Abstract
Nucleotide excision repair (NER) removes UV-induced photoproducts and numerous other DNA lesions in a highly conserved ‘cut-and-paste’ reaction that involves approximately 25 core components. In addition, several other proteins have been identified which are dispensable for NER in vitro but have an undefined role in vivo and may act at the interface of NER and other cellular processes. An intriguing example is the Saccharomyces cerevisiae Mms19 protein that has an unknown dual function in NER and RNA polymerase II transcription. Here we report the cloning and characterization of a human homolog, designated hMMS19, that encodes a 1030 amino acid protein with 26% identity and 51% similarity to S.cerevisiae Mms19p and with a strikingly similar size. The expression profile and nuclear location are consistent with a repair function. Co-immunoprecipitation experiments revealed that hMMS19 directly interacts with the XPB and XPD subunits of NER-transcription factor TFIIH. These findings extend the conservation of the NER apparatus and the link between NER and basal transcription and suggest that hMMS19 exerts its function in repair and transcription by interacting with the XPB and XPD helicases.
INTRODUCTION
To prevent the detrimental effects of gene damage and maintain the integrity of genetic information a number of DNA repair systems have evolved as part of an intricate network of genome care-taking mechanisms. The most versatile and complex repair pathway is nucleotide excision repair (NER), which removes a remarkably wide range of structurally diverse DNA lesions, including UV-induced photoproducts, bulky chemical adducts and intra-strand cross-links (1). During eukaryotic evolution the basic ‘cut-and-paste’ reaction mechanism of NER has been strongly conserved with striking parallels between humans and yeast and, to a lesser degree, even with prokaryotes. Thus, Escherichia coli uses a set of six proteins (UvrA–D, DNA polymerase I and ligase) to eliminate NER lesions (2,3 and references therein) while in eukaryotes 25–30 proteins are implicated (4–6). Genome-wide lesion detection is carried out by the XPC–hHR23B complex (7), probably assisted by the UV-DDB (XPE) dimer for the recognition of UV-induced cyclobutane pyrimidine dimers (CPD) (8). Lesions in the transcribed strand of active genes, that block ongoing transcription, are likely signaled by an elongating RNA polymerase II complex (9). Subsequently, the multi-subunit TFIIH factor, the damage verifier XPA and the single-stranded DNA binding protein trimer RPA are recruited to the site of the lesion (4,10–12). The bi-directional helicase activity of TFIIH involving the XPB and XPD helicases facilitates formation of a locally opened repair intermediate (13–16). A dual single-strand incision is made around the lesion by XPG at the 3′ (17), and the ERCC1–XPF complex at the 5′ site (18) allowing excision of a 24–32mer oligonucleotide containing the damage (19). The last step of gap-filling is accomplished by general replication factors PCNA, RPA, RFC, DNA polymerase δ and/or ɛ and ligase sealing the final nick. The efficiency with which DNA injuries are excised is lesion- and location-dependent. Lesions for which the global genome NER is slow (such as CPDs) are preferentially removed when present in the transcribed strand of active genes (20,21). This points to a direct link between the transcription apparatus and repair. Cells carrying mutations in the CSA and CSB genes suffer from a selective defect in this transcription-coupled NER (TC-NER) subpathway (22–24).
Intriguingly, deficiencies in NER genes are associated with three distinct genetic disorders: xeroderma pigmentosum (XP), Cockayne syndrome (CS) and trichothiodystrophy (TTD) (25,26). XP is mainly characterized by sun (UV)-hypersensitivity and a strong predisposition to skin cancer. Frequently accelerated neurodegeneration occurs. CS and TTD patients are not particularly cancer-prone but present sun sensitivity, pronounced developmental abnormalities and very severe neurological problems due to neurodysmyelination. The additional hallmarks of TTD include brittle hair and nails and ichthyosis. The heterogeneous spectrum of clinical manifestations present in CS and TTD, but absent in XP, is difficult to attribute to deficiencies in NER which in general are more severe in XP. Since specifically the NER genes implicated in TTD encode subunits of the repair-transcription factor TFIIH and thus have a dual involvement in NER and transcription initiation (15,16,18,27), we have proposed that part of the clinical features are the consequence of a viable defect in basal transcription (28,29).
As the basic reaction of NER is highly conserved in eukaryotes, each yeast NER gene has in general at least one structural human counterpart. In recent years functions in the reaction mechanism have been assigned to all core NER factors (4). However, an additional category of UV-sensitive yeast mutants discloses genes that appear to be involved in NER in vivo, but are partly dispensable for the NER reaction in vitro. This class is comprised of RAD7, RAD16, RAD23 (30) and MMS19 (31) and is functionally poorly understood. Except for Rad23p (32) no human homologs are known suggesting that these genes may be less well conserved during eukaryotic evolution. Here we focus on MMS19. The mms19Δ mutant exhibits a unique phenotype for NER mutants: moderate UV-sensitivity due to a defect affecting both NER subpathways, global genome repair and TC-NER (31,33), thermosensitive transcriptional activity as well as methionine auxotrophy. Short treatment of a total cell extract at non-permissive temperature leads to a strong reduction of RNA polymerase II transcription activity. Remarkably, the transcription defect can be corrected by addition of purified TFIIH (31) although Mms19p appears not to be a subunit of this repair-transcription factor. Thus, the protein may not be directly involved in these two processes but influence them via TFIIH as an upstream regulatory factor. To examine whether this intriguing factor is conserved in evolution and to permit studies in higher species we have searched for homologs in mammals. Here we report the identification and partial characterization of a presumed human homolog of Mms19p.
