Abstract
Most deep-ocean life relies on organic carbon from the surface ocean. While settling primary production rapidly attenuates in the water column, pulses of organic material can be quickly transported to depth in the form of food falls. One example of fresh material that can reach great depths across the tropical Atlantic Ocean and Caribbean Sea is the pelagic macroalgae Sargassum. However, little is known about the deep-ocean organisms able to use this food source. Here, we encountered the isopod Bathyopsurus nybelini at depths 5002–6288 m in the Puerto Rico Trench and Mid-Cayman Spreading Center using the Deep Submergence Vehicle Alvin. In most of the 32 observations, the isopods carried fronds of Sargassum. Through an integrative suite of morphological, DNA sequencing, and microbiological approaches, we show that this species is adapted to feed on Sargassum by using a specialized swimming stroke, having serrated and grinding mouthparts, and containing a gut microbiome that provides a dietary contribution through the degradation of macroalgal polysaccharides and fixing nitrogen. The isopod’s physiological, morphological, and ecological adaptations demonstrate that vertical deposition of Sargassum is a direct trophic link between the surface and deep ocean and that some deep-sea organisms are poised to use this material.
Keywords: Munnopsidae, macroalgae, carbon sequestration, gut microbiome, deep sea, Alvin
1. Introduction
In the planet’s largest habitat—the deep sea—most life relies on fixed organic carbon and energy that originates in the surface ocean. Because only a small fraction of surface-derived primary production makes it to the deep seafloor, with most consumed by pelagic organisms throughout the water column (e.g. [1]), deep-ocean fauna are often energy and carbon limited. However, some organic material can sink quickly, bypassing pelagic processing and degradation. This rapidly transported carbon includes episodic deposits of microbial phytoplankton (e.g. [2,3]) and dense, high biomass material, such as whale falls and other carrion (e.g. [4,5]). To consume episodic, localized, and often recalcitrant material, deep-sea taxa have developed an array of specialized adaptations. Well-studied organisms that capitalize on these food sources include the bone-eating polychaete Osedax [6], the wood-boring mollusc Xylophaga [7], and the many taxa that rely on chemosynthesis at hydrothermal vents and cold seeps (e.g. [8]). These trophic specialists forge important connections in global ocean food webs, contributing to energy and nutrient cycling (e.g. [9]).
Another alternative food source for deep-ocean fauna is plant and macroalgal debris. This material can reach abyssal (4000−6000 m) and hadal (6000−11 000 m) habitats and may serve as an important trophic link between the surface and organisms at great depths (e.g [10–13]). For example, the holopelagic macroalgae Sargassum spp. has been observed on the seafloor in and around the western Atlantic (e.g [10,11,14–16]). At the surface, the quantity and distribution of Sargassum have increased since 2011, expanding from the Sargasso Sea to the central Atlantic Ocean and Caribbean Sea (e.g. [17–19]), causing ecosystem and economic impacts on coastal waters through large beaching events [20]. While Sargassum could be a growing source of labile carbon to the deep ocean (e.g. [21,22]), the fate of this organic material and the organisms that consume it have not been fully investigated. This is in part due to the technical challenges of studying abyssal and hadal ecosystems, as most studies use untethered landers baited with carrion to collect video and physical samples from a single location (e.g. [23–25]).
In this study, we opportunistically encountered the swimming isopod Bathyopsurus nybelini Nordenstam, 1955 (Suborder Asellota, Family Munnopsidae Lilljeborg, 1864) carrying Sargassum at abyssal and hadal depths in the western Atlantic Ocean and Caribbean Sea. Previous lander-based pictures [15] and gut contents from trawled specimens collected in poor condition during the 1948 Swedish Deep-Sea Expedition to the Puerto Rico Trench [26,27] suggested this organism may consume Sargassum. We leveraged the newly refit Deep Submergence Vehicle (DSV) Alvin, which as of 2022 can now reach depths exceeding 6000 m [28,29], to explore the importance of surface-derived macroalgae on the lifestyle of Bathyopsurus using in situ video, morphological analysis, and DNA sequencing of the gut microbiome. We provide conclusive evidence that Bathyopsurus nybelini is specialized to feed on Sargassum, forming a trophic link between the surface and organisms at the deepest ocean depths across the tropical Atlantic and Caribbean Sea.
2. Results
(a). Distribution of macroalgae and observation of isopods at abyssal and hadal depths
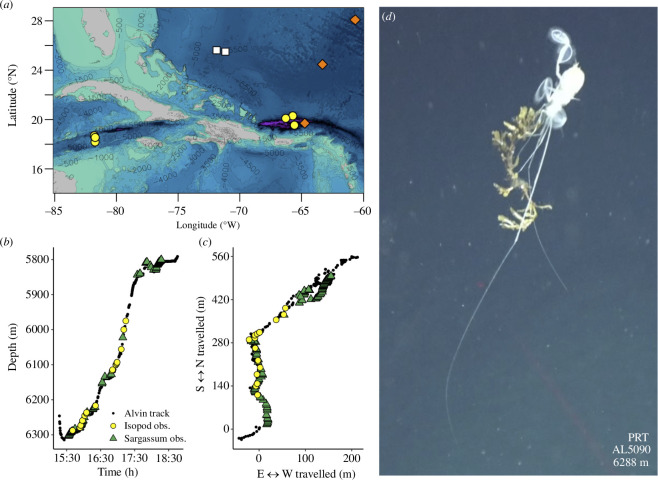
We explored the Puerto Rico Trench, Mid-Cayman Spreading Center, and surrounding waters on 14 dives with DSV Alvin. Video from three dives in the Puerto Rico Trench (depths 5605−6303 m), each with 2–3 h of bottom time and covering <2 km, showed that macroalgae was abundant on the seafloor (figure 1, electronic supplementary material, figures S1 and S2). Macroalgae was observed >300 times on each dive at an average interval of less than 4 m between observations. Much of this material was visually identifiable as Sargassum. Falls ranged in size from small pieces to large piles >50 cm in length and were commonly identified on soft sediment. Macroalgae was seen floating past the submersible while being carried by the current and often became entangled around benthic organisms, including glass sponges and anemones (electronic supplementary material, figure S1). Hundreds of macroalgal fragments <3 cm were observed when the camera zoomed in on the seafloor but were not quantified.
Figure 1.
The swimming isopod Bathyopsurus nybelini was consistently observed carrying Sargassum at abyssal and hadal depths. (a) A map showing observations of B. nybelini during this study and historical records. Yellow circles, this study; orange diamonds, 1948 Swedish Deep-Sea Expedition collections [26,27]; white squares, lander observations [15]. (b,c) Isopod and macroalgae observations along the seafloor during Alvin dive AL5090 as a function of depth, time, and relative distance travelled. Data begin shortly before the sub arrives on the seafloor. The legend is the same for both panels. (d) Representative image of B. nybelini carrying Sargassum. PRT, Puerto Rico Trench.
Swimming isopods were consistently observed interacting with Sargassum (electronic supplementary material, videos S1–S4). Thirty-two distinct individuals were seen at six locations (depths 5002−6288 m), three within the Puerto Rico Trench (Central Ridge, North Wall, and South Wall) and three near the Mid-Cayman Spreading Center (Axial Volcanic Ridge, North Ridge, and Beebe Woods; figure 1, table 1, electronic supplementary material, table S1). Nineteen individuals were filmed carrying Sargassum; in some cases, the frond was longer than the body length of the isopod. Isopods swam by paddle stroking above the seafloor with their two natatory legs (pereopods V–VI). Individuals swam backwards, and in some instances upside down, either travelling parallel to or vertically away from the seafloor. Isopods swam continuously, with a mean frequency of 1.27 ± 0.38 strokes s−1 (range 0.60–2.15 strokes s−1). The isopods carried Sargassum off the seafloor by pinching gnathopod 1.
Table 1.
