Abstract
The 3′-termini of maize mitochondrial RNAs were characterized by ligation of an anchor oligonucleotide, reverse transcription and amplification. DNA sequence analysis of cDNA clones for tRNASer and 18S rRNA confirmed the expected 3′-terminal nucleotides and demonstrated the accuracy and fidelity of the protocol. Analysis of cDNAs for rps12, cox2 and atp9 indicated that non-genomically encoded nucleotides were present at the 3′-terminus. rps12 cDNAs exhibited the highest degree of modification, with 94% of 35 cDNA clones analyzed containing one to four non-genomically encoded C or A residues; 83% of these cDNAs terminated with the trinucleotide CCA. DNA sequence and transcript mapping analyses demonstrated that four positions exhibited modified 3′-termini within a small region of the 3′ flank of rps12 transcripts. These transcript termini represented low abundance, truncated forms of rps12 mRNAs which may be intermediates in degradation. cox2 mRNAs are also modified at a truncated position. Sixty percent of the cox2 cDNAs were modified with 1–5 nt that most frequently included A and C residues, but also included a few G and T residues. Non-genomically encoded nucleotides were detected in 27% of the atp9 cDNAs as a single C or A residue.
INTRODUCTION
3′ Modification is a common post-transcriptional modification of mRNAs and 3′ polyadenylation is typically required for efficient translation of cytosolic mRNAs. In addition, 3′ polyadenylation of bacterial and certain organellar mRNAs is an important post-transcriptional regulatory mechanism to mark transcripts for degradation (1–3). The sequence of events in prokaryotic RNA degradation is thought to involve endonucleolytic cleavage that makes an initial cleavage in the 3′ stem–loop region, followed by polyadenylation and 3′→5′ degradation (1). Transcript turnover is thought to involve a ‘degradosome’ (4) that includes: RNase E, an endonuclease (5,6); a DEAD-box RNA helicase that can unwind RNA substrates (7); a 3′→5′ polynucleotide phosphorylase (PNPase) with 3′→5′ exonuclease activity (8).
The adenylation of mitochondrial mRNAs in animal mitochondria has also been well characterized. Mammalian transcripts have long been recognized to be adenylated and post-transcriptional adenylation may complete the UAA termination codon in some cases (9). Trypanosome and yeast mitochondria also contain polyadenylated mRNAs (10,11).
In plant mitochondria 3′ modification of mRNAs is less well characterized. Plant mitochondrial transcripts typically have 3′ inverted repeats that are predicted to fold into stem and loop structures (12). Transcript stability in plant mitochondria has been shown to depend on the structure at the 3′-terminus (13,14). Polyadenylated RNAs have recently been reported for plant mitochondrial mRNAs (15,16). In cytoplasmic male sterile (cms) sunflower, transcripts from a cms-related gene (atpA-orf522) are polyadenylated and the level of polyadenylation correlates with transcript instability (16). Polyadenylation has also been shown to occur in cox2 mRNAs in maize mitochondria (17).
In this study the nature of the 3′-termini of plant mitochondrial transcripts has been examined by an anchor ligation strategy. T4 RNA ligase catalyzes the ATP-dependent ligation of 3′-OH and 5′-phosphorylated DNA or RNA and the ligation junction between the anchor oligonucleotide and mitochondrial RNA should reflect the 3′-terminus of the transcript. This reaction has been used rather extensively, especially for the creation of circular RNAs (18) and for the analysis of 3′-termini (19). Using this approach, we report the presence of non-genomically encoded nucleotides at the 3′-end of plant mitochondrial mRNAs.
MATERIALS AND METHODS
Isolation of maize mitochondrial nucleic acids
Mitochondria were isolated from 5–7-day-old etiolated seedlings of Zea mays (B37N) (Pioneer Hi-Bred International, Johnston, IA) and purified by centrifugation on a discontinuous sucrose density gradient (20).
Maize mitochondria were lysed with 0.5% SDS and total nucleic acids were extracted with phenol/chloroform followed by ethanol precipitation. Mitochondrial RNA was prepared by digesting total nucleic acids with RNase-free DNase (Boehringer Mannheim) for 1 h at 37°C, followed by phenol/chloroform extraction and ethanol precipitation.
Anchor ligation, reverse transcription, amplification and cloning
One to two micrograms of maize mitochondrial RNA was ligated with 40 pmol of anchor oligonucleotide BW (TTTAGTGAGGGTTAATAAGCGGCCGCGTCG). The anchor oligonucleotide contained a 5′-phosphate and a 3′-dideoxy A (cordycepin). The ligation reactions contained 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 0.2 mg/ml BSA, 1 mM hexamine cobalt chloride, 20 µM ATP, 12.5% PEG 8000 and 15 U T4 RNA ligase (New England Biolabs). Following a 2 h incubation at room temperature, the nucleic acids were directly precipitated with ethanol and ammonium acetate.
To examine the 3′-ends of endogenous rps12, atp9 and cox2 transcripts, cDNA clones were produced. cDNAs were prepared by reverse transcription (RT) using anchor-ligated maize mitochondrial RNA and 240 ng oligo GO (CGACGCGGCCGCTTATTAACCCTC) as previously described (21). A typical RT reaction contained 2 µg anchor-ligated RNA and was incubated at 42°C for 15–30 min. The RT reaction was terminated by heating at 99°C for 5 min and the entire reaction was used in a PCR reaction. cDNAs were amplified with 120 ng of gene-specific oligonucleotides and anti-anchor oligonucleotide as described (21). Amplification reactions were performed as follows: 3 min denaturation at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50 or 55°C and 1 min at 72°C, followed by 10 min at 72°C. PCR products were cloned into pCR2.1 (Invitrogen).
