Abstract
Detecting chemical signals is important for identifying food sources and avoiding harmful agents. Like many animals, C. elegans use olfaction to chemotax towards their main food source, bacteria. However, little is known about the bacterial compounds governing C. elegans attraction to bacteria and the physiological importance of these compounds to bacteria. Here, we address these questions by investigating the function of a small RNA, P11, in the pathogen, Pseudomonas aeruginosa, that was previously shown to mediate learned pathogen avoidance. We discovered that this RNA also affects the attraction of untrained C. elegans to P. aeruginosa and does so by controlling production of ammonia, a volatile odorant produced during nitrogen assimilation. We describe the complex regulation of P. aeruginosa nitrogen assimilation, which is mediated by a partner-switching mechanism involving environmental nitrates, sensor proteins, and P11. In addition to mediating C. elegans attraction, we demonstrate that nitrogen assimilation mutants perturb bacterial fitness and pathogenesis during C. elegans infection by P. aeruginosa. These studies define ammonia as a major mediator of trans-kingdom signaling, implicate nitrogen assimilation as important for both bacteria and host organisms, and highlight how a bacterial metabolic pathway can either benefit or harm a host in different contexts.
Subject terms: Bacteriology, Bacteria, Bacterial infection
Ammonia production enzymes mediate the initial attraction of C. elegans towards P. aeruginosa. Here, Marogi et al show that the bacterial small RNA, P11, affects worm attraction by modulating nitrogen assimilation.
Introduction
Animals depend on sensory information to promote survival by detecting and responding to a variety of environmental stimuli, such as moving toward nutrient sources or avoiding harmful pathogens. Within their natural environment of soil and rotting vegetation, Caenorhabditis elegans are surrounded by diverse bacterial species, which constitute their major food source1–3. C. elegans respond and chemotaxis toward volatile compounds using olfactory neurons that control their locomotion4. A number of C. elegans attractants and repellents have been defined, and some of these attractants and repellants have been shown to be produced by bacteria4–9. However, the contributions of the production of specific bacterially produced molecules for the chemotaxis of C. elegans towards specific bacterial species have remained largely elusive.
One example of the complex responses of C. elegans to its environment is its multifaceted behaviors in the presence of the bacterium, Pseudomonas aeruginosa8–13. Pseudomonas species represent the most common bacteria found in the C. elegans microbiome, suggesting that C. elegans encounters these bacteria frequently in nature3. P. aeruginosa has a complex regulatory network that controls its pathogenesis such that in some contexts, it is non-pathogenic and a beneficial food source for C. elegans. But in other contexts, such as higher temperatures or upon association with rigid surfaces, P. aeruginosa produces virulence factors that make it pathogenic towards C. elegans14–16. Many of P. aeruginosa’s effects on C. elegans are dynamic, as are C. elegans’ responses to P. aeruginosa. Naive C. elegans that have never previously encountered P. aeruginosa are attracted towards P. aeruginosa, and even prefer P. aeruginosa over other bacteria like Escherichia coli that are constitutively non-pathogenic towards C. elegans10,12,13. The specific molecule that mediates the attraction of C. elegans to P. aeruginosa was previously unknown. Here, we identify volatile ammonia, a byproduct of Pseudomonas aeruginosa nitrogen assimilation, as a key mediator of C. elegans attraction.
Our studies highlight the importance of nitrogen metabolism in P. aeruginosa’s interactions with C. elegans. Nitrogen is essential for amino acid synthesis and also plays important roles in a variety of other processes17–19. Like all animals, C. elegans cannot directly utilize inorganic nitrogen sources like ammonium and must, therefore, obtain processed organic nitrogen from the proteins and amino acids in their bacterial food sources17,20. In contrast, bacteria like Pseudomonas species can process inorganic environmental ammonium into organic nitrogen21–24. Inorganic nitrogen is typically found in two forms: ammonium and nitrate. When ammonium is abundant, it is the preferred nitrogen source and is reduced into glutamine and glutamate, which subsequently function as intracellular organic nitrogen donors19. When ammonium availability is limited, most Pseudomonas species induce a process known as nitrogen assimilation, where they reduce environmental nitrate into ammonium, which can then be further reduced into glutamine and glutamate21–24.
Although many studies have interrogated nitrogen assimilation pathways across diverse bacterial species, its importance in trans-kingdom communication and bacterial host interactions remains largely unknown. Here, we combine P. aeruginosa and C. elegans genetics to help bridge a gap in trans-kingdom signaling by linking animal chemosensation with bacterial metabolism and physiology. Our data show that volatile ammonia is produced by P. aeruginosa nitrogen assimilation, which is regulated by a small RNA (sRNA)-mediated transcriptional termination and anti-termination mechanism. Ammonia produced by P. aeruginosa is an attractant for C. elegans and causes them to specifically prefer P. aeruginosa over E. coli. Finally, we found that nitrogen assimilation affects the ability of P. aeruginosa to colonize C. elegans, which affects pathogenesis and demonstrates the importance of metabolism in trans-kingdom signaling and host-microbe interactions.
Results
Disrupting the P11 sRNA perturbs the naive attraction of C. elegans to PA14
Within their natural habitat, C. elegans are surrounded by diverse bacterial species1–3. Previous studies have demonstrated that naive, untrained C. elegans are attracted to P. aeruginosa PA14 (PA14) and prefer it to E. coli OP50 (OP50)10,12,13. P11, also known as NalA in PA0124, is a sRNA previously shown to be necessary for a process in which trained C. elegans use transgenerational epigenetic inheritance to learn to avoid PA14 for four generations10,25. The role of P11 in the attraction of naive C. elegans to PA14 has not been previously examined. In the PA14 genome, P11 is found immediately upstream of an operon that mediates the first step of nitrogen assimilation, including the two subunits of nitrite reductase (NirB and NirD), and nitrate reductase (NasC). Given that P11 is upstream of the nitrogen assimilation operon that produces ammonium and that ammonium-related compounds have been previously shown to affect worm chemotaxis7, we sought to interrogate P11 function in naive worm chemotaxis.
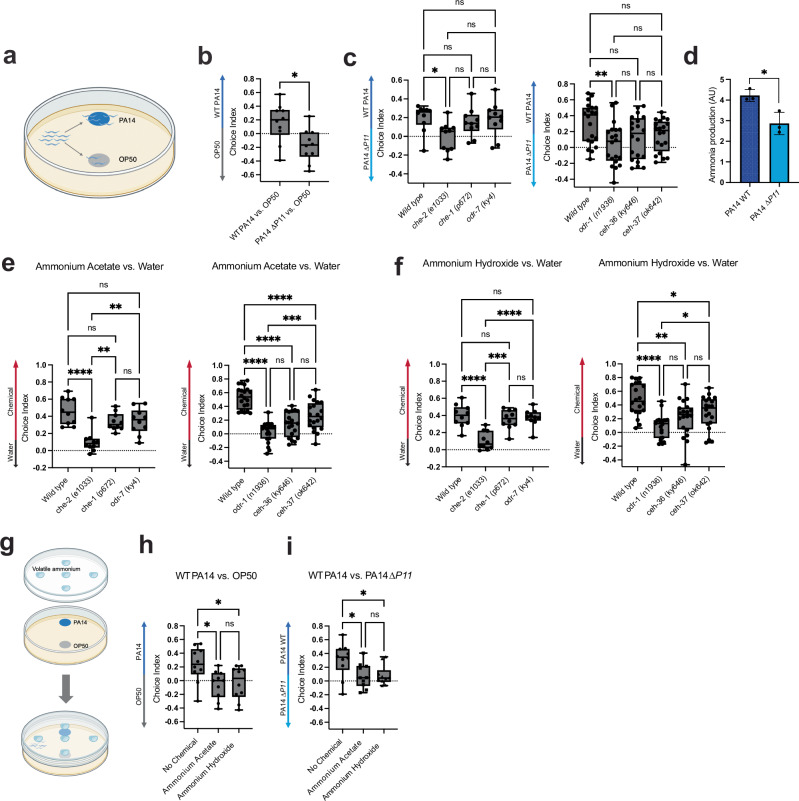
We examined the role of P11 in mediating the attraction of C. elegans to PA14 using an OP50 vs. PA14 bacterial choice assay (Fig. 1a, b). We first confirmed the previous finding that naive C. elegans prefer PA14 to OP50. We then performed choice assays between a PA14 strain with an ~150 base pair deletion surrounding P11 (ΔP11) and OP50. We found that ΔP11 eliminated the preference of PA14 over OP50 (Fig. 1b), suggesting that in the absence of P11, PA14 lacks the attractant that mediates preference over OP50.