Observations of Bathyopsurus nybelini in the Puerto Rico Trench and Mid-Cayman Spreading Center on the Alvin Science Verification Expedition (AT50−02). Date and encounter time are reported in GMT. Individuals that were observed carrying Sargassum are indicated with a Y (yes), while individuals that were not carrying Sargassum are shown with an N (no). Observations where it was not possible to determine if the isopod was carrying Sargassum due to distance or focus are blank. Individual 5089–05, denoted by an asterisk, was seen six times over a short period, with the initial encounter time and location reported here. Example videos and paddle stroke frequencies are available in the electronic supplementary material.
| Location | Alvin dive ID | Encounter ID | Dive site | Date | Encounter time (GMT) | Latitude (°N) | Longitude (°W) | Depth (m) | Carrying Sargassum |
|---|---|---|---|---|---|---|---|---|---|
| Puerto Rico Trench | 5089 | 5089-01 | Central Ridge | 7.29.22 | 16:38:46 | 19.53967 | −65.57386 | 6103.5 | N |
| Puerto Rico Trench | 5089 | 5089-02 | Central Ridge | 7.29.22 | 16:47:22 | 19.54050 | −65.57327 | 6095.1 | Y |
| Puerto Rico Trench | 5089 | 5089-03 | Central Ridge | 7.29.22 | 16:47:40 | 19.54054 | −65.57324 | 6094.4 | N |
| Puerto Rico Trench | 5089 | 5089-04 | Central Ridge | 7.29.22 | 16:59:19 | 19.54188 | −65.57231 | 6069.3 | Y |
| Puerto Rico Trench | 5089 | 5089-05* | Central Ridge | 7.29.22 | 17:01:12 | 19.54194 | −65.57227 | 6067.6 | N |
| Puerto Rico Trench | 5089 | 5089-06 | Central Ridge | 7.29.22 | 17:27:08 | 19.54355 | −65.57149 | 6034.8 | N |
| Puerto Rico Trench | 5090 | 5090-01 | North Wall | 7.30.22 | 15:40:32 | 20.29516 | −65.70579 | 6287.6 | Y |
| Puerto Rico Trench | 5090 | 5090-02 | North Wall | 7.30.22 | 15:53:31 | 20.29542 | −65.70582 | 6279.2 | N |
| Puerto Rico Trench | 5090 | 5090-03 | North Wall | 7.30.22 | 15:54:56 | 20.29549 | −65.70579 | 6273.1 | Y |
| Puerto Rico Trench | 5090 | 5090-04 | North Wall | 7.30.22 | 15:58:01 | 20.29576 | −65.70579 | 6259.2 | Y |
| Puerto Rico Trench | 5090 | 5090-05 | North Wall | 7.30.22 | 16:01:55 | 20.29596 | −65.70572 | 6245.3 | Y |
| Puerto Rico Trench | 5090 | 5090-06 | North Wall | 7.30.22 | 16:04:55 | 20.29614 | −65.70576 | 6238.9 | |
| Puerto Rico Trench | 5090 | 5090-07 | North Wall | 7.30.22 | 16:05:09 | 20.29617 | −65.70576 | 6236.3 | Y |
| Puerto Rico Trench | 5090 | 5090-08 | North Wall | 7.30.22 | 16:20:55 | 20.29653 | −65.70581 | 6216.8 | |
| Puerto Rico Trench | 5090 | 5090-09 | North Wall | 7.30.22 | 16:50:23 | 20.29683 | −65.70581 | 6117.0 | Y |
| Puerto Rico Trench | 5090 | 5090-10 | North Wall | 7.30.22 | 16:51:03 | 20.29678 | −65.70593 | 6114.3 | Y |
| Puerto Rico Trench | 5090 | 5090-11 | North Wall | 7.30.22 | 16:56:49 | 20.29691 | −65.70581 | 6100.2 | Y |
| Puerto Rico Trench | 5090 | 5090-12 | North Wall | 7.30.22 | 16:58:15 | 20.29695 | −65.70576 | 6096.8 | Y |
| Puerto Rico Trench | 5090 | 5090-13 | North Wall | 7.30.22 | 16:59:02 | 20.29699 | −65.70571 | 6093.1 | Y |
| Puerto Rico Trench | 5090 | 5090-14 | North Wall | 7.30.22 | 16:59:02 | 20.29699 | −65.70571 | 6093.1 | |
| Puerto Rico Trench | 5090 | 5090-15 | North Wall | 7.30.22 | 16:59:02 | 20.29699 | −65.70571 | 6093.1 | |
| Puerto Rico Trench | 5090 | 5090-16 | North Wall | 7.30.22 | 17:06:22 | 20.29735 | −65.70536 | 6055.9 | Y |
| Puerto Rico Trench | 5090 | 5090-17 | North Wall | 7.30.22 | 17:10:51 | 20.29750 | −65.70520 | 5999.8 | Y |
| Puerto Rico Trench | 5090 | 5090-18 | North Wall | 7.30.22 | 17:14:25 | 20.29769 | −65.70516 | 5974.9 | Y |
| Puerto Rico Trench | 5091 | 5091-01 | South Wall | 7.31.22 | 17:55:01 | 20.09438 | −66.27648 | 5868.6 | Y |
| Puerto Rico Trench | 5091 | 5091-02 | South Wall | 7.31.22 | 18:38:42 | 20.09536 | −66.27606 | 5806.2 | |
| Puerto Rico Trench | 5091 | 5091-03 | South Wall | 7.31.22 | 18:45:27 | 20.09584 | −66.27616 | 5753.9 | Y |
| Mid-Cayman Spreading Center | 5094 | 5094-01 | Axial volcanic Ridge | 8.8.22 | 17:48:23 | 18.17784 | −81.72858 | 5191.7 | Y |
| Mid-Cayman Spreading Center | 5097 | 5097-01 | North Ridge | 8.11.22 | 16:19:13 | 18.68616 | −81.78744 | 6026.5 | N |
| Mid-Cayman Spreading Center | 5097 | 5097-02 | North Ridge | 8.11.22 | 16:25:55 | 18.68445 | −81.78717 | 6040.5 | Y |
| Mid-Cayman Spreading Center | 5101 | 5101-01 | Beebe Woods | 8.15.22 | 18:10:49 | 18.54712 | −81.71882 | 5001.9 | N |
| Mid-Cayman Spreading Center | 5101 | 5101-02 | Beebe Woods | 8.15.22 | 18:10:57 | 18.54713 | −81.71881 | 5001.9 | N |
(b). Integrative taxonomy of Bathyopsurus nybelini
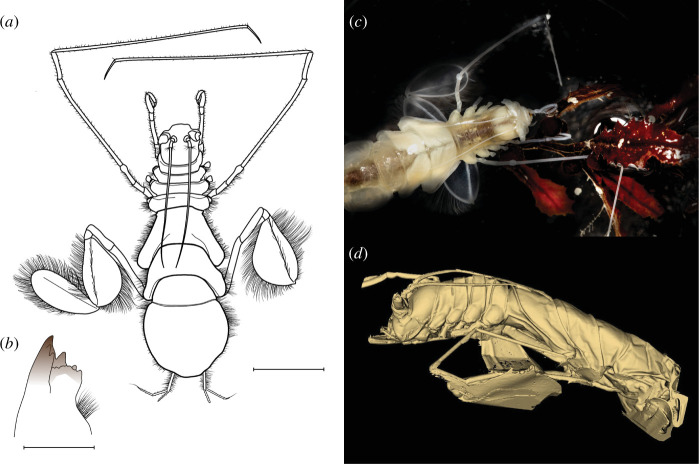
Two isopods were collected from depths of 6100 and 6114 m within the Puerto Rico Trench (Dive AL5090; figure 2, table 1). Specimens are female, with total body lengths of 33.6 and 35.2 mm. Their bodies are transparent and thin, of parchment-like consistency, with highly fragile appendages. We identify these individuals as Bathyopsurus nybelini due to (i) the separation of the first four pereion segments, of equal length and width, from the expanded and evenly rounded pleotelson, (ii) the lack of molar process and palp on the mandibles, (iii) a left mandible having five teeth on the incisor, and (iv) the operculum being broader than long with near parallel front and hind margins. This new material informed a redescription of characters that were absent or damaged during previous trawl-based collections (electronic supplementary material; [26,27]). Ribosomal 18S and 28S RNA gene sequencing placed B. nybelini within the Bathyopsurinae sub-family and sister to Paropsurus giganteus Wolff, 1962 [27] (electronic supplementary material, figures S3 and S4).
Figure 2.
Bathyopsurus nybelini is morphologically adapted to feed on Sargassum using large paddles for a specialized swimming stroke and mouthparts for tearing. (a) Whole body dorsal illustration of collected female specimen with 2 cm scale. (b) Left mandible with 0.5 mm scale. Credit: Johanna Weston. (c) Isopod staged with associated Sargassum. Credit: Daniel Hentz. (d) Micro-CT scan showing lateral morphology at 19.4 µm resolution. Credit: Mackenzie Gerringer. All images are specimen AT50−02−017.
Both individuals were carrying Sargassum at the time of collection. The macroalgae was morphologically identified as Sargassum natans VIII (electronic supplementary material; [30]). Fronds hosted an epiphyte community that covered <20% of the surface area. Epibiont species present included the hydroid Aglaophenia latecarinata, the polychaete Spirorbis spirorbis, and a filamentous alga, suggesting rapid deposition from the surface and a lack of degradation (electronic supplementary material). Sargassum was nitrogen poor. Carbon (C) and nitrogen (N) reflected 28.3% and 0.59% of the biomass dry weight, respectively, yielding a C:N molar ratio of 56.3.
