Abstract
The generation of new neurons at the hippocampal neurogenic niche, known as adult hippocampal neurogenesis (AHN), and its impairment, have been implicated in Alzheimer’s disease (AD). MicroRNA-132 (miR-132), the most consistently downregulated microRNA (miRNA) in AD, was recently identified as a potent regulator of AHN, exerting multilayered proneurogenic effects in adult neural stem cells (NSCs) and their progeny. Supplementing miR-132 in AD mouse brain restores AHN and relevant memory deficits, yet the exact mechanisms involved are still unknown. Here, we identify NACC2 as a novel miR-132 target implicated in both AHN and AD. miR-132 deficiency in mouse hippocampus induces Nacc2 expression and inflammatory signaling in adult NSCs. We show that miR-132-dependent regulation of NACC2 is involved in the initial stages of human NSC differentiation towards astrocytes and neurons. Later, NACC2 function in astrocytic maturation becomes uncoupled from miR-132. We demonstrate that NACC2 is present in reactive astrocytes surrounding amyloid plaques in mouse and human AD hippocampus, and that there is an anticorrelation between miR-132 and NACC2 levels in AD and upon induction of inflammation. Unraveling the molecular mechanisms by which miR-132 regulates neurogenesis and cellular reactivity in AD, will provide valuable insights towards its possible application as a therapeutic target.
Subject terms: Molecular biology, Neuroscience
Introduction
Alzheimer’s disease (AD) is the most common form of dementia, causing progressive decline in cognitive abilities1. Novel genetic and molecular insights have challenged the unidirectional linearity of the pathogenic cascade in AD, which was long thought to primarily consist of the accumulation of extracellular beta amyloid (Aβ) plaques and intracellular hyperphosphorylated TAU tangles2–4. The critical contribution of complex, intertwined cellular pathways going beyond amyloid and TAU pathologies to also involve -among others- astrogliosis, synaptic damage, neuronal loss and neuroinflammation, and differentially impacting distinct AD endophenotypes, is increasingly appreciated3–7. Currently, an effective disease-modifying treatment is still lacking8, partially attributable to the multifactorial nature of the disease. One of the cellular processes that has more recently been associated with AD is the generation of new neurons at the adult hippocampal neurogenic niche, also referred to as adult hippocampal neurogenesis (AHN). Even though the existence of AHN in humans is still under debate, there is evidence suggesting that AHN may be altered in the AD brain, in both rodents and humans9–12. Additionally, boosting AHN in mouse models of AD was shown to beneficially impact both pathology and cognition12,13.
MicroRNAs (miRNAs) are pleiotropic cellular regulators of gene expression, which can typically target multiple sensitive nodes of various molecular cascades deregulated in disease conditions in the AD brain4. Under physiological conditions, miRNAs in the central nervous system (CNS) control gene expression in various cell types and in a highly regulated time-, space-, and neuronal activity- dependent manner, via mRNA degradation and/or translational inhibition14–20.
This functional pleiotropy makes them attractive candidates to treat multifactorial diseases, such as AD, as they can simultaneously regulate numerous targets in diverse cell types4,21,22. microRNA-132 (miR-132), one of the most consistently downregulated microRNAs in AD22–25, was recently identified as a potent regulator of AHN, exerting multilayered proneurogenic effects in adult neural stem cells and their progeny12. Supplementing miR-132 in the brain of an AD mouse model could rescue AHN and relevant memory deficits12. Additionally, miR-132 has been linked to the amelioration of multiple other aspects of AD pathology, including amyloidosis, TAU pathology and cognitive deficits, highlighting its potential as a putative therapeutic target for AD12,23,26–32. Understanding the interplay between AD, AHN and miR-132 has implications for understanding disease progression and identifying potential therapeutic targets.
The goal of this study was to identify the molecular effectors of miR-132 regulation at the interface of AHN and AD. Employing a systematic meta-analysis of previously published datasets, we identify NACC2 as a putative miR-132 target implicated in both adult neurogenesis and AD. We show that NACC2 becomes upregulated in response to miR-132 knockdown, as expected for a miRNA target, both in radial glia-like cells (RGLs) isolated from the adult mouse dentate gyrus and in human neural stem cells (NSCs) in vitro. We report that NACC2 regulates cell cycle exit and neural stem cell differentiation under physiological conditions and is expressed in reactive astrocytes in the AD brain.