Fig. 1. C. elegans are attracted to PA14-produced ammonia.
a Schematic of worm bacterial choice assay between PA14 and OP50. Worms were hatched and grown on OP50 for 2 days prior to bacterial choice assays. Worms were given 1 hour to chemotax to bacteria and paralyzed at first choice. ~30–100 worms were analyzed per plate, and precise numbers of worms used for each assay are noted in the source data file for each experiment. Created with BioRender.com. b Worm choice assay between PA14 WT/∆P11 and OP50. Choice index (CI) = (# on PA14–OP50)/(Total). p = 0.0117. c Choice assays between WT PA14 and ∆P11 with Wild type, che-2(e1033), che-1(p627), and odr-7(ky4) worm mutants (left) or odr-1(n1936), ceh-36(ky646), and ceh-37(ok642) (right). CI = (# on WT PA14–PA14 ∆P11)/(Total). d PA14 WT and ∆P11 ammonia production measured using an ammonia colorimetric assay. Colorimetric fluorescence intensity (ex = 360 nm/em = 450 nm) normalized to CFU and multiplied by 105. p = 0.0305. Chemical choice assays between water and ammonium acetate (e) and ammonium hydroxide (f) with Wild type, che-2(e1033), che-1(p627), and odr-7(ky4) worm mutants (left) or odr-1(n1936), ceh-36(ky646), and ceh-37(ok642) (right). CI = (# on chemical - # on water)/(# on chemical + # on water). g Schematic of bacterial choice assay with saturating concentrations of volatile ammonia. Bacteria spotted on NGM agar and chemical spotted on lid (5 spots of 10 µl chemical, 2.5 M each). Created with BioRender.com. h Addition of ammonia (ammonium acetate and ammonium hydroxide) to choice assay between PA14 WT and OP50. (CI) = (# on PA14–OP50)/(Total). i Addition of volatile ammonia (ammonium acetate and ammonium hydroxide) to choice assay between PA14 WT and ∆P11. CI = (# on PA14 WT– PA14 ∆P11)/(Total). Box Plots: center line = median, box range 25th–75th percentile, minimum/maximum denoted by whiskers, individual choice assay plates are represented by dots and are technical replicates (10 plates/technical replicates for (b, c (left), e (left), f (left), h, i) and 20 plates/technical replicates for c(right), e(right), f(right)). 3 Biological replicates were performed per choice assay and graphs represent one replicate. Bar graph error bars denote SD, center denotes mean, and each dot represents a biological replicate (3 total). One-Way ANOVA (c, e, f, h, i) Tukey’s multiple comparison test. Unpaired T-test, two-sided (b, d). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant (Prism 9). For exact sample size (n), all biological replicates, and p values see source data file. Figure 1a, g were created with BioRender.com and released under a CC BY-NC-ND lisence.
Worms are attracted to the PA14 nitrogen assimilation metabolite, ammonia
To gain insight into the nature of the P11-dependent signal produced by PA14, we first interrogated the mechanism by which C. elegans responds to this signal. Both the AWA and AWC neurons facilitate worm chemotaxis to volatile compounds, whereas the ASE neuron is involved in taste sensation4,26. To determine which neuron mediates attraction to PA14, we subjected worms with individual mutants that affect these neurons to choice assays between wild-type PA14 (WT) and ΔP11. Like wild-type C. elegans, worms defective in the function of ASE (che-1(p672))27 or AWA (odr-7(ky4))28 preferred WT PA14 to PA14 ΔP11. However, che-2(e1033) mutants, which are defective in the function of ciliated neurons including AWA, AWB, AWC, and others29, no longer exhibited a preference (Fig. 1c, left). Because che-2(e1033) disrupts the function of all ciliated neurons29, we examined additional mutants defective in the functions of specific subsets of these neurons. Worm mutants defective in AWB (ceh-37(ok642))30,31 or AWC and ASE (ceh-36(ky646))30 still preferred WT PA14 over PA14 ΔP11. However, a worm mutant defective in the functions of AWB and AWC (odr-1(n1936))31 did not demonstrate a preference (Fig. 1c, right). These results suggest that P11-dependent attraction to PA14 is mediated by a volatile odorant and are consistent with a previous study that did not identify the specific attractant C. elegans use for naive chemotaxis to P. aeruginosa but did show that this preference is mediated by the AWB-AWC circuit13.
Nitrogen assimilation produces ammonium, which can spontaneously convert to a volatile form, ammonia32. Thus, if the deletion surrounding P11 disrupts nitrogen assimilation, it might reduce ammonia production. To determine if the P11 deletion disrupted nitrogen assimilation, we assayed the ability of ΔP11 to grow in nitrogen-free media supplemented with inorganic nitrates. WT PA14 grew robustly in these conditions, but ΔP11 mutants did not (Supplementary Fig. 1a). We confirmed that this growth defect was due to an inability to reduce nitrate by demonstrating that adding the products of the nitrogen assimilation reactions (ammonium, Glu, or Gln) rescued PA14 ΔP11 growth (Supplementary Fig. 1b–e). We also confirmed that the ΔP11 mutant lacked expression of the downstream nitrogen assimilation genes, providing a molecular explanation for its nitrogen assimilation defect (Supplementary Fig. 1f).
Given that the P11 deletion disrupts nitrogen assimilation, we next sought to directly determine the effect of this mutant on ammonia production. For this purpose, we adapted a colorimetric ammonia assay so that it could be used in the surface-associated conditions employed for C. elegans choice assays. We found that ΔP11 produced statistically significantly less ammonia than WT PA14 in the conditions in which we assayed bacterial choice preference (Fig. 1d). We also demonstrated that ammonia is sufficient to promote C. elegans attraction by confirming previous findings that C. elegans is attracted to ammonia and that this attraction requires the function of the ciliated sensory neurons that detect volatile odorants7 (Fig. 1e, f). To determine if ammonia sensing is necessary for C. elegans’ PA14 preference, we exposed C. elegans to saturating levels of exogenous ammonia, which should eliminate any ammonia gradients produced by bacteria in this environment. In the presence of saturating concentrations of ammonia, we found that C. elegans no longer preferred WT PA14 to OP50 or ΔP11 (Fig. 1g–i). Therefore, our data suggest that P11-dependent ammonia production is necessary and sufficient to explain naive C. elegans’ attraction to PA14.
A complex regulatory mechanism controls PA14 nitrogen assimilation
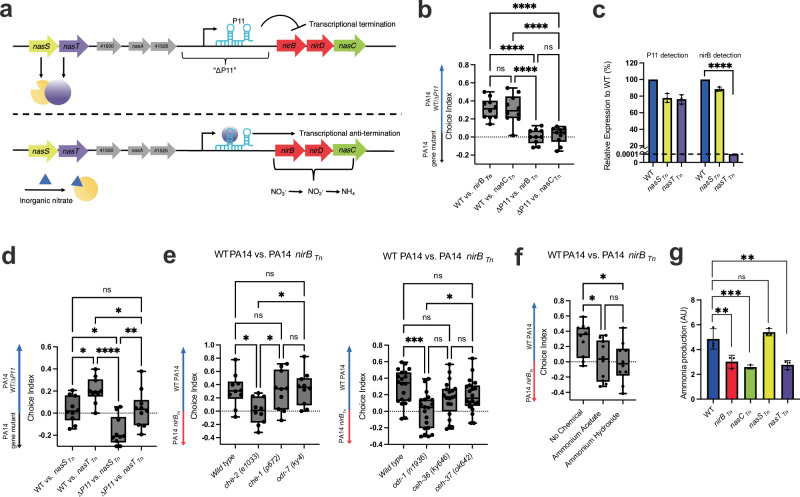
The mutant referred to as “ΔP11” both above and in prior studies10 deletes >150 bp of the genomic region that resides between two operons implicated in nitrogen assimilation (Fig. 2a). We thus examined mutants in these operons to dissect their functions in nitrogen assimilation. As expected, transposon insertions in either nitrite reductase, nirB, or nitrate reductase, nasC, rendered PA14 unable to grow in conditions in which nitrates are the sole nitrogen source (Supplementary Fig. 2a). These mutants also affected C. elegans attraction: C. elegans preferred WT PA14 to nirBTn or nasCTn. Loss of nirB or nasC was also sufficient to completely explain the ΔP11 C. elegans attraction defect, as C. elegans showed no preference between ΔP11 and nirBTn or nasCTn (Fig. 2b).