In vitro processing assays
Mitochondrial protein extracts were prepared by resuspension of a mitochondrial pellet, typically 100–200 µl packed volume, with 1 vol of lysis buffer (30 mM HEPES, pH 7.7, 3 mM MgOAc, 45 mM KOAc, 1 mM DTT, 10% glycerol and 5 µg/ml of each of the following protease inhibitors: leupeptin, amino caproic acid, antipain, aprotinin and pepstatin). The diluted mitochondria were lysed in 0.5% Triton X-100 for 5 min on ice and insoluble components were removed by centrifugation at 60 000 g for 2 h. The supernatant was dialyzed for 2 h against two changes each of 0.5 l of lysis buffer. Aliquots were rapidly frozen in liquid nitrogen and stored at –80°C. Protein concentrations were determined with the Bradford protein assay (Bio-Rad).
In vitro processing assays were performed in a total reaction volume of 20 µl and contained the following components: 100–200 ng in vitro transcribed RNA, 20 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 40 mM KCl, 1 mM DTT, 0.1 EDTA, 0.1 mM ATP, 0.1 mM CTP, 10 µCi 32P-radiolabeled nucleotide, 10 U RNasin (Gibco BRL) and 20–40 µg mitochondrial protein extract. Reactions were incubated for 30 min at room temperature, followed by phenol/chloroform extraction and ethanol precipitation. After resuspension in 5 µl of formamide loading buffer (USB), processing reaction products were electrophoresed on 7% denaturing polyacrylamide gels and visualized by autoradiagraphy.
The plasmid used to produce PCR products to generate substrate RNAs for processing reactions was an rps12 clone (pCL/CK in BsSK+) that extended into the 3′-UTR. This plasmid was used as a template for PCR amplification using oligo GZ (GCGGAGCTCAATACGACTCACTATAG), which contained the 18 nt T7 RNA polymerase promoter, and either oligo HU (GGCATCTTCCATTCATTTAGATTT), for the cleaved RNA substrate, oligo GY (TGGGGCATCTTCCATTCATTTAGA) for the CCA RNA substrate, or oligo CL (CCGGATCCCATCTCATAGTTACTC), for the long 3′ RNA substrate. The reaction products were purified on a 1% agarose gel using a QIA quick gel extraction kit (Qiagen).
RNAs were transcribed directly from the T7 promoter-containing PCR products using T7 RNA polymerase (Promega) and the manufacturer’s protocol. Transcription products were purified on a 1% agarose gel using a QIAquick gel extraction kit (Qiagen).
DNA sequencing analysis
DNA sequencing was performed by the chain termination method with an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). DNA sequence analysis was performed with an ABI model 370 automated DNA sequencer. DNA sequences were also determined with [35S]dATP as previously described (21).
RESULTS
Anchor ligation and RT–PCR accurately amplifies the 3′-ends of cDNAs
An anchor oligonucleotide was ligated to total maize mitochondrial RNA with T4 RNA ligase and first strand cDNA synthesis was performed by hybridization of an anti-anchor oligonucleotide and reverse transcription. Subsequent amplification was performed by PCR with a gene-specific oligonucleotide and the anti-anchor oligonucleotide. Amplification of anchor-ligated cDNAs was shown to be dependent on the addition of T4 RNA ligase for atp9 (Fig. 1A), cox2 (Fig. 1B), rps12 (Fig. 1C, lanes 5 and 6), trnS (Fig. 1C, lanes 7 and 8) and rrn18 (data not shown).
Figure 1.
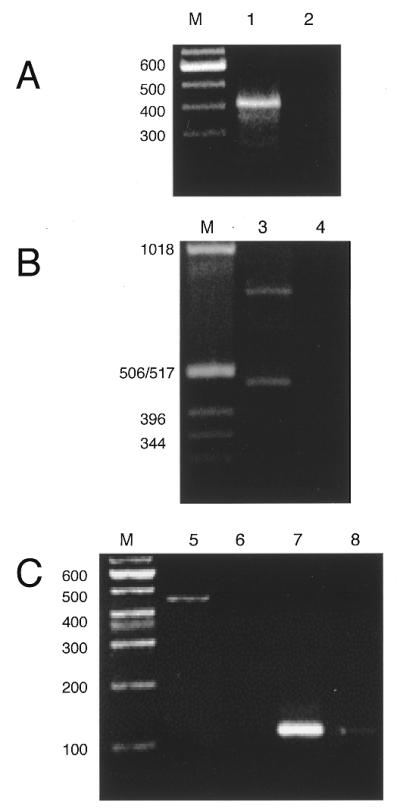
Amplification of anchor-ligated cDNAs is dependent on T4 RNA ligase. Maize mitochondrial RNA (1–2 µg) was incubated with 40 pmol of anchor oligonucleotide in the presence or absence of T4 RNA ligase. Anchor-ligated RNAs were reverse transcribed and amplified by PCR and the cDNA products were electrophoresed on agarose gels. Lanes marked M show the migration of commercial DNA size markers. PCR products for the following cDNAs are shown: (A) atp9, lanes 1 and 2; (B) cox2, lanes 3 and 4; (C) rps12, lanes 5 and 6, and trnS, lanes 7 and 8. Amplification of anchor-ligated atp9. Odd numbered lanes (1, 3, 5 and 7) included T4 RNA ligase and even numbered lanes (2, 4, 6 and 8) omitted T4 RNA ligase.