The gross morphology from both direct examination and micro-CT scanning revealed key functional adaptations to support the isopod’s feeding on macroalgae (figure 2). The specimens appear to lack eyes; the long flagella of the antennule and sensory setae may allow the isopod to find macroalgae on the seafloor by touch or chemosensation. Individuals carried Sargassum by compressing the dactylus and propodus on the carpus in pereopod I (electronic supplementary material, figure S5). The serrated pereopod I dactylus and mandible incisor and robust and knotty left lacinia mobilis aid in the tearing and grinding of Sargassum (electronic supplementary material, figure S6). The large dual paddles support the isopod’s characteristic swimming gait, which involves alternating power strokes of adjacent appendages. In addition to the broadening and separating of the two paddles on each appendage during the power stroke, setae provide additional surface area for propulsion power to carry large fronds of Sargassum. These paddles were missing in the original collections [26,27].
(c). Gut content metagenomic sequencing
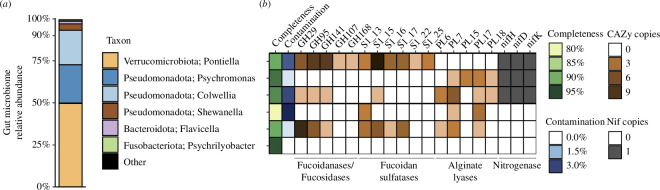
Shotgun metagenomic sequencing of the isopod’s gut contents indicated a diet containing macroalgae. At least 30 marker genes related to Sargassum were present within the gut, including rRNA genes, cytochrome c oxidases, and ribosomal proteins (electronic supplementary material, figure S7). Most of these genes were identical to published sequences belonging to Sargassum fluitans III. Phylogenetically informative single-copy marker genes revealed the gut microbiome is composed primarily (>90%) of the genera Pontiella (phylum Verrucomicrobiota in GTDB-Tk and Kiritimatiellota in NCBI; electronic supplementary material, figure S8), Psychromonas, and Colwellia (figure 3). Members of the genera Shewanella, Flavicella, and Psychrilyobacter together represented approximately 6% of the remaining community. To determine if these microorganisms contribute to the digestion of macroalgal polysaccharides, we explored their genomic functional potential using metagenome-assembled genome (MAG) binning (electronic supplementary material, table S2). The MAGs within the gut belonging to Pontiella and Flavicella have putative genes for the degradation of fucose-containing sulfated fucoidans and polysaccharides, including fucosidases (GH29, GH95, GH141), fucanoidases (GH107, GH168), and fucoidan sulfatases (S1_13, S1_15, S1_16, S1_17, S1_22, S1_25; e.g. [31,32]). MAGs related to Psychromonas, Colwellia, and Shewanella have genes for the use of alginate (PL6, PL7, PL15, PL17, PL18; e.g. [33,34]). The three most abundant organisms in the gut—Pontiella, Psychromonas, and Colwellia—all contain the nitrogenase nif operon for N2 fixation, suggesting these organisms may provide the host with bioavailable nitrogen. While the Psychromonas and Colwellia MAGs are related to other deep-ocean lineages (electronic supplementary material, figures S9 and S10), the ability to fix nitrogen is absent in other deep-sea members of these genera (electronic supplementary material, figure S11).
Figure 3.
The gut microbiome of Bathyopsurus nybelini can degrade sulfated polysaccharides and fix nitrogen. (a) The composition of the gut microbiome based on the averaged relative abundances of ten single-copy marker genes. (b) Genome completeness, contamination, and gene copy numbers from gut metagenome-assembled genomes. The rows in (b) are aligned with their corresponding taxa in (a). CAZy, carbohydrate-active enzymes; Nif, nitrogenase genes.
3. Discussion
We provide conclusive evidence that the isopod Bathyopsurus nybelini is adapted to feed on surface-derived macroalgae in abyssal and hadal ecosystems of the Atlantic Ocean and Caribbean Sea using a specialized swimming style, serrated mouthparts, and a diazotrophic gut microbiome. Our work shows that Sargassum is a direct trophic link between the surface and deep ocean and that organisms at great depths are using this material. The isopod appears to lack eyes, likely relying instead on specialized antennule and sensory setae to chemosense macroalgal odours (e.g. [35]). Video and morphological evidence highlight that this isopod has a specialized method of locomotion—swimming upside down and backward with large paddles—that allows it to carry Sargassum off the seafloor. Munnopsidae are commonly observed swimming in the deep ocean (e.g. [36–38]), although many swim only in short bursts to avoid predation (e.g. [39–41]). We hypothesize that Bathyopsurus may carry Sargassum into the water column to avoid predation at the seafloor. Future work to determine where this organism carries this algal material would reveal further details on the evolutionary significance of this behaviour. Together, our findings demonstrate that Bathyopsurus is specifically adapted to use a rather unconventional source of food at abyssal depths, illustrating the importance of food falls to deep-sea specialists.
In addition to the isopod’s morphological adaptations, we show that Bathyopsurus has a unique gut microbiome specialized in the degradation of macroalgal material. Brown algal cell walls are composed of sulfated fucoidans and alginates, polysaccharides that are recalcitrant and require highly specialized microorganisms for degradation (e.g. [31,42,43]). The high abundances of Pontiella that contain genes for the degradation of sulfated fucoidans (e.g. [44–46]), along with Psychromonas and Colwellia that can degrade alginate (e.g. [46,47]), are consistent with the isopod consuming macroalgae. These findings also agree with hypotheses from terrestrial isopods suggesting that gut microorganisms play an important role in host nutrition (e.g. [48,49]). While members of the genera Psychromonas and Colwellia are commonly reported on sinking particles in the deep sea [50] and within the gut microbiomes of hadal animals (e.g. [51–53]), the presence of Pontiella here reveals a distinction from the gut microbiomes of other deep-ocean organisms that feed on carrion, which instead have high abundances of Mycoplasmataceae (e.g. [53,54]). All three of these MAGs reported here are capable of fixing N2 gas into biologically available N, a finding not previously observed in microbiomes of abyssal and hadal animals. Because Sargassum is a carbon-rich, stoichiometrically imbalanced food source with a C:N ratio above 20:1 (our data; e.g. [18,22,55]), N2 fixation may provide a source of supplemental N for both the microorganism and host. The association of diazotrophs with animals occurs across many marine and terrestrial symbioses (e.g. [56,57]), especially those which feed on carbon-rich substrates, including urchins [58], wood-boring bivalves [59,60], and insects [61–63]. Therefore, our findings support the hypothesis that microorganisms form an important trophic link between detritus and larger organisms at great depth (e.g. [11,22,64,65]), with herbivorous detritivores supported by a distinct gut microbiome capable of degrading algal carbon-rich polysaccharides and supplementing N content via N2 fixation. Our results provide a unique example of how some deep-sea megafauna rely on specialized microbial communities to access varied nutrient and energy sources. While the importance of chemosynthetic microorganisms to higher trophic levels has been well documented (e.g. [66]), a complete picture of global ocean food webs requires interdisciplinary investigations that integrate across taxa and scales to understand these vital connections.
From a biogeographic perspective, our observations in both the Atlantic and Caribbean at distances more than 1500 km apart indicate this isopod species is widely distributed and consuming Sargassum at depths of 5000−7900 m across the Sargassum belt. Bathyopsurus nybelini has also been reported from the abyssal plains near the Kermadec Trench in the Southwestern Pacific and Tasman Sea (depths 4400−5900 m, [27]), albeit without molecular identification, and the genus was found in the Southern Ocean [67]. Isopods are among the most abundant and diverse fauna in the deep ocean [68]. Both shallow- and deep-sea isopods have been observed associated with macroalgae [11,69–71], although many species are generalists and detritivores, consuming a wide range of food sources (e.g. [39,72,73]). Previous analyses of B. nybelini gut contents documented both macroalgae and rare remnants of other organisms [27], suggesting macroalgae may not be its only food source. Episodic pulses, seasonal cycles, and climatic changes in primary production can impact organismal distributions and physiology in the deep ocean (e.g. [74,75]). Given that Sargassum abundances at the surface can show seasonality, patchy distributions, and have been increasing over the last decade (e.g. [16]), future work should link B. nybelini distributions and behaviour to estimates of Sargassum biomass at the surface and its depositional flux using satellites, sediment traps, and transects along the seafloor. Overall, we hypothesize that Bathyopsurus and related herbivorous isopods may be prominent members of benthopelagic megafauna communities across the world’s oceans in areas with macroalgal deposition.