Fig. 2. The regulation of ammonia production and C. elegans chemoattraction in PA14.
a Model of PA14 nitrogen assimilation mechanism. When preferred nitrogen sources are limited, the two-component NtrCB system is activated, thus promoting expression at the start of P11. NasS binds to inorganic nitrates, and NasT inhibition is relieved. NasT binds to P11, allowing for anti-termination at the leader sequence and expression of the nitrogen assimilation operon. b Choice assays between WT PA14 ΔP11 and PA14 mutants in nitrite reductase (nirBTn) and nitrate reductase (nasCTn). c Relative expression of P11 and nirB in PA14 nasSTn/nasTTn to WT PA14. Normalized to 5S expression. The dotted line represents the limit of detection. P = < 0.001. d Choice assays between WT PA14ΔP11 and PA14 mutants in the partner-switching genes (nasSTn and nasTTn). CI = (# on WT PA14 or ∆P11 PA14 gene mutants)/(Total). e Choice assays between WT PA14 and nirBTn with Wild type, che-2(e1033), che-1(p627), and odr-7(ky4) worm mutants (left) or odr-1(n1936), ceh-36(ky646), and ceh-37(ok642) (right). CI = (# on WT PA14–PA14 nirBTn)/(Total). f Addition of ammonia (ammonium acetate and ammonium hydroxide) to choice assay between WT PA14 and nirBTn. CI = (# on WT PA14–PA14 nirBTn)/(Total). g WT PA14 and gene mutants ammonia production measured using an ammonia colorimetric assay. Colorimetric fluorescence intensity (ex = 360 nm/em = 450 nm) normalized to CFU and multiplied by 105. p values using WT as control: nirBTn = 0.0032, nasCTn = 0.0007, nasTTn = 0.0013. Box Plots: center line = median, box range 25th–75th percentile, minimum/maximum denoted by whiskers, individual choice assay plates are represented by dots and are technical replicates (10 plates/technical replicates for (b, d, e (left), f) and 20 plates/technical replicates for (e, right). Bar graph error bars denote SD, center denotes mean, and each dot represents a biological replicate (3 total). One-Way ANOVA, (b, d–f) Tukey’s multiple comparison test, c, g Dunett’s multiple comparison test using WT as control. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant (Prism 9). For exact sample size (n), all biological replicates, and p values see source data file.
Upstream of P11 is another operon containing two regulatory genes, nasS and nasT. In a closely related Pseudomonas strain, PA01, it has been shown that NasT can bind to either the P11-containing 5’ leader region of the nirB gene (P11) or to NasS in a mutually exclusive manner24. P11 contains a predicted hairpin that terminates transcription when unbound, and NasT binding to the leader is required for downstream transcription of nirB, nirD, and nasC. Meanwhile, NasS mutually exclusively binds to either NasT or to nitrate. Thus, in the absence of nitrates, NasS binds to NasT, which leaves P11 unbound and results in termination that prevents nirBD nasC expression. When nitrates are present, nitrates bind to NasS, freeing NasT to bind to P11 and promote nirBD nasC expression24 (Fig. 2a). To determine if PA14 nitrogen assimilation is also regulated by this partner-switching regulatory mechanism24, we assayed PA14 nasS and nasT mutants for their ability to grow in nitrogen-free media supplemented with nitrates. Whereas nasSTn grew as well as WT PA14, nasTTn was unable to grow (Supplementary Fig. 2a). We also analyzed the expression of both the P11 and nirB RNA’s by qRT-PCR in surface-attached conditions mimicking those used during C. elegans choice assays. We found that nasTTn dramatically reduced the downstream nirB gene to undetectable levels but retained relatively high P11 levels. While nasSTn exhibited similar P11 levels to those seen in nasTTn, the levels of nirB were significantly higher in nasSTn than in nasTTn (Fig. 2c). These results suggest that the NasT-NasS partner-switching mechanism regulates nitrate/nitrite reductase expression in PA14, providing a molecular explanation for their effects on nitrogen assimilation.
Next, we tested the function of nasS and nasT in C. elegans chemotaxis through choice assays against WT PA14 or ΔP11 and mutants in both genes. Worms demonstrated a preference for WT over nasTTn but did not prefer WT to nasSTn (Fig. 2d). Conversely, ΔP11 was less attractive to worms than nasSTn, but worm attraction to ΔP11 and nasTTn was equal (Fig. 2d). To confirm that mutants in nirB, nasC, and nasT interfere with C. elegans attraction by disrupting the production of a volatile molecule, we assayed C. elegans chemotaxis to WT PA14 and mutants in these three genes with the same neuronal mutants as previously described. Worms defective in the function of ASE (che-1(p672))27, AWA (odr-7(ky4))28, AWB (ceh-37(ok642))30, or AWC/ASE (ceh-36(ky646))30 preferred WT PA14 to nirBTn, nasCTn, or nasTTn. Meanwhile, worms defective in the function of ciliated sensory neurons (che-2(e1033))29 or AWB and AWC (odr-1(n1936))31 no longer exhibited a preference (Fig. 2e, Supplementary Fig. 2b, c). We further confirmed the identity of the signal as ammonia by introducing exogenous ammonia to choice assays between WT PA14 and nirBTn, nasCTn, or nasTTn. In the presence of saturating concentrations of ammonia, C. elegans were no longer able to distinguish different PA14 strains (Fig. 2f, Supplementary Fig. 2d, e).
Finally, we determined whether our PA14 nitrogen assimilation pathway mutants directly affect ammonia production. Specifically, we assayed ammonia production in surface-attached conditions by PA14 mutants in nirB, nasC, nasS, and nasT. Mutants that render PA14 unable to assimilate nitrates (nirBTn, nasCTn, nasTTn) showed a significant decrease in ammonia production, whereas nasSTn produced WT ammonia levels (Fig. 2g). Altogether, these data support our hypothesis that NasS functions as a nitrate sensor and, with NasT, regulates nitrogen assimilation by a partner-switching mechanism. Additionally, all disruptions of the nitrogen assimilation operon affect PA14 ammonia production and consequently interfere with C. elegans attraction to PA14. Furthermore, while it is possible that our transposon mutants could have polar and off-target effects, the phenotypic agreement amongst all of the independent transposon mutant studies suggests that these mutants affect these behaviors through their predicted functions in nitrogen assimilation.
The specificity and relevance of worm attraction to Pseudomonas-produced ammonia
While our results thus far are consistent with bacterially produced ammonia representing the attractant that drives worm chemoattraction to P. aeruginosa, the specificity and relevance of this behavior remained unclear. For example, saturating ammonia disrupted the preference for P. aeruginosa, but it is formally possible that saturating ammonia nonspecifically disrupts all C. elegans chemoattraction. To address this possibility, we assessed the effect of saturating ammonia on worm attraction towards benzaldehyde, another known worm attractant31. Regardless of the presence of saturating ammonia, worms were still able to sense and locomote toward benzaldehyde, suggesting that sensing and chemotaxis towards other attractants is not disrupted by saturating levels of ammonia (Fig. 3a). As another test of odorant specificity, we challenged worms to a bacterial choice assay between WT PA14 and ∆P11 or nirBTn in the presence of saturating concentrations of benzaldehyde. Worms still preferred WT PA14 over PA14 mutants defective in nitrogen assimilation expression (Fig. 3b), demonstrating that worms can specifically identify differential levels of ammonia produced by PA14 in the presence of saturating levels of another attractant.
Fig. 3. C. elegans attraction to ammonia is independent of other chemotactic responses and conserved in multiple Pseudomonas species.
a Chemical choice assay between benzaldehyde and water with saturating concentrations of volatile ammonium acetate (left) or ammonium hydroxide (right). CI = (# on chemical - # on water)/(# on chemical + # on water). b Bacterial choice assays between WT PA14 and ∆P11 (left) or nirBTn (right) in the presence of saturating concentrations of benzaldehyde. CI = (# on WT PA14–PA14 mutant)/(Total). c Naive avoidance of WT, ∆P11, and nirBTn PA14 lawns over a time course of 12 h. Percent worms on lawn = ((number of worms on lawn/total)*100). d Bacterial choice assay between OP50 and wild Pseudomonas isolates P. vranovensis (left) and P. mendocina (right) with saturating concentrations of volatile ammonia. CI = (# on Pseudomonas–OP50)/(Total). Box Plots: center line = median, box range 25th–75th percentile, minimum/maximum denoted by whiskers, individual choice assay plates are represented by dots and are technical replicates (10 plates/technical replicates for a, b, d). Line graph error bars denote SD and center denotes mean (3 plates/technical replicates per condition). Unpaired T test, two-sided (a, b). One-Way ANOVA (c, d) Tukey’s multiple comparison test. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant (Prism 9). For exact sample size (n), all biological replicates, and p values see source data file.