In order to document the fidelity of the method, the 3′-ends of cDNAs with known 3′-termini were characterized. cDNAs for tRNASer and 18S rRNA were amplified and cloned from anchor-ligated total mitochondrial RNA. The 3′-termini of 15 18S rRNA cDNAs were determined by this method (Fig. 2A). The sequences of 14 18S rRNA cDNAs terminated at the expected 3′-terminus of 18S rRNA (22) and this technique amplified cDNAs with the expected sequence at the 3′-terminus of 18S rRNA. One of the 15 cDNAs reflected a cDNA that was truncated 155 nt upstream of the 3′-terminus. The sequence of this cDNA read TTTCGCCCCA-anchor oligonucleotide (genomic sequence underlined, 3′ non-encoded nucleotides bold), such that a non-encoded CA was introduced at the 3′-terminus of the cleaved 18S rRNA. The cDNA could reflect a truncated 18S rRNA that is engaged in degradation.
Figure 2.
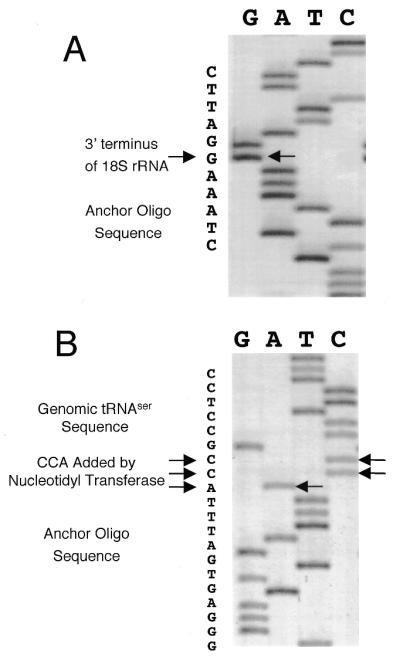
Anchor-ligated cDNAs for 18S rRNA and tRNASer show the expected 3′-termini. (A) DNA sequence of an anchor-ligated cDNA for 18S rRNA demonstrates that no non-genomically encoded nucleotides are introduced at the 3′-terminus of mature 18S rRNA. The arrow indicates the expected 3′-terminus of 18S rRNA. (B) DNA sequence of an anchor-ligated cDNA for tRNASer shows the presence of the CCA post-transcriptionally added to the 3′-terminus. Arrows indicate the non-genomically encoded nucleotides.
The 3′-ends of tRNASer cDNAs were determined to include the post-transcriptional addition of CCA. Ten of the 11 clones sequenced included the genomically encoded DNA sequence, the CCA added by tRNA nucleotidyltransferase and the anchor oligonucleotide sequence as shown in Figure 2B. The DNA sequence of one of the 11 clones had a single C at the 3′-terminus (TCCTCCGC-anchor oligo) and may reflect an incompletely modified tRNA molecule. Thus, this experiment confirmed that the anchor ligation and amplification method accurately detected the 3′-terminus of a post-transcriptionally modified tRNA, as well as the unmodified 18S rRNA.
Maize mitochondrial mRNAs have non-genomically encoded nucleotides at their 3′-termini
Thirty-five rps12 cDNAs were obtained by anchor ligation and PCR and 33 of these cDNAs were determined to have a 3′ modification that involved addition of cytidine or adenine residues. The last genomically encoded nucleotide of these cDNAs was identified at position 1, 2, 3 or 4 (Fig. 3A). Transcript mapping experiments described in the next section demonstrated that rps12 transcripts exist with termini at these positions, but they are low in abundance and apparently represent 3′ truncated forms of rps12 RNAs. The last genomically-encoded nucleotide of these rps12 transcripts was typically followed by 1–4 nt composed of C and A residues (Fig. 3A). The 3′-termini of 29 of the 35 cDNAs ended with the nucleotide sequence CCA; in clones D, E and J the trinucleotide was partially genomically encoded. Figure 3B–D shows DNA sequences of rps12 cDNAs with examples of non-genomically encoded nucleotides at the 3′-termini.
Figure 3.
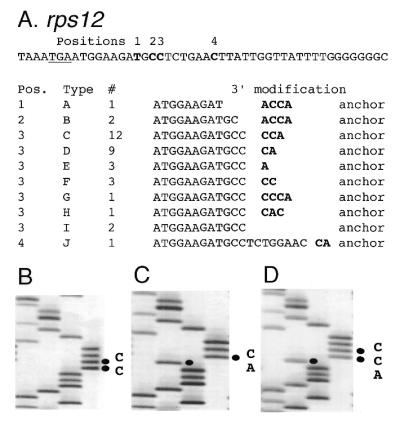
The 3′ modification of rps12 mRNAs. (A) Sequences of the 3′-terminus of the rps12 mRNAs. The genomic sequence of downstream of rps12 is shown above with the stop codon (TGA) underlined. Bold letters indicate the modified sites. The modification patterns of the 3′-termini of rps12 mRNAs are shown in the table below. Bold letters show non-genomically encoded nucleotides. (B–D) Autoradiograms of the sequences at the 3′-terminus of the rps12 mRNAs. DNA sequence reactions for G, A, T and C were loaded from left to right. (B) DNA sequence of clone F. (C) DNA sequence of clone D. (D) DNA sequence of clone C.
