Abstract
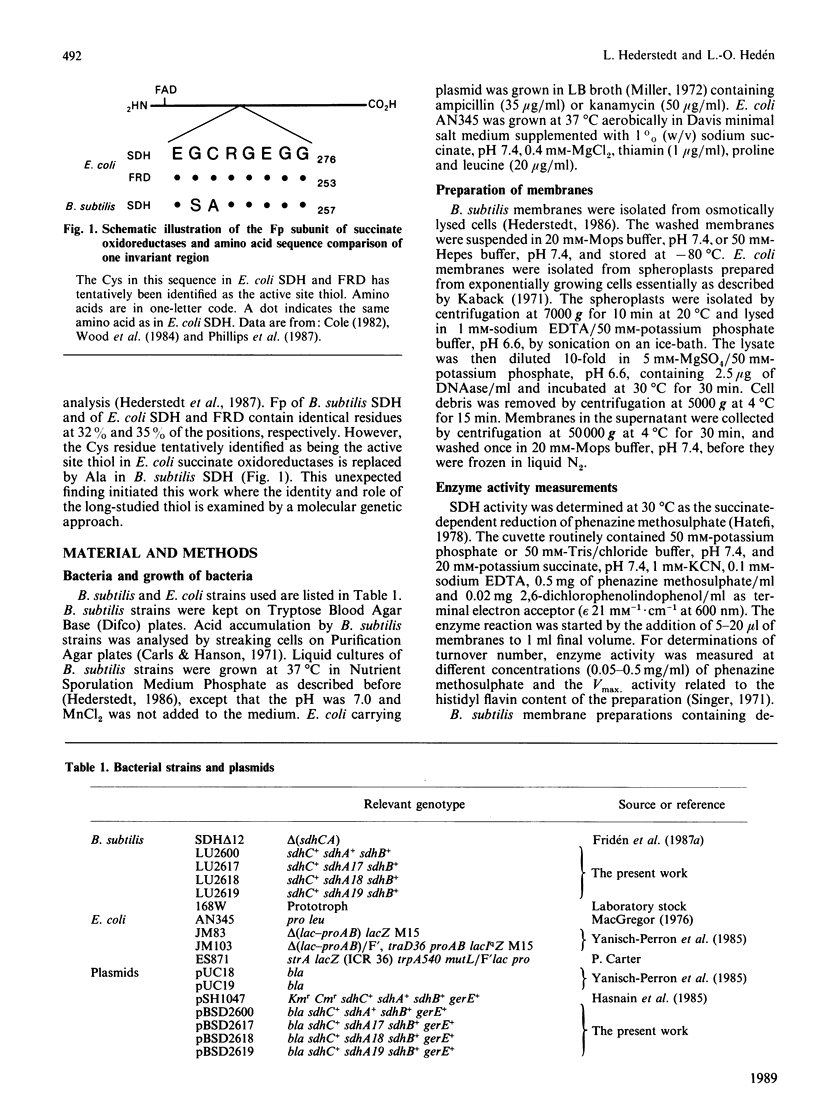
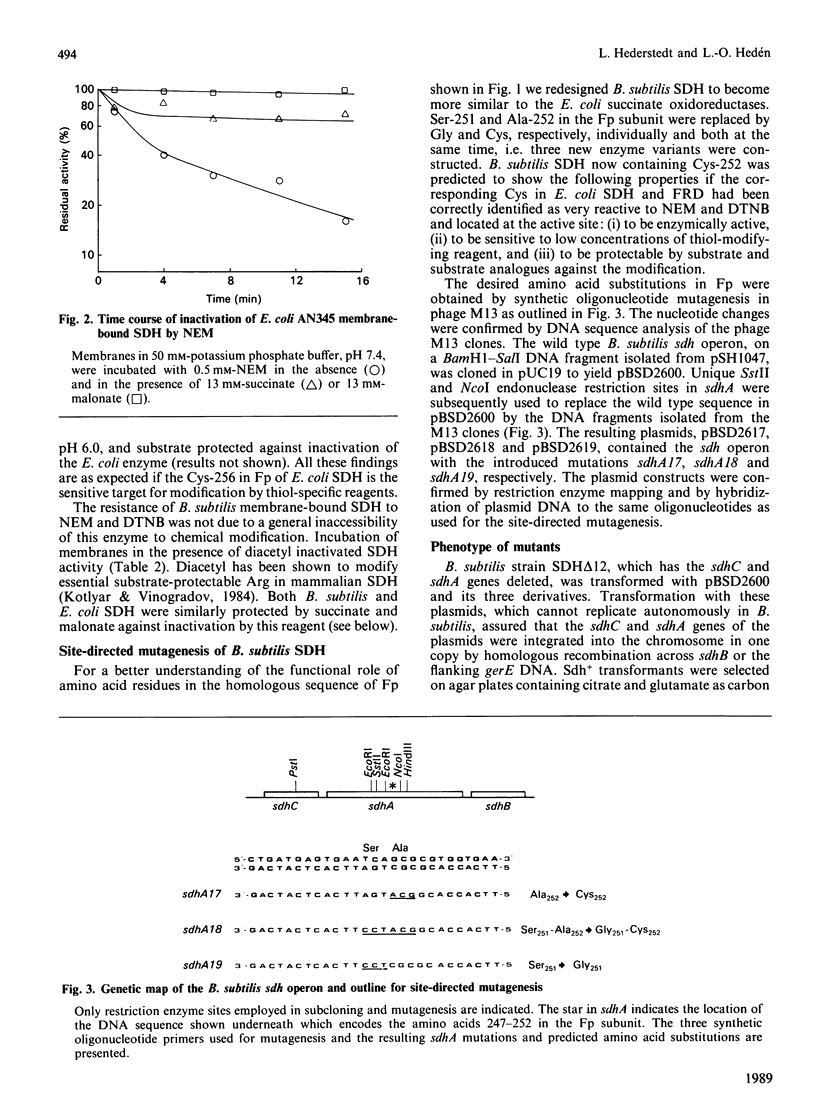
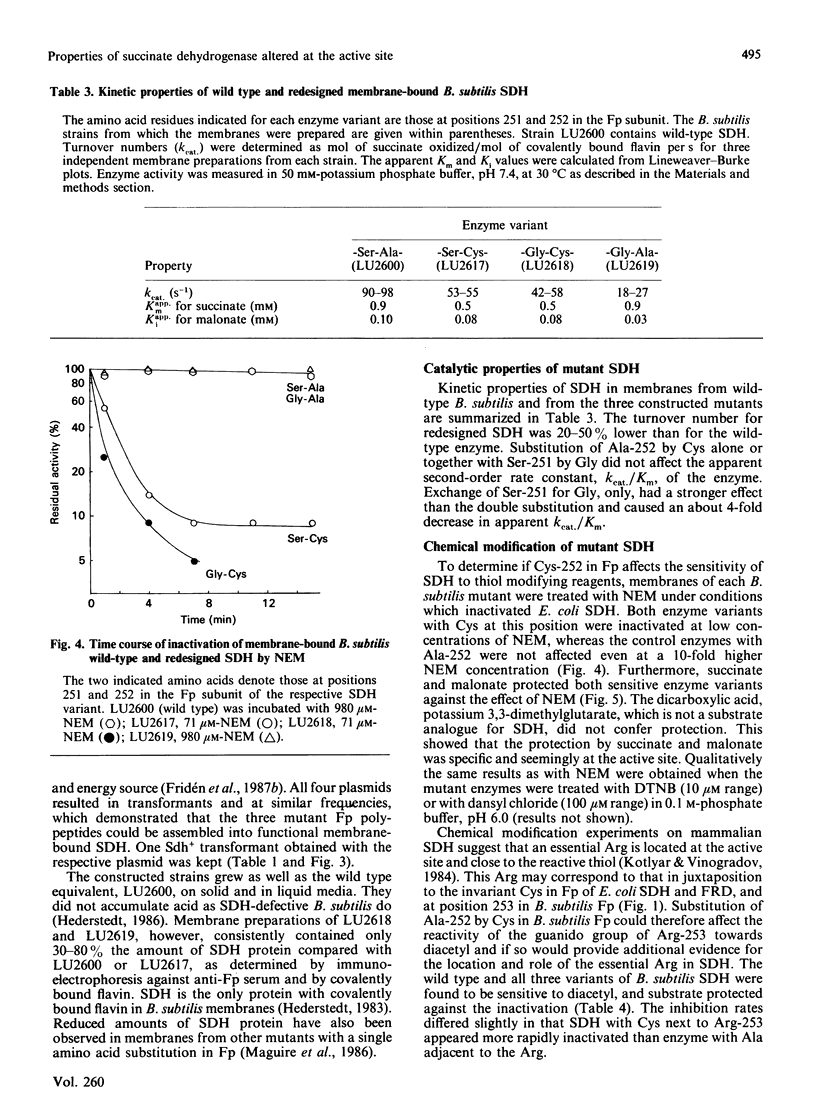
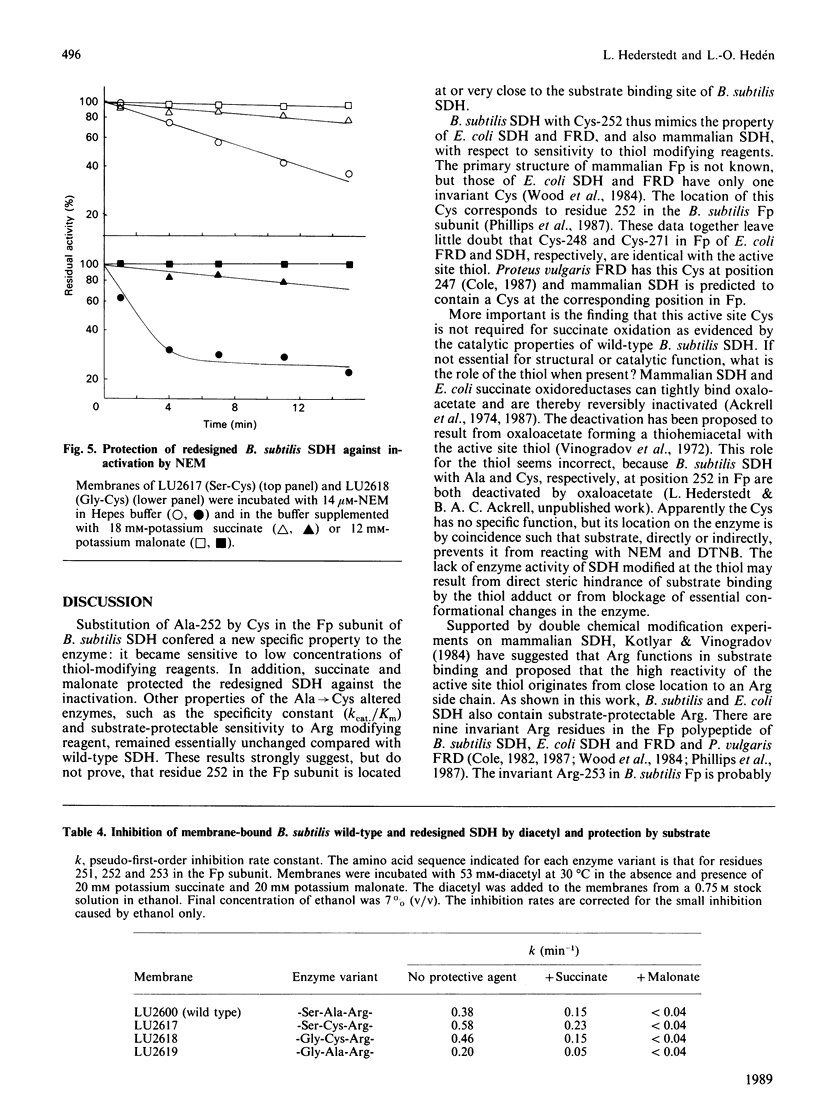
Mammalian and Escherichia coli succinate dehydrogenase (SDH) and E. coli fumarate reductase apparently contain an essential cysteine residue at the active site, as shown by substrate-protectable inactivation with thiol-specific reagents. Bacillus subtilis SDH was found to be resistant to this type of reagent and contains an alanine residue at the amino acid position equivalent to the only invariant cysteine in the flavoprotein subunit of E. coli succinate oxidoreductases. Substitution of this alanine, at position 252 in the flavoprotein subunit of B. subtilis SDH, by cysteine resulted in an enzyme sensitive to thiol-specific reagents and protectable by substrate. Other biochemical properties of the redesigned SDH were similar to those of the wild-type enzyme. It is concluded that the invariant cysteine in the flavoprotein of E. coli succinate oxidoreductases corresponds to the active site thiol. However, this cysteine is most likely not essential for succinate oxidation and seemingly lacks an assignable specific function. An invariant arginine in juxtaposition to Ala-252 in the flavoprotein of B. subtilis SDH, and to the invariant cysteine in the E. coli homologous enzymes, is probably essential for substrate binding.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackrell B. A., Kearney E. B., Mayr M. Role 3f oxalacetate in the regulation of mammalian succinate dehydrogenase. J Biol Chem. 1974 Apr 10;249(7):2021–2027. [PubMed] [Google Scholar]
- Arwert F., Venema G. Transformation in Bacillus subtilis. Fate of newly introduced transforming DNA. Mol Gen Genet. 1973;123(2):185–198. doi: 10.1007/BF00267334. [DOI] [PubMed] [Google Scholar]