It was previously shown that PA14 can produce cues such as 1-undecene and blue light (pyocyanin) that are avoided by naive worms8,33. Although our data suggest that PA14 nitrogen assimilation is involved in naive worm attraction to PA14, it is plausible that nitrogen assimilation can also affect avoidance behaviors in naive worms. To assess the specificity of ammonia for worm attraction, we employed worm avoidance assays to WT PA14 and PA14 strains defective in nitrogen assimilation (PA14 ∆P11 and nirBTn). Our data show that worms avoid PA14 lawns at the same rate regardless of a functional PA14 nitrogen assimilation pathway (Fig. 3c). Thus, nitrogen assimilation appears to affect C. elegans attraction to P. aeruginosa, but does not affect repulsion from other previously described cues.
PA14 is a human clinical isolate that is not an abundant Pseudomonas species in the natural soil environments typically encountered by C. elegans. Therefore, we sought to determine if ammonia’s role as a chemoattractant is specific to PA14 or if ammonia might also play a role in worm attraction to soil-resident Pseudomonas species that might be more physiologically relevant for C. elegans’ natural environment. We performed choice assays between OP50 and Pseudomonas vranovensis or Pseudomonas mendocina, both of which are soil-resident Pseudomonas species and are preferred by naive worms over OP503,34,35 (Fig. 3d). Similar to the behavior we observed for PA14, worm preference towards each of these Pseudomonas species was abolished by the presence of saturating ammonia, suggesting that worms also use ammonia sensing to chemotax to natural isolates of Pseudomonas (Fig. 3d).
Two P11 stem-loops inversely regulate nitrogen assimilation expression
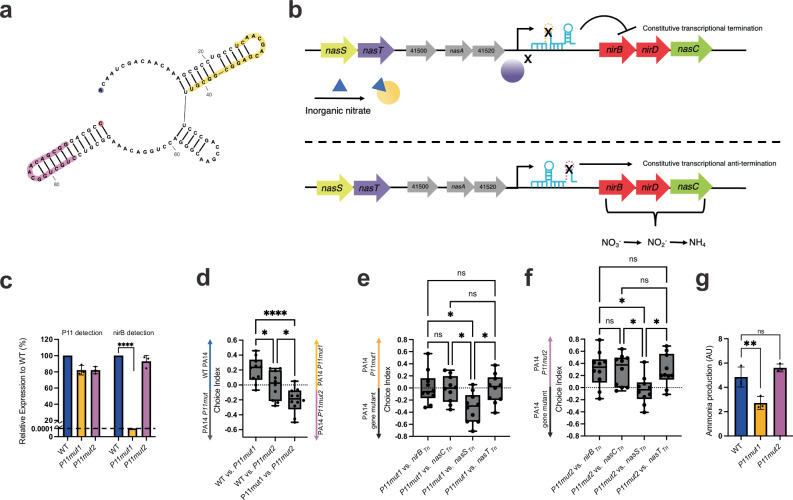
RNA secondary structure formation is important for numerous RNA functions. The predicted secondary structure of P11 contains stem loops (Fig. 4a), which often facilitate protein-RNA interactions or function as transcriptional terminators36–38. To determine the functions of the P11 stem-loops and to better understand the mechanism by which P11 regulates the nitrogen assimilation operon, we made two mutants that each specifically deleted one of the stem-loops (P11mut1 and P11mut2) (Fig. 4a, b). Unlike the large ΔP11 deletion described above, these small mutations do not extend upstream into the operon promoter. PA14 P11mut1 failed to grow in nitrogen-free media supplemented with nitrate (Supplementary Fig. 2f), suggesting that deletion of the first stem-loop leads to a loss of expression of the nitrogen assimilation enzymes. To understand the cause of this nitrogen assimilation phenotypic defect, we examined the expression of both the P11 leader region and the downstream nirB gene. P11mut1 retained robust P11 expression, indicating that the operon promoter remained intact. However, nirB mRNA expression was so low as to remain undetectable in P11mut1 (Fig. 4c). These results suggest that P11mut1 results in transcriptional termination between P11 and nirB, which could be explained by loss of NasT binding, resulting in a loss of anti-termination activity. Conversely, P11mut2 grew robustly in nitrogen-free media supplemented with nitrate (Supplementary Fig. 2f) and retained relatively high levels of both P11 and nirB expression (Fig. 4c). These data suggest that disrupting the second stem-loop leads to constitutive expression of nirB. The second P11 stem-loop is followed by a poly-T sequence, a hallmark indicator of a rho-independent terminator38. Thus, our results support the hypothesis that within P11, the first stem-loop is the site of anti-termination by NasT, and the second stem-loop functions as a rho-independent terminator that leads to transcriptional termination when NasT is not bound.
Fig. 4. The roles of P11 stem-loops in C. elegans attraction and nitrogen assimilation.
a Diagram showing putative P11 secondary structure. Yellow: NasT binding site (P11mut1) deletion; purple: rho-independent terminator (P11mut2) deletion; blue: 5’ nucleotide; red: 3’ nucleotide followed by …UUUUUUGUUU, poly-U sequence following the rho-independent terminator. Structure created in RNAfold and visually optimized in rnacanvas.app52. b Model showing the effects of P11mut1 and P11mut2 on nitrogen assimilation operon expression. P11mut1 prevents NasT binding to P11 and transcriptional anti-termination through the leader sequence (top). P11mut1 prevents transcriptional termination and therefore, causes constitutive expression of the nitrogen assimilation operon (bottom). c Relative expression of P11 and nirB in PA14 P11mut1/2 to PA14 WT. Normalized to 5S expression. Dotted line represents limit of detection. p = <0.0001. d Choice assays between PA14 WT and p11mut1/2. CI = (# on PA14 WT PA14 P11 mutants)/(Total). Right-hand CI = (# on PA14 P11mut1–PA14 P11mut2)/(Total) represents last box and whicker plot. e Choice assays between PA14 P11mut1 and nitrogen assimilation/partner-switching gene mutants. CI = (# on PA14 P11mut1–PA14 gene mutant)/(Total). f Choice assays between PA14 P11mut2 and nitrogen assimilation/partner-switching gene mutants. CI = (# on PA14 P11mut2–PA14 gene mutant)/(Total). g Ammonia production of P11 mutants. Colorimetric fluorescence intensity normalized to CFU and multiplied by 105. p = 0.0025. Box Plots: center line = median, box range 25th–75th percentile, minimum/maximum denoted by whiskers, individual choice assay plates are represented by dots and are technical replicates (10 plates/technical replicates for d–f). Bar graph error bars denote SD, center denotes mean, and each dot represents a biological replicate (3 total). One-Way ANOVA, (c–f), Tukey’s multiple comparison test, (g) Dunett’s multiple comparison test using WT as control. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001, ns = not significant (Prism 9). For exact sample size (n), all biological replicates, and p values see source data file.
To examine the phenotypic consequences of stem-loop mutations, we subjected C. elegans to choice assays between WT PA14 and the two P11 stem-loop mutants. WT PA14 was preferred over P11mut1, whereas no preference was exhibited between WT PA14 and P11mut2 (Fig. 4d). When given the choice between P11mut1 and P11mut2, P11mut2 was preferred (Fig. 4d). We also tested worm preference between the P11 mutants and nirBTn, nasCTn, nasSTn, and nasTTn. Worms did not prefer P11mut1 over nirBTn, nasCTn, or nasTTn (Fig. 4e). However, nasSTn was more attractive than P11mut1 (Fig. 4e). The opposite effect was observed when comparing mutants in all these genes to P11mut2, i.e., worms preferred P11mut2 over nirBTn, nasCTn, and nasTTn, but did not prefer P11mut2 to nasSTn (Fig. 4f). Finally, we tested ammonia production in the P11 mutants and found that ammonia levels were reduced in P11mut1 but were normal in P11mut2 (Fig. 4g). These results confirm the functions of the two stem-loops of P11 in regulating the expression of the nitrogen assimilation enzymes and the correlations between ammonia production and chemosensation provide further support for the central role for P11-dependent ammonia production in mediating the attraction of C. elegans to P. aeruginosa.