MATERIALS AND METHODS
Isolation of a cDNA encoding a putative human homolog of yeast MMS19
A HeLa poly(A)+ cDNA library constructed in λgt10 with relatively long inserts (32) was screened with a probe described below. Approximately 600 000 recombinant bacteriophage plaques were transferred to Hybond-N membranes in duplicate. The probe used to screen the library was obtained by RT–PCR using random synthetic oligonucleotides for cDNA synthesis from HeLa total RNA. The oligonucleotide primer pairs used for amplification were: p274 5′-CATCCTGTTATTTCCCTATCG-3′ and p276 5′-ACAAACACGCAGTCAGGGAG-3′. The 345 bp RT–PCR product was partially sequenced and confirmed to correspond to the amino acid sequence of EST c18436, purified from a 2% agarose gel, and labeled with [32P]dATP and random hexamers. Hybridization was carried out in a solution of 10× Denhardt’s (1×: 0.02% bovine serum albumin, 0.02% Ficoll, 0.02% polyvinylpyrolidone), 3× SSC (1×: 0.15 M NaCl, 15 mM Na-citrate), 0.1% SDS, 50 µg/ml salmon sperm DNA, 9.0% dextran sulphate overnight at 65°C. Membranes were washed for 20 min in 3× SSC/0.1% SDS, 1× SSC/0.1% SDS and 0.3× SSC/0.1% SDS at 65°C and exposed to autoradiographic films with intensifying screens at –70°C.
Computer analysis
Various sequences in database with homology to the hMMS19 ORF were identified using the TBLASTN algorithm (34). Reconstruction of the most likely ORF deduced from various exons was done with the help of Netgene2, Genescan, Grail, BCM genefinder, EST partial cDNA sequences and by visual inspection of the sequence. Protein alignments were carried out using ClustalX and printed in boxshade. Phylogenetic trees were generated with the aid of the Phylip package.
RNA isolation and northern blot analysis
Total RNA was isolated by the LiCl–urea method described (35). RNA samples were loaded onto a 0.9% agarose–formaldehyde gel, separated by electrophoresis and transferred onto a nitrocellulose membrane. The blot was probed with 32P-labeled random-primed DNA corresponding to the ORF of the cDNA isolated above. Hybridization of the filter was performed at 42°C in 50% formamide, 5× SSPE, 1% SDS, 5× Denhardts solution and 100 µg/ml denatured salmon sperm DNA (36). The blot was washed twice for 30 min in 1× SSC containing 0.1% SDS at 65°C and exposed for autoradiography.
Fluorescent in situ hybridization (FISH)
Human lymphocyte metaphase spreads were processed prior to hybridization as described (37). Briefly, the metaphase spreads were treated with 100 µg/ml RNAse A in 2× SSC during 1 h at 37°C, rinsed and incubated with proteinase K (20 mg/100 ml at 37°C) for 1 h. The chromosomes were fixed with 1% formaldehyde, washed, dehydrated in ethanol and air-dried. The slides were hybridized in a mixture containing 50% formamide, 2× SSC, 40 mM Na-phosphate pH 7.0, 10% dextran sulfate, 0.1 mg/ml sonicated salmon sperm DNA, 0.1 mg/ml yeast tRNA and 2 ng/ml labeled probe. The biotin-labeled complete ORF used as a probe was denatured at 80°C for 5 min and hybridized to the chromosomes overnight at 37°C. After a wash with 50% formamide in 2× SSC at 45°C followed by a second wash with 2× SSC containing 0.05% Tween-20 at room temperature, the slides were incubated with 5 mg/ml avidin D-FITC (Vector). To increase the sensitivity of the D-FITC signals, one round of amplification was performed using biotinylated goat anti-avidin D antibody. To generate clear reverse bands, metaphase chromosomes were either counterstained with propidium iodide in antifade medium or banded with DAPI and actinomycin D.
Baculovirus infection
Baculoviruses allowing the expression of TFIIH subunits were constructed as previously described (38,39). The cDNA encoding the hMMS19 protein was inserted into pFastBac vector (Gibco BRL). The resulting vector was recombined with baculovirus DNA using DH10 Bac E.coli cells (Gibco BRL). Spodoptera frugiperda 9 (Sf9) cells were transfected with the recombinant Bacmid DNA to prepare a viral stock. Sf9 cells were infected with hMMS19 baculovirus in combination with each individual core TFIIH subunit or with either core, CAK or TFIIH complex subunits. Cells were collected 48 h after infection, washed once in 1× PBS 30% glycerol before lysate preparation.
Preparation of HeLa whole cell extracts and Sf9 cell lysates
Whole cell extract (WCE) was prepared from HeLa cells as previously described (40) and dialyzed in buffer A (50 mM Tris–HCl pH 7.8, 10% glycerol, 0.1 mM EDTA, 0.5 mM DTT) containing 50 mM KCl. Sf9 cell extracts were prepared by homogenization in buffer B (20 mM Tris–HCl pH 7.8, 150 mM NaCl, 20% glycerol, 0.1% Nonidet P-40, 5 mM 2-mercaptoethanol, 0.5 mM PMSF, 1× protease inhibitor cocktail) and centrifugation at 14 000 r.p.m. for 30 min at 4°C (39).