Our finding that surface-derived macroalgae is common at the abyssal-hadal boundary and actively eaten by deep-ocean taxa may have implications for carbon cycling and storage at great depth. Exposure of macroalgae to turbulence and deep-mixing events that exceed depths of 130 m (1.3 Megapascals), which can occur during storms [76,77], are enough to make Sargassum negatively buoyant and rapidly sink to the abyssal seafloor in approximately 40 h (sinking speeds of approx. 3000 m d−1 [78]). The many Sargassum observations here suggest this fixed carbon source is readily available and may be abundant in the abyssal ocean (e.g. [15,21,22]). On a global scale, total carbon sequestration from macroalgae is estimated to be about 173 Tg C yr−1, 88% of which is in the deep oceans [21]. The fate of Sargassum is important as its abundance and distribution in the tropical Atlantic and Gulf of Mexico have been increasing, with important ramifications for both the ecology and economies in the Atlantic (e.g. [20,79,80]). Furthermore, it has been suggested to combat climate change by deliberately sinking macroalgae into the deep sea to store carbon (e.g. [81,82]). Bathyopsurus nybelini is not the only species that consumes macroalgae; pulses of organic matter can elicit responses by sea cucumbers, crustaceans, polychaetes, cumaceans, and microorganisms (e.g. [11,12,70,83–86]). Given that numerous organisms consume this material, increasing biomass flux to the seafloor will have downstream impacts of largely unknown consequence on the ecology of deep-sea communities (e.g. [87,88]). Future work will be needed to quantitatively evaluate the flux of Sargassum and its relative importance to the deep-ocean food web.
While interest in the deep ocean has increased over the last few decades [89], sampling at abyssal and hadal depths remains constrained by technology that is limited to only a few nations and institutions [90]. Following its upgrade, DSV Alvin is now one of only a few human-occupied vehicles capable of reaching depths exceeding 6000 m (e.g. [91,92]), providing increased opportunity to actively image and sample the deep abyssal and hadal seafloor. Our observations using Alvin follow previous lander-based images [15] and gut content morphological analysis of trawled specimens [27] that suggested this organism consumes Sargassum. Our study highlights how the ability to selectively target and collect specimens from the deep sea fills gaps in knowledge about the fate of fixed carbon and nutrients in surface waters and uncovers novel adaptations that link oceanic food webs. The deep ocean—the largest biome on the planet—is commonly deemed ‘extreme.’ We add to a growing body of evidence that abyssal and hadal communities are directly impacted by conditions at the surface, including through the deposition of pollutants and plastics (e.g. [93–95]). Further exploration will only continue to reveal that the fate of organisms at the greatest ocean depths is inexorably connected to human activities far above.
4. Methods
(a). Study location and methodology
Samples were collected aboard the R/V Atlantis during the Alvin Science Verification Expedition (AT50−02) to the Puerto Rico Trench (26 July–2 August 2022) and the Mid-Cayman Spreading Center (3−19 August 2022). Video and sampling were performed using Alvin during five scientific dives surrounding the Puerto Rico Trench (AL5088−5092; depths approx. 5600–6300 m) and nine dives at the Mid-Cayman Spreading Center (AL5093−5101; depths approx. 2300–6000 m). Average bottom time for each dive was 3 h. Alvin was equipped with two 4K ultra-high-definition Optim SeaCam cameras (Deep-Sea Power and Light, San Diego, CA) on pan and tilt platforms. Lighting was provided by ten LSL-2000 SeaLights (Deep-Sea Power and Light).
(b). Estimating Bathyopsurus and macroalgae abundances on the seafloor
The isopod Bathyopsurus nybelini was encountered during six dives (table 1). Macroalgae pieces bigger than approximately 3 cm were quantified during three dives (dives AL5089, AL5090, AL5091) within the Puerto Rico Trench along sedimented seafloor where isopods were also observed. Dives had multiple objectives, including verification of navigation systems, collections, and other protocols; therefore, straight transects were not typically conducted. Due to the lack of fixed transects and non-calibrated laser scale indicators, quantitative abundances of isopods and macroalgae were not determined. Instead, we report the total number of observations on each dive relative to the duration of bottom time and total distance travelled. Observations were plotted using the R [96] packages marmap [97] and rgdal v. 1.5-28.
(c). Sample collection
Two individual isopod specimens carrying Sargassum were collected via a suction sampler into the same chamber, approximately four minutes apart, from 6114 and 6100 m depth (dive AL5090; 20.29679°N, 65.70597°W and 20.29694°N, 65.70575°W). Upon return to the surface, the specimens were kept in cold seawater on ice and moved to a 4°C cold room for processing. One isopod (AT50-02-017) and one frond of Sargassum were preserved in 95% ethanol as vouchers for morphological taxonomy. The second isopod (AT50-02-018) was sampled for phylogenetic and microbiological analyses as described below and then preserved in 95% ethanol. The second piece of Sargassum was subsampled for chemical content. All sub-samples were flash-frozen in liquid N2 and stored at −80°C.
(d). Isopod and Sargassum morphology
The collected isopod and Sargassum material were morphologically examined with a Zeiss Axio Zoom V16 stereo microscope (Oberkochen, Germany). Images were digitally inked using Inkscape v.1.2.2. For the isopod, terminology, measurements, and identifications follow Wolff, 1962 [27], Wilson, 1989 [98], and Osborn, 2009 [37]. Total body length was measured medially from the anterior edge of the cephalon to the posterior tip of the pleotelson. The length of segments was measured medially or laterally from the anterior margin to the posterior margin. For the Sargassum, terminology and morphotype determination follow Parr, 1939 [30] and Schell et al., 2015 [17]. Further details can be found in the electronic supplementary material.
(e). 3D imaging using micro-computed tomography
One specimen (AT50-02-017) was imaged using micro-computed tomography (micro-CT) scanning using a Bruker SkyScan 1173 (Karel F. Liem Bioimaging Center, Friday Harbor Laboratories, University of Washington, USA). The isopod was wrapped in plastic with a minimal quantity of 75% ethanol to keep the specimen wet during scanning. Two scans were conducted at 50 kV and 145 µA, with no metal filter, optimized for low-density invertebrates. Voxel size was 19.4 µm resolution for the first, full body scan and 7.1 µm for the second, head-specific scan. After scanning, two-dimensional images were reconstructed using NRecon (Bruker, 2005−2011). During reconstruction, we followed a standard protocol for optimizing x/y alignment, reducing ring artefacts generated by variations in sensitivity on the scanner’s detector, correcting for beam hardening, and post-aligning the scan (e.g. [99]). To reduce scan size for analysis, the reconstructed scans were segmented in DataViewer (Bruker, 2004−2011). Images for this manuscript were rendered using the open-source image computing platform 3D Slicer via the extension SlicerMorph [100,101].
(f). Isopod DNA barcoding
Total genomic DNA was extracted from one isopod (AT50-02-018) ventral pleotelson using a standard phenol-chloroform approach. Two regions for PCR amplification were targeted using primers previously used on the Munnopsidae [37]: approximately 1800 bp of the nuclear small-subunit 18S rRNA gene with mitchA and mitchB [102] and approximately 1100 bp surrounding the D1−D3 region of the nuclear large-subunit 28S rRNA gene with LSUD1F and D3AR [103]. PCR products were purified using a QIAquick PCR Purification kit (Qiagen) and sequenced by Sanger sequencing on an ABI 3730XL capillary sequencer (Eurofins Genomics, Louisville, KY, USA). Electropherograms were manually inspected, and ambiguous base calls were denoted with N. Primers were trimmed in MEGA X [104]. Phylogenetic analyses are described in the electronic supplementary material.
(g). Sargassum biomass carbon and nitrogen content
Approximately 1 g of flash-frozen Sargassum, which was actively carried by an isopod during collection, was stored at −80°C. This material included epiphytic polychaetes that were not removed prior to analysis as we have no evidence they are selectively removed by the isopod prior to consumption. Sargassum was freeze-dried, homogenized, and subsampled. Carbon and nitrogen content were determined using an Exeter Analytical CE-440. The %C and %N data were used to calculate molar C:N ratios.
(h). Gut content metagenomic sequencing
For gut content metagenomic shotgun sequencing, gut contents were aseptically obtained from one isopod (AT50-02-018) by placing a syringe needle through the abdomen wall. DNA was extracted using a Qiagen DNeasy Blood and Tissue Kit (Hilden, Germany) and shotgun metagenomic sequencing was performed on an Illumina Novaseq (Novogene, Sacramento, CA). Raw reads were quality trimmed using Trimmomatic v. 0.39 [105] and assembled with MEGAHIT v. 1.2.9 [106]. The depth of coverage of contigs was estimated using Bowtie 2 v. 2.3.5.1 [107] and SAMtools v. 1.10 [108].