Nitrogen assimilation is important for PA14 fitness during worm infections
Our findings demonstrate that the nitrogen assimilation pathway affects the attraction of C. elegans towards PA14, but a previous study also demonstrated that the large ΔP11 deletion affects the pathogenesis of PA14 towards C. elegans10. To determine if the pathogenesis defect of ΔP11 is due to this mutant’s defect in nitrogen assimilation, we assayed C. elegans survival in the presence of a specific mutant in nitrite reductase, nirBTn, as well as in a P11 mutant that does not disrupt the nitrogen assimilation operon promoter, P11mut1. C. elegans infected by nirBTn and P11mut1 survived significantly longer than worms infected by WT PA14 (Fig. 5a). To assess whether the pathogenesis defect is specifically due to nitrogen assimilation, we supplemented infected worms with glutamine, a preferred nitrogen source that eliminates the need for nitrogen assimilation. We found that glutamine supplementation fully rescued the pathogenesis defect of nirBTn and P11mut1. These results suggest that the nitrogen assimilation pathway is important for optimal pathogenesis of C. elegans by PA14.
Fig. 5. PA14 nitrogen assimilation impacts bacterial growth and C. elegans pathogenesis.
a Survival of worms infected by PA14 WT and PA14 nirBTn (left) or P11mut1 (right). Worms were counted every 10 hours and were declared dead if they were unresponsive to mechanical agitation. Glutamine supplementation (10 mM) decreased worm survival (%) infected by PA14 nirBTn (left) or P11mut1 (right) to survival (%) by PA14 WT infections. P value calculated using a Mantel-Cox test compared to PA14 WT (P = < 0.0001). b PA14 CFU’s quantified from worm guts 30 h and 50 h post infection. At 30 h, bacterial load does not differ between strains. At 50 h, PA14 nirBTn (left) or P11mut1 (right) is ~2× less abundant than PA14 WT and is rescued by glutamine supplementation (10 mM). P = 0.0007 (left) and P = 0.0002 (right). Bar graph error bars denote SD, center denotes mean, and each dot represents a technical replicate (5 total). One-Way ANOVA analysis was performed compared to WT. Dunett’s multiple comparison test using WT PA14 as control. *p ≤ 0.05. (Prism 9). c RNA sequencing results from PA14 ∆P11 or d PA14 nirBTn relative to WT PA14. 3 Biological replicates used per strain. (c, d: left) Volcano plots were generated from all identified genes in the RNA sequencing results. X axis denotes Log2(Fold Change). Y axis denotes −Log10(P adjusted) with horizontal dotted line denoting Padj = 0.05. Significantly differentially expressed gene in PA14 nirBTn were identified based on Log2(Fold Change) values ≥ 2 or ≤−2 with P adjusted values ≤ 0.05. P values calculated using two-sided quasi-likelihood F test. (c, right) Gene Ontology enrichment analysis was performed to identify differentially expressed gene functional groups. For exact sample size (n), all biological replicates, p values, and full list of quantified genes from RNA sequencing results see source data file.
Nitrogen assimilation could influence pathogenesis by affecting the ability of the bacteria to grow within the host (the number of bacteria present), or by affecting the virulence potential of individual bacteria (how harmful each bacterial cell is towards C. elegans). To differentiate these possibilities, we assessed the number of bacteria present within C. elegans at different time points after infection with WT PA14, nirBTn, or P11mut1. Specifically, we killed all bacteria outside the worms, used mechanical disruption to release the bacteria found inside the worm, and then quantified intrahost bacterial load by quantifying colony- forming units (CFU’s) from the contents released from the worms.
After 30 hours of infection, we observed no difference in either C. elegans survival or intrahost PA14 CFU between WT and nirBTn or P11mut1 (Fig. 5a, b). However, after 50 hours of infection, we observed that worms survived significantly less in WT than in nirBTn, and that intrahost PA14 CFUs were ~30% greater for WT than for nirBTn or P11mut1 (Fig. 5b). Supplementing the infections with glutamine, which rescued the survival defect of nirBTn and P11mut1, also rescued the intrahost CFU phenotype, restoring the number of nirBTn and P11mut1 bacteria inside the worms to WT levels (Fig. 5b). This inverse correlation between bacterial numbers and worm survival are consistent with the nitrogen assimilation pathway affecting worm pathogenesis by primarily influencing the growth of P. aeruginosa within C. elegans.
To further explore whether the effects of nirBTn and ΔP11 might be due to effects on virulence factor regulation, we performed RNA-seq analysis on WT, nirBTn, and ΔP11 PA14 RNA isolated from PA14 grown in the same conditions in which we performed survival assays (see methods). We found relatively few genes whose levels consistently changed in these mutants (Fig. 5c, d). Gene ontology enrichment analysis on genes with Log2(Fold Change) values ≤ −2 and ≥2 and P-adjusted values ≤0.05 failed to identify any major virulence networks upregulated in either PA14 ΔP11 (Fig. 5c) or nirBTn (Fig. 5d). Consistent with these genes playing a fairly minor role in gene regulation, we found no significantly downregulated gene classes in either mutant and no significantly upregulated gene classes in nirBTn (Fig. 5c, d). Finally, we directly examined whether individual virulence factors were differentially regulated but found that the only virulence factors whose levels changed, such as the phenazine biosynthesis genes phzA/B, were expressed higher in the nirBTn and ΔP11 mutants than WT PA14 (Fig. 5c, d, Supplementary Table 1). This result is the opposite of what one would expect to account for the reduced virulence of these mutants. Together, our data suggest that at later stages of P. aeruginosa infection, the metabolic function of the nitrogen assimilation pathway becomes important for virulence. We suggest that at these stages the internal environment of C. elegans becomes limited for preferred nitrogen sources, making the ability to assimilate nitrogen important for intrahost bacterial growth and pathogenesis.
Discussion
Here, we interrogated the functions of nitrogen assimilation in PA14 and how its sRNA-mediated regulation produces volatile ammonia that is attractive to C. elegans. C. elegans depend on olfactory neurons to sense ammonia from bacteria, which in turn governs their preference towards pathogenic PA14 over less pathogenic PA14 or E. coli. Additionally, our data show that PA14 nitrogen assimilation promotes the growth and pathogenesis of PA14 during the later stages of C. elegans infection.
Our findings demonstrate that the P11 sRNA, which was previously shown to mediate learned avoidance of C. elegans by downregulating a specific mRNA in the worm10, functions in PA14 as a major regulator of nitrate assimilation. Nitrogen assimilation is an energetically expensive pathway19, which may explain why P. aeruginosa PA14 has evolved multiple regulatory components to tightly control its expression. NasS and NasT function in a partner-switching mechanism in response to intracellular inorganic nitrates. By binding to P11, NasT facilitates transcriptional anti-termination of the nitrogen assimilation operon, allowing PA14 to reduce inorganic nitrates into ammonium and, subsequently, glutamine and glutamate. This multifaceted approach to regulating nitrogen assimilation ensures a proper balance between energy conservation and growth in a specific environment. Interestingly, our results demonstrate that one such environment where nitrogen assimilation is important for bacterial growth is within the C. elegans body at later stages of infection. While it is impossible to rule out all secondary functions, our data suggest that P11’s roles in regulating nitrogen assimilation and bacterial growth are sufficient to explain the pathogenesis defect associated with the loss of P11. These data highlight the importance of considering environmental regulation and metabolism in the context of understanding bacterial fitness and pathogenesis during infections. In the future, assaying bacterial load and metabolism within animals may help to untangle the roles of other factors in contributing to either bacterial growth in that environment or specific virulence pathways.
In addition to shedding light on the role of P11 in bacteria, our findings provide insights into how C. elegans interact with bacteria in their environment. Previous studies examining molecules produced by P. aeruginosa and sensed by C. elegans identified several compounds that are repulsive to C. elegans and one molecule that is attractive, but without being able to eliminate production of these molecules from the bacteria the actual activity of these molecules remained unclear8,9,11,39. Excitingly, we identified the nitrogen assimilation product, ammonia, as the major attractant that causes C. elegans to prefer P. aeruginosa over E. coli. In addition to confirming that ammonia is produced when P. aeruginosa attracts C. elegans, we demonstrate that ammonia sensing is necessary for C. elegans’ preference for P. aeruginosa, as well as other Pseudomonas species that are naturally encountered by C. elegans, over E. coli. Specifically, disrupting ammonia gradients by either saturating the environment with excess ammonia or using P. aeruginosa mutants defective in ammonia production, inhibits C. elegans attraction. A recent study identified a second nitrite reductase gene, nirA, in P. aeruginosa PA0140. It is possible that the residual ammonia we observed to be produced by nirB mutants could be due to the activity of nirA24,39. Future work to untangle the conditions in which nirA expression occurs and its impact on P. aeruginosa ammonia production and C. elegans interactions could further illuminate the complex mechanisms by which P. aeruginosa and C. elegans interact.