- Carls R. A., Hanson R. S. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J Bacteriol. 1971 Jun;106(3):848–855. doi: 10.1128/jb.106.3.848-855.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T., Condon C., Lemire B. D., Weiner J. H. Molecular biology, biochemistry and bioenergetics of fumarate reductase, a complex membrane-bound iron-sulfur flavoenzyme of Escherichia coli. Biochim Biophys Acta. 1985 Dec;811(4):381–403. doi: 10.1016/0304-4173(85)90008-4. [DOI] [PubMed] [Google Scholar]
- Cole S. T. Nucleotide sequence and comparative analysis of the frd operon encoding the fumarate reductase of Proteus vulgaris. Extensive sequence divergence of the membrane anchors and absence of an frd-linked ampC cephalosporinase gene. Eur J Biochem. 1987 Sep 15;167(3):481–488. doi: 10.1111/j.1432-1033.1987.tb13362.x. [DOI] [PubMed] [Google Scholar]
- Cole S. T. Nucleotide sequence coding for the flavoprotein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Mar 1;122(3):479–484. doi: 10.1111/j.1432-1033.1982.tb06462.x. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Djavadi-Ohaniance L., Friguet B., Goldberg M. E. Conformational effects of ligand binding on the beta 2 subunit of Escherichia coli tryptophan synthase analyzed with monoclonal antibodies. Biochemistry. 1986 May 6;25(9):2502–2508. doi: 10.1021/bi00357a033. [DOI] [PubMed] [Google Scholar]
- Fridén H., Rutberg L., Magnusson K., Hederstedt L. Genetic and biochemical characterization of Bacillus subtilis mutants defective in expression and function of cytochrome b-558. Eur J Biochem. 1987 Nov 2;168(3):695–701. doi: 10.1111/j.1432-1033.1987.tb13471.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hasnain S., Sammons R., Roberts I., Thomas C. M. Cloning and deletion analysis of a genomic segment of Bacillus subtilis coding for the sdhA, B, C (succinate dehydrogenase) and gerE (spore germination) loci. J Gen Microbiol. 1985 Sep;131(9):2269–2279. doi: 10.1099/00221287-131-9-2269. [DOI] [PubMed] [Google Scholar]