Results
In silico identification of putative miR-132 targets at the interface between adult neurogenesis and Alzheimer’s disease
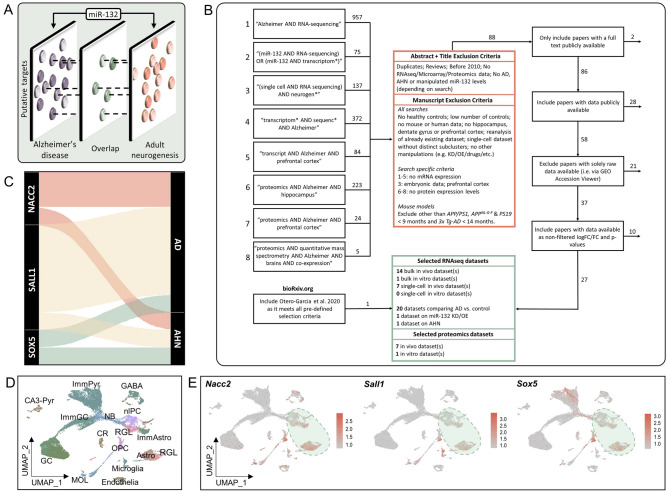
To screen for putative downstream molecular targets of miR-132 involved in both AHN and AD, we employed a systematic meta-analysis of previously published transcriptomic and proteomic datasets (Fig. 1A,B). We considered datasets containing information on differentially expressed genes or proteins in both human and mouse AD brain, a study involving experimental manipulation of miR-132 levels in mice27, as well as one single-cell RNA sequencing dataset with annotated marker genes of neurogenic populations in the adult mouse brain33 (Supplementary Table 1). We first constructed a list of genes showing anticorrelated expression levels to miR-132, i.e. upregulated in AD brain or upregulated in a miR-132 knockout mouse model. These were compared to genes previously identified at the adult hippocampal neurogenic niche in astrocytes, RGLs, neuronal intermediate progenitor cells and neuroblasts33. Lastly, this list was compared to genes predicted to be putative miR-132 targets by three miRNA prediction algorithms (Targetscan, DIANA MicroT-CDS and miRDB). This approach yielded three genes which were upregulated in AD, anticorrelated to experimental miR-132 manipulation, expressed in resident neurogenic cells in the adult hippocampus and predicted to be targeted by miR-132 (Supplementary Table 2). Consequently, we focused on these 3 predicted targets that were identified in more than one dataset and linked to miR-132, AD and AHN, namely SALL1, NACC2 and SOX5 (Fig. 1C).
Fig. 1.
Meta-analysis identifying putative miR-132 targets involved in both AD and AHN. (A) Meta-analysis strategy to identify targets of miR-132 involved in both AHN and AD. (B) Eight independent PubMed searches yielded a total of 1877 manuscripts, which were screened on whether papers fulfilled all the inclusion and exclusion criteria based on the abstract and title, and the manuscript details, followed by filtering based on dataset availability. A final selection of 27 articles with one or more proteomic or transcriptomic datasets each were included for further analysis. One extra article from bioRxiv.org was included meeting all criteria. Further information on the datasets and the corresponding articles can be found in Supplementary Table 1. (C) Alluvial diagram depicting three predicted miR-132 targets selected for further experimental validation and their links to AHN and AD as identified in the analyzed datasets. The weight of association reflects the number of datasets a gene was identified in. (D) UMAP visualization of 24.185 isolated mouse dentate gyrus single-cell transcriptomes33. Cells are colored by identified cell type. Immature pyramidal neurons, ImmPyr; CA3-pyramidal neurons, CA3-Pyr; Granule cells, GC; Immature granule cells, ImmGC; Neuroblasts, NB; Cajal-Retzius cells, CR; Myelinating oligodendrocytes, MOL; GABAergic neurons, GABA; Neural intermediate progenitor cells, nIPC; Radial glia-like cells, RGL; Oligodendrocyte precursor cells, OPC; Immature astrocytes, ImmAstro; Astrocytes, Astro. (E) UMAP visualizations indicating the expression of the three predicted miR-132 targets selected for further experimental validation on the dataset depicted in (D).
