild-type U3 served as an internal positive control in (A)–(D). Wild-type U1 was included in (A) while a variant U1 RNA (U1sm– RNA), lacking a functional Sm binding site and unable to undergo cap hypermethylation, was included in (B)–(D).
DISCUSSION
We have determined the cis-acting elements required for 5′-cap hypermethylation of U3 and U14 Box C/D snoRNAs. A systematic mutational analysis of conserved elements demonstrated that the Box C/D motif, including Box C (C′ in U3), Box D and the terminal stem, is responsible for m2,2,7G cap formation in these RNAs. Two experimental lines of evidence support our conclusion that the Box C/D motif is both necessary and sufficient for 5′-cap hypermethylation. First, for both U3 and U14, 5′-cap hypermethylation was prevented when mutations were introduced in Box C or Box D or when the terminal stem was disrupted (Figs 2 and 3; 32) but not when mutations were present in other conserved elements (Figs 2 and 4). Second, deletion analysis showed that minimal RNAs consisting essentially of the Box C/D motif were efficient substrates for 5′-cap hypermethylation (Fig. 4). Our results indicate that a common mechanism exists for hypermethylation of m7G-capped Box C/D snoRNAs in many diverse organisms.
It was over 30 years ago when it was first discovered that certain nuclear RNAs (including both snRNAs and snoRNAs) contain 5′-trimethylguanosine cap structures (9,10). Since that time, significant advances have been made, primarily in understanding how the spliceosomal snRNAs (U1, U2, U4 and U5) become hypermethylated in the cell and what role the trimethylguanosine 5′-cap plays in the biogenesis of these RNAs. Studies performed mainly with the Xenopus oocyte system have shown that m7G-capped precursor spliceosomal snRNAs must first be exported to the cytoplasm to acquire a trimethylguanosine cap structure (11,21). The exact cytoplasmic events leading to the conversion of an m7G to an m2,2,7G cap structure are not fully understood but binding of the Sm proteins to a conserved RNA motif consisting of the Sm site [consensus PuA(U)4–6GPu] and flanking sequences (17) present within each snRNA is crucial for cap hypermethylation (11). Sm protein binding apparently generates a suitable RNP substrate for hypermethylation. The methyltransferase responsible for cap hypermethylation is not included among the bound Sm proteins (58) and remains unidentified. The trimethylguanosine cap together with bound Sm proteins provide the signals required for import of the cytoplasmic snRNPs to the nucleus (20,21), where they function in pre-mRNA splicing. The trimethylguanosine cap of snRNAs serves as a binding site for a protein known as snurportin that acts as an adaptor to link the snRNP to a nuclear import receptor (19). Interestingly, the trimethylguanosine cap structure is not essential for import of all snRNA species and its relative importance to snRNP import is cell-type dependent (59–61).
Interesting similarities and differences between the 5′-cap hypermethylation of snRNAs and snoRNAs are becoming apparent. While snRNAs receive their hypermethylated 5′-caps within the cytoplasm, snoRNAs are not exported and undergo 5′-cap hypermethylation within the nucleus (32,36,62). Unlike snRNAs, snoRNAs lack Sm sites and fail to bind Sm proteins. Instead, our data indicate that the Box C/D motif, which is common to this class of snoRNAs, provides the cis-acting information that directs cap hypermethylation of these RNAs. Previous studies revealed that variant U3 RNAs containing two mutated box elements (i.e. Boxes B + C′, B + D, C + C′ and C + D) failed to be hypermethylated in vivo, raising the possibility that each Box B, Box C, Box C′ and Box D may be required for cap hypermethylation (36). It is clear from the present study that Box B and Box C of U3 are not important for cap hypermethylation. The earlier results are explained since each of the four double Box mutants contained a mutation in either the Box C′ or Box D elements.