Immunoprecipitations of recombinant and endogenous proteins
Recombinant proteins from baculovirus-infected Sf9 cell lysates were immunoprecipitated for 4 h at 4°C in buffer B with either Ab-hMMS19 (2G2, 3H10, 3H1, 3B7), Ab-XPB (1B3), Ab-XPD (2F6), Ab-p62 (3c9), Ab-p52 (1D11), Ab-p44 (1H5), Ab-p34 (2B1), Ab-cdk7 (2F8), Ab-cyclin H (2DA) or a control antibody (Ab-GST: 1D10) crosslinked to protein G–Sepharose beads. Endogenous proteins were immunoprecipitated from WCE for 4 h at 4°C in buffer A containing 0.01% Nonidet P-40 using protein G–Sepharose linked antibodies raised either against XPD or hMMS19. Beads were extensively washed with buffer A containing 150 or 250 mM KCl in the presence of 0.1% Nonidet P-40. The immunoprecipitated polypeptides were resolved on SDS–PAGE and probed with antibodies raised against TFIIH subunits or hMMS19.
RESULTS
Identification, cloning and sequence analysis of a human homolog of S.cerevisiae MMS19
The S.cerevisiae Mms19 protein sequence was used to systematically screen public databases using the TBLASTN algorithm (34). A computer translated partial cDNA clone from human placenta (GenBank accession number c18436) attracted our attention because of potentially significant homology with a portion of Mms19p at amino acid positions 216–377. To see whether the homology extended, an RT–PCR probe was used to screen a HeLa poly(A)+ cDNA library. The nucleotide sequence of the longest cDNA clone isolated (3.4 kb) showed that it was derived from the expected mRNA (see Materials and Methods for details).
The sequence predicts a protein of 1030 amino acids (Fig. 1A) with a calculated molecular weight of 113 kDa consistent with the observed mobility on SDS–PAGE of the in vitro translated protein and with the normal gene product in WCE of HeLa cells detected using mono-specific antibodies (data not shown).
Figure 1.
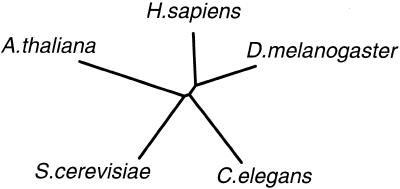
(A, opposite) Comparison of amino acid sequences of the human hMMS19 with its yeast counterpart and homologous translated ESTs of different species. Highly conserved amino acids are boxed in black. Related residues are shaded in gray. The putative nuclear localization signal is underlined. Accession numbers: S.cerevisiae (U70559), C.elegans [tentative exon identification from cosmid C24G6 (AF067936) also using various partial cDNA sequences], D.melanogaster [ORF tentatively identified from genomic sequence AC007532, with exon sequence determined using various programmes and using partial cDNAs: LP10101 (AI296246), LD30346 (AA950519)] and A.thaliana [g4220638 (AB023039) genomic sequence, ORF tentatively deduced from intron/exon border identification]. In addition to the above genes the Schizosaccharomyces pombe homolog has also been identified in the genome sequencing projects. (B, above) Phylogenetic tree constructed with the aid of the Phylip package using the alignment in (A). The distance matrix (Dayhoff PAM 001 matrix) is calculated for the 164 amino acids in the first part of the ORF using the PROTDIST software. An unrooted tree is determined from this with the help of NEIGHBOR.
The hMMS19 polypeptide has a very alanine-rich N-terminus and is particularly rich in hydrophobic amino acids (17% leucines) with highly hydrophobic regions in the N- and C-terminal parts, containing 18 trimers of hydrophobic residues frequently preceded and/or followed by proline, glutamic acid or histidine. Using a 3D structure prediction program amphiphatic helical wheel-like motifs with one side mainly polar and the opposite side hydrophobic residues were found mostly concentrated in the 360 C-terminal amino acids. Also, a putative nuclear localization signal, KKRXXRK, is present in the C-terminus. No other known motifs were found except for several potential casein kinase II and protein kinase C phosphorylation sites.
Comparison with yeast Mms19p revealed a striking correspondence in size (1030 versus 1032 amino acids for the human and yeast products, respectively) and highly significant overall homology: 26% identity and 51% similarity (when aligned with each other). The N- and C-termini are better conserved than the central part (Fig. 1A). Leucine-rich motifs in Mms19p as defined by Lauder et al. (31) appear poorly conserved. Screening of the complete human ORF against the entire S.cerevisiae protein database using the BLAST algorithm (34) identified Mms19p as the by far most homologous protein (p = 2 × 10–41) indicating that within the yeast genome Mms19p is the only counterpart. On the basis of the above and the findings and arguments below we designate the gene as ‘hMMS19’, although until completion of the human genome sequence it is not excluded that additional human homologs exist defining a gene family. Using the human hMMS19 sequence we identified DNA fragments of various other species allowing a tentative reconstruction of the gene from separate exons for Drosophila melanogaster, Caenorhabditis elegans and Arabidopsis thaliana compiled in Figure 1A. This enabled the definition of several highly conserved sequence motifs in the N- and C-termini such as DGEKDPRNL and EDLFEVX2CYFPIXF (in addition to the putative NLS). However, screening of various databases failed to identify any significant similarity with other proteins that would hint to a function suggesting that these motifs are MMS19-specific. The alignment of the various homologous proteins enabled us to deduce a phylogenetic tree (Fig. 1B). The degree of similarity between the different proteins matches well with the overall evolutionary relatedness of the species involved. These findings led us to conclude that in all species there is (at least) one clear homolog of Mms19p.