Worms rely on sensory information to find vital resources like nitrogen-based compounds, and bacteria provide rich nutrient sources20,31. Pseudomonads comprise the largest subgroup of the bacterial genre within the C. elegans natural microbiome1–3. Many Pseudomonads produce ammonia but are not harmful to C. elegans, and many other non-pathogenic soil-based bacteria also possess nitrogen assimilation machinery3,21–23,41–44. Therefore, we hypothesize that the benefits of being attracted to nutrient sources signaled by ammonia production may generally outweigh the risk to C. elegans of becoming infected by a pathogen. If C. elegans does encounter a pathogen like P. aeruginosa, pathogenesis is regulated in a complex manner such that the bacteria could be present in a non-pathogenic state14–16. And even when P. aeruginosa is pathogenic, it often takes several days to kill C. elegans, during which time C. elegans can continue to produce offspring. Those offspring will have been taught to move away from P. aeruginosa due to the P11-mediated and other learned avoidance mechanisms10,12,45, giving them a chance to survive and find alternative food sources in the next generation. Future studies interrogating nitrogen assimilation and ammonia production in different bacteria and how they influence C. elegans chemotaxis will shed further light on the generality of these processes. For example, while our studies suggest that the pathogenesis defect associated with nitrogen assimilation mutants is due to the impact of these processes on bacterial growth, it remains possible that they also influence virulence pathways more directly.
Our work highlights the complexity of trans-kingdom signaling: the same bacterial pathways that are important for C. elegans to acquire essential nutrients appear to also be important for the growth of harmful bacteria within C. elegans. The fact that untrained C. elegans prefer PA14 suggests that in the short term, the need to find nutrients is prioritized over the risk of encountering a pathogen. However, C. elegans have also evolved mechanisms to avoid harmful bacteria in future generations after ingesting them10,12,25,35. Interestingly, in PA14 these behaviors are coupled in that the same sRNA that mediates transgenerationally inherited-learned avoidance of PA14 is also required for naive C. elegans attraction to PA14 by regulating ammonia production. In this manner, C. elegans may use chemotaxis as a simple form of an adaptive immune system that specifically learns to avoid harmful pathogens only once they are encountered. This system of combining ammonia sensing and sRNA-mediated adaptive avoidance enables C. elegans to identify food reservoirs and then avoid pathogens with specificity.
Methods
Worm strains
Worm strains were gifted by the Murphy lab, which received them from the C. elegans Genetics Center (CGC). N2, che-2(e1033) X, che-1(p672) I, odr-7(ky4) X, odr-1(n1936), ceh-36(ky646), and ceh-37(ok642). Hermaphrodites were used in all experiments.
Bacterial strains
OP50 was from the CGC. P. vranovensis GRb0427 and P. mendocina MSPm1 were from Dr. Buck Samuel and MSPm1 from the CGC. PA14 mutants obtained from a transposon library46. The following mutants were used: nirB::MAR2xT7, nasC::MAR2xT7, nasS::MAR2xT7, nasT::MAR2xT7. In the text, mutants are denoted by the Tn subscript.
General worm maintenance
Worm strains were maintained at 20 °C on Nematode Growth Media (NGM) ((3 g/L NaCl, 2.5 g/L Bacto-peptone, 17 g/L Bacto-agar in distilled water, with 1 mL/L cholesterol (5 mg/mL in ethanol), 1 mL/L 1 M CaCl2, 1 mL/L 1 M MgSO4, and 25 mL/L 1 M potassium phosphate buffer (pH 6.0) added to molten agar after autoclaving) plates with E. coli OP50 using standard methods.
General bacterial cultivation
OP50 and PA14 were cultured overnight in Luria Broth (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl in distilled water) shaking (250 rpm) at 37 °C.
Bacterial cultivation to assay nitrogen assimilation
PA14 strains were grown overnight in LB. 1 mL of overnight culture was washed 2× in minimal salts media (40 mM K2HPO4, 22 mM KH2PO4, 0.5 mM MgSO4, 10 mM FeSO4, pH 7.0) before back-diluting 1:100 in 200 µL minimal salts media supplemented with 20 mM sodium succinate for a carbon source and 10 mM potassium nitrate, ammonium chloride, ammonium sulfate, glutamine, and glutamate for nitrogen sources. OD600 readings were performed every 12 minutes for 16 hours in a plate reader.
Bacterial choice assays
Eggs from young hermaphrodites were obtained by bleaching and hatched on NGM plates. Worms were grown at 20 °C for 2 days to achieve synchronized L4 populations. Overnight bacterial cultures were diluted to optical density (OD600) = 1 and 25 µl were spotted on opposite sides of 60 mm NGM plates and incubated for 2 days at 25 °C. Plates were incubated at room temperature 1 hour before use, and 1 µl of 400 mM sodium azide was spotted on each bacteria spot to paralyze worms at first choice. L4’s were washed off of OP50 NGM plates with 1 mL M9 buffer and pelleted with gravity for 5 minutes. M9 supernatant was removed, and worms were washed again with 1 mL M9 to remove external OP50. After pelleting with gravity for 5 minutes, 5 µL of worms were spotted on choice assay plates (Fig. 1a), and worms were given 1 hour at room temperature to chemotax to bacteria before counted at first choice. ~30–100 worms were used per plate and 10 or 20 plate replicates total were used per condition.
Chemical choice assays
Method as described7. Briefly, 10 µL of 2.5 M volatile ammonia (Ammonium acetate or ammonium hydroxide) or 1 µL of 10% benzaldehyde were spotted on 100 mm NGM plates opposite of ddH2O control immediately before choice assay. L4’s were washed as described above and spotted in the center of the plate between the two choices. Worms were given 1 hour of chemotaxis to choice and paralyzed with sodium azide (1 µL of 400 mM sodium azide on each choice). For chemical choice assays in the presence of saturating levels of a different attractan, 1 µL spots (5) of 10% benzaldehyde were evenly placed on the lids of the plate. ~30–100 worms were used per plate, and 10 or 20 plate replicates total were used per condition.
Bacterial choice assays with saturating concentrations of volatile chemical: Bacterial choice assay plates and worm preparation were prepared as previously described. Immediately before worms were spotted on choice assay plate, 10 µL spots (5) of 2.5 M volatile ammonia (ammonium acetate or ammonium hydroxide) or 1 µL spots (5) of 10% benzaldehyde were placed on the lid of the plate. Worms were given 1 hour at room temperature to chemotax to bacteria and paralyzed at first choice with sodium azide. ~30–100 worms were used per plate and 10 plate replicates total were used per condition.
Naive worm avoidance assay
Method as described8. Briefly, 50 µL of overnight PA14 cultures were spotted on the center of NGM plates and incubated at 37 °C for 12 h. ~50 worms were placed in the center of the bacterial lawns before incubating at 25 °C. Worms were scored for positioning on or off the lawns every 4 h for 12 h. ~50–60 worms were used per plate, and 3 plate replicates were used per condition.
Bacterial RNA isolation
RNA was isolated from PA14 strains grown on NGM plates. Overnight PA14 cells grown in LB were backdiluted to OD600 = 1 and 500 µL of cells were spread on NGM plates, dried, and incubated for 2 days at 25 °C. Bacterial cells were isolated from NGM plates by applying 1 mL of M9 buffer and suspending them with a cell scraper. Cells were pelleted at 5000 × g for 10 min at 4 °C. After removing the supernatant, cells were suspended in 1 mL Trizol, vortexed, and incubated at 65 °C for 10 minutes. Samples were removed and brought to room temperature before adding 200 µL of chloroform, vortexing for 1 minute, and centrifuging at 12,000 × g for 10 minutes at 4 °C. The aqueous phase was inputted for RNA purification using mirVana RNA isolation kit according to manufacturer’s instructions for total RNA. RNA was stored at −80 °C.
DNase treatment
100 µg RNA per sample was treated with DNase according to the Invitrogen DNA-free kit manufacturer’s instructions to remove DNA contamination.
qRT-PCR
Two-Step qRT-PCR was performed. First, 500 ng of RNA was used as input for cDNA synthesis. Reverse transcription was performed using a LunaScript RT Supermix Kit according to manufacturer’s instructions. cDNA was diluted 1:10, and 3 µl of diluted cDNA was used as input for qPCR using Luna by NEB master mix for a 20 µL total reaction. 40 cycles were performed for PCR at an annealing temperature of 55 °C and extension at 68 °C for 20 seconds. For P11 detection, primer pair P11.1/2 was used, and nirB.1/2 for nirB detection. P11 and nirB expression normalized to 5S expression (primer pair 5S.1/2). Three biological replicates were used per condition.