We characterized the microbial community and its functional potential. Open reading frames were identified with Prodigal v. 2.6 [109] and Prokka v. 1.14.6 [110]. Gene annotation and the identification of carbohydrate active enzymes and sulfatases were performed using GhostKOALA [111], dbCAN3 [112] and SulfAtlas [113], respectively. Ten microbial single-copy marker genes (rps2, rps7, rps8, rps15, rpl3, rpl5, rpl16, rpoB, recA, 16S rRNA) were used to characterize the prokaryotic community based on their depth of coverage. Single-copy marker genes related to Sargassum were identified by blastp [114] comparisons against published Sargassum mitochondrial genomes. Trees were created by sequence alignment with MUSCLE [115], built using FastTree [116] and visualized using iTOL [117].
Metagenome-assembled genomes (MAGs) were obtained using MetaBAT 2 v. 2.11.1 [118]. Contigs longer than 5 kb were retained. The quality of each MAG was evaluated using CheckM v. 1.0.13 [119]. MAGs related to Colwellia and Psychromonas were further refined manually, including through blastn similarity against published genomes of these genera. Genomes were taxonomically classified with GTDB-Tk v. 1.6.0 [120] using Kbase [121]. Average nucleotide identity (ANI) comparisons were performed using OrthoANI [122]. Whole-genome trees of selected MAGs were created using concatenated single-copy marker genes obtained and aligned using CheckM, constructed using FastTree, and visualized using iTOL. Further comparative analyses can be found in the electronic supplementary material.
Acknowledgements
We are grateful to the captain and crew of the R/V Atlantis for supporting this expedition. The authors extend their sincere gratitude to the US National Deep Submergence Facility and the DSV Alvin team both at sea and on shore, including Bruce Strickrott, Bob Waters, Danik Forsman, Kaitlyn Beardshear, John Dymek, Benen ElShakhs, Nick Osadcia, Randy Holt, and Matt Skorina (WHOI). We thank the Alvin Science Verification Expedition Science Party for their contributions to dive planning, video data collection, and expedition success. Thank you to Joe Garcia, Audrey Mickle, and Tina Haskins (WHOI) for their data management work that supports Alvin operations and research. We thank Adam Summers and the Karel F. Liem Bioimaging Center, Friday Harbor Laboratories, University of Washington for access to CT Scanning facilities and Louis Kerr at the Marine Biological Laboratory’s Central Microscopy Facility. Amy Suida (Eckard College) provided helpful advice on Sargassum identification. Thank you to Adam Baumann, Sydni Racki, and Miranda Seixas (Flathead Lake Biological Station, University of Montana) for technical support. Robert Hall Jr. (Flathead Lake Biological Station) provided helpful feedback on previous versions of this manuscript.
Contributor Information
Logan M. Peoples, Email: logan.peoples@flbs.umt.edu.
Mackenzie E. Gerringer, Email: gerringer@geneseo.edu.
Johanna N. J. Weston, Email: johanna.weston@whoi.edu.
Rosa León-Zayas, Email: rleonzayas@willamette.edu.
Abisage Sekarore, Email: ans100@geneseo.edu.
Grace Sheehan, Email: gasheehan@willamette.edu.
Matthew J. Church, Email: matt.church@flbs.umt.edu.
Anna P. M. Michel, Email: amichel@whoi.edu.
S. Adam Soule, Email: adamsoule@uri.edu.
Timothy M. Shank, Email: tshank@whoi.edu.
Ethics
Sampling in the Cayman Islands was carried out with permission of the Cayman Island authorities and the Ocean Policy Unit (Legal Directorate) of the Foreign, Commonwealth & Development Office, London (Reference 468/2022).
Data accessibility
Isopod specimens have been deposited at the Smithsonian Institution National Museum of Natural History (Suitland, MD, USA) under accession numbers USNM 1716401 and 1716402. CT-scan reconstructions are available on MorphoSource (Duke University) under accession number 000640894. Isopod and A. latecarinata rRNA gene sequences are available at NCBI under accession numbers PP351657, PP351679, and PP351649. The gut metagenome and MAGs are available at NCBI BioProject accession number PRJNA1075769. Raw video data are available from the Woods Hole Oceanographic Institution.
Supplementary material is available online [123].
Declaration of AI use
We have not used AI-assisted technologies in creating this article.
Authors’ contributions
L.M.P.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; M.E.G.: conceptualization, data curation, formal analysis, investigation, methodology, supervision, writing—original draft, writing—review and editing; J.N.J.W.: conceptualization, data curation, formal analysis, investigation, methodology, writing—original draft, writing—review and editing; R.L.Z.: conceptualization, investigation, project administration, supervision, writing—review and editing; A.S.: formal analysis, investigation, writing—review and editing; G.S.: formal analysis, investigation, writing—review and editing; M.J.C.: formal analysis, funding acquisition, investigation, methodology, resources, supervision, writing—review and editing; A.P.M.M.: conceptualization, funding acquisition, investigation, project administration, resources, supervision, writing—review and editing; S.A.S.: conceptualization, funding acquisition, investigation, project administration, resources, supervision, writing—review and editing; T.M.S.: conceptualization, formal analysis, investigation, methodology, project administration, resources, supervision, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Funding for this work was provided by the National Science Foundation (NSF grant #2129431; S.A.S.), the Simons Collaboration on Ocean Processes and Ecology (SCOPE grant #721221; L.M.P., M.J.C.), the State University of New York at Geneseo (M.E.G., A.S.), and the Woods Hole Oceanographic Institution (J.N.J.W., A.P.M.M.).
References
- 1. Martin JH, Knauer GA, Karl DM, Broenkow WW. 1987. VERTEX: carbon cycling in the northeast Pacific. Deep Sea Res. Oceanogr. Res. Pap. 34 , 267–285. ( 10.1016/0198-0149(87)90086-0) [DOI] [Google Scholar]
- 2. Thiel H, et al. 1989. Phytodetritus on the deep-sea floor in a central oceanic region of the northeast Atlantic. Biol. Oceanogr. 6 , 203–239. ( 10.1080/01965581.1988.10749527) [DOI] [Google Scholar]
- 3. Smith KL, Ruhl HA, Huffard CL, Messié M, Kahru M. 2018. Episodic organic carbon fluxes from surface ocean to abyssal depths during long-term monitoring in NE pacific. Proc. Natl Acad. Sci. USA 115 , 12235–12240. ( 10.1073/pnas.1814559115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith CR, Baco AR. 2003. Ecology of whale falls at the deep-sea floor. Oceanogr. Mar. Biol. Annu. Rev. 41 , 311–354. [Google Scholar]
- 5. Robison BH, Reisenbichler KR, Sherlock RE. 2005. Giant larvacean houses: rapid carbon transport to the deep sea floor. Science 308 , 1609–1611. ( 10.1126/science.1109104) [DOI] [PubMed] [Google Scholar]
- 6. Rouse GW, Goffredi SK, Vrijenhoek RC. 2004. Osedax: bone-eating marine worms with dwarf males. Science 305 , 668–671. ( 10.1126/science.1098650) [DOI] [PubMed] [Google Scholar]
- 7. Voight JR. 2015. Xylotrophic bivalves: aspects of their biology and the impacts of humans. J. Molluscan Stud. 81 , 175–186. ( 10.1093/mollus/eyv008) [DOI] [Google Scholar]
- 8. Tunnicliffe V, Juniper SK, Sibuet M. 2003. Reducing environments of the deep-sea floor. Ecosyst. World 81–110. [Google Scholar]
- 9. Levin LA, et al. 2016. Hydrothermal vents and methane seeps: rethinking the sphere of influence. Front. Mar. Sci. 3 , 195812. ( 10.3389/fmars.2016.00072) [DOI] [Google Scholar]
- 10. George RY, Higgins RP. 1979. Eutrophic hadal benthic community in the Puerto Rico Trench. Ambio. Spec. Rep. 51–58. [Google Scholar]
- 11. Wolff T. 1979. Magrofaunal utilization of plant remains in the deep sea. Sarsia 64 , 117–143. ( 10.1080/00364827.1979.10411373) [DOI] [Google Scholar]
- 12. Gallo ND, Cameron J, Hardy K, Fryer P, Bartlett DH, Levin LA. 2015. Submersible- and lander-observed community patterns in the Mariana and New Britain trenches: influence of productivity and depth on epibenthic and scavenging communities. Deep Sea Res. Oceanogr. Res. Papers 99 , 119–133. ( 10.1016/j.dsr.2014.12.012) [DOI] [Google Scholar]
