Abstract
Neural EGFL like 1 (NELL-1), is a secreted glycoprotein and stimulates osteogenic cell differentiation and bone mineralization. This study aimed to explore the relationship between NELL-1 and Trabecular Bone Score (TBS) as a novel tool for the evaluation of osteoporosis in an elderly population-based cohort study in Iran. A single-locus analysis was performed on TBS using data from 2,071 participants in the Bushehr Elderly Health (BEH) Program. The study investigated 376 independent single nucleotide polymorphisms (SNPs) within the NELL-1 on chromosome 11p15.1. The association between SNPs and the mean TBS L1 to L4 was analyzed through an additive model. Significant variants in the additive model (PFDR<0.05) were further examined within dominant, recessive, over-dominant, and co-dominant models. Multiple linear regression was employed to assess the relationship between the genetic risk score (GRS) derived from significant SNPs and TBS. Three SNPs within the NELL-1 showed a statistically significant association with TBS after adjusting for age and sex. The associations for rs1901945 (β = 0.013, PFDR = 0.0007), rs1584851 (β = -0.011, PFDR = 0.0003), and rs58028601 (β = 0.011, PFDR = 0.0003) were significant in the additive model. Additionally, significant results were observed for rs1901945 and rs58028601 in the dominant model (P<0.05). The GRS showed a statistically significant relationship with TBS, considering adjustments for age, sex, Body Mass Index, type 2 diabetes, and smoking (β = 0.077, P = 1.7×10−5). This study highlights the association of NELL-1 with TBS, underscoring its potential as a candidate for further research and personalized medicine concerning the impact of this gene on bone quality.
Introduction
Osteoporosis is a systemic skeletal disease defined as a bone mineral density (BMD) 2.5 Standard deviation or more below that of young, healthy women in lumbar spine, total hip, or femoral neck assessed by DXA [1, 2]. The global burden of osteoporosis is increasing; its Worldwide prevalence in women and men is estimated at 23.1% and 11.7%, respectively [3, 4]. The age-standardized prevalence of osteoporosis among Iranian men and women aged 60 years or older, was 24.6% and 62.7%, respectively [5]. Additionally, the estimated annual cumulative incidence of hip fractures within this population is reported at 138 per 100,000 in men and 157 per 100,000 in women [6]. The economic ramifications of osteoporosis are substantial, with the economic burden in Iran reaching an estimated US$ 393.24 million in 2020 [7].
A significant number of osteoporotic fractures occur in individuals with normal BMD scores, indicating that factors beyond bone density, such as bone microarchitecture, play a significant role in fracture risks [8–11]. The Trabecular Bone Score (TBS) is a novel and complementary index that evaluates bone microarchitecture based on pixel variation in gray levels in the lumbar spine dual-energy X-ray absorptiometry (DXA) image [12–18]. TBS and BMD are highly correlated and share similar heritability with genetic factors accounting for around 45% of the variance in TBS. This indicates that genetics play an important role in determining both bone mineral density and microarchitecture [19]. A low TBS is related to an increased fracture risk, independent of BMD, and combining these two parameters improves the fracture risk prediction process [14–17].
The development of osteoporosis results from the interaction between environmental, metabolic and genetic factors, each exerting modest effects on bone metabolism and fracture susceptibility [20–24]. Although osteoporosis is in the early stages of personalized medicine implementation, polygenic risk scores based on multiple genetic variants have allowed for the assessment of an individual’s genetic susceptibility to the condition [25, 26].
Neural EGFL like 1 (NELL-1) is a protein-coding gene that encodes a cytoplasmic protein and is involved in regulating cell growth and differentiation [27]. NELL-1 promotes osteoblast cell and osteoblastic mineralization and proliferation [28]. Recent studies have revealed the role of NELL-1 in bone health. Researchers have found, for the first time, that NELL-1 expression is increased in cranial intramembranous bone from patients with craniosynostosis [29]. Several investigations have consistently shown that NELL-1 plays a crucial role in the osteogenic differentiation of osteoblasts, and it has been found to interact synergistically with other genes implicated in the development of osteoporosis, such as Bone Morphogenetic Protein 2 (BMP2) [30–32]. NELL-1 has been identified as a promising therapeutic target for treating osteoporosis in animal studies. The findings revealed that the groups treated with NELL-1 indicated increased bone formation and enhanced new bone growth compared to the control group [33, 34].
Genetic risk factors can be varied across different human populations [35]. Given the novelty of TBS, no prior studies have investigated genes potentially influencing TBS in the Iranian population. The investigation of genetic loci associated with TBS is crucial for personalized medicine, specifically in identifying high-risk elderly individuals at high risk with low TBS. Our study was the first to demonstrate the genetic risk of NELL-1 on TBS within the Iranian population.
Materials and methods
Participants
The present study was conducted on 2,071 participants of the Bushehr Elderly Health (BEH) study. This population-based prospective cohort study initiated in 2013, comprises a total of 3,000 adults, including 1,000 men (48.2%), all aged 60 years or older, selected through a multistage cluster random sampling method [36]. The main objective of the second measurement of the first phase was to investigate musculoskeletal health and the related risk factors and consequences in 2,772 participants who were under follow-up in 2015 [37].
Genotyping and quality control
Blood samples obtained from the participants in the BEH study were genotyped using the Infinium Global Screening Array (GSA) from Illumina. A total of 47,109,443 autosomal SNPs were imputed, and the quality of the imputation was assessed by means of MACH R-square. A systematic and comprehensive approach was adopted to implement genomic quality control (QC) [38]. Good quality was defined as a MACH R-square above 0.3 and SNPs with a Hardy–Weinberg equilibrium P<10−6, and minor allele frequency ≤0.01 were excluded from the analysis. All SNPs and gene locations were relative to the GRCh37 genome assembly.