To better profile the expression of these predicted miR-132 targets in neurogenesis-relevant cellular populations in the adult brain, we visualized their levels in the single-cell RNA sequencing dataset from mouse dentate gyrus previously employed in our meta-analysis33. All three miR-132 predicted targets were highly expressed in astrocytes, adult neural stem cells (NSCs), also known as RGLs, and oligodendrocyte precursor cells (OPCs), confirming their expression in the neurogenic niche (Fig. 1D,E), whereas lower levels were overall observed in mature neuronal populations. Taken together, these data suggest that SALL1, NACC2 and SOX5 may be among the miR-132 targets mediating AD-relevant effects in neurogenic populations of the adult brain.
NACC2 is a putative target of miR-132 in adult neural stem cells in vivo
In order to validate SALL1, NACC2 and SOX5 as miR-132 targets in the adult neurogenic niche, we employed a nestin promoter-based green fluorescent protein (nestin-GFP) reporter mouse line34 (MGI:5523870, source indicated in Supplementary Table 3), in which GFP labels -among others- all the neurogenic cell types in the dentate gyrus, but not the mature neurons, thereby allowing for the enrichment of putatively rare populations, like the RGLs, upon Fluorescence-Activated Cell (FAC) sorting of GFP+ cells12. Mice were injected intracerebroventricularly with either a control or an antisense oligonucleotide against miR-132 (Ant-132) to mimic the deficiency of miR-132 that we and others previously observed in AD22–25. Nestin-GFP+ niche cells were isolated from the dentate gyri and single-cell cDNA libraries were prepared as discussed in Materials and Methods (Fig. 2A). A total of 723 nestin-GFP+ single niche cells were sequenced using the Smart-seq2 protocol. Unsupervised clustering of the dataset using the Seurat algorithm yielded 7 populations, which were projected on a Uniform Manifold Approximation and Projection (UMAP) map for visualization (Fig. 2B). Considering well-characterized marker genes33,35, we defined cluster identity (Fig. 2B), annotating the subpopulations as RGLs, neuronal precursor cells (NPCs), astrocytes, OPCs, myelin-forming oligodendrocytes (MFOLs), endothelial cells, and pericytes. The extent of miR-132 knockdown was assessed in nestin-GFP+ and nestin-GFP− sorted cells (Fig. 2C).
Fig. 2.
Single-cell transcriptomic profiling of the adult neurogenic niche to validate miR-132 predicted targets in vivo. (A) Schematic representation of the experimental procedure employed for the isolation and analysis of adult nestin-GFP+ niche cells derived from the mouse dentate gyrus upon miR-132 knockdown. n = 6 mice per group. (B) Unsupervised clustering of the dataset, defined cluster identity and normalized expression of cell type-specific markers. (C) Semi-quantitative real-time PCR of miR-132 levels in GFP+ and GFP- sorted cells from the dentate gyrus of nestin-GFP mice. n = 6 mice per group; Student’s t-test was used for statistical analysis. (D) Differential expression analysis between miR-132 knockdown and control group in astrocytes, NPCs and RGLs. Results for Nacc2, Sall1 and Sox5 are depicted. For p-value adjustments the Bonferroni correction was employed. The full list of differentially expressed genes can be found in Supplementary Table 4. (E and F) GOrilla biological pathways enriched among the downregulated (E) or upregulated (F) differentially expressed genes between miR-132 knockdown and control group within the RGL cluster, with color indicating significance.
Focusing on the neurogenesis-relevant populations, we noticed that within the astrocytic and NPC clusters, none of the top 3 predicted miR-132 targets exhibited a significant response upon miR-132 knockdown (Fig. 2D). However, in RGLs, Nacc2 was the only miR-132 predicted target significantly upregulated, as expected for a miRNA target, suggesting that under physiological conditions, miR-132 regulation over Nacc2 might be critical during earlier stages of stem cell activation, but less so upon further commitment of the RGL progeny towards neurons or astrocytes. Pathway enrichment analysis among the significantly downregulated genes upon miR-132 knockdown in the RGL cluster suggested that signaling related to cell proliferation and differentiation might be impeded by decreased levels of miR-132 in this cellular population, confirming earlier findings12. Surprisingly, miR-132 deficiency in RGLs induced molecular pathways related to the immune response, possibly indicating that miR-132 regulation is required for keeping the inflammatory signaling in check at the adult neurogenic niche, a regulatory layer that has been previously shown to be required for the neurogenic process36 (Fig. 2E,F).