By analogy with the snRNPs we propose that nuclear proteins interact with the Box C/D motif to convert the m7G-capped snoRNAs into RNP substrates (snoRNPs) recognized by a nuclear methyltransferase enzyme. Structural probing of snoRNP complexes has provided evidence that the Box C/D motif indeed serves as a phylogentically conserved protein binding site (47,49,53,63) and multiple common Box C/D snoRNP proteins have been identified (28,64–70). While the identity of the nuclear methyltransferase responsible for 5′-cap hypermethylation of Box C/D snoRNAs is not yet known, numerous candidate genes encoding putative S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases exist (71). In fact, fibrillarin/Nop1p, Nop2p, Ncl1p, Spblp and Dim1p are each nucleolar proteins important in pre-rRNA processing that have either been demonstrated to be or are suspected to be methyltransferases (72–78). In most cases the full spectrum of methylation substrates of these enzymes is unknown.
Dissection of the cis-acting sequences that direct hypermethylation of Box C/D snoRNAs has provided important information toward understanding how these RNAs acquire their hypermethylated 5′-cap structures. Further studies are required to identify and characterize the role of trans-acting factors (including the 5′-cap methyltransferase) in this process. A detailed mechanistic understanding of the reaction should be aided by the development of an accurate in vitro snoRNA 5′-cap hypermethylation system (32). Finally, a conserved function for the trimethylguanosine cap in snoRNP biogenesis and/or function has not yet been found. The observations that an m7G cap was required for nucleolar localization of Box C/D snoRNAs U3 and U8 and that nucleolar localization was competed by excess m7G cap dinucleotide (31) are consistent with a role of hypermethylation in the nucleolar targeting of these snoRNAs. However, several studies utilizing Xenopus oocytes clearly show that a hypermethylated cap structure is not essential for nucleolar localization (33–35). Furthermore, the nucleolar targeting of numerous Box C/D snoRNAs which are processed from pre-mRNA introns is clearly cap-independent. The available evidence also indicates that the hypermethylated cap structure may not be serving a critical functional role within the nucleolus during rRNA processing (34).
Hypermethylation of snoRNAs is likely an early event in the biogenesis of snoRNP complexes and occurs prior to functional localization of snoRNPs to nucleoli. Biochemical evidence indicates that cap hypermethylation takes place in the nucleoplasm rather than nucleoli (32). Other data indicate that cap hypermethylation may be essential for the localization of U3 and U8 snoRNAs to nucleoli in injected somatic cells (31). Our analysis reveals that snoRNA 5′-cap hypermethylation and snoRNA function are not obligatorily coupled, since functionally incompetent mutant snoRNAs are nevertheless capable of becoming hypermethylated in vivo. Specifically, our results show that Box A′, Box A and the hinge region of U3 (Figs 2 and 4 and data not shown), which are essential for the base pairing of U3 snoRNA to pre-rRNA and for the function of U3 in pre-rRNA processing (44–48), are not required for cap hypermethylation. In addition, Box B and Box C, the key sequence elements of the functionally important B/C motif of U3 (36,52), are also not required for 5′-cap hypermethylation. Together these findings indicate that cap hypermethylation occurs independent of localization to nucleoli and of ability to function.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Reinhard Luhrmann (anti-m2,2,7G antibodies), Elsebet Lund and James Dahlberg (anti-m7G antibodies), Stu Maxwell (U14 constructs) and Ram Reddy (me-pppG U6 cap dinucleotide) for providing valuable reagents. This work was supported by a grant from the National Institutes of Health (GM54682) to M.P.T.