Gene selection
The NELL-1 is a protein-coding gene located on chromosome 11 (11p15.1), with its genomic region spanning from position 20,691,097 to 21,597,232 (GRCh37) on the chromosome (27). To ensure comprehensive coverage of potential primer regions, we considered a genomic region spanning approximately ±5 kb surrounding the gene. Following quality control procedures, the dataset contained 3,451 SNPs related to the NELL-1 gene. Out of the extracted SNPs, 376 independent SNPs with a low linkage disequilibrium (r2≤0.2) were selected.
Outcome measures
The Trabecular Bone Score (TBS) of each lumbar spine was assessed using TBS iNsight® software version 2.2, installed on a dual X-ray absorptiometry (DXA) machine (Discovery WI, Hologic Inc., USA). The mean L1 to L4 lumbar spine TBS was considered as the outcome of this study. Higher TBS values indicate a more robust trabecular structure, whereas lower TBS values are associated with an increased risk of osteoporosis and osteoporotic fractures.
Statistical analysis
A generalized linear model (GLM) was applied to examine various genetic models, including additive, dominant, recessive, over-dominant and co-dominant models. Initially, an analysis was conducted on the 376 NELL-1 gene variants to explore their relationship with TBS using the additive model, with a significance threshold set at a False Discovery Rate (PFDR) <0.05. Subsequently, SNPs identified as significant in the additive model (PFDR <0.05) were further evaluated across dominant, recessive, over-dominant, and co-dominant models. Different genetic models for a SNP were defined as demonstrated in the following example. For rs1901945 with alleles C/G, where C is the minor allele, the coding labels for the additive model were defined based on the minor allele count as GG = 0, CG = 1, and CC = 2. Additionally, the dominant model (CC + CG versus GG), recessive model (CC versus CG + GG), co-dominant model (CC and CG versus GG), and over-dominant model (CG versus CC + GG) were investigated.
A genetic risk score (GRS) was calculated for each individual through regression coefficients and allele values derived from significant genetic variants. To calculate the GRS for an individual, we assigned values to the alleles based on whether or not it is the effect allele (0, 1, and 2). Then, we multiplied these allele values by the corresponding beta coefficients for each genetic variant. The sum of these products provides the GRS for that individual. The relationship between GRS and TBS was investigated using simple and multiple linear regression analyses, adjusting for age, sex, Body Mass Index (BMI), type 2 diabetes and smoking status. Associations between GRS quartiles and TBS were also investigated by Analysis of Variance (ANOVA).
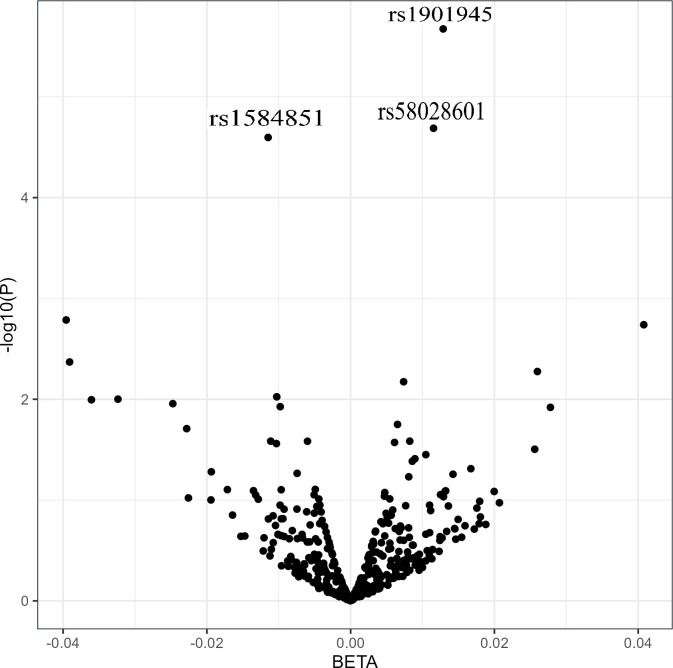
A volcano plot was utilized to visually represent the results of the additive model for all the 376 investigated SNPs. This plot typically displays the negative logarithm (base 10) of the p-value on the y-axis and regression coefficients on the x-axis for each SNP. Each point on the plot corresponds to a specific SNP. The interpretation involves assessing the position of points relative to the axes. SNPs positioned higher on the y-axis signify a stronger relationship with TBS, while those located towards the edges on the x-axis indicate larger expression differences, suggesting biological relevance.
The statistical significance threshold for the additive model was considered to be PFDR<0.05. Additionally, the threshold for the GRS was set at P<0.05.
Quality control processes were performed using PLINK version 2 software, and the relationships of SNPs with TBS in different genetic models were investigated using R version 4.4.0 (SNPassoc package) [39]. In addition, simple and multiple linear regression models, as well as data visualization were conducted using R version 4.4.0.
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of the Tehran University of Medical Sciences under code IR.TUMS.SPH.REC.1400.237. Participants were recruited in the BEH Program only after obtaining written informed consent. Data was accessed for research purposes on November 9, 2021. Authors did not have access to information that could identify individual participants during or after data collection.
Results
The participants’ characteristics are presented in Table 1. Among the participants, 51.7% were women, and the average age was 69.4 years. The mean BMI among older adults was 27.53±4.87, with 31.3% of them having been diagnosed with Type 2 diabetes. The mean Trabecular Bone Score (TBS) for the lumbar spine region (L1-L4) was 1.296 ± 0.105 across both sexes. Specifically, the mean TBS values for men and women were 1.353 ± 0.091 and 1.241 ± 0.087, respectively. Additionally, the mean TBS values for the individual lumbar segments (L1, L2, L3, and L4) were estimated as follows: 1.261 ± 0.132, 1.310 ± 0.122, 1.315 ± 0.116, and 1.299 ± 0.125, respectively.