Physiological expression of NACC2 along neuronal differentiation and in the neurogenic niche
To gain insight into the physiological context of the interaction between miR-132 and NACC2 during neurogenesis, we assessed the endogenous expression profiles of miR-132 and NACC2 upon human stem cell differentiation into neurons in vitro. We employed an immortalized human NSC and progenitor line (ReNcell VM), which gives rise to both astrocytes and neurons, and a human induced pluripotent stem cell (iPSC) line, that can be reprogrammed primarily towards neurons37 (Fig. 3A,B). In both cell lines, an upregulation of miR-132 levels upon induction of neuronal differentiation was observed (Fig. 3A,B). Differentiation efficiency of iPSCs to neurons was confirmed by downregulation of a pluripotency marker (OCT4) and upregulation of a mature neuronal marker (MAP2) (Supplementary Fig. 1). The differentiation efficiency of ReNcells was assessed by monitoring proliferation (MKI67), neuronal differentiation (MAP2) and astrocytic differentiation (AQP4). MAP2 expression already increased four days (D4) after induction of differentiation, whereas AQP4 significantly increased only from two weeks on (D14), suggesting that astrocytic differentiation is delayed compared to neuronal differentiation in ReNcells (Fig. 3C). Interestingly, NACC2 expression initially decreased upon induction of neuronal differentiation at D7, after which it became upregulated at D30 (Fig. 3D). This reverse association with miR-132 selectively during the early differentiation stages suggests that miR-132 may target NACC2 upon initial neuronal commitment of the NSCs, and that there may be decoupling of miR-132 regulation over NACC2 at later stages of neuronal differentiation and once astrogliogenesis commences. NACC2 expression was detected with immunostaining in non-differentiated NSCs, and in both neurons and astrocytes in more differentiated cultures (Fig. 3F). Upon neuronal differentiation of human iPSCs, NACC2 levels exhibited a consistent decrease all along the neurogenic trajectory, in consensus with previously published data38–47 and remained anticorrelated with miR-132 (Fig. 3E). Together, these results underscore a possible requirement for NACC2 expression to be -at least initially- kept under the control of miR-132 for neurogenesis -but not astrogliogenesis- to proceed.
Fig. 3.
In vitro and in vivo assessment of physiological NACC2 expression. (A, B) Semi-quantitative real-time PCR of miR-132 levels during neuronal differentiation of ReNcells (A) and human iPSCs (B). (A) n = 12 technical replicates; (B) n = 3 technical replicates. One-way ANOVA with Bonferroni correction was used for statistical analysis. (C, D) Semi-quantitative real-time PCR of MKI67, MAP2, AQP4 (C) and NACC2 (D) upon induction of differentiation of ReNcells. n = 4 technical replicates. One-way ANOVA with Bonferroni correction was used. (E) Semi-quantitative real-time PCR of NACC2 levels during neuronal differentiation of human iPSCs. n = 3 technical replicates. One-way ANOVA with Bonferroni correction was used for statistical analysis. (F) Immunolabeling of NACC2+, MAP2+ and GFAP+ cells in ReNcells at D0, D7 and D30 of neuronal differentiation. Scale bar, 50 μm. The green arrowhead indicates a NACC2+ neuron and the red arrow indicates a NACC2+ astrocyte. (G) Immunolabeling of NACC2+, GFP+ and GFAP+ cells in the dentate gyrus of nestin-GFP mice at 3 months and 9 months of age. Scale bar, 200 μm. Green arrowheads indicate NACC2- and nestin-GFP-positive cells, red arrowheads indicate NACC2- and GFAP-positive cells. Lower panels in 3- and 9-months groups are magnified inlets of the areas indicated in dashed-line boxes in the upper panels. Dashed line color corresponds to arrowhead color. MCL, molecular cell layer; GCL, granular cell layer; SGZ, subgranular zone.