The nature of the 3′-termini of 26 atp9 cDNAs was also analyzed by the anchor ligation and amplification technique (Fig. 4A). Nineteen of 26 cDNAs (73%) had unmodified 3′-termini at one of two adjacent nucleotides. Seven of the 26 cDNAs (27%) were modified with a single A or C residue. Nuclease S1 protection analyses (data not shown) demonstrated that positions 1 and 2 represent the major transcript terminus for atp9.
Figure 4.
The 3′ modification of atp9 and cox2 mRNAs. Sequences of the 3′-terminus of the (A) atp9 and (B) cox2 cDNAs. The genomic sequences are shown above and bold letters indicate the modified sites. The modification patterns of the 3′-termini of mRNAs are shown in the tables below. Bold letters show non-genomically encoded nucleotides.
The nature of the 3′-termini of 22 cox2 cDNAs was also analyzed by the anchor ligation and amplification technique (Fig. 4B). Nine of the 22 cDNAs (41%) were unmodified, while the remaining 13 cDNAs (59%) included non-genomically encoded nucleotides. In the case of cox2 mRNAs the non-encoded nucleotides included not only C and A residues but also an occasional G or T residue. The last genomically encoded nucleotide of these cox2 cDNAs was within a 6 nt region. Thus 95% of the rps12 cDNAs included non-genomically encoded nucleotides at the 3′-terminus, while only 59 and 27% of the cox2 and atp9 cDNAs, respectively, were modified at their 3′-termini.
A detailed analysis of the extensively modified rps12 transcripts was performed. Nuclease S1 protection analyses were performed to determine the abundance and position of 3′-termini of rps12 transcripts. A genomic DNA clone was labeled by fill-in reaction at the EcoRI site and released from the plasmid by digestion at the XbaI site in the polylinker to produce a 320 nt fragment (lane 5). This probe protected a very abundant RNA species of ∼310 nt (lanes 1, 2) and the size shift between the 320 nt probe and the protected fragment of 310 nt reflects the removal of polylinker sequences by nuclease S1. The intensity of the 310 nt protected fragment indicated that a very abundant transcript extended downstream of the probe that probably represents the mature form of rps12 mRNAs. Within the proximal 3′-region of the rps12 coding sequence a number of less abundant transcript 3′-termini were detected (Fig. 5A). The shortest fragment detected at 80 nt corresponds to positions 1, 2 and 3 in Figure 3A, while the protected fragment at ∼88 nt corresponds to position 4 in Figure 3A. In addition, several additional protected fragments correspond to 3′-termini in the next 50 nt. Control reactions that omitted mitochondrial RNA (lanes 3 and 4) failed to protect any fragments and the omission of nuclease S1 (lane 5) failed to produce smaller fragments from the DNA probe.
Figure 5.
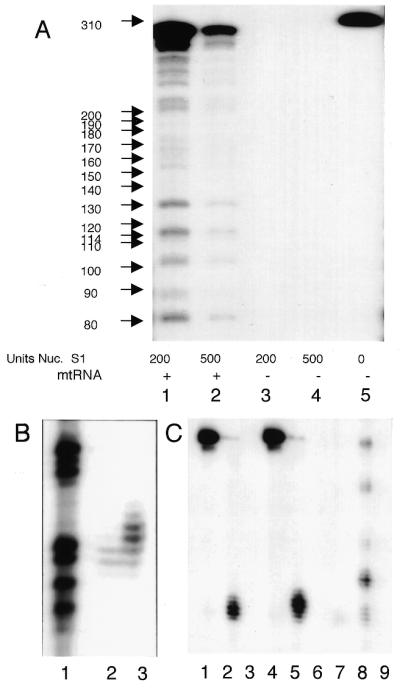
Transcript mapping of 3′-termini of rps12 mRNAs. (A) Nuclease S1 protection with a genomic probe shows an abundant RNA at ∼310 nt that extends through the probe and several hundred nucleotides beyond (lanes 1 and 2). Additional transcripts at relatively low abundance are evident in the gel. (B) cDNA clone C (3′-CCA) (A) was used as probe in a nuclease S1 protection experiment (lane 3) and demonstrates that the cDNA clone protects a few additional nucleotides relative to the genomic clone (lane 2). (C) cDNA clones D (3′-CA, lanes 1–3) and F (3′-CC, lanes 4–6) and the genomic clone (lanes 7–9) were used as probes in nuclease S1 protection assays. Lanes 1 and 4 are probe only. Lanes 2 and 5 show the DNA fragment protected from nuclease S1 digestion compared to the fragments protected by the genomic clone (lane 8). Mitochondrial RNA was omitted from lanes 3, 6 and 9.