Table 1. Baseline and TBS characteristics of study population.
| Variable | Men N = 1000 (48.3%) |
Women N = 1071(51.7) |
Total N = 2071 |
|---|---|---|---|
| Age, Mean (SD*) (yr) | 69.6 (6.4) | 69.2 (6.4) | 69.4 (6.4) |
| BMI, Mean (SD) (kg/m2) | 26.33 (3.97) | 28.65 (5.34) | 27.53 (4.87) |
| Type 2 Diabetes, Frequency (%) | |||
| Yes | 274 (27.6) | 371 (34.8) | 645 (31.3) |
| No | 720 (72.4) | 694 (65.2) | 1414 (68.7) |
| Smoking status, Frequency (%) | |||
| Never | 417 (42) | 500 (46.9) | 917 (44.5) |
| Current smoker | 346 (34.8) | 374 (35) | 720 (34.9) |
| Former smoker | 231 (23.2) | 193 (18.1) | 424 (20.6) |
| TBS L1**, Mean (SD) | 1.312 (0.120) | 1.213 (0.124) | 1.261 (0.132) |
| TBS L2, Mean (SD) | 1.364 (0.108) | 1.258 (0.111) | 1.31 (0.122) |
| TBS L3, Mean (SD) | 1.374 (0.103) | 1.258 (0.098) | 1.315 (0.116) |
| TBS L4, Mean (SD) | 1.364 (0.102) | 1.236 (0.112) | 1.299 (0.125) |
| TBS L1-L4, Mean (SD) | 1.353 (0.091) | 1.241 (0.087) | 1.296 (0.105) |
SD: Standard deviation, TBS: Trabecular bone score
Fig 1 displays a volcano plot representing the p-values versus regression coefficients of 376 analyzed SNPs. From the analysis, three SNPs were identified to be significantly associated with TBS adjusted for age and sex in the additive model. The specific characteristics of these significant SNPs are presented in Table 2.
Fig 1. Volcano plot of the relationship between 376 NELL-1 SNPs and trabecular bone score.
The x-axis represents the -log10 of the p-value. The y-axis represents the regression coefficients, reflecting the magnitude and direction of the effect size.
Table 2. Characteristics of the statistically significant genetic variants of NELL-1 related to trabecular bone score.
| rsID | Position (GRCh37) | Allele1/Allele2 | MAF* | Functional consequence |
|---|---|---|---|---|
| rs1901945 | 21285923 | C/G | 0.48 | Intron |
| rs1584851 | 21301343 | T/C | 0.49 | Intron |
| rs58028601 | 21312971 | A/G | 0.48 | Intron |
*Minor Allele Frequency
The relationships between the genetic variants rs1901945-C (β = 0.013, P = 2.12×10−6, PFDR = 0.0007), rs1584851- T (β = -0.011, P = 2.52×10−5, PFDR = 0.0003), and rs58028601-A (β = 0.11, P = 2.04×10−5, PFDR = 0.0003), with TBS were found to be statistically significant in the additive model. For rs1901945-C, each copy of the C allele, on average, increased the TBS by 0.013. Further analyses in different genetic models revealed additional insights: in the dominant model, two SNPs, rs1901945-C (β = 0.02, P = 2.73×10−6, PFDR = 0.001), and rs58028601-A (β = 0.018, P = 4.47×10−5, PFDR = 0.008) showed a statistically significant association with TBS. As a result, for rs1901945-C, compared to the GG genotype, having one or two copies of the C allele increased TBS by 0.02. Under the co-dominant model, rs1901945-C was related to TBS. For this variant, CG (β = 0.017, P = 9×10–5, PFDR = 0.001) and CC (β = 0.025, P = 3.24×10–6, PFDR = 0.0008) genotypes were statistically significant compared to GG. Significant results for rs1584851-T and rs58028601-A were observed only for the homozygous genotypes with two effect alleles. No significant relationships between the variants and TBS were found in the recessive and over-dominant genetic models (PFDR >0.05) (Table 3).
Table 3. Association of genetic variants with the Trabecular bone score, adjusted for age and sex.
| rsID | Effect allele | Beta* | CI, 95%* | P-value | PFDR *** |
|---|---|---|---|---|---|
| Additive | |||||
| rs1901945(C/G) | C | 0.013 | (0.008, 0.018) | 2.12×10−6 | 0.0007 |
| rs1584851(T/C) | T | -0.011 | (-0.017, -0.006) | 2.52×10−5 | 0.003 |
| rs58028601(A/G) | A | 0.011 | (0.006, 0.017) | 2.04×10−5 | 0.003 |
| Dominant | |||||
| rs1901945(C/G) | C | 0.02 | (0.01, 0.3) | 2.73e×10−6 | 0.001 |
| rs1584851(T/C) | T | -0.016 | (-0.024, -0.007) | 0.0004 | 0.055 |
| rs58028601(A/G) | A | 0.018 | (0.009, 0.026) | 4.47×10−5 | 0.008 |
| Recessive | |||||
| rs1901945(C/G) | C | 0.013 | (0.005, 0.023) | 0.002 | 0.2 |
| rs1584851(T/C) | T | -0.015 | (-0.024, -0.006) | 0.0007 | 0.2 |
| rs58028601(A/G) | A | 0.013 | (0.004, 0.022) | 0.004 | 0.3 |
| Over-dominant | |||||
| rs1901945(C/G) | C | 0.006 | (-0.001, 0.014) | 0.1 | 0.7 |
| rs1584851(T/C) | T | -0.001 | (-0.008, 0.007) | 0.8 | 0.99 |
| rs58028601(A/G) | A | 0.005 | (-0.003, 0.012) | 0.2 | 0.9 |
| Co-dominant (Reference) | |||||
| rs1901945 (GG) | |||||
| CG | 0.017 | (0.008, 0.026) | 9×10−5 | 0.001 | |
| CC | 0.025 | (0.014, 0.035) | 3.24×10−6 | 0.0008 | |
| rs1584851 (CC) | |||||
| TC | -0.011 | (-0.021, -0.002) | 0.01 | 0.8 | |
| TT | -0.02 | (-0.033, -0.012) | 2.50×10−5 | 0.001 | |
| rs58028601 (GG) | |||||
| AG | 0.015 | (0.006, 0.024) | 0.001 | 0.3 | |
| AA | 0.022 | (0.012, 0.033) | 2.7×10−5 | 0.0002 |
*Adjusted for age and sex
**95% Confidence interval
***False discovery rate
The risk score for the significant SNPs was computed based on the number of risk alleles present, taking into account the respective regression coefficients. The risk scores of all three SNPs were associated with TBS (P<0.05). The results of both simple and multiple linear regression analyses can be found in Table 4.