- 13. Leduc D, Rowden AA. 2018. Not to be sneezed at: does pollen from forests of exotic pine affect deep oceanic trench ecosystems? Ecosystems 21 , 237–247. ( 10.1007/s10021-017-0146-8) [DOI] [Google Scholar]
- 14. Schoener A, Rowe GT. 1970. Pelagic Sargassum and its presence among the deep-sea benthos. Deep Sea Res. Oceanogr. Abstr. 17 , 923–925. ( 10.1016/0011-7471(70)90010-0) [DOI] [Google Scholar]
- 15. Fleury AG, Drazen JC. 2013. Abyssal scavenging communities attracted to Sargassum and fish in the Sargasso Sea. Deep Sea Res. Oceanogr. Res. Papers 72 , 141–147. ( 10.1016/j.dsr.2012.11.004) [DOI] [Google Scholar]
- 16. Wang M, Hu C, Barnes BB, Mitchum G, Lapointe B, Montoya JP. 2019. The great Atlantic Sargassum belt. Science 365 , 83–87. ( 10.1126/science.aaw7912) [DOI] [PubMed] [Google Scholar]
- 17. Schell JM, Goodwin DS, Siuda ANS. 2015. Recent Sargassum inundation events in the Caribbean: shipboard observations reveal dominance of a previously rare form. Oceanography 28 , 8–10. ( 10.5670/oceanog.2015.70) [DOI] [Google Scholar]
- 18. Lapointe BE, Brewton RA, Herren LW, Wang M, Hu C, McGillicuddy DJ, Lindell S, Hernandez FJ, Morton PL. 2021. Nutrient content and stoichiometry of pelagic Sargassum reflects increasing nitrogen availability in the Atlantic Basin. Nat. Commun. 12 , 3060. ( 10.1038/s41467-021-23135-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGillicuddy DJ, Morton PL, Brewton RA, Hu C, Kelly TB, Solow AR, Lapointe BE. 2023. Nutrient and arsenic biogeochemistry of Sargassum in the western Atlantic. Nat. Commun. 14 , 6205. ( 10.1038/s41467-023-41904-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. United Nations Environment Programme – Caribbean Environment Programme . 2021. Sargassum white paper – turning the crisis into an opportunity. Ninth meeting of the Scientific and Technical Advisory Committee (STAC) to the protocol concerning Specially Protected Areas and Wildlife (SPAW) in the wider Caribbean region. Kingston, Jamaica [Google Scholar]
- 21. Krause-Jensen D, Duarte CM. 2016. Substantial role of macroalgae in marine carbon sequestration. Nat. Geosci. 9 , 737–742. ( 10.1038/ngeo2790) [DOI] [Google Scholar]
- 22. Baker P, et al. 2018. Potential contribution of surface-dwelling Sargassum algae to deep-sea ecosystems in the southern North Atlantic. Deep Sea Res. Part II: Top. Stud. Oceanogr. 148 , 21–34. ( 10.1016/j.dsr2.2017.10.002) [DOI] [Google Scholar]
- 23. Isaacs JD, Schwartzlose RA. 1975. Active animals of the deep-sea floor. Sci. Am. 233 , 84–91. ( 10.1038/scientificamerican1075-84) [DOI] [Google Scholar]
- 24. Bailey DM, King NJ, Priede IG. 2007. Cameras and carcasses: historical and current methods for using artificial food falls to study deep-water animals. Mar. Ecol. Prog. Ser. 350 , 179–191. ( 10.3354/meps07187) [DOI] [Google Scholar]
- 25. Linley TD, Stewart AL, McMillan PJ, Clark MR, Gerringer ME, Drazen JC, Fujii T, Jamieson AJ. 2017. Bait attending fishes of the abyssal zone and hadal boundary: community structure, functional groups and species distribution in the Kermadec, New Hebrides and Mariana trenches. Deep Sea Res. Oceanogr. Res. Papers 121 , 38–53. ( 10.1016/j.dsr.2016.12.009) [DOI] [Google Scholar]
- 26. Nordenstam A. 1955. A new isopod from the deep sea. Reports of the Swedish Deep-Sea Expedition 2 , 203–212. [Google Scholar]
- 27. Wolff T. 1962. The systematics and biology of bathyal and abyssal Isopoda Asellota. Galathea Rep. 6 , 1–320. [Google Scholar]
- 28. Walden BB, Brown RS. 2004. A replacement for the Alvin submersible. Mar. Technol. Soc. J. 38 , 85–91. ( 10.4031/002533204787522721) [DOI] [Google Scholar]
- 29. Humphris SE, German CR, Hickey JP. 2014. Fifty years of deep ocean exploration with the DSV Alvin. EOS 95 , 181–182. ( 10.1002/2014EO220001) [DOI] [Google Scholar]
- 30. Parr AE. 1939. Quantitative observations on the pelagic Sargassum vegetation of the western North Atlantic. Bull. Bingham Oceanogr. Collect. 6 , 1–94. [Google Scholar]
- 31. Sichert A, et al. 2020. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5 , 1026–1039. ( 10.1038/s41564-020-0720-2) [DOI] [PubMed] [Google Scholar]
- 32. Orellana LH, Francis TB, Ferraro M, Hehemann JH, Fuchs BM, Amann RI. 2022. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 16 , 630–641. ( 10.1038/s41396-021-01105-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, Pascual-García A, Cordero OX. 2019. Modular assembly of polysaccharide-degrading marine microbial communities. Curr. Biol. 29 , 1528–1535.( 10.1016/j.cub.2019.03.047) [DOI] [PubMed] [Google Scholar]
- 34. Bunse C, Koch H, Breider S, Simon M, Wietz M. 2021. Sweet spheres: succession and CAZyme expression of marine bacterial communities colonizing a mix of alginate and pectin particles. Environ. Microbiol. 23 , 3130–3148. ( 10.1111/1462-2920.15536) [DOI] [PubMed] [Google Scholar]
- 35. Thomson M, Robertson K, Pile A. 2009. Microscopic structure of the antennulae and antennae on the deep-sea isopod Bathynomus pelor. J. Crust. Biol. 29 , 302–316. ( 10.1651/08-3083.1) [DOI] [Google Scholar]
- 36. Marshall N, Diebel C. 1995. “Deep-sea spiders” that walk through the water. J. Exp. Biol. 198 , 1371–1379. ( 10.1242/jeb.198.6.1371) [DOI] [PubMed] [Google Scholar]
- 37. Osborn KJ. 2009. Relationships within the Munnopsidae (Crustacea, Isopoda, Asellota) based on three genes. Zool. Scr. 38 , 617–635. ( 10.1111/j.1463-6409.2009.00394.x) [DOI] [Google Scholar]
- 38. Bober S, Brix S, Riehl T, Schwentner M, Brandt A. 2018. Does the Mid-Atlantic Ridge affect the distribution of abyssal benthic crustaceans across the Atlantic Ocean? Deep Sea Res. Part II: Top. Stud. Oceanogr. 148 , 91–104. ( 10.1016/j.dsr2.2018.02.007) [DOI] [Google Scholar]
- 39. Hessler RR, Strömberg JO. 1989. Behavior of janiroidean isopods (Asellota), with special reference to deep-sea genera. Sarsia. 74 , 145–159. ( 10.1080/00364827.1989.10413424) [DOI] [Google Scholar]
- 40. Hessler RR. 1993. Swimming morphology in Eurycope cornuta (Isopoda: Asellota). J. Crust. Biol. 13 , 667–674. ( 10.1163/193724093X00237) [DOI] [Google Scholar]
- 41. Jamieson AJ, Fujii T, Priede IG. 2012. Locomotory activity and feeding strategy of the hadal munnopsid isopod Rectisura cf. herculea (Crustacea: Asellota) in the Japan Trench. J. Exp. Biol. 215 , 3010–3017. ( 10.1242/jeb.067025) [DOI] [PubMed] [Google Scholar]
- 42. Deniaud-Bouët E, Kervarec N, Michel G, Tonon T, Kloareg B, Hervé C. 2014. Chemical and enzymatic fractionation of cell walls from Fucales: insights into the structure of the extracellular matrix of brown algae. Ann. Bot. 114 , 1203–1216. ( 10.1093/aob/mcu096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Podell S, et al. 2023. Herbivorous fish microbiome adaptations to sulfated dietary polysaccharides. Appl. Environ. Microbiol. 89 , e0215422. ( 10.1128/aem.02154-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Vliet DM, Lin Y, Bale NJ, Koenen M, Villanueva L, Stams AJM, Sánchez-Andrea I. 2020. Pontiella desulfatans gen. nov., sp. nov., and Pontiella sulfatireligans sp. nov., two marine anaerobes of the Pontiellaceae fam. nov. producing sulfated glycosaminoglycan-like exopolymers. Microorganisms 8 , 920. ( 10.3390/microorganisms8060920) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu N, et al. 2024. Pontiella agarivorans sp. nov., a novel marine anaerobic bacterium capable of degrading macroalgal polysaccharides and fixing nitrogen. Appl. Environ. Microbiol. 90 , e0091423. ( 10.1128/aem.00914-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang YS, et al. 2024. Metagenomic insights into the dynamic degradation of brown algal polysaccharides by kelp-associated microbiota. Appl. Environ. Microbiol. 90 , e0202523. ( 10.1128/aem.02025-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu XA, et al. 2024. Low-level resource partitioning supports coexistence among functionally redundant bacteria during successional dynamics. ISME J. 18 , wrad013. ( 10.1093/ismejo/wrad013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kostanjšek R, Štrus J, Lapanje A, Avguštin G, Rupnik M, Drobne D. 2006. Intestinal microbiota of terrestrial isopods. In Intestinal Microorganisms of Termites and Other Invertebrates. Soil Biology (eds König H, Varma A), vol. 6. Berlin, Heidelberg: Springer. ( 10.1007/3-540-28185-1_5) [DOI] [Google Scholar]