In order to independently demonstrate the presence of 3′ modified transcripts without the use of PCR, we produced 3′-end-labeled probes from rps12 cDNA clones C, D and F (Fig. 3A). The cDNA probes were labeled by fill-in of the same EcoRI site as the genomic probe and the sizes of the protected fragments are therefore directly comparable. Probes from the rps12 cDNAs should protect one to several additional nucleotides longer than the genomic rps12 probe if RNAs are present with additional 3′ nucleotides. rps12 cDNA clone C has a CCA trinucleotide added at position 3 (Fig. 3A) and this clone represented one third of the rps12 cDNAs analyzed. Figure 5B shows the DNA fragments protected from nuclease S1 digestion with the genomic rps12 probe (lane 2) resolved on a DNA sequencing gel; lane 1 shows the G track from a dideoxy sequencing reaction with an unrelated control template. Mitochondrial RNA protected DNA fragments produced from rps12 cDNA clone C (lane 3) by an additional 1 and 2 nt and somewhat more weakly protected additional fragments of 3 and 4 nt longer than the genomic probe. These results directly confirm the occurrence and relatively high abundance of RNAs with these 3′ modifications.
rps12 cDNA clones D (3′-CC) and F (3′-CA) were also well represented in the analysis of rps12 cDNAs and end-labeled DNA probes were produced by fill-in reaction at the EcoRI site and used in nuclease S1 protection analyses. Figure 5C shows the protected fragments from rps12 cDNA clones D and F (lanes 2 and 5, respectively) and these probes protected an additional few nucleotides longer than the genomic probe (lane 8). Lanes 1, 4 and 7 show the DNA probe (omission of nuclease S1) and lanes 3, 6 and 9 are controls for the omission of mitochondrial RNA. Thus, nuclease S1 protection experiments independently verify the existence and document that 3′ modified rps12 RNAs are a substantial fraction of these truncated rps12 transcripts.
In vitro RNA processing reactions
An in vitro labeling system was developed to study the biochemistry of the post-transcriptional modification. A mitochondrial lysate was prepared that catalyzed incorporation of [α-32P]NTP into exogenous RNA substrates. A S-60 fraction was obtained by lysis of intact maize mitochondria with 0.5% Triton X-100 and removal of insoluble components by centrifugation at 60 000 g for 2 h. This fraction was active in the incorporation of [α-32P]ATP into exogenous substrate RNAs (Fig. 6). Three RNAs were tested for RNA specificity. rps12 RNA substrates were produced from PCR products that incorporated the promoter for T7 RNA polymerase into the 5′-end and utilized various 3′-ends: (i) a ‘cleaved RNA’ substrate was produced with a 3′ nucleotide at position 3 (Fig. 3); (ii) a ‘3′-CCA RNA’ was produced with a 3′-CCA at position 3, identical to clone C (Fig. 3); (iii) a ‘long 3′ RNA’ was prepared that had an additional 200 nt of 3′-UTR from rps12. Incubation of these substrates with the S-60 mitochondrial lysate and [α-32P]ATP resulted in the incorporation of label into the exogenous RNAs (lanes 1, 4 and 7). The incorporation of label was dependent on the exogenous substrate (exogenous RNA omitted, lanes 2, 5 and 8) and was also dependent on the S-60 lysate (mitochondrial lysate omitted, lanes 3, 6 and 9). Thus, an in vitro labeling reaction is catalyzed by this fraction, but it is apparently not specific for RNA substrate, at least among these RNAs tested.
Figure 6.
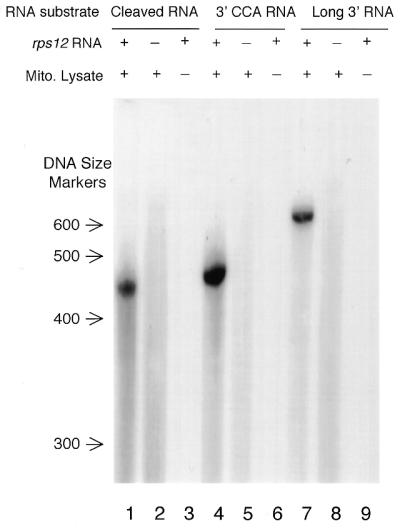
Nucleotide addition assay with different RNA substrates. Three RNA substrates were used that differed at the 3′-termini as described in the text. rps12 RNAs were incubated with [α-32P]ATP and the S-60 mitochondrial lysate (lanes 1, 4 and 7). In control reactions the substrate RNA (lane 2, 5 and 8) or the S-60 mitochondrial lysate (lane 3, 6 and 9) was omitted. Substitution of radiolabeled CTP for ATP gave similar results (data not shown).
The nucleotide specificity of the labeling reaction was examined by incubation of the cleaved rps12 RNA (3′-terminus at position 3, Fig. 3). Both CTP and ATP were readily incorporated into the exogenous RNA (Fig. 7, lanes 1 and 3), but GTP and UTP were much less efficiently incorporated (lanes 5 and 7). Thus, the nucleotide specificity of the in vitro reaction reflected the non-genomically encoded nucleotides that were detected by DNA sequence analysis in Figure 3. In addition, it is noteworthy that the gels are not ‘smeary’, as would be expected for a polyadenylation type of modification; in contrast the labeled RNAs ran essentially as a single band within the limits of resolution of these slab gels. Also, other RNAs present in the S-60 fraction were radiolabeled by CTP or ATP and these were mostly in the <100 nt size range and probably reflect labeling of endogenous tRNAs and possibly other RNAs. Thus, the 3′ modification detected in rps12 mRNAs frequently resulted in the termini of the transcript bearing a CCA (Fig. 3) and the nucleotide specificity indicated that C and A residues were most efficiently incorporated into the exogenous rps12 RNAs.
Figure 7.