Table 4. Relationship between SNP risk scores and the trabecular bone score.
| Simple linear regression | Multiple linear regression* | ||||||
|---|---|---|---|---|---|---|---|
| Effect allele | Standardized Coefficient | CI, 95%* | P-value | Beta | CI, 95% | P-value | |
| Risk score for the additive model | |||||||
| rs1901945 (C/G) | C | 0.092 | (0.049, 0.135) | 2.9×10 −5 | 0.082 | (0.047, 0.117) | 5.0×10 −6 |
| rs1584851 (T/C) | T | 0.088 | (0.045, 0.131) | 6.5×10 −5 | 0.073 | (0.038, 0.108) | 5.2×10 −5 |
| rs58028601 (A/G) | A | 0.084 | (0.041, 0.127) | 1.4×10 −4 | 0.071 | (0.036, 0.106) | 8.3×10 −5 |
| Risk score for the dominant model | |||||||
| rs1901945 (C/G) | C | 0.096 | (0.053, 0.139) | 1.2×10 −5 | 0.083 | (0.048, 0.118) | 3×10 −6 |
| rs1584851 (T/C) | T | 0.068 | (0.025, 0.111) | 0.001 | 0.061 | (0.025, 0.096) | 6.1×10 −4 |
| rs58028601 (A/G) | A | 0.095 | (0.052, 0.138) | 1.3×10 −5 | 0.07 | (0.035, 0.105) | 9.9×10 −5 |
†Adjusted for age, sex, BMI, type 2 diabetes and smoking status
*95% Confidence interval
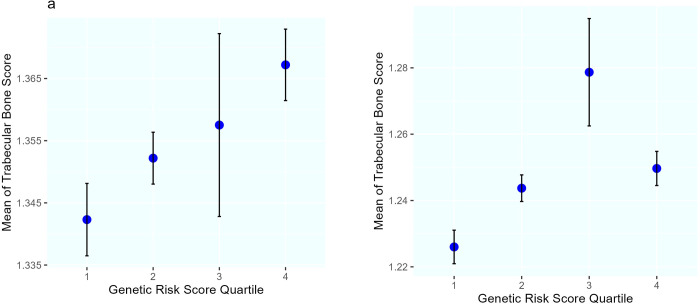
In the subsequent analysis, a Genetic Risk Score (GRS) was calculated by summing up the effect of the three SNPs related to TBS. The Correlation between risk scores derived from the additive genetic model (r = 0.09, P = 4×10−5) and the dominant genetic model (r = 0.1, P = 4×10−6) with TBS was found to be statistically significant. Further exploration involved investigating the associations of GRS quartiles in both additive and dominant models with TBS using ANOVA. The differences between TBS means were statistically significant in four quartile groups of GRS in both men and women (P<0.05). Fig 2 presents the mean TBS in quartiles of GRS for men and women, illustrating an increasing trend across the GRS quartiles. It is noted that the higher mean TBS in the third quartile compared to the fourth in women could be attributed to the smaller sample size, as indicated by the confidence intervals.
Fig 2.
Mean trabecular bone scores (95% confidence interval for the standard error) among quartile categories of the genetic risk score derived from the additive model in a) men and b) women.
Standardized regression coefficients were used to investigate the effect size corresponding to GRS for TBS in older adults. The GRS derived from the additive model showed a statistically significant relationship with TBS even after adjusting for age, sex, BMI, type 2 diabetes, and smoking status. The results indicated that a one standard deviation increase in the GRS corresponds to a 0.086 standard deviation increase in TBS (p = 2×10–6). Similarly, the GRS obtained from the dominant model also demonstrated a significant relationship with TBS (β = 0.086, P = 2×10–6) (Table 5).
Table 5. Relationship between genetic risk score (weighted sum scores of risk alleles) and TBS.
| Additive genetic model | Dominant genetic model | |||||||
|---|---|---|---|---|---|---|---|---|
| Genetic risk score | Standardized Coefficient | CI, 95%* | P-value | Adjusted R-Square | Standardized Coefficient | CI, 95% | P-value | R-Square |
| Model 1 | 0.09 | (0.047, 0.133) | 4×10 −5 | 0.008 | 0.1 | (0.062, 0.148) | 2×10 −6 | 0.01 |
| Model 2 | 0.083 | (0.047, 0.119) | 5×10 −6 | 0.31 | 0.091 | (0.055, 0.126) | 7.02×10 −7 | 0.31 |
| Model 3 | 0.077 | (0.042, 0.112) | 1.7×10 −5 | 0.34 | 0.086 | (0.051, 0.126) | 2×10 −6 | 0.34 |
| Model 4 | - | - | - | 0.30 | - | - | - | 0.30 |
| Model 5 | - | - | - | 0.33 | - | - | - | 0.33 |
Model 1: Crude, Model 2: Age, sex, and GRS, Model 3: Age, sex, BMI, type 2 diabetes, smoking status, and GRS, Model 4: Age, sex, Model 5: Age, sex, BMI, type 2 diabetes, and smoking status
*95% Confidence interval
Furthermore, adjusted R-squared (R2) values were calculated for various regression models to evaluate the proportion of the variance in TBS explained by the independent variables. The R2 values for the five regression models, including: 1) the crude model, 2) the model with Age, sex, and GRS, 3) Age, sex, BMI, type 2 diabetes, smoking status, and GRS, Model, 4) Age, sex, 5) Age, sex, BMI, type 2 diabetes, and smoking status, were determined to be 0.008, 0.31, 0.34, 0.3, and 0.339, respectively. It was observed that the models incorporating GRS showed a slight increase in R2 values compared to the models without GRS (Model 2 vs Model 4 and Model 3 vs Model 5).