Expression pattern and subcellular localization of hMMS19
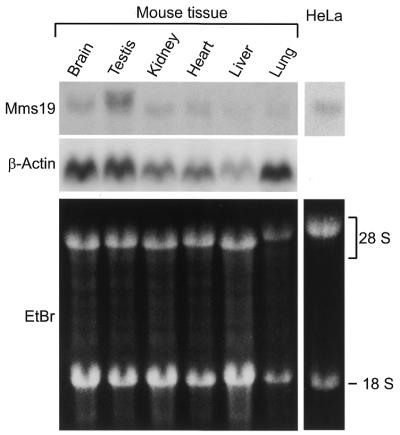
Northern blot analysis revealed a single transcript of 4 kb in various mouse tissues and in human cells (Fig. 2). With the exception of testis the gene appears to be expressed at a rather low level in all organs and tissues examined. ESTs from various databases confirmed that the gene is widely expressed with hMMS19 mRNA present in skin, placenta, cerebellum, mammary gland, parathyroid gland, myotubes, uterus and at all stages of development (blastocyst, embryo, fetus as well as adult tissues). To detect the protein in cells and on blots and to enable immunoprecipitation experiments we generated monoclonal antibodies. These tools were used to investigate the subcellular localization of hMMS19 protein after transfection of Cos-1 cells with a pcDNA3–hMMS19 construct. An anti-hMMS19 monoclonal antibody raised against an N-terminal peptide revealed a clear nuclear localization confirmed by a second antibody raised against the C-terminus (data not shown).
Figure 2.
Northern blot analysis of hMMS19. Hybridization was achieved with a specific hMMS19 or β-actin (as an internal control) cDNA probe, as described in Materials and Methods.
Chromosomal localization
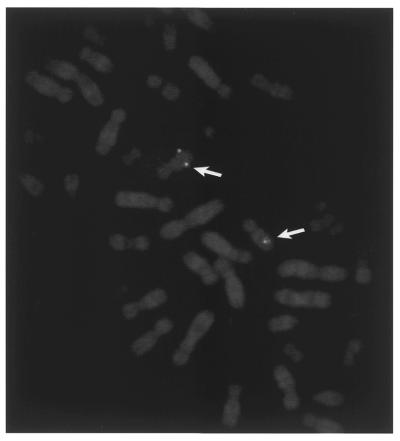
To further characterize hMMS19 and its potential involvement in human disorders, chromosomal localization was carried out. The cDNA encompassing the complete ORF was biotinylated and used as a probe for in situ hybridization to human metaphase spreads. A single site of hybridization on the human karyotype was found located on the long arm of chromosome 10 at the junction of q24 and q25 (Fig. 3).
Figure 3.
Chromosomal localization of hMMS19 by fluorescent in situ hybridization. The biotinylated hMMS19 cDNA was hybridized with a spread of human metaphase chromosomes that were thereafter counterstained with propidium iodide. The arrows indicate the fluorescent spots present in the region 10q24–10q25 of the R-banding pattern.
Interspecies complementation
In order to know whether the human protein is able to complement the yeast defect, an mms19Δ strain (MGSC217) in which almost the complete ORF of MMS19 is replaced by the URA3 gene, was transformed with the hMMS19 cDNA subcloned in the yeast expression vector, pYET2 (33). UV-sensitivity, thermosensitivity as well as methionine auxotrophy of this transformed strain were examined. We failed to observe any significant rescue of the cells for any of the defects, although the human protein could be detected as a weak band in the transformed yeast extract by immunoblot analysis using the monoclonal antibodies of above (data not shown). These findings suggest that the human protein has diverged too far from its yeast counterpart to be able to complement the various mms19Δ deficiencies.