- 49. Bouchon D, Zimmer M, Dittmer J. 2016. The terrestrial isopod microbiome: an all-in-one toolbox for animal-microbe interactions of ecological relevance. Front. Microbiol. 7 , 1472. ( 10.3389/fmicb.2016.01472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boeuf D, Edwards BR, Eppley JM, Hu SK, Poff KE, Romano AE, Caron DA, Karl DM, DeLong EF. 2019. Biological composition and microbial dynamics of sinking particulate organic matter at abyssal depths in the oligotrophic open ocean. Proc. Natl Acad. Sci. USA 116 , 11824–11832. ( 10.1073/pnas.1903080116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. León-Zayas R, Novotny M, Podell S, Shepard CM, Berkenpas E, Nikolenko S, Pevzner P, Lasken RS, Bartlett DH. 2015. Single cells within the Puerto Rico Trench suggest hadal adaptation of microbial lineages. Appl. Environ. Microbiol. 81 , 8265–8276. ( 10.1128/AEM.01659-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang W, et al. 2018. Genome reduction in Psychromonas species within the gut of an amphipod from the ocean’s deepest point. mSystems 3 , 10–1128. ( 10.1128/mSystems.00009-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Blanton JM, et al. 2022. Microbiomes of hadal fishes across trench habitats contain similar taxa and known piezophiles. mSphere 7 , e0003222. ( 10.1128/msphere.00032-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheng X, Wang Y, Li J, Yan G, He L. 2019. Comparative analysis of the gut microbial communities between two dominant amphipods from the Challenger Deep, Mariana Trench. Deep Sea Res. Part I: Oceanogr. Res. Papers 151 , 103081. ( 10.1016/j.dsr.2019.103081) [DOI] [Google Scholar]
- 55. Lapointe BE. 1995. A comparison of nutrient‐limited productivity in Sargassum natans from neritic vs. oceanic waters of the western North Atlantic Ocean. Limnol. Oceanogr. 40 , 625–633. ( 10.4319/lo.1995.40.3.0625) [DOI] [Google Scholar]
- 56. Kneip C, Lockhart P, Voss C, Maier UG. 2007. Nitrogen fixation in eukaryotes - new models for symbiosis. BMC Evol. Biol. 7 , 1–12. ( 10.1186/1471-2148-7-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fiore CL, Jarett JK, Olson ND, Lesser MP. 2010. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 18 , 455–463. ( 10.1016/j.tim.2010.07.001) [DOI] [PubMed] [Google Scholar]
- 58. Guerinot ML, Patriquin DG. 1981. The association of N2-fixing bacteria with sea urchins. Mar. Biol. 62 , 197–207. ( 10.1007/BF00388183) [DOI] [Google Scholar]
- 59. Waterbury JB, Calloway CB, Turner RD. 1983. A cellulolytic nitrogen-fixing bacterium cultured from the gland of Deshayes in shipworms (Bivalvia: Teredinidae). Science 221 , 1401–1403. ( 10.1126/science.221.4618.1401) [DOI] [PubMed] [Google Scholar]
- 60. Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. 2002. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int. J. Syst. Evol. Microbiol. 52 , 2261–2269. ( 10.1099/00207713-52-6-2261) [DOI] [PubMed] [Google Scholar]
- 61. Breznak JA, Brill WJ, Mertins JW, Coppel HC. 1973. Nitrogen fixation in termites. Nature 244 , 577–580. ( 10.1038/244577a0) [DOI] [PubMed] [Google Scholar]
- 62. Pinto-Tomás AA, Anderson MA, Suen G, Stevenson DM, Chu FST, Cleland WW, Weimer PJ, Currie CR. 2009. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science 326 , 1120–1123. ( 10.1126/science.1173036) [DOI] [PubMed] [Google Scholar]
- 63. Bar-Shmuel N, Behar A, Segoli M. 2020. What do we know about biological nitrogen fixation in insects? Evidence and implications for the insect and the ecosystem. Insect Sci. 27 , 392–403. ( 10.1111/1744-7917.12697) [DOI] [PubMed] [Google Scholar]
- 64. Deming JW, Colwell RR. 1982. Barophilic bacteria associated with digestive tracts of abyssal holothurians. Appl. Environ. Microbiol. 44 , 1222–1230. ( 10.1128/aem.44.5.1222-1230.1982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Romero-Romero S, Miller EC, Black JA, Popp BN, Drazen JC. 2021. Abyssal deposit feeders are secondary consumers of detritus and rely on nutrition derived from microbial communities in their guts. Sci. Rep. 11 , 12594. ( 10.1038/s41598-021-91927-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dubilier N, Bergin C, Lott C. 2008. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat. Rev. Microbiol. 6 , 725–740. ( 10.1038/nrmicro1992) [DOI] [PubMed] [Google Scholar]
- 67. Brandt A, Brix S, Brökeland W, Choudhury M, Kaiser S, Malyutina M. 2007. Deep-sea isopod biodiversity, abundance, and endemism in the Atlantic sector of the Southern Ocean—results from the ANDEEP I–III expeditions. Deep Sea Res. Part II: Top. Stud. Oceanogr. 54 , 1760–1775. ( 10.1016/j.dsr2.2007.07.015) [DOI] [Google Scholar]
- 68. Hessler RR, Sanders HL. 1967. Faunal diversity in the deep-sea. Deep Sea Res. Oceanogr. Abstr. 14 , 65–78. ( 10.1016/0011-7471(67)90029-0) [DOI] [Google Scholar]
- 69. Carefoot TH. 1973. Feeding, food preference, and the uptake of food energy by the supralittoral isopod Ligia pallasii. Mar. Biol. 18 , 228–236. ( 10.1007/BF00367989) [DOI] [Google Scholar]
- 70. Wolff T. 1976. Utilization of seagrass in the deep sea. Aquat. Bot. 2 , 161–174. ( 10.1016/0304-3770(76)90017-6) [DOI] [Google Scholar]
- 71. Jormalainen V, Honkanen T, Heikkilä N. 2001. Feeding preferences and performance of a marine isopod on seaweed hosts: cost of habitat specialization. Mar. Ecol. Prog. Ser. 220 , 219–230. ( 10.3354/meps220219) [DOI] [Google Scholar]
- 72. Storch V, Strus J, Brandt A. 2002. Microscopic anatomy and ultrastructure of the digestive system of Natatolana obtusata (Vanhöffen, 1914) (Crustacea, Isopoda). Acta Zool. 83 , 1–14. ( 10.1046/j.1463-6395.2002.00093.x) [DOI] [Google Scholar]
- 73. Brökeland W, Guðmundsson G, Svavarsson J. 2010. Diet of four species of deep-sea isopods (Crustacea: Malacostraca: Peracarida) in the South Atlantic and the Southern Ocean. Mar. Biol. 157 , 177–187. ( 10.1007/s00227-009-1308-9) [DOI] [Google Scholar]
- 74. Ruhl HA, Smith KL. 2004. Shifts in deep-sea community structure linked to climate and food supply. Science 305 , 513–515. ( 10.1126/science.1099759) [DOI] [PubMed] [Google Scholar]
- 75. Smith KL, Ruhl HA, Kahru M, Huffard CL, Sherman AD. 2013. Deep ocean communities impacted by changing climate over 24 y in the abyssal northeast Pacific Ocean. Proc. Natl Acad. Sci. USA 110 , 19838–19841. ( 10.1073/pnas.1315447110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sosa-Gutierrez R, Chevalier C. 2022. Impact of tropical cyclones on pelagic Sargassum. Geophys. Res. Lett. ( 10.1029/2021GL097484) [DOI] [Google Scholar]
- 77. Putman NF, Hu C. Sinking Sargassum. Geophys. Res. Lett. e2022GL100189. ( 10.1029/2021GL097484) [DOI] [Google Scholar]
- 78. Johnson DL, Richardson PL. 1977. On the wind-induced sinking of Sargassum. J. Exp. Mar. Biol. Ecol. 28 , 255–267. ( 10.1016/0022-0981(77)90095-8) [DOI] [Google Scholar]
- 79. Chávez V, et al. 2020. Massive influx of pelagic Sargassum spp. on the coasts of the Mexican Caribbean 2014–2020: challenges and opportunities. Water 12 , 2908. ( 10.3390/w12102908) [DOI] [Google Scholar]
- 80. Oxenford HA, Cox SA, van Tussenbroek BI, Desrochers A. 2021. Challenges of turning the Sargassum crisis into gold: current constraints and implications for the Caribbean. Phycol. 1 , 27–48. ( 10.3390/phycology1010003) [DOI] [Google Scholar]
- 81. Gouvêa LP, et al. 2020. Golden carbon of Sargassum forests revealed as an opportunity for climate change mitigation. Sci. Total Environ. 729 , 138745. ( 10.1016/j.scitotenv.2020.138745) [DOI] [PubMed] [Google Scholar]