Discussion
The Current study has identified a relationship between TBS and three specific SNPs located in the NELL-1 gene (rs1901945, rs1584851, and rs58028601) under the additive, dominant, and co-dominant genetic models.
NELL-1 was found to be associated with TBS in our study. Only a limited number of studies have been conducted to identify genes that influence TBS. Among the genes that have been found to be related to TBS are GREM2 and CYP24A1 [40, 41], while a genome-wide association study identified two loci near IRX3 and MAP2K5 that may also have a role in TBS [42]. Given that TBS is a relatively recent tool, the absence of prior reports of NELL-1 as a gene associated with TBS is not unexpected. Nonetheless, numerous studies in recent decades have demonstrated the functional impact of NELL-1 on bone formation.
Previous studies have identified the functional impact of the NELL-1 gene on bone health. Ting et al. in 1999, showed that NELL-1 is expressed and up-regulated in cranial intramembranous bone in craniosynostosis patients [29]. A study conducted in 2006 revealed that decreased expression of NELL-1 affected osteogenesis in mice, leading to vertebral and cranial defects due to the down-regulation of crucial extracellular matrix proteins essential for bone formation [43].
The synergistic effects of NELL-1 and bone morphogenetic proteins (BMPs) have been confirmed. Cowan et al. in 2007 investigated the synergistic effects of BMP-2 and NELL-1 on bone formation, demonstrating a robust synergistic impact on myoblasts both in vitro and in vivo. The results suggest that simultaneously stimulating both NELL-1 and BMP-2 could have greater therapeutic benefits and fewer negative side effects compared to only one pathway [32]. Another study also revealed this synergic effect in calvarial bone regeneration in vivo [44]. Moreover, research on the coding region of human BMP9 and human NELL-1, revealed that NELL-1 can be upregulated by BMP9, leading to the acceleration and enhancement of mineralization and maturity of BMP9-induced bone formation [45]. Considering that BMPs are crucial genes for bone and cartilage development [46], leveraging the synergistic effect of these genes with NELL-1 could hold significant promise in the treatment of osteoporosis.
Gene therapy involving the use of NELL-1 has shown promising outcomes in promoting bone regeneration. Li et al. investigated the treatment effects of NELL-1 in a femoral defect model in rats. Three groups of treatments were established, each with eight rats, receiving either 1.5 mg/ml NELL-1, 0.6 mg/ml NELL-1, or phosphate-buffered saline as a Nell-free control. The results showed that both NELL-1 -treated groups had more bone formation compared to the control group, and higher concentrations of NELL-1 led to greater bone volume [33]. Similarly, Guo et al. investigated the treatment effect of NELL-1 on osteolysis caused by polyethylene using a mouse model, finding that NELL-1 promoted new bone growth, compensating for osteolysis better than control group [34].
NELL-1 promotes osteoblast differentiation and mineralization, leading to increased bone formation. This is achieved by stimulating the expression of key osteogenic genes like RUNX2 and Osterix, which are essential for osteoblast function. An investigation on human osteogenic sarcoma and primary human osteoblast cells the findings indicate that Osterix functions as a direct transcriptional regulator, suppressing NELL-1 gene expression. This contributes to a fine-tuned balance of regulatory effects on NELL-1 transcription in coordination with Runx2, potentially playing a critical role in osteoblast differentiation and mineralization [47].
NELL-1 significantly increased the expression levels of various osteogenic genes and proteins, such as ALP, OCN, Runx2, OPG, Col-I, and Osterix. NELL-1 can enhance the osteogenic differentiation of pre-osteoblasts on titanium surfaces by activating the mitogen-activated protein kinase/extracellular signal-regulated kinase signaling Pathway [30]. The activation of the NELL-1 gene also promotes osteogenesis by regulating the Runx2/Osterix [31]. The binding between NELL-1 and APR3 is another pathway to enhance human osteoblast differentiation and mineralization [28].
In our study, rs1901945, rs1584851, and rs58028601 in the NELL-1 were related to TBS. Several genome-wide association studies have identified SNPs within or near to this gene related to bone mineral density and osteoporosis [48–50]. These signals, along with the research findings that reveal the biological effect of the NELL-1 gene on bone, indicate the potential impact of this gene on BMD and TBS.
We found that the GRS derived from SNPs of NELL-1 was related to TBS. This suggests that GRS contributes significantly to explaining the variability in TBS levels beyond the other variables considered in the analysis. It is reasonable that the R2 values for the GRS in our study were relatively smaller compared to the other variables in the models. Despite this, the consistent higher R2 values in the models, including GRS underline its significance as a contributing factor to TBS. While the impact of GRS was small compared to other factors, it is still a significant factor for TBS.
Our study had some limitations. In this study, GRS was calculated using the three SNPs located on a single gene. Other limitation was the inability to compare our results with other studies due to the limited evidence about genes related to TBS, especially in a similar population. However, the present study was the first genetic investigation focusing on the TBS phenotype within the Iranian population.
Conclusion
We investigated genetic variants of NELL-1 to test their relationships with TBS. Three SNPs were associated to TBS after adjusting for age and sex. NELL-1 is primarily expressed in osteoblasts, the cells responsible for creating bone tissue. This gene plays a critical role in bone formation by promoting osteoblast differentiation and mineralization. Given that NELL-1 is primarily expressed in osteoblasts, targeting this gene for therapeutic interventions could hold promise for treating bone-related conditions such as TBS and osteoporosis. Utilizing genetic information for personalized medicine interventions based on individual genetic profiles could lead to more targeted and effective prevention, diagnosis, and treatment strategies to enhance bone health outcomes. Further research is essential to gain a deeper understanding of the genetic factors influencing TBS and to uncover additional genes that may be involved in regulating bone quality.