hMMS19 associates with TFIIH
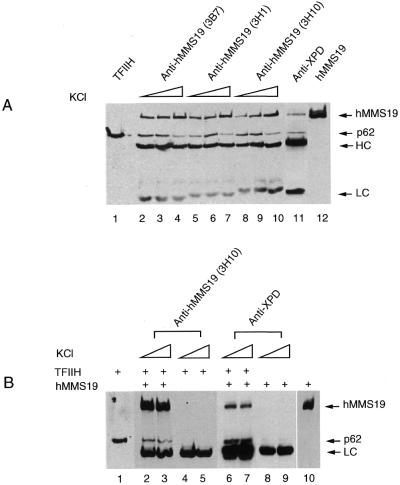
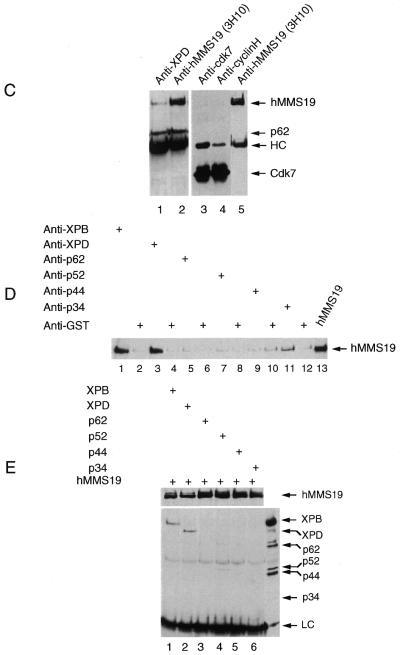
Several observations (31,33) suggest that Mms19p affects NER and transcription by influencing the activity or stability of TFIIH. How Mms19 exerts this effect is currently unknown. To find out whether hMMS19 affects TFIIH via direct or indirect interactions the different polypeptides present in a fraction of TFIIH purified from HeLa cells were resolved by SDS–PAGE and probed with antibodies against hMMS19. The p62 antibody indicated that TFIIH subunits could be easily visualized. We failed to observe detectable quantities of hMMS19 indicating that it is probably not a stable component of TFIIH (data not shown). To investigate the possibility of a weak interaction we performed immunoprecipitation from HeLa WCE using three different monoclonal antibodies raised against hMMS19. Interestingly, in addition to hMMS19 itself the p62 subunit of TFIIH was found to precipitate (Fig. 4A, lanes 2–10). Conversely, monoclonal antibodies against another component of TFIIH, XPD, not only precipitated the p62 subunit of TFIIH but also significant quantities of hMMS19.
Figure 4.
hMMS19 interacts with TFIIH via the XPB and XPD helicase subunits. (A) HeLa WCE was immunoprecipitated with either various monoclonal antibodies raised against hMMS19 (lanes 2–10) or antibodies against XPD (lane 11) linked to protein G–Sepharose beads. Beads were washed with buffers containing 50–250 mM KCl (lanes 2–4, 5–7 and 8–10) or 150 mM KCl (lane 11) in the presence of 0.1% NP-40. After immunoprecipitation, proteins remaining attached to the beads were revealed by immunoblotting using appropriate antibodies as indicated on the side of each panel. (B) Lysates of Sf9 cells infected with either baculoviruses expressing each of the nine TFIIH subunits (lanes 4 and 5) or these in combination with hMMS19 baculovirus (lanes 2, 3, 6 and 7) or hMMS19 alone (lanes 8 and 9) were immunoprecipitated with protein G–Sepharose beads containing antibodies against XPD or hMMS19. The beads were washed with buffers containing 150–250 mM KCl and 0.1% NP-40. (C) Sf9 cells infected with hMMS19 baculovirus in combination with baculoviruses expressing the CAK subunits (lanes 3–5) or the TFIIH core subunits (lanes 1 and 2) were lysed and immunoprecipitated with antibodies against XPD (lane 1), cdk7 (lane 3) or cyclin H (lane 4) or hMMS19 (lanes 2 and 5) bound to protein G–Sepharose beads. The beads were subsequently washed with a buffer containing 150 mM KCl as well as 0.1% NP-40. (D and E) Lysates of co-infected Sf9 cells with hMMS19 baculovirus in addition to baculoviruses for either XPB (lanes 1 and 2) or XPD (lanes 3 and 4) or p62 (lanes 5 and 6) or p52 (lanes 7 and 8) or p44 (lanes 9 and 10) or p34 (lanes 11 and 12) were immunoprecipitated with either antibodies against hMMS19 or anti-GST (as a control) as indicated in (D) or antibodies against each of the TFIIH core subunits as indicated in (E). The washing of the beads was done with a 150 mM KCl buffer containing 0.1% NP-40. After immunoprecipitation, proteins remaining attached to the beads were revealed by immunoblotting using appropriate antibodies as indicated on the side of each panel.
To examine whether hMMS19 interacts with TFIIH directly or via a partner protein we infected Sf9 insect cells with recombinant baculoviruses containing the cDNAs of all nine TFIIH subunits as well as hMMS19 to produce recombinant TFIIH and hMMS19 proteins as described by Tirode et al. (38). Immunoprecipitation of lysates of the infected Sf9 cells revealed that hMMS19 coprecipitated specifically with TFIIH (Fig. 4B: compare lanes 6 and 7 with lanes 8 and 9) and vice versa (lanes 2–5). This interaction is highly significant as it was resistant to stringent washing conditions (250 mM salt buffer containing 0.1% Nonidet P-40). These observations strongly suggest that hMMS19 interacts physically with TFIIH.
To determine whether the physical interaction between hMMS19 and TFIIH has functional implications the reaction mixture of an in vitro reconstituted transcription assay was supplemented with the recombinant hMMS19–TFIIH complex. We observed a similar transcriptional activity in the presence and in the absence of hMMS19 in the transcription assay (data not shown). Therefore, in spite of its physical interaction with TFIIH, hMMS19 neither up- nor downregulates the in vitro transcriptional activity of TFIIH. Perhaps other factors are required in addition to hMMS19, or the effect of the protein is not visible in the in vitro system, that reflects only part of the in vivo complexity.
hMMS19 interacts via the XPB and XPD subunits with TFIIH
TFIIH is comprised of two subcomplexes: core TFIIH–XPD containing XPB, p62, p52, p44, p34 and less tightly associated XPD, and the CAK complex consisting of cdk7, cyclin H and Mat1. To determine which subcomplex interacts with hMMS19, immunoprecipitations were performed with extracts of Sf9 cells simultaneously expressing either hMMS19 together with the six core subunits of TFIIH or hMMS19 together with the three CAK subunits. Figure 4C shows that hMMS19 interacts with core TFIIH–XPD but not with the CAK complex (Fig 4C, compare lanes 1 and 2 with lanes 3–5).