5S.1: GAACCACCTGATCCCTTCCC
5S.2: TAGGAGCTTGACGATGACCT
P11.1: CGACAACAAAGCGCCTG
P11.2: GCTTGCCCTGGACCTG
nirB.1: GCGCAAGTTGGCACG
nirB.2: GCCGACCATTACCAGCTTG
RNA sequencing and Gene Ontology analysis
RNA samples were sent to SeqCenter for sequencing using the Illumina Stranded RNA library preparation with RiboZero plus rRNA depletion. Gene Enrichment analysis performed from the Gene Ontology data base47–49, GO Ontology database DOI: 10.5281/zenodo.10536401 Released 2024-01-17. Analysis was performed on significantly differentially expressed genes that were identified based on Log2(Fold Change) values ≥ 2 or ≤−2 with P adjusted values ≤ 0.05.
Ammonia production assay
200 µL NGM agar was aliquoted into each well of a flat bottom 96 well plate. Overnight PA14 strains were backdiluted to OD600 = 1. 25 µL of culture were spotted in wells and left to dry at room temperature before incubating at 25 °C for 2 days. Plates were equilibrated to room temperature for 15 min. Master mix of ammonia colorimetric assay was spotted on bacteria for 15 min, removed by pipetting, and centrifuged at 10,000 × g for 2 minutes to remove debris before fluorescent intensity measures (all according to Sigma-Aldrich Ammonia Assay Kit manufacturer’s instructions). CFU’s were quantified by resuspending bacteria grown in the same conditions in LB. Fluorescent intensity normalized to CFU to get final relative differences in ammonia production. 3 biological replicates were used per condition.
P11 mutants strain construction
P11 mutants were constructed by a two-step allelic exchange using plasmid pEXG2. ~500 bp upstream and downstream of the deletion were amplified from genomic DNA using primer pairs P11mut1-1/2 and P11mut1-3/4 for P11mut1 and P11mut2-1/2 and P11mut2-3/4 for P11mut2. Fragments were fused by overlap extension PCR and inserted by Gibson assembly into pEXG2 (PCR linearized with primers pEXG2-1/2). The pEXG2 plasmid was conjugated and integrated into PA14 genome from donor cells E. coli S17. Exconjugates were selected on gent 30 µg/mL and irg 100 µg/mL, and mutants were counterselected on sucrose 15%. Mutants were verified using Seq1/2 primers.
pEXG2-1: AATTAATTTCCACGGGTGCGCATG
pEXG2-2: CTTTACATTTATGCTTCCGGCTCGTA
P11mut1-1: CGCACCCGTGGAAATTAATTGCTATCCCTATGGCGAGATCG
P11mut1-2: TTCAGCATGCTTGCGGCTCGAGTTGGCAGGCGCTTTGTTGT
P11mut1-3: AACTCGAGCCGCAAGCATGCTGAACGTTTCCCGACCGAACGGG
P11mut1-4: CCGGAAGCATAAATGTAAGCTTGAGGCCGTTGGCC
P11mut2-1: CGCACCCGTGGAAATTAATTGCTATCCCTATGGCGAGATCG
P11mut2-2: TTCAGCATGCTTGCGGCTCGAGTTGACGCCTTTGTCCAGGTC
P11mut2-3: AACTCGAGCCGCAAGCATGCTGAAGACGCCTTTTTTGTTTGCGC
P11mut2-4: CCGGAAGCATAAATGTAAGCTTGAGGCCGTTGGCC
Seq1: GCTATCCCTATGGCGAGATCG
Seq2: GCTTGAGGCCGTTGGCC
Survival assay
Wild-type worms were maintained on E. coli OP50-coated (NGM) plates prior to experiments. For P. aeruginosa-coated plates, overnight cultures were diluted to OD600 = 1, spread onto NGM plates, incubated overnight at 37 °C, and equilibrated to 25 °C. 10 mM glutamine was supplemented into backdiluted cultures before plating on NGM. For virulence assays, synchronized L4 worms were transferred to P. aeruginosa plates. Worms were counted at time t = 0, 30, 40, 50, and 60 hours to assess viability and were declared dead if they were unresponsive to mechanical agitations. ~30–35 worms were used per plate, and five plates were used per condition. P values were calculated using a Mantel-Cox test comparing all conditions to WT PA14 (Prism 9).
CFU quantification inside worms
CFU’s of PA14 inside of worms were analyzed as described50,51. Briefly, PA14 was allowed to infect C. elegans for either 30 or 50 hours. At the relevant timepoints, external bacteria were eliminated by paralyzing worms in 25 mM levamisole to reduce internalization of additional bacteria, placed on NGM plates with 1 mg/mL carbenicillin and 1 mg/mL gentamycin for 15 min, and then moved to fresh NGM plates supplemented with same antibiotics for 30 min to kill external bacteria. Worms were then resuspended in 1 mL M9 and mechanically lysed to release internal bacteria. Lysates were serially diluted in M9 and plated on Pseudomonas isolation agar plates. Plates were incubated overnight at 37 °C before CFU quantification. Ten worms were used for each CFU quantification, with five replicates in total.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We appreciate the training received from Rachel Kaletsky, Jasmine Ashraf, and Renee Seto on work with C. elegans and Christopher Guan on qRT-PCR. Funding was provided in part by NIH (1R01AT011963-01 to Z.G. and C.T.M. and R21 AI168808-01 to C.M.) and CDC (75D30122C15113 to C.M.). We thank Dr. Cori Bargmann for helpful comments on the preprint. The opinions, findings, and conclusions are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
Author contributions
Conceptualization, methodology, writing–review & editing: Z.G., J.M., C.M., C.T.M. Validation, formal analysis, visualization, and data collection: J.M. Writing–original draft, supervision, and funding acquisition: Z.G., C.M.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
Metadata accession number GSE273751. Source data are provided with this paper.
Competing interests
Z.G. has a competing interest as founder of ArrePath, Inc. C.M. is a cofounder of Carver Biosciences, a startup company developing Cas13-based antivirals, and holds equity in Carver Biosciences. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52227-3.
References
- 1.Berg, M. et al. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J.10, 1998–2009 (2016). 10.1038/ismej.2015.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dirksen, P. et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol.14, 38 (2016). 10.1186/s12915-016-0258-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samuel, B. S., Rowedder, H., Braendle, C., Félix, M. A. & Ruvkun, G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl Acad. Sci. USA113, E3941–E3949 (2016). 10.1073/pnas.1607183113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bargmann, C. I., Hartwieg, E. & Robert Horvitz, H. Odorant-selective genes and neurons mediate olfaction in C elegans. Cell74, 515–527 (1993). 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- 5.Worthy, S. E. et al. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLoS One13, 7 (2018). 10.1371/journal.pone.0201158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werner, K. M., Perez, L. J., Ghosh, R., Semmelhack, M. F. & Bassler, B. L. Caenorhabditis elegans recognizes a bacterial quorum-sensing signal molecule through the AWCON neuron. J. Biol. Chem.289, 26566–26573 (2014). 10.1074/jbc.M114.573832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frøkjær-Jensen, C., Ailion, M. & Lockery, S. R. Ammonium-acetate is sensed by gustatory and olfactory neurons in Caenorhabditis elegans. PLoS One3, 6 (2008). 10.1371/journal.pone.0002467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prakash, D. et al. 1‐Undecene from Pseudomonas aeruginosa is an olfactory signal for flight‐or‐fight response in Caenorhabditis elegans. EMBO J.40, 13 (2021). 10.15252/embj.2020106938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De-Souza, E. A., Thompson, M. A. & Taylor, R. C. Olfactory chemosensation extends lifespan through TGF-β signaling and UPR activation. Nat. Aging3, 938–947 (2023). 10.1038/s43587-023-00467-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaletsky, R. et al. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature586, 445–451 (2020). 10.1038/s41586-020-2699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meisel, J. D., Panda, O., Mahanti, P., Schroeder, F. C. & Kim, D. H. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell159, 267–280 (2014). 10.1016/j.cell.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, Y., Lu, H. & Bargmann, C. I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature438, 179–184 (2005). 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