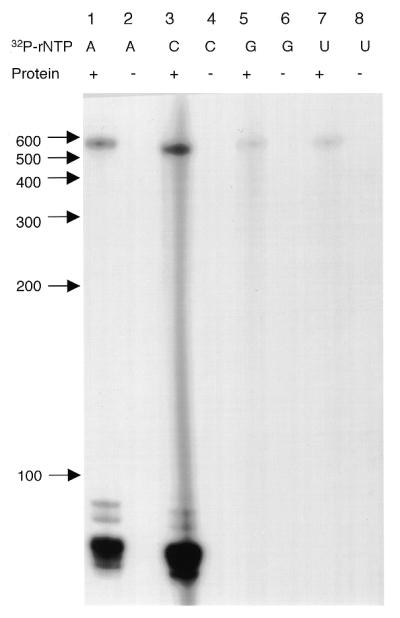
The nucleotide specificity of the labeling reaction for cleaved rps12 RNA. The cleaved rps12 RNA (position 3, Fig. 3A) was incubated with all four rNTPs and one [α-32P]rNTP as indicated. Lanes 1, 3, 5 and 7 included both radiolabeled nucleotide and mitochondrial lysate; lanes 2, 4, 6 and 8 were control reactions with the omission of mitochondrial lysate.
DISCUSSION
3′ Addition of non-genomically encoded nucleotides to plant mitochondrial mRNAs
The addition of non-genomically encoded nucleotides to the 3′-termini of maize mitochondrial mRNAs is a novel observation. The extent of modification differed substantially between transcripts for the three genes surveyed in this paper. rps12 exhibited the highest degree of modification, with 95% of the cDNAs possessing between one and four non-genomically encoded residues. In addition, 83% of the cDNAs analyzed terminated with the trinucleotide CCA, identical to the 3′ processing reaction that occurs in the maturation of tRNA. Nuclease S1 protection experiments confirmed the presence of low abundance transcripts from this region, which is 8–19 nt downstream of the stop codon. Additional transcript termini existed in this region from other low abundance transcripts; however, a much higher abundance transcript that probably represents the mature form of the rps12 mRNA extended downstream. Thus, the low abundance transcripts that exhibit a very high degree of 3′ modification apparently represent RNAs involved in the degradation process.
cox2 cDNAs exhibited a high degree of modification over a 4 nt region. The modifications were characterized by the addition of 1–5 nt that included mostly A and C residues and an occasional G or T residue. cox2 mRNAs from maize mitochondria have also been reported to be polyadenylated in this region of the 3′-UTR (17). These RNAs were characterized as low abundance RNAs that probably represented intermediates in RNA turnover (17).
atp9 cDNAs also exhibited non-genomically encoded nucleotides, but to a much lower extent, and the number of non-genomically encoded residues was limited to a single A or C in a relatively small fraction of the atp9 cDNAs. The 3′ modification of atp9 mRNAs was at one of two adjacent nucleotides of the mature mRNA.
An important consideration in the interpretation of these data is the specificity and reactivity of T4 RNA ligase with various 3′-termini. Both sequence and secondary structure could influence the ligation efficiency and thereby bias the cloning of cDNAs. T4 RNA ligase is reported to require a single unpaired nucleotide at the 3′-terminus. The use of a complementary oligonucleotide facilitates ligation (23) and it seems that a strictly single-stranded structure is not required. In our results atp9 cDNA clones were obtained that utilized a ligation site which was 2 or 3 nt immediately downstream of an extensive stem–loop structure (Fig. 8B) and these results are consistent with the specificity of T4 RNA ligase.
Figure 8.
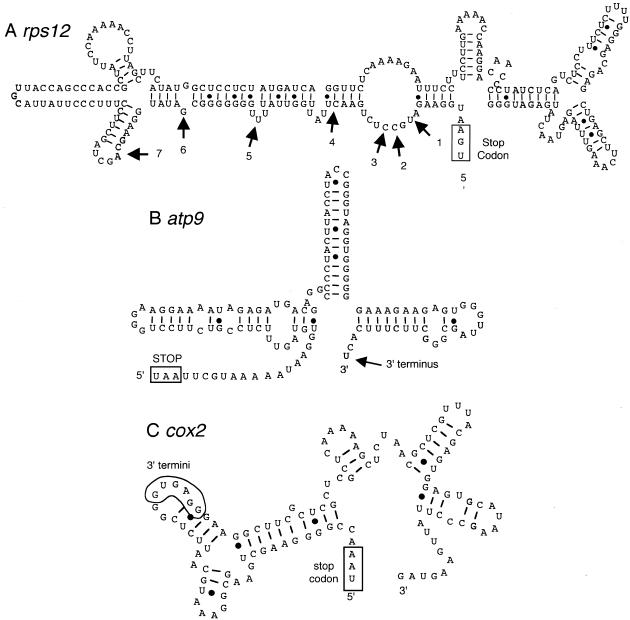
Predicted secondary structure of the 3′-regions of rps12, atp9 and cox2 mRNAs. Secondary structures were predicted by the Mfold program accessed on Prof. M. Zukers’ web site at Washington University (http://mfold2.wustl.edu/∼mfold/rna/form1.cgi ). The stop codons are indicated by a box and are labeled. The positions of transcript 3′-termini are indicated with arrows. (A) Secondary structure prediction of the 3′-UTR of rps12. The positions of low abundance transcript termini are shown by numbered arrows. Transcripts with highly modified termini were detected at positions 1–4. (B) Secondary structure prediction of the 3′-UTR of atp9. The arrow indicates the position of the 3′-terminus of the mature mRNA. (C) Secondary structure prediction of the 3′-UTR of cox2. The position of modified 3′-termini was within the 6 nt enclosed in the polygon.