To establish which core subunit was responsible for the interaction, Sf9 cells were infected with hMMS19 baculovirus in combination with each individual core TFIIH subunit. Immunoprecipitation using antibodies specific for the corresponding TFIIH component demonstrates that hMMS19 specifically coprecipitated with XPB and XPD and to a lesser extent with the p34 subunit (Fig. 4D, lanes 1, 3 and 11). hMMS19 did not coprecipitate with any of the three other core subunits nor with GST alone (Fig. 4D, lanes 5, 7 and 9). Conversely, anti hMMS19 antibodies coprecipitated XPB and XPD as well as trace amounts of p52 (Fig. 4E, lanes 1 and 2). Taken together these data indicate that, in contrast to what is reported in yeast (31), hMMS19 interacts with TFIIH as detected in WCE using natural expression conditions. Interaction specifically involves the XPB and XPD helicases establishing a direct link between hMMS19 and these NER/transcription factors. In agreement with the absence of an effect on the in vitro transcription function of TFIIH we failed to find a significant effect of the addition of hMMS19 on the in vitro helicase activity of both XPD and XPB (data not shown).
DISCUSSION
Phenotypic characterization of UV-sensitive S.cerevisiae mutants revealed the existence of a large number of genes involved in NER, collectively assigned to the RAD3 epistasis group. Similarly, studies on three human genetic disorders, XP, CS, TTD and a series of UV-sensitive Chinese hamster mutants uncovered a large set of mammalian NER factors. Analysis of the genes highlighted the structural and functional conservation of the core of the NER machinery from yeast to mammals. Here we report the isolation of a unique human cDNA, the deduced protein sequence of which shows a clear overall co-linear homology and similarity in size with Mms19p. MMS19 is the only gene in the entire yeast genome that exhibits such a highly significant homology with the human cDNA we have isolated.
The ubiquitous expression, the elevated mRNA level in testis as well as the nuclear localization of the protein and the interaction with NER transcription factor TFIIH are entirely consistent with a function in DNA repair and transcription. Taken together these findings strongly support the idea that the isolated cDNA reflects the human homolog of yeast MMS19.
hMMS19 was localized to chromosome 10q24–25, a region with a relatively high density of genes but where no genetic disorders with significance in relation to hMMS19 have been assigned.
Despite extensive efforts we failed to find significant complementation of the yeast mutant by the human gene although expression of the human protein could be detected. These negative findings are consistent with the notion that only few human NER genes have been reported to correct (part of) the corresponding yeast mutant phenotype (41,42).
The function of Mms19 in transcription and NER has been enigmatic in view of the very pleiotropic phenotype of the mutant. This includes temperature sensitivity and methionine auxotrophy, and a partial defect in NER reflected by an intermediate UV sensitivity. Remarkably, the compromised repair function affects both NER subpathways global genome repair as well as transcription-coupled repair (33). On the other hand, the connections with TFIIH are multiple. Extracts of mms19Δ mutants exhibit a temperature-sensitive transcription defect that can be corrected by addition of purified TFIIH, but not by purified Mms19p (31). The status of TFIIH in the treated extract was not determined. The NER defect present in cell-free extracts prepared from an mms19Δ mutant can be complemented in vitro by mixing with extracts of other non-TFIIH NER mutants (e.g. by rad4), but not by a TFIIH NER mutant (e.g. rad3) (33). However, a direct interaction between Mms19 and TFIIH was not demonstrated (31). Importantly, here we identify a physical association between hMMS19 and the two helicase subunits of TFIIH, XPB and XPD. These two proteins have a direct engagement in NER as well as in basal transcription initiation. The binding of Mms19 apparently has important biological consequences as in its absence the complex becomes functionally unstable and cannot optimally exert its role in NER. Similar to the p44 subunit of TFIIH (43,44), hMMS19 interacts with both helicases. In addition to the fundamental role of p44 in maintaining the architecture of TFIIH holo-complex by anchoring the CAK–XPD subcomplex to the five subunit TFIIH core, p44 acts as a regulator of the XPD helicase activity (44,45). Since no separate subcomplex of TFIIH containing both helicases has been reported in vivo so far, the binding of hMMS19 to both helicases probably occurs within the context of the holo-complex, as p44, through two distinct domains. The effect of the binding of hMMS19 to both TFIIH helicases is different to that of p44 since we failed to find a direct influence of hMMS19 on the helicase as well as the transcriptional activity of TFIIH in vitro. hMMS19 might therefore participate in the stabilisation of TFIIH upstream of the transcription process or may exert its effect only in the more complex in vivo context.