- Hatefi Y. Resolution of complex II and isolation of succinate dehydrogenase (EC 1.3.99.1). Methods Enzymol. 1978;53:27–35. doi: 10.1016/s0076-6879(78)53009-7. [DOI] [PubMed] [Google Scholar]
- Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- Hederstedt L., Bergman T., Jörnvall H. Processing of Bacillus subtilis succinate dehydrogenase and cytochrome b-558 polypeptides. Lack of covalently bound flavin in the Bacillus enzyme expressed in Escherichia coli. FEBS Lett. 1987 Mar 23;213(2):385–390. doi: 10.1016/0014-5793(87)81527-2. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. Cytochrome b reducible by succinate in an isolated succinate dehydrogenase-cytochrome b complex from Bacillus subtilis membranes. J Bacteriol. 1980 Dec;144(3):933–940. doi: 10.1128/jb.144.3.933-940.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hederstedt L., Hatefi Y. Modification of bovine heart succinate dehydrogenase with ethoxyformic anhydride and rose bengal: evidence for essential histidyl residues protectable by substrates. Arch Biochem Biophys. 1986 Jun;247(2):346–354. doi: 10.1016/0003-9861(86)90593-x. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 1986;126:399–414. doi: 10.1016/s0076-6879(86)26040-1. [DOI] [PubMed] [Google Scholar]
- Hederstedt L. Succinate dehydrogenase mutants of Bacillus subtilis lacking covalently bound flavin in the flavoprotein subunit. Eur J Biochem. 1983 May 16;132(3):589–593. doi: 10.1111/j.1432-1033.1983.tb07404.x. [DOI] [PubMed] [Google Scholar]
- Hopkins F. G., Morgan E. J., Lutwak-Mann C. The influence of thiol groups in the activity of dehydrogenases. II: With an addendum on the location of dehydrogenases in muscle. Biochem J. 1938 Oct;32(10):1829–1848. doi: 10.1042/bj0321829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins F. G., Morgan E. J. The influence of thiol-groups in the activity of dehydrogenases. Biochem J. 1938 Mar;32(3):611–620. doi: 10.1042/bj0320611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney K. B., Ackrell B. A., Mayr M., Singer T. P. Activation of succinate dehydrogenase by anions and pH. J Biol Chem. 1974 Apr 10;249(7):2016–2020. [PubMed] [Google Scholar]