The nucleotide specificity at the 3′-end of the acceptor nucleic acid in the ligation reaction has been systematically investigated (24,25). The minimum chain length for the 5′ substrate is 3 nt and longer chain length does not substantially affect activity. Any of the four dNTPs in the three 3′-terminal positions will support ligation activity, however, the ligation efficiency is dependent on the nucleotides in this region. The order of reactivity is dependent on position, but is roughly in the order A > C = G > U (24,25). Examination of the ligation sites reported in Figures 3 and 4 are consistent with the expected order of ligation efficiency. Fifty-four percent of the 3′ nucleotides were A residues, while 20% of 3′ residues were U. Thus, although we did recover cDNAs with all nucleotides utilized at the ligation site, the specificity of the enzyme probably had an influence on the reactivity of different RNAs. This sort of selectivity may explain why some transcript termini were better represented in the cDNA clones than others. For example, rps12 cDNAs with 3′ positions at 1, 2, 3 or 4 were readily recovered, but no cDNAs with 3′-termini in the U-rich region around position 5 were obtained (Fig. 8A).
No polyadenylated cDNAs were detected in these analyses, although polyadenylation of plant mitochondrial mRNAs have been reported in sunflower atpA-orf522 and maize cox2 (15,16). In these studies polyadenylation was detected by reverse transcription with an oligo(dT)-adapter primer and amplified with a gene-specific primer and an anti-adapter oligonucleotide; thus, this approach would selectively amplify polyadenylated mRNAs. The ligation approach used in this report may select and amplify cDNAs based on the reactivity of T4 RNA ligase, although polyadenylated transcripts would appear to be excellent substrates for the ligation reaction. Further characterization of the relative abundance of the CA-modified and polyadenylated mRNAs would be useful to understand how these processes are involved in gene expression and RNA metabolism.
Mechanism of 3′ modification
The prevalence of 3′ modifications of rps12 transcripts that exhibited the 3′ trinucleotide CCA is strikingly similar to the post-transcriptional maturation of tRNAs. The in vitro modification assays suggested that the system was not highly specific for the rps12 substrate RNA within the range of substrates that were tested. However, the modification assay clearly utilized CTP and ATP much better than GTP or UTP, a result that is consistent with the prevalence of non-genomically encoded C and A residues detected by DNA sequence analyses. Taken together, these results suggest that the 3′ modification of rps12 mRNAs might be directed by a nucleotidyltransferase similar to the CCA:tRNA nucleotidyl transferase. In addition, the rps12 substrate RNAs labeled by the in vitro assay electrophoresed as a discrete single band; in contrast, polyadenylation reactions produce heterogeneous reaction products that run as smears on gels (26–28). Thus, both the 3′ modifications detected and the in vitro reactions appear to reflect the activity of a tRNA nucleotidyltransferase-like activity.
Both the CCA-adding enzyme [ATP (CTP):tRNA nucleotide transferase] and poly(A) polymerase are members of the nucleotidyltransferase (Ntr) superfamily that includes many enzymes that catalyze nucleotide transfer reactions to a variety of substrates (29–31). tRNA nucleotidyltransferase and eubacterial poly(A) polymerase are both class II nucleotidyltransferases and show substantial similarities at the level of characteristic amino acid sequence motifs (29,31).
Secondary structure and modification
The 3′-termini of plant mitochondrial mRNAs typically possess an inverted repeat that can create a relatively stable stem–loop (12) and the secondary structure correlates with transcript stability (13,14). Secondary structure predictions for the 3′-termini of rps12, atp9 and cox2 RNAs are shown in Figure 8. The positions of the 3′-termini of rps12 RNAs are indicated in Figure 8A by arrows. Positions 1–4 were highly modified by addition of between one and four C or A residues and these sequences failed to fold into secondary structures with substantial stability (ΔG values were approximately –10 kCal/mol); however, the 3′-UTR does fold into a reasonable stable secondary structure with approximately –70 kCal/mol for the 250 nt region. In addition, the relatively high abundance of RNA with a long 3′-UTR and relatively low abundance of the transcripts indicated at positions 1–7 suggests that these RNAs may be in the process of degradation.
The 3′-terminus of atp9 transcripts (Fig. 8B) has three short and nearly perfect inverted repeats that are predicted to form a very stable and compact multiple stem–loop structure of approximately –44 kCal/mol for the 125 nt sequence. The 3′-terminus of atp9 mRNA was represented as the 3′-terminal or penultimate nucleotide shown in Figure 8B and a single non-genomically encoded C or A residue was detected at these two positions in a fraction of the cDNAs. These nucleotides corresponded to the most abundant 3′-terminus by nuclease S1 protection assays; thus, this structure apparently represents the mature form of the mRNA.
The predicted secondary structure of the cox2 mRNA is shown in Figure 8C. There are several relatively complex inverted repeat sequences which are predicted to fold into a structure with a ΔG of approximately –33 kCal/mol for the 118 nt region. 3′ Modifications were detected at the 6 nt enclosed by the polygon. Transcripts with termini in this region represent truncated forms of cox2 mRNAs (17) and one of the two most abundant polyadenylation sites was detected in this region.