In favor of a possible in vivo function of hMMS19 in stabilization of TFIIH it is worth noting that we have recently obtained evidence that in all cases of TTD the overall levels of TFIIH are substantially reduced (45). Thus, instability of TFIIH occurs in vivo adding a clinical dimension to the potential hMMS19 function towards the severe symptoms of this pleiotropic human NER/transcription disorder which is associated with TFIIH defects (26). The above intricate link with TFIIH prompted us to investigate whether this protein is implicated in TTD. One genetic form of this syndrome is not due to mutations in the XPB or XPD genes (26) nor to alterations in any of the other TFIIH core subunits (45). This is complementation group TTD-A, which was originally represented by a single case (46). We searched for abnormalities in hMMS19 but no obvious ORF mutations could be detected in fibroblasts of this patient, nor did we find correction of the TTD-A NER defect upon microinjection of hMMS19 cDNA. Nevertheless, the existence of a class of genes that, like hMMS19, directly interacts with TFIIH and affects its functioning in repair and transcription provides a plausible source for mutations causing TTD-like syndromes, a number of which have been described in the literature (26 and references therein).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professor D. Bootsma for continuous support and stimulating interest, T. Verkerk for assistance with database screenings, Mrs Cathy Braun for technical help and H. Bluyssen for very fruitful discussions. G.S.W. was supported by the Dutch Science Foundation NWO, division Medical Sciences (NWO grant 901-501-151). J.H.J.H. is recipient of the Spinoza Award of NWO. We also acknowledge support by the Dutch Cancer Society (EUR project 1774 and 2004), the EC (QLRT-1999-02002), the Louis Jeantet Foundation, the INSERM, the CNRS and the Université Louis Pasteur of Strasbourg.
REFERENCES
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis, American Society for Microbiology Press, Washington,DC.
- 2.Hoeijmakers J.H.J. (1993). Trends Genet., 9, 173–177. [DOI] [PubMed] [Google Scholar]
- 3.Verhoeven E.E., van Kesteren,M., Moolenaar,G.F., Visse,R. and Goosen,N. (2000) J. Biol. Chem., 275, 5120–5123. [DOI] [PubMed] [Google Scholar]
- 4.de Laat W.L., Jaspers,N.G.J. and Hoeijmakers,J.H.J. (1999) Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 5.Sancar A. (1996) Annu. Rev. Biochem., 65, 43–82. [DOI] [PubMed] [Google Scholar]
- 6.Wood R.D. (1997) J. Biol. Chem., 272, 23465–23468. [DOI] [PubMed] [Google Scholar]
- 7.Sugasawa K., Ng,J.M.Y., Masutani,C., Iwai,S., van der Spek,P.J., Eker,A.P.M., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H.J. (1998) Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- 8.Hwang B.J., Ford,J.M., Hanawalt,P.C. and Chu,G. (1999) Proc. Natl Acad. Sci. USA, 96, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanawalt P.C. (1994) Science, 266, 1957–1958. [DOI] [PubMed] [Google Scholar]
- 10.He Z., Henricksen,L.A., Wold,M.S. and Ingle,C.J. (1995) Nature 374, 566–569. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Lu,X., Peterson,C.A. and Legerski,R.J. (1995) Mol. Cell. Biol., 15, 5396–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuda T., Saijo,M., Kuraoka,I., Kobayashi,T., Nakatsu,Y., Nagai,A., Enjoji,T., Masutani,C., Sugasawa,K., Hanaoka,F., Yasui,A. and Tanaka,K. (1995) J. Biol. Chem., 270, 4152–4157. [DOI] [PubMed] [Google Scholar]
- 13.Drapkin R., Reardon,J.T., Ansari,A., Huang,J.C., Zawel,L., Ahn,K., Sancar,A. and Reinberg,D. (1994) Nature, 368, 769–772. [DOI] [PubMed] [Google Scholar]
- 14.Evans E., Moggs,J.G., Hwang,J.R., Egly,J.M. and Wood,R.D. (1997) EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaeffer L., Roy,R., Humbert,S., Moncollin,V., Vermeulen,W., Hoeijmakers,J.H.J., Chambon,P. and Egly,J.M. (1993) Science, 260, 58–63. [DOI] [PubMed] [Google Scholar]
- 16.van Vuuren A.J., Vermeulen,W., Ma,L., Weeda,G., Appeldoorn,E., Jaspers,N.G.J., van der Eb,A.J., Bootsma,D., Hoeijmakers,J.H.J., Humbert,S., Schaeffer,L. and Egly,J.M. (1994) EMBO J., 13, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donovan A., Davies,A.A., Moggs,J.G., West,S.C. and Wood,R.D. (1994) Nature, 371, 432–435. [DOI] [PubMed] [Google Scholar]
- 18.Sijbers A.M., De Laat,W.L., Ariza,R.R., Biggerstaff,M., Wei,Y.-F., Moggs,J.G., Carter,K.C., Shell,B.K., Evans,E., De Jong,M.C., Rademakers,S., De Rooij,J., Jaspers,N.G.J., Hoeijmakers,J.H.J. and Wood,R.D. (1996) Cell, 86, 811–822. [DOI] [PubMed] [Google Scholar]
- 19.Huang J.C., Svoboda,D.L., Reardon,J.T. and Sancar,A. (1992) Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohr V.A., Smith,C.A., Okumoto,D.S. and Hanawalt,P.C. (1985) Cell, 40, 359–369. [DOI] [PubMed] [Google Scholar]
- 21.Mellon I., Spivak,G. and Hanawalt,P.C. (1987) Cell, 51, 241–249. [DOI] [PubMed] [Google Scholar]
- 22.Henning K.A., Li,L., Iyer,N., McDaniel,L., Reagan,M.S., Legerski,R., Schultz,R.A., Stefanini,M., Lehmann,A.R., Mayne,L.V. and Friedberg,E.C. (1995) Cell, 82, 555–564. [DOI] [PubMed] [Google Scholar]
- 23.Troelstra C., van Gool,A., de Wit,J., Vermeulen,W., Bootsma,D. and Hoeijmakers,J.H.J. (1992) Cell, 71, 939–953. [DOI] [PubMed] [Google Scholar]
- 24.Venema J., Mullenders,L.H.F., Natarajan,A.T., van Zeeland,A.A. and Mayne,L.V. (1990) Proc. Natl Acad. Sci. USA, 87, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bootsma D., Kraemer,K.H., Cleaver,J. and Hoeijmakers,J.H.J. (1998) In Vogelstein,B. and Kinzler,K.W. (eds), The Genetic Basis of Human Cancer. McGraw-Hill, New York, pp. 245–274.