REFERENCES
- 1.Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- 2.Caponigro G. and Parker,R. (1996) Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty S.M., Fortes,P., Izaurralde,E., Mattaj,I.W. and Gilmartin,G.M. (1997) Proc. Natl Acad. Sci. USA, 94, 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamm J. and Mattaj,I.W. (1990) Cell, 63, 109–118. [DOI] [PubMed] [Google Scholar]
- 5.Gallie D.R. (1991) Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 6.Izaurralde E., Lewis,J., McGuigan,C., Jankowska,M., Darzynkiewicz,E. and Mattaj,I.W. (1994) Cell, 78, 657–668. [DOI] [PubMed] [Google Scholar]
- 7.Izaurralde E., Lewis,J., Gamberi,C., Jarmolowski,A., McGuigan,C. and Mattaj,I.W. (1995) Nature, 376, 709–712. [DOI] [PubMed] [Google Scholar]
- 8.Terns M.P., Dahlberg,J.E. and Lund,E. (1993) Genes Dev., 7, 1898–1908. [DOI] [PubMed] [Google Scholar]
- 9.Reddy R., Ro,C.T., Henning,D., Shibata,H., Choi,Y. and Busch,H. (1972) J. Biol. Chem., 247, 7245–7250. [PubMed] [Google Scholar]
- 10.Saponara A.G. and Enger,M.D. (1969) Nature, 223, 1365–1366. [DOI] [PubMed] [Google Scholar]
- 11.Mattaj I.W. (1986) Cell, 46, 905–911. [DOI] [PubMed] [Google Scholar]
- 12.Branlant C., Krol,A., Ebel,J.P., Lazar,E., Haendler,B. and Jacob,M. (1982) EMBO J., 1, 1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liautard J.P., Sri-Widada,J., Brunel,C. and Jeanteur,P. (1982) J. Mol. Biol., 162, 623–643. [DOI] [PubMed] [Google Scholar]
- 14.Sauterer R.A., Goyal,A. and Zieve,G.W. (1990) J. Biol. Chem., 265, 1048–1058. [PubMed] [Google Scholar]
- 15.Raker V.A., Plessel,G. and Luhrmann,R. (1996) EMBO J., 15, 2256–2269. [PMC free article] [PubMed] [Google Scholar]
- 16.Raker V.A., Hartmuth,K., Kastner,B. and Luhrmann,R. (1999) Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarmolowski A. and Mattaj,I.W. (1993) EMBO J., 12, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattaj I.W. and De Robertis,E.M. (1985) Cell, 40, 111–118. [DOI] [PubMed] [Google Scholar]
- 19.Huber J., Cronshagen,U., Kadokura,M., Marshallsay,C., Wada,T., Sekine,M. and Luhrmann,R. (1998) EMBO J., 17, 4114–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamm J., Darzynkiewicz,E., Tahara,S.M. and Mattaj,I.W. (1990) Cell, 62, 569–577. [DOI] [PubMed] [Google Scholar]
- 21.Fischer U. and Luhrmann,R. (1990) Science, 249, 786–790. [DOI] [PubMed] [Google Scholar]
- 22.Fischer U., Sumpter,V., Sekine,M., Satoh,T. and Luhrmann,R. (1993) EMBO J., 12, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell E. and Fournier,M. (1995) Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 24.Smith C. and Steitz,J. (1997) Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 25.Weinstein L.B. and Steitz,J.A. (1999) Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 26.Venema J. and Tollervey,D. (1999) Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 27.Tollervey D. and Kiss,T. (1997) Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 28.Tyc K. and Steitz,J.A. (1989) EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sollner-Webb B. (1993) Cell, 75, 403–405. [DOI] [PubMed] [Google Scholar]
- 30.Balakin A.G., Lempicki,R.A., Huang,G.M. and Fournier,M.J. (1994) J. Biol. Chem., 269, 739–746. [PubMed] [Google Scholar]
- 31.Jacobson M.R. and Pederson,T. (1998) Nucleic Acids Res., 26, 756–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terns M.P., Grimm,C., Lund,E. and Dahlberg,J.E. (1995) EMBO J., 14, 4860–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lange T.S., Borovjagin,A.V. and Gerbi,S.A. (1998) RNA, 4, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peculis B. and Steitz,J. (1994) Genes Dev., 8, 2241–2255. [DOI] [PubMed] [Google Scholar]
- 35.Narayanan A., Speckmann,W., Terns,R. and Terns,M.P. (1999) Mol. Biol Cell., 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speckmann W., Narayanan,A., Terns,R. and Terns,M.P. (1999) Mol. Cell. Biol., 19, 8412–8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanab G.M. and Maxwell,E.S. (1992) Eur. J. Biochem., 206, 391–400. [DOI] [PubMed] [Google Scholar]
- 38.Watkins N., Leverette,R., Xia,L. andrews,M. and Maxwell,E. (1996) RNA, 2, 118–133. [PMC free article] [PubMed] [Google Scholar]
- 39.Singh R., Gupta,S. and Reddy,R. (1990) Mol. Cell. Biol., 10, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terns M.P. and Goldfarb,D.S. (1998) Methods Cell Biol., 53, 559–589. [DOI] [PubMed] [Google Scholar]