- 13.Ha, Hick et al. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron68, 1173–1186 (2010). 10.1016/j.neuron.2010.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siryaporn, A., Kuchma, S. L., O’Toole, G. A., Gitai, Z. & Ausubel, F. M. Surface attachment induces Pseudomonas aeruginosa virulence. Proc. Natl Acad. Sci. USA111, 16860–16865 (2014). 10.1073/pnas.1415712111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persat, A., Inclan, Y. F., Engel, J. N., Stone, H. A. & Gitai, Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA112, 7563–7568 (2015). 10.1073/pnas.1502025112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosso-Becerra, M. V. et al. Regulation of pseudomonas aeruginosa virulence factors by two novel RNA thermometers. Proc. Natl Acad. Sci. USA111, 15562–15567 (2014). 10.1073/pnas.1402536111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu, G. Amino acids: metabolism, functions, and nutrition. Amino Acids37, 1–17 (2009). 10.1007/s00726-009-0269-0 [DOI] [PubMed] [Google Scholar]
- 18.Gusarov, I. et al. Bacterial nitric oxide extends the lifespan of C. elegans. Cell152, 818–830 (2013). 10.1016/j.cell.2012.12.043 [DOI] [PubMed] [Google Scholar]
- 19.van Heeswijk, W. C., Westerhoff, H. V. & Boogerd, F. C. Nitrogen assimilation in escherichia coli: putting molecular data into a systems perspective. Microbiol. Mol. Biol. Rev.77, 628–695 (2013). 10.1128/MMBR.00025-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zečić, A., Dhondt, I. & Braeckman, B. P. The nutritional requirements of Caenorhabditis elegans. Genes Nutr.14, 15 (2019). 10.1186/s12263-019-0637-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, C. M., Macdonald, D. S. & Stanley, S. O. The mechanisms of nitrogen assimilation in pseudomonads. Antonie van. Leeuwenhoek39, 89–98 (1973). 10.1007/BF02578844 [DOI] [PubMed] [Google Scholar]
- 22.Betlach, M. R., Tiedje, J. M. & Firestone, R. B. Assimilatory nitrate uptake in pseudomonas fluorescens studied using nitrogen-13. Arch. Microbiol129, 2 (1981). 10.1007/BF00455349 [DOI] [PubMed] [Google Scholar]
- 23.Hervás, A. B., Canosa, I., Little, R., Dixon, R. & Santero, E. NtrC-dependent regulatory network for nitrogen assimilation in Pseudomonas putida. J. Bacteriol.191, 6123–6135 (2009). 10.1128/JB.00744-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romeo, A. et al. Transcriptional regulation of nitrate assimilation in Pseudomonas aeruginosa occurs via transcriptional antitermination within the nirBD-PA1779-cobA operon. Microbiology158, 1543–1552 (2012). 10.1099/mic.0.053850-0 [DOI] [PubMed] [Google Scholar]
- 25.Moore, R. S., Kaletsky, R. & Murphy, C. T. Piwi/PRG-1 argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell177, 1827–1841.e12 (2019). 10.1016/j.cell.2019.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bargmann, C. I. & Horvitz, H. R. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron7, 729–742 (1991). 10.1016/0896-6273(91)90276-6 [DOI] [PubMed] [Google Scholar]
- 27.Uchida, O., Nakano, H., Koga, M., Ohshima, Y. & The, C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development130, 1215–1224 (2003). 10.1242/dev.00341 [DOI] [PubMed] [Google Scholar]
- 28.Sengupta, P., Colbert, H. A. & Bargmann, C. I. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell79, 971–980 (1994). 10.1016/0092-8674(94)90028-0 [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara, M., Ishihara, T. & Katsura, I. A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development126, 4839–4848 (1999). 10.1242/dev.126.21.4839 [DOI] [PubMed] [Google Scholar]
- 30.Lanjuin, A., Vanhoven, M. K., Bargmann, C. I., Thompson, J. K. & Sengupta, P. Otx/Otd homeobox genes specify distinct sensory neuron identities in C. Elegans. Dev. Cell5, 4 (2003). 10.1016/S1534-5807(03)00293-4 [DOI] [PubMed] [Google Scholar]
- 31.Bargmann, C. I., Hartwieg, E. & Robert Horvitz, H. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell74, 515–527 (1993). 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- 32.Freney, J. R., Simpson, J. R. & Denmead, O. T. Ammonia volatilization. Ecol. Bull.291, 302 (1981). [Google Scholar]
- 33.Ghosh, D. D., Lee, D., Jin, X., Horvitz, H. R. & Nitabach, M. N. C. Elegans discriminates colors to guide foraging. Science371, 1059–1063 (2021). 10.1126/science.abd3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, A. J., Zuazo, C., Mellman, K., Chandra, R. & L’etoile, N. C. Elegans show preference for Pseudomonas mendocina (MSPm1) and proteus mirabilis (P. Mirabilis Sp?), and repulsion to pseudomonas lurida (MYb11); growth on Pseudomonas mendocina (MSPm1) increases attraction to 2-nonanone. MicroPubl. Biol.2022, 10.17912/micropub.biology.000535 (2022). [DOI] [PMC free article] [PubMed]
- 35.Sengupta, T. et al. A natural bacterial pathogen of C. elegans uses a small RNA to induce transgenerational inheritance of learned avoidance. PLoS Genet.20, 3 (2024). 10.1371/journal.pgen.1011178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Małecka, E. M., Strózecka, J., Sobańska, D. & Olejniczak, M. Structure of bacterial regulatory RNAS determines their performance in competition for the chaperone protein HFQ. Biochemistry54, 1157–1170 (2015). 10.1021/bi500741d [DOI] [PubMed] [Google Scholar]
- 37.Pichon, C. & Felden, B. Proteins that interact with bacterial small RNA regulators. FEMS Microbiol. Rev.31, 614–625 (2007). 10.1111/j.1574-6976.2007.00079.x [DOI] [PubMed] [Google Scholar]
- 38.Wilsont, K. S. & Hippel Von, P. H. Transcription termination at intrinsic terminators: the role of the RNA hairpin (Escherichia Coli/RNA polymerase/rho-independent termination). Biochemistry92, 19 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prakash, D., Siddiqui, R., Chalasani, S. H. & Singh, V. Pyrrole produced by Pseudomonas aeruginosa influences olfactory food choice of Caenorhabditis elegans. 10.1101/2022.01.27.477966 (2022).
- 40.Fenn, S. et al. NirA is an alternative nitrite reductase from pseudomonas aeruginosa with potential as an antivirulence target. mBio12, 2 (2021). 10.1128/mBio.00207-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramos, F., Blanco, G., Gutiérrez, J. C., Luque, F. & Tortolero, M. Identification of an operon involved in the assimilatory nitrate‐reducing system of Azotobacter vineiandii. Mol. Microbiol.8, 1145–1153 (1993). 10.1111/j.1365-2958.1993.tb01659.x [DOI] [PubMed] [Google Scholar]
- 42.Wu, Q. & Stewart, V. NasFED proteins mediate assimilatory nitrate and nitrite transport in klebsiella oxytoca (Pneumoniae) M5al. J. Bacteriol.180, 5 (1998). 10.1128/JB.180.5.1311-1322.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa, K.-I. et al. The NasB operon and NasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J. Bacteriol.177, 5 (1995). 10.1128/jb.177.5.1409-1413.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren, J., Bai, X., Liu, Y. & Huang, X. Simultaneous nitrification and aerobic denitrification by a novel isolated Ochrobactrum anthropi HND19. Bioresour. Technol.340, 125582 (2021). 10.1016/j.biortech.2021.125582 [DOI] [PubMed] [Google Scholar]
- 45.Filipowicz, A., Lalsiamthara, J. & Aballay, A. Trpm channels mediate learned pathogen avoidance following intestinal distention. Elife10, e65935 (2021). 10.7554/eLife.65935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liberati, N. T. et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl Acad. Sci. USA103, 2833–2839 (2006). 10.1073/pnas.0511100103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium*. Nat. Genet.25, 25–9 (2000). 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aleksander, S. A. et al. The gene ontology knowledgebase in 2023. Genetics224, iyad031 (2023). 10.1093/genetics/iyad031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, P. D. et al. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci.31, 8–22 (2022). 10.1002/pro.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker, A. C., Bhargava, R., Vaziriyan-Sani, A. S., Brust, A. S. & Czyz, D. M. Quantification of bacterial loads in Caenorhabditis elegans. Bio Protoc.12, e4291 (2022). 10.21769/BioProtoc.4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, Y., Zhi, L., Wu, Q., Jing, L. & Wang, D. NPR-9 regulates the innate immune response in Caenorhabditis elegans by antagonizing the activity of AIB interneurons. Cell Mol. Immunol.15, 27–37 (2018). 10.1038/cmi.2016.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson, P. Z. & Simon, A. E. RNAcanvas: interactive drawing and exploration of nucleic acid structures. Nucleic Acids Res.51, W501–W508 (2023). 10.1093/nar/gkad302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metadata accession number GSE273751. Source data are provided with this paper.