- 82. Bach LT, Tamsitt V, Gower J, Hurd CL, Raven JA, Boyd PW. 2021. Testing the climate intervention potential of ocean afforestation using the Great Atlantic Sargassum Belt. Nat. Commun. 12 , 2556. ( 10.1038/s41467-021-22837-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bernardino AF, Smith CR, Baco A, Altamira I, Sumida PYG. 2010. Macrofaunal succession in sediments around kelp and wood falls in the deep NE Pacific and community overlap with other reducing habitats. Deep Sea Res. Part I: Oceanogr. Res. Papers 57 , 708–723. ( 10.1016/j.dsr.2010.03.004) [DOI] [Google Scholar]
- 84. Hoffmann K, Hassenrück C, Salman-Carvalho V, Holtappels M, Bienhold C. 2017. Response of bacterial communities to different detritus compositions in Arctic deep-sea sediments. Front. Microbiol. 8 , 266. ( 10.3389/fmicb.2017.00266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Harbour R, Smith C, Simon-Nutbrown C, Cecchetto M, Young E, Coral C, Sweetman A. 2021. Biodiversity, community structure and ecosystem function on kelp and wood falls in the Norwegian deep sea. Mar. Ecol. Prog. Ser. 657 , 73–91. ( 10.3354/meps13541) [DOI] [Google Scholar]
- 86. Nomaki H, et al. 2021. In situ experimental evidences for responses of abyssal benthic biota to shifts in phytodetritus compositions linked to global climate change. Glob. Chang. Biol. 27 , 6139–6155. ( 10.1111/gcb.15882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ricart AM, Krause-Jensen D, Hancke K, Price NN, Masqué P, Duarte CM. 2022. Sinking seaweed in the deep ocean for carbon neutrality is ahead of science and beyond the ethics. Environ. Res. Lett. 17 , 081003. ( 10.1088/1748-9326/ac82ff) [DOI] [Google Scholar]
- 88. Levin LA, et al. 2023. Deep-sea impacts of climate interventions. Science 379 , 978–981. ( 10.1126/science.ade7521) [DOI] [PubMed] [Google Scholar]
- 89. Weston JNJ, Jamieson AJ. 2022. Exponential growth of hadal science: perspectives and future directions identified using topic modelling. ICES J. Mar. Sci. 79 , 1048–1062. ( 10.1093/icesjms/fsac074) [DOI] [Google Scholar]
- 90. Bell KLC, et al. 2023. Exposing inequities in deep-sea exploration and research: results of the 2022 global deep-sea capacity assessment. Front. Mar. Sci. 10 , 1217227. ( 10.3389/fmars.2023.1217227) [DOI] [Google Scholar]
- 91. Kohnen W. 2013. Review of deep ocean manned submersible activity in 2013. Mar. Technol. Society J. 47 , 56–68. ( 10.4031/MTSJ.47.5.6) [DOI] [Google Scholar]
- 92. Jamieson AJ, Ramsey J, Lahey P. 2019. Hadal manned submersible. Sea. Technol. 60 , 22–24. [Google Scholar]
- 93. Fischer V, Elsner NO, Brenke N, Schwabe E, Brandt A. 2015. Plastic pollution of the Kuril–Kamchatka Trench area (NW Pacific). Deep Sea Res. Part II: Top. Stud. Oceanogr. 111 , 399–405. ( 10.1016/j.dsr2.2014.08.012) [DOI] [Google Scholar]
- 94. Jamieson AJ, Malkocs T, Piertney SB, Fujii T, Zhang Z. 2017. Bioaccumulation of persistent organic pollutants in the deepest ocean fauna. Nat. Ecol. Evol. 1 , 51. ( 10.1038/s41559-016-0051) [DOI] [PubMed] [Google Scholar]
- 95. Jamieson AJ, Brooks LSR, Reid WDK, Piertney SB, Narayanaswamy BE, Linley TD. 2019. Microplastics and synthetic particles ingested by deep-sea amphipods in six of the deepest marine ecosystems on Earth. R. Soc. Open Sci. 6 , 180667. ( 10.1098/rsos.180667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. R Core Team . 2024. R: A language and environment for statistical computing. R Foundation for Statistical Computing Vienna, Austria. https://www.R-project.org/
- 97. Pante E, Simon-Bouhet B. 2013. Marmap: a package for importing, plotting and analyzing bathymetric and topographic data in R. PLoS One 8 , e73051. ( 10.1371/journal.pone.0073051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wilson GDF. 1989. A systematic revision of the deep-sea subfamily Lipomerinae of the isopod crustacean family Munnopsidae. Univ. of Calif. Press 27 . [Google Scholar]
- 99. Gerringer ME, Dias AS, von Hagel AA, Orr JW, Summers AP, Farina S. 2021. Habitat influences skeletal morphology and density in the snailfishes (family Liparidae). Front. Zool. 18 , 16. ( 10.1186/s12983-021-00399-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fedorov A, et al. 2012. 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 30 , 1323–1341. ( 10.1016/j.mri.2012.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rolfe S, et al. 2021. SlicerMorph: an open and extensible platform to retrieve, visualize and analyse 3D morphology. Methods Ecol. Evol. 12 , 1816–1825. ( 10.1111/2041-210X.13669) [DOI] [Google Scholar]
- 102. Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71 , 491–499. ( 10.1016/0378-1119(88)90066-2) [DOI] [PubMed] [Google Scholar]
- 103. Lenaers G, Maroteaux L, Michot B, Herzog M. 1989. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J. Mol. Evol. 29 , 40–51. ( 10.1007/BF02106180) [DOI] [PubMed] [Google Scholar]
- 104. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35 , 1547–1549. ( 10.1093/molbev/msy096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 , 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31 , 1674–1676. ( 10.1093/bioinformatics/btv033) [DOI] [PubMed] [Google Scholar]
- 107. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 , 357–359. ( 10.1038/nmeth.1923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25 , 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11 , 119. ( 10.1186/1471-2105-11-119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30 , 2068–2069. ( 10.1093/bioinformatics/btu153) [DOI] [PubMed] [Google Scholar]
- 111. Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428 , 726–731. ( 10.1016/j.jmb.2015.11.006) [DOI] [PubMed] [Google Scholar]
- 112. Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y. 2023. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 51 , W115–W121. ( 10.1093/nar/gkad328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Stam M, Lelièvre P, Hoebeke M, Corre E, Barbeyron T, Michel G. 2023. SulfAtlas, the sulfatase database: state of the art and new developments. Nucleic Acids Res. 51 , D647–D653. https://doi.org/10.1093%2Fnar%2Fgkac977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215 , 403–410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 115. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 , 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Price MN, Dehal PS, Arkin AP. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5 , e9490. ( 10.1371/journal.pone.0009490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Letunic I, Bork P. 2021. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 , W293–W296. ( 10.1093/nar/gkab301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7 , e7359. ( 10.7717/peerj.7359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25 , 1043–1055. ( 10.1101/gr.186072.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36 , 1925–1927. ( 10.1093/bioinformatics/btz848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Arkin AP, et al. 2018. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 36 , 566–569. ( 10.1038/nbt.4163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Yoon SH, Ha SM, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110 , 1281–1286. ( 10.1007/s10482-017-0844-4) [DOI] [PubMed] [Google Scholar]
- 123. Peoples LM, Gerringer ME, Weston JN, León-Zayas R, Sekarore A, Sheehan Get al. 2024. Data from: A deep-sea isopod that consumes Sargassum sinking from the ocean’s surface. Figshare. ( 10.6084/m9.figshare.c.7412468) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Isopod specimens have been deposited at the Smithsonian Institution National Museum of Natural History (Suitland, MD, USA) under accession numbers USNM 1716401 and 1716402. CT-scan reconstructions are available on MorphoSource (Duke University) under accession number 000640894. Isopod and A. latecarinata rRNA gene sequences are available at NCBI under accession numbers PP351657, PP351679, and PP351649. The gut metagenome and MAGs are available at NCBI BioProject accession number PRJNA1075769. Raw video data are available from the Woods Hole Oceanographic Institution.
Supplementary material is available online [123].