Supporting information
(DOCX)
Acknowledgments
The authors are thankful to the researchers and staff of the BEH program.
Data Availability
Data cannot be shared publicly because of ethical considerations. Data are available from the School of Public Health & Allied Medical Sciences- Tehran University of Medical Sciences Ethics Committee (contact via research ethics Committees of School of Public Health & Allied Medical Sciences- Tehran University of Medical Sciences, Email: research@tums.ac.ir & Ethics@sina.tums.ac.ir, Tel: +98-81633619) for researchers who meet the criteria for access to confidential data.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.International Osteoporosis Foundation. Osteoporosis, diagnosis 2022. [cited 2022 December 29]. Available from: https://www.osteoporosis.foundation/health-professionals/diagnosis#ref_1.
- 2.Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group. World Health Organ Tech Rep Ser. 1994;843:1–129. PubMed PMID: 7941614. [PubMed]
- 3.Shen Y, Huang X, Wu J, Lin X, Zhou X, Zhu Z, et al. The Global Burden of Osteoporosis, Low Bone Mass, and Its Related Fracture in 204 Countries and Territories, 1990–2019. Front Endocrinol (Lausanne). 2022;13:882241. Epub 20220520. doi: 10.3389/fendo.2022.882241 ; PubMed Central PMCID: PMC9165055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salari N, Ghasemi H, Mohammadi L, Behzadi MH, Rabieenia E, Shohaimi S, et al. The global prevalence of osteoporosis in the world: a comprehensive systematic review and meta-analysis. J Orthop Surg Res. 2021;16(1):609. Epub 20211017. doi: 10.1186/s13018-021-02772-0 ; PubMed Central PMCID: PMC8522202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahimfar N, Noorali S, Yousefi S, Gharibzadeh S, Shafiee G, Panahi N, et al. Prevalence of osteoporosis among the elderly population of Iran. Archives of Osteoporosis. 2021;16(1):16. doi: 10.1007/s11657-020-00872-8 [DOI] [PubMed] [Google Scholar]
- 6.Tanha K, Fahimfar N, Nematollahi S, Sajjadi-Jazi SM, Gharibzadeh S, Sanjari M, et al. Annual incidence of osteoporotic hip fractures in Iran: a systematic review and meta-analysis. BMC Geriatrics. 2021;21(1):668. doi: 10.1186/s12877-021-02603-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostovar A, Mousavi A, Sajjadi-Jazi SM, Rajabi M, Larijani B, Fahimfar N, et al. The economic burden of osteoporosis in Iran in 2020. Osteoporosis International. 2022;33(11):2337–46. doi: 10.1007/s00198-022-06484-x [DOI] [PubMed] [Google Scholar]
- 8.Hans D, Goertzen AL, Krieg MA, Leslie WD. Bone microarchitecture assessed by TBS predicts osteoporotic fractures independent of bone density: the Manitoba study. J Bone Miner Res. 2011;26(11):2762–9. doi: 10.1002/jbmr.499 . [DOI] [PubMed] [Google Scholar]
- 9.Compston JE. SP0201 Usefulness and Limitations of DXA for Diagnosing Osteoporosis. Annals of the Rheumatic Diseases. 2014;73(Suppl 2):52-. doi: 10.1136/annrheumdis-2014-eular.6150 [DOI] [Google Scholar]
- 10.Pothuaud L, Barthe N, Krieg MA, Mehsen N, Carceller P, Hans D. Evaluation of the potential use of trabecular bone score to complement bone mineral density in the diagnosis of osteoporosis: a preliminary spine BMD-matched, case-control study. J Clin Densitom. 2009;12(2):170–6. Epub 20090131. doi: 10.1016/j.jocd.2008.11.006 . [DOI] [PubMed] [Google Scholar]
- 11.Panahi N, Ostovar A, Fahimfar N, Aghaei Meybodi HR, Gharibzadeh S, Arjmand B, et al. Factors associated with TBS worse than BMD in non-osteoporotic elderly population: Bushehr elderly health program. BMC Geriatrics. 2021;21(1):444. doi: 10.1186/s12877-021-02375-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pothuaud L. Process of Interpretation of Two-Dimensional Densitometry Images for the Prediction of Bone Mechanical Strength. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2004. 2004:1079–80. [Google Scholar]
- 13.Pothuaud L, Carceller P, Hans D. Correlations between grey-level variations in 2D projection images (TBS) and 3D microarchitecture: applications in the study of human trabecular bone microarchitecture. Bone. 2008;42(4):775–87. Epub 20080129. doi: 10.1016/j.bone.2007.11.018 . [DOI] [PubMed] [Google Scholar]
- 14.McCloskey EV, Odén A, Harvey NC, Leslie WD, Hans D, Johansson H, et al. A Meta-Analysis of Trabecular Bone Score in Fracture Risk Prediction and Its Relationship to FRAX. J Bone Miner Res. 2016;31(5):940–8. Epub 20151119. doi: 10.1002/jbmr.2734 . [DOI] [PubMed] [Google Scholar]
- 15.Krueger D, Fidler E, Libber J, Aubry-Rozier B, Hans D, Binkley N. Spine trabecular bone score subsequent to bone mineral density improves fracture discrimination in women. J Clin Densitom. 2014;17(1):60–5. Epub 20130613. doi: 10.1016/j.jocd.2013.05.001 . [DOI] [PubMed] [Google Scholar]