- Kenney W. C., Mowery P. C., Seng R. L., Singer T. P. Localization of the substrate and oxalacetate binding site of succinate dehydrogenase. J Biol Chem. 1976 Apr 25;251(8):2369–2373. [PubMed] [Google Scholar]
- Kenney W. C. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975 Apr 25;250(8):3089–3094. [PubMed] [Google Scholar]
- Kotlyar A. B., Vinogradov A. D. Evidence for an essential arginine residue in the substrate binding site of the mammalian succinate dehydrogenase. Biochem Int. 1984 Apr;8(4):545–552. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- MacGregor C. H. Biosynthesis of membrane-bound nitrate reductase in Escherichia coli: evidence for a soluble precursor. J Bacteriol. 1976 Apr;126(1):122–131. doi: 10.1128/jb.126.1.122-131.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson K., Philips M. K., Guest J. R., Rutberg L. Nucleotide sequence of the gene for cytochrome b558 of the Bacillus subtilis succinate dehydrogenase complex. J Bacteriol. 1986 Jun;166(3):1067–1071. doi: 10.1128/jb.166.3.1067-1071.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Molecular genetic aspects of the citric acid cycle of Escherichia coli. Biochem Soc Symp. 1987;54:45–65. [PubMed] [Google Scholar]
- Phillips M. K., Hederstedt L., Hasnain S., Rutberg L., Guest J. R. Nucleotide sequence encoding the flavoprotein and iron-sulfur protein subunits of the Bacillus subtilis PY79 succinate dehydrogenase complex. J Bacteriol. 1987 Feb;169(2):864–873. doi: 10.1128/jb.169.2.864-873.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. J., Weiner J. H. Molecular properties of fumarate reductase isolated from the cytoplasmic membrane of Escherichia coli. Can J Biochem. 1982 Aug;60(8):811–816. doi: 10.1139/o82-101. [DOI] [PubMed] [Google Scholar]