- 41.Birkenmeier E.H., Brown,D.D. and Jordan,E. (1978) Cell, 15, 1077–1086. [DOI] [PubMed] [Google Scholar]
- 42.Munns T.W., Liszewski,M.K., Tellam,J.T., Sims,H.F. and Rhoads,R.E. (1982) Biochemistry, 21, 2922–2928. [DOI] [PubMed] [Google Scholar]
- 43.Bringmann P., Rinke,J., Appel,B., Reuter,R. and Luhrmann,R. (1983) EMBO J., 2, 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beltrame M. and Tollervey,D. (1995) EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borovjagin A.V. and Gerbi,S.A. (2000) J. Mol. Biol., 300, 57–74. [DOI] [PubMed] [Google Scholar]
- 46.Hughes J. (1996) J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 47.Mereau A., Fournier,R., Gregoire,A., Mougin,A., Fabrizio,P., Luhrmann,R. and Branlant,C. (1997) J. Mol. Biol., 273, 552–571. [DOI] [PubMed] [Google Scholar]
- 48.Sharma K. and Tollervey,D. (1999) Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeppesen C., Stebbins,B.B. and Gerbi,S. (1988) Nucleic Acids Res., 16, 2127–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiss T. and Solymosy,F. (1990) Nucleic Acids Res., 18, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porter G.L., Brennwald,P.J., Holm,K.A. and Wise,J.A. (1988) Nucleic Acids Res., 16, 10131–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samarsky D. and Fournier,M. (1998) Mol. Cell. Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hartshorne T. and Agabian,N. (1994) Nucleic Acids Res., 22, 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baserga S.J., Gilmore-Hebert,M. and Yang,X.W. (1992) Genes Dev., 6, 1120–1130. [DOI] [PubMed] [Google Scholar]
- 55.Leverette R.D. andrews,M.T. and Maxwell,E.S. (1992) Cell, 71, 1215–1221. [DOI] [PubMed] [Google Scholar]
- 56.Samarsky D., Fournier,M., Singer,R. and Bertrand,E. (1998) EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiss-Laszlo Z., Henry,Y., Bachellerie,J., Caizergues-Ferrer,M. and Kiss,T. (1996) Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 58.Plessel G., Fischer,U. and Luhrmann,R. (1994) Mol. Cell. Biol., 14, 4160–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fischer U., Darzynkiewicz,E., Tahara,S.M., Dathan,N.A., Luhrmann,R. and Mattaj,I.W. (1991) J. Cell Biol., 113, 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer U., Heinrich,J., van,Z.K., Fanning,E. and Luhrmann,R. (1994) J. Cell Biol., 125, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marshallsay C. and Luhrmann,R. (1994) EMBO J., 13, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terns M.P. and Dahlberg,J.E. (1994) Science, 264, 959–961. [DOI] [PubMed] [Google Scholar]
- 63.Parker K. and Steitz,J. (1987) Mol. Cell. Biol., 7, 2899–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caffarelli E., Losito,M., Giorgi,C., Fatica,A. and Bozzoni,I. (1998) Mol. Cell. Biol., 18, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lafontaine D.L. and Tollervey,D. (2000) Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lafontaine D.L. and Tollervey,D. (1999) RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E. (1989) EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watkins N., Newman,D., Kuhn,J. and Maxwell,E. (1998) RNA, 4, 582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu P., Brockenbrough,J., Metcalfe,A., Chen,S. and Aris,J. (1998) J. Biol. Chem., 273, 16453–16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niewmierzycka A. and Clarke,S. (1999) J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- 72.Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.H. (2000) EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu P., Brockenbrough,J.S., Paddy,M.R. and Aris,J.P. (1998) Gene, 220, 109–117. [DOI] [PubMed] [Google Scholar]
- 74.Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E.C. (1993) Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- 75.Pintard L., Kressler,D. and Lapeyre,B. (2000) Mol. Cell. Biol., 20, 1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lafontaine D., Vandenhaute,J. and Tollervey,D. (1995) Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- 77.Hong B., Brockenbrough,J., Wu,P. and Aris,J. (1997) Mol. Cell. Biol., 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.de B.E., Brockenbrough,J., Hong,B. and Aris,J. (1994) J. Cell Biol., 127, 1799–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shanab G.M. and Maxwell,E.S. (1991) Nucleic Acids Res., 19, 4891–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]