- 16.Sakulpisuti C, Sritara C, Kositwattanarerk A, Fuangfa P, Suppasilp C, Vathesatogkit P, et al. Bone Mineral Density and Trabecular Bone Score in Predicting Vertebral Fractures in Male Employees of the Electricity Generating Authority of Thailand. Journal of Osteoporosis. 2022;2022:6832166. doi: 10.1155/2022/6832166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benhamou CL, Poupon S, Lespessailles E, Loiseau S, Jennane R, Siroux V, et al. Fractal analysis of radiographic trabecular bone texture and bone mineral density: two complementary parameters related to osteoporotic fractures. J Bone Miner Res. 2001;16(4):697–704. doi: 10.1359/jbmr.2001.16.4.697 . [DOI] [PubMed] [Google Scholar]
- 18.Harvey NC, Glüer CC, Binkley N, McCloskey EV, Brandi ML, Cooper C, et al. Trabecular bone score (TBS) as a new complementary approach for osteoporosis evaluation in clinical practice. Bone. 2015;78:216–24. Epub 20150516. doi: 10.1016/j.bone.2015.05.016 ; PubMed Central PMCID: PMC4538791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho-Pham LT, Hans D, Doan MC, Mai LD, Nguyen TV. Genetic determinant of trabecular bone score (TBS) and bone mineral density: A bivariate analysis. Bone. 2016;92:79–84. Epub 20160820. doi: 10.1016/j.bone.2016.08.015 . [DOI] [PubMed] [Google Scholar]
- 20.Sonoda T, Takada J, Iba K, Asakura S, Yamashita T, Mori M. Interaction between ESRα polymorphisms and environmental factors in osteoporosis. J Orthop Res. 2012;30(10):1529–34. Epub 20120210. doi: 10.1002/jor.22083 . [DOI] [PubMed] [Google Scholar]
- 21.Ongphiphadhanakul B. Osteoporosis: the role of genetics and the environment. Forum Nutr. 2007;60:158–67. doi: 10.1159/000107166 . [DOI] [PubMed] [Google Scholar]
- 22.Kelly PJ, Eisman JA, Sambrook PN. Interaction of genetic and environmental influences on peak bone density. Osteoporos Int. 1990;1(1):56–60. doi: 10.1007/BF01880417 . [DOI] [PubMed] [Google Scholar]
- 23.Panahi N, Arjmand B, Ostovar A, Kouhestani E, Heshmat R, Soltani A, et al. Metabolomic biomarkers of low BMD: a systematic review. Osteoporos Int. 2021;32(12):2407–31. Epub 20210726. doi: 10.1007/s00198-021-06037-8 . [DOI] [PubMed] [Google Scholar]
- 24.Panahi N, Fahimfar N, Roshani S, Arjmand B, Gharibzadeh S, Shafiee G, et al. Association of amino acid metabolites with osteoporosis, a metabolomic approach: Bushehr elderly health program. Metabolomics. 2022;18(8):63. Epub 20220801. doi: 10.1007/s11306-022-01919-2 . [DOI] [PubMed] [Google Scholar]
- 25.Yalaev B, Tyurin A, Prokopenko I, Karunas A, Khusnutdinova E, Khusainova R. Using a Polygenic Score to Predict the Risk of Developing Primary Osteoporosis. Int J Mol Sci. 2022;23(17). Epub 20220902. doi: 10.3390/ijms231710021 ; PubMed Central PMCID: PMC9456390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, Jung J. Genome-wide polygenic risk score for major osteoporotic fractures in postmenopausal women using associated single nucleotide polymorphisms. J Transl Med. 2023;21(1):127. Epub 20230216. doi: 10.1186/s12967-023-03974-2 ; PubMed Central PMCID: PMC9933300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Center for Biotechnology Information. NELL1 neural EGFL like 1 [Homo sapiens (human) ] 2024. [cited 2023 May 10]. Available from: https://www.ncbi.nlm.nih.gov/gene/4745.
- 28.Zou X, Shen J, Chen F, Ting K, Zheng Z, Pang S, et al. NELL-1 binds to APR3 affecting human osteoblast proliferation and differentiation. FEBS Lett. 2011;585(15):2410–8. Epub 20110626. doi: 10.1016/j.febslet.2011.06.024 ; PubMed Central PMCID: PMC3209538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ting K, Vastardis H, Mulliken JB, Soo C, Tieu A, Do H, et al. Human NELL-1 expressed in unilateral coronal synostosis. J Bone Miner Res. 1999;14(1):80–9. doi: 10.1359/jbmr.1999.14.1.80 . [DOI] [PubMed] [Google Scholar]
- 30.Shen MJ, Wang GG, Wang YZ, Xie J, Ding X. Nell-1 Enhances Osteogenic Differentiation of Pre-Osteoblasts on Titanium Surfaces via the MAPK-ERK Signaling Pathway. Cell Physiol Biochem. 2018;50(4):1522–34. Epub 20181025. doi: 10.1159/000494651 . [DOI] [PubMed] [Google Scholar]
- 31.Lai K, Xi Y, Du X, Jiang Z, Li Y, Huang T, et al. Activation of Nell-1 in BMSC Sheet Promotes Implant Osseointegration Through Regulating Runx2/Osterix Axis. Front Cell Dev Biol. 2020;8:868. Epub 20200922. doi: 10.3389/fcell.2020.00868 ; PubMed Central PMCID: PMC7536315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowan CM, Jiang X, Hsu T, Soo C, Zhang B, Wang JZ, et al. Synergistic effects of Nell-1 and BMP-2 on the osteogenic differentiation of myoblasts. J Bone Miner Res. 2007;22(6):918–30. doi: 10.1359/jbmr.070312 ; PubMed Central PMCID: PMC2866074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Zara JN, Siu RK, Lee M, Aghaloo T, Zhang X, et al. Nell-1 enhances bone regeneration in a rat critical-sized femoral segmental defect model. Plast Reconstr Surg. 2011;127(2):580–7. doi: 10.1097/PRS.0b013e3181fed5ae ; PubMed Central PMCID: PMC3089952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo X, Peng J, Wang Y, Wang A, Zhang X, Yuan M, et al. NELL1 promotes bone regeneration in polyethylene particle-induced osteolysis. Tissue Eng Part A. 2012;18(13–14):1344–51. Epub 20120625. doi: 10.1089/ten.TEA.2011.0578 . [DOI] [PubMed] [Google Scholar]