- 26.Hoeijmakers J.H.J. (1994) Eur. J. Cancer, 30A, 1912–1921. [DOI] [PubMed] [Google Scholar]
- 27.Feaver W.J., Svejstrup,J.Q., Bardwell,L., Bardwell,A.J., Buratowski,S., Gulyas,K.D., Donahue,T.F., Friedberg,E.C. and Kornberg,R.D. (1993) Cell, 75, 1379–1387. [DOI] [PubMed] [Google Scholar]
- 28.Vermeulen W., van Vuuren,A.J., Chipoulet,M., Schaeffer,L., Appeldoorn,E., Weeda,G., Jaspers,N.G.J., Priestley,A., Arlett,C.F., Lehmann,A.R., Stefanini,M., Mezzina,M., Sarasin,A., Bootsma,D., Egly,J.M. and Hoeijmakers,J.H.J. (1994) Cold Spring Harb. Symp. Quant. Biol., 59, 317–329. [DOI] [PubMed] [Google Scholar]
- 29.Bootsma D. and Hoeijmakers,J.H.J. (1993) Nature, 363, 114–115. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., Wei,S., Reed,S.H., Wu,X., Svejstrup,J.Q., Feaver,W.J., Kornberg,R.D. and Friedberg,E.C. (1997) Mol. Cell. Biol., 17, 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauder S., Bankmann,M., Guzder,S.N., Sung,P., Prakash,L. and Prakash,S. (1996) Mol. Cell. Biol., 16, 6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masutani C., Sugasawa,K., Yanagisawa,J., Sonoyama,T., Ui,M., Enomoto,T., Takio,K., Tanaka,K., van der Spek,P.J., Bootsma,D. Hoeijmakers,J.H.J. and Hanaoka,F. (1994) EMBO J., 13, 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombaerts M., Tijsterman,M., Verhage,R.A. and Brouwer,J. (1997) Nucleic Acids Res., 25, 3974–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 35.Auffray C. and Rougeon,F. (1980) Eur. J. Biochem., 107, 303–314. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 37.Weeda G., Wiegant,J., van der Ploeg,M., Geurts van Kessel,A.H., van der Eb,A.J. and Hoeijmakers,J.H.J. (1991) Genomics, 10, 1035–1040. [DOI] [PubMed] [Google Scholar]
- 38.Tirode F., Busso,D., Coin,F. and Egly,J.M. (1999) Mol. Cell, 3, 87–95. [DOI] [PubMed] [Google Scholar]
- 39.Rossignol M., Kolb-Cheynel,I. and Egly,J.M. (1997) EMBO J., 16, 1628–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manley J.L., Fire,A., Samuels,M. and Sharp,P.A. (1983) Methods Enzymol., 101, 568–582 [DOI] [PubMed] [Google Scholar]
- 41.Sung P., Bailly,V., Weber,C., Thompson,L.H., Prakash,L. and Prakash,S. (1993) Nature, 365, 852–855. [DOI] [PubMed] [Google Scholar]
- 42.Rodel C., Jupitz,T. and Schmidt,H. (1997) Nucleic Acids Res., 25, 2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz P., Fribourg,S., Poterszman,A., Mallouh1,V., Moras,D. and Egly,J.M. (2000) Cell, 102, 599–607. [DOI] [PubMed] [Google Scholar]
- 44.Seroz T., Perez,C., Bergmann,E., Bradsher,J. and Egly,J.M. (2000) J. Biol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 45.Vermeulen W., Bergmann,E., Auriol,J., Rademakers,S., Frit,P., Appeldoorn,E., Hoeijmakers,J.H.J. and Egly,J.M. (2000) Nature Genet., in press. [DOI] [PubMed] [Google Scholar]
- 46.Stefanini M., Lagomarsini,P., Giliani,S., Nardo,T., Botta,E., Peserico,A., Kleijer,W.J., Lehmann,R., and Sarasin,A. (1993) Carcinogenesis, 14, 1101–1105. [DOI] [PubMed] [Google Scholar]