Polyadenylation of bacterial and plastid transcripts has been well documented to play a role in RNA turnover. Plant mitochondria also have polyadenylated mRNAs that show some similarities and some differences to the bacterial and plastid systems. The stability of polyadenylated and non-polyadenylated RNAs has been compared by in vitro degradation assays (15,16). Enhanced degradation of polyadenylated substrates was detected (16) and polyadenylation may be involved in RNA turnover metabolism. Since the same sites are reported to be polyadenylated and 3′ modified in maize cox2 transcripts, it is possible that both polyadenylation and short CA modification are involved in RNA metabolism. Since these cox2 transcripts either exist as polyadenylated or as short CA-modified RNAs, it seems possible that these modifications could regulate RNA metabolism, perhaps in an antagonistic manner. Additional studies on the nature and extent of polyadenylation and 3′ modification will be necessary to establish the roles of these processes in mitochondrial gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
The maize seed was a gift from Pioneer Hi-bred Seed Co. (Johnston, IA). The authors would like to thank Drs Daniel P. Morse and Brenda L. Bass of the University of Utah for helpful suggestions in the development of the RNA ligation assay. This research was supported by USDA grant 98-35301-6043.
REFERENCES
- 1.Coburn G.A. and Mackie,G.A. (1999) Prog. Nucleic Acid Res. Mol. Biol., 62, 55–108. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar N. (1996) Microbiology, 142, 3125–3133. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar N. (1997) Annu. Rev. Biochem., 66, 173–197. [DOI] [PubMed] [Google Scholar]
- 4.Carpousis A.J., Vanzo,N.F. and Raynal,L.C. (1999) Trends Genet., 15, 24–28. [DOI] [PubMed] [Google Scholar]
- 5.Mackie G.A. (1998) Nature, 395, 720–723. [DOI] [PubMed] [Google Scholar]
- 6.Vanzo N.F., Li,Y.S., Py,B., Blum,E., Higgins,C.F., Raynal,L.C., Krisch,H.M. and Carpousis,A.J. (1998) Genes Dev., 12, 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Py B., Higgins,C.F., Krisch,H.M. and Carpousis,A.J. (1996) Nature, 381, 169–172. [DOI] [PubMed] [Google Scholar]
- 8.Cao G.J., Kalapos,M.P. and Sarkar,N. (1997) Biochimie, 79, 211–220. [DOI] [PubMed] [Google Scholar]
- 9.Ojala D., Merkel,C., Gelfand,R. and Attardi,G. (1980) Cell, 22, 393–403. [DOI] [PubMed] [Google Scholar]
- 10.Koslowsky D.J. and Yahampath,G. (1997) Mol. Biochem. Parasitol., 90, 81–94. [DOI] [PubMed] [Google Scholar]
- 11.Hendler F.J., Padmanaban,G., Patzer,J., Ryan,R. and Rabinowitz,M. (1975) Nature, 258, 357–359. [DOI] [PubMed] [Google Scholar]
- 12.Schuster W., Hiesel,R., Isaac,P.G., Leaver,C.J. and Brennicke,A. (1986) Nucleic Acids Res., 14, 5943–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaleikau E.K., Andre,C.P. and Walbot,V. (1990) Nucleic Acids Res., 18, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellaoui M., Pelletier,G. and Budar,F. (1997) EMBO J., 16, 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupold D.S., Caoile,A. and Stern,D.B. (1999) J. Biol. Chem., 274, 3897–3903. [DOI] [PubMed] [Google Scholar]
- 16.Gagliardi D. and Leaver,C.J. (1999) EMBO J., 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupold D.S., Caoile,A. and Stern,D.B. (1999) Plant Cell, 11, 1565–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L. and Ruffner,D.E. (1998) Nucleic Acids Res., 26, 2502–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali Ansari-Lari M., Jones,S.N., Timms,K.M. and Gibbs,R.A. (1996) Biotechniques, 21, 34–36, 38. [DOI] [PubMed] [Google Scholar]
- 20.Mulligan R.M., Leon,P. and Walbot,V. (1991) Mol. Cell. Biol., 11, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phreaner C.G., Williams,M.A. and Mulligan,R.M. (1996) Plant Cell, 8, 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloney A.P., Traynor,P.L., Levings,C.S.,III and Walbot,V. (1989) Curr. Genet., 15, 207–212. [DOI] [PubMed] [Google Scholar]
- 23.Morse D.P. and Bass,B.L. (1997) Biochemistry, 36, 8429–8434. [DOI] [PubMed] [Google Scholar]
- 24.England T.E. and Uhlenbeck,O.C. (1978) Biochemistry, 17, 2069–2076. [DOI] [PubMed] [Google Scholar]
- 25.Romaniuk E., McLaughlin,L.W., Neilson,T. and Romaniuk,P.J. (1982) Eur. J. Biochem., 125, 639–643. [DOI] [PubMed] [Google Scholar]
- 26.Lisitsky I., Klaff,P. and Schuster,G. (1996) Proc. Natl Acad. Sci. USA, 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisitsky I., Kotler,A. and Schuster,G. (1997) J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- 28.Kudla J., Hayes,R. and Gruissem,W. (1996) EMBO J., 15, 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 29.Yue D., Maizels,N. and Weiner,A.M. (1996) RNA, 2, 895–908. [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L. and Sander,C. (1995) Trends Biochem. Sci., 20, 345–347. [DOI] [PubMed] [Google Scholar]
- 31.Raynal L.C., Krisch,H.M. and Carpousis,A.J. (1998) J. Bacteriol., 180, 6276–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]