- 35.Kim MS, Patel KP, Teng AK, Berens AJ, Lachance J. Genetic disease risks can be misestimated across global populations. Genome Biol. 2018;19(1):179. Epub 20181114. doi: 10.1186/s13059-018-1561-7 ; PubMed Central PMCID: PMC6234640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostovar A, Nabipour I, Larijani B, Heshmat R, Darabi H, Vahdat K, et al. Bushehr Elderly Health (BEH) Programme, phase I (cardiovascular system). BMJ Open. 2015;5(12):e009597. Epub 20151216. doi: 10.1136/bmjopen-2015-009597 ; PubMed Central PMCID: PMC4691780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafiee G, Ostovar A, Heshmat R, Darabi H, Sharifi F, Raeisi A, et al. Bushehr Elderly Health (BEH) programme: study protocol and design of musculoskeletal system and cognitive function (stage II). BMJ Open. 2017;7(8):e013606. Epub 20170804. doi: 10.1136/bmjopen-2016-013606 ; PubMed Central PMCID: PMC5577871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, et al. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27(2):e1608. Epub 20180227. doi: 10.1002/mpr.1608 ; PubMed Central PMCID: PMC6001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.González JR, Armengol L, Solé X, Guinó E, Mercader JM, Estivill X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23(5):644–5. Epub 20070131. doi: 10.1093/bioinformatics/btm025 . [DOI] [PubMed] [Google Scholar]
- 40.Pineda-Moncusí M, Rodríguez-Sanz M, Servitja S, Díez-Pérez A, Tusquets I, Nogués X, et al. Study of the genetic basis of Trabecular Bone Score reduction related to aromatase inhibitors. Revista de Osteoporosis y Metabolismo Mineral. 2018;10(2):82–7. [Google Scholar]
- 41.Paternoster L, Lorentzon M, Lehtimäki T, Eriksson J, Kähönen M, Raitakari O, et al. Genetic determinants of trabecular and cortical volumetric bone mineral densities and bone microstructure. PLoS Genet. 2013;9(2):e1003247. Epub 20130221. doi: 10.1371/journal.pgen.1003247 ; PubMed Central PMCID: PMC3578773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong SH, Yoon JW, Kim JH, Park J, Choi J, Lee JH, et al. Identification of Novel Genetic Variants Related to Trabecular Bone Score in Community-Dwelling Older Adults. Endocrinol Metab (Seoul). 2020;35(4):801–10. Epub 20201124. doi: 10.3803/EnM.2020.735 ; PubMed Central PMCID: PMC7803610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai J, Shannon ME, Johnson MD, Ruff DW, Hughes LA, Kerley MK, et al. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15(8):1329–41. Epub 20060314. doi: 10.1093/hmg/ddl053 . [DOI] [PubMed] [Google Scholar]
- 44.Aghaloo T, Cowan CM, Zhang X, Freymiller E, Soo C, Wu B, et al. The effect of NELL1 and bone morphogenetic protein-2 on calvarial bone regeneration. J Oral Maxillofac Surg. 2010;68(2):300–8. doi: 10.1016/j.joms.2009.03.066 ; PubMed Central PMCID: PMC3113462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Liao J, Zhang F, Song D, Lu M, Liu J, et al. NEL-Like Molecule-1 (Nell1) Is Regulated by Bone Morphogenetic Protein 9 (BMP9) and Potentiates BMP9-Induced Osteogenic Differentiation at the Expense of Adipogenesis in Mesenchymal Stem Cells. Cell Physiol Biochem. 2017;41(2):484–500. Epub 20170130. doi: 10.1159/000456885 . [DOI] [PubMed] [Google Scholar]
- 46.National Center for Biotechnology Information. BMP2 bone morphogenetic protein 2 [Homo sapiens (human) ]. 2024. [cited 2023 May 10]. Available from: https://www.ncbi.nlm.nih.gov/gene/650.
- 47.Chen F, Zhang X, Sun S, Zara JN, Zou X, Chiu R, et al. NELL-1, an osteoinductive factor, is a direct transcriptional target of Osterix. PLoS One. 2011;6(9):e24638. Epub 20110913. doi: 10.1371/journal.pone.0024638 ; PubMed Central PMCID: PMC3172249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inaba H, Cao X, Han AQ, Panetta JC, Ness KK, Metzger ML, et al. Bone mineral density in children with acute lymphoblastic leukemia. Cancer. 2018;124(5):1025–35. Epub 20171219. doi: 10.1002/cncr.31184 ; PubMed Central PMCID: PMC5821586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He D, Liu H, Wei W, Zhao Y, Cai Q, Shi S, et al. A longitudinal genome-wide association study of bone mineral density mean and variability in the UK Biobank. Osteoporos Int. 2023;34(11):1907–16. Epub 20230727. doi: 10.1007/s00198-023-06852-1 . [DOI] [PubMed] [Google Scholar]
- 50.Karasik D, Hsu YH, Zhou Y, Cupples LA, Kiel DP, Demissie S. Genome-wide pleiotropy of osteoporosis-related phenotypes: the Framingham Study. J Bone Miner Res. 2010;25(7):1555–63. doi: 10.1002/jbmr.38 ; PubMed Central PMCID: PMC3153998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
Data cannot be shared publicly because of ethical considerations. Data are available from the School of Public Health & Allied Medical Sciences- Tehran University of Medical Sciences Ethics Committee (contact via research ethics Committees of School of Public Health & Allied Medical Sciences- Tehran University of Medical Sciences, Email: research@tums.ac.ir & Ethics@sina.tums.ac.ir, Tel: +98-81633619) for researchers who meet the criteria for access to confidential data.