- Rutberg B., Hederstedt L., Holmgren E., Rutberg L. Characterization of succinic dehydrogenase mutants of Bacillus subtilis by crossed immunoelectrophoresis. J Bacteriol. 1978 Oct;136(1):304–311. doi: 10.1128/jb.136.1.304-311.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T. P. Determination of the activity of succinate, NADH, choline, and alpha-glycerophosphate dehydrogenases. Methods Biochem Anal. 1974;22:123–175. doi: 10.1002/9780470110423.ch3. [DOI] [PubMed] [Google Scholar]
- Singer T. P., Kearney E. B., Kenney W. C. Succinate dehydrogenase. Adv Enzymol Relat Areas Mol Biol. 1973;37:189–272. doi: 10.1002/9780470122822.ch4. [DOI] [PubMed] [Google Scholar]
- Unden G., Kröger A. An essential sulfhydryl group at the substrate site of the fumarate reductase of Vibrio succinogenes. FEBS Lett. 1980 Aug 11;117(1):323–326. doi: 10.1016/0014-5793(80)80972-0. [DOI] [PubMed] [Google Scholar]
- Vik S. B., Hatefi Y. Possible occurrence and role of an essential histidyl residue in succinate dehydrogenase. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6749–6753. doi: 10.1073/pnas.78.11.6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov A. D., Gavrikova E. V., Zuevsky V. V. Reactivity of the sulfhydryl groups of soluble succinate dehydrogenase. Eur J Biochem. 1976 Apr 1;63(2):365–371. doi: 10.1111/j.1432-1033.1976.tb10238.x. [DOI] [PubMed] [Google Scholar]
- Vinogradov A. D. Suktsinat-ubikhinon reduktaznyi uchastok dykhatel'noi tsepi. Biokhimiia. 1986 Dec;51(12):1944–1973. [PubMed] [Google Scholar]
- Vinogradov A. D., Winter D., King T. E. The binding site for oxaloacetate on succinate dehydrogenase. Biochem Biophys Res Commun. 1972 Oct 17;49(2):441–444. doi: 10.1016/0006-291x(72)90430-5. [DOI] [PubMed] [Google Scholar]
- Wood D., Darlison M. G., Wilde R. J., Guest J. R. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Sep 1;222(2):519–534. doi: 10.1042/bj2220519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