To profile NACC2 expression in the microenvironment of the adult neurogenic niche in vivo, we performed NACC2 immunolabeling in the dentate gyrus of young (3-month-old) nestin-GFP mice. NACC2 was expressed in nestin-GFP+ RGLs (adult NSCs) in the subgranular layer of the dentate gyrus, and in astrocytes throughout the dentate gyrus (Fig. 3G). A similar overall expression pattern was observed in old (9-month-old) mice (Fig. 3G). NACC2 immunopositivity was also detected in neurons in the dentate gyrus of 9-month-old mice, albeit to a much lesser extent (Supplementary Fig. 2). Interestingly, fewer NACC2+/GFP+ cells were found in the subgranular layer of old animals, possibly due to a general trend for fewer GFP+ RGLs (Supplementary Fig. 3). In addition, a significant increase in NACC2+/GFAP+ cells was observed with age, after normalizing for the total number of GFAP+ astrocytes (Supplementary Fig. 3), suggesting increased astrocytic expression of NACC2 in old dentate gyrus. Considering its spatial and temporal physiological expression profile in vitro and in vivo, NACC2 may be part of different miR-132-dependent and -independent, cell-autonomous and non-cell-autonomous, cellular programs regulating the neurogenic process.
NACC2 is a functional miR-132 target regulating cell cycle exit and neural stem cell differentiation in human NSCs
To directly probe the regulatory relationship between miR-132 and NACC2 and its temporal determinants, we knocked down miR-132 in non-differentiated human NSCs (ReNcells) and in differentiated human iPSC-derived neurons using cholesterol-conjugated miR-132 antisense oligonucleotides (Fig. 4A,B). In ReNcells, expression levels of NACC2 significantly increased upon miR-132 knockdown, confirming NACC2 as a target of miR-132 in human NSCs (Fig. 4C). However, no change in NACC2 was detected upon miR-132 knockdown in differentiated iPSC-derived neurons and differentiated ReNcells (Fig. 4D, Supplementary Fig. 4), suggesting that NACC2 repression by miR-132 is essential for the neurogenic process before lineage commitment, in agreement with our miR-132 knockdown data in vivo (Fig. 2D).
Fig. 4.
Functional significance of NACC2 suppression by miR-132 during neurogenesis. (A, B) Schematic representation of neuronal differentiation and timing of miR-132 knock down in ReNcells (A) and human iPSCs (B). (C, D) Semi-quantitative real-time PCR of miR-132 levels and NACC2 levels upon miR-132 knockdown in ReNcells at D0 (C) and human iPSC-derived neurons at D47 (D). n = 4 independent experiments (C); n = 10 technical replicates from 2 independent experiments (D). Student’s t-test was used for statistical analysis. (E–J) Semi-quantitative real-time PCR of NACC2 (E), MKI67 (F), DCX (G), AQP4 (H), MDM2 (I) and CASP3 (J) levels upon NACC2 knockdown in ReNcells at D0. n = 5 independent experiments. Student’s t-test was used for statistical analysis.
To test the functional significance of NACC2 suppression by miR-132 during the initial stages of neurogenesis, a NACC2 gene silencing approach by means of siRNA transfection was used in ReNcells (Fig. 4E). NACC2 knockdown led to a significant decrease of NSC proliferation as measured by MKI67 (Fig. 4F). In addition, the astrocytic marker AQP4 and the early neuronal marker DCX were significantly upregulated (Fig. 4G,H), suggesting that NACC2 suppression may direct proliferating progenitors to exit the cycle and start differentiating.
Previous work has shown that NACC2 can affect apoptotic signaling by repressing the MDM2 pathway48. Given the involvement of apoptosis in the neurogenic process49, we next determined MDM2 levels and also measured caspase-3 (CASP3), since CASP3 is a key effector in the execution phase of apoptosis50. While MDM2 levels did not change in response to NACC2 knockdown (Fig. 4I), CASP3 was significantly downregulated (Fig. 4J), suggesting that the effects of NACC2 on cell proliferation and differentiation may be—at least partially- mediated by its impact on apoptosis in an MDM2-independent manner.
NACC2 may be involved in the response of astrocytes and NSCs to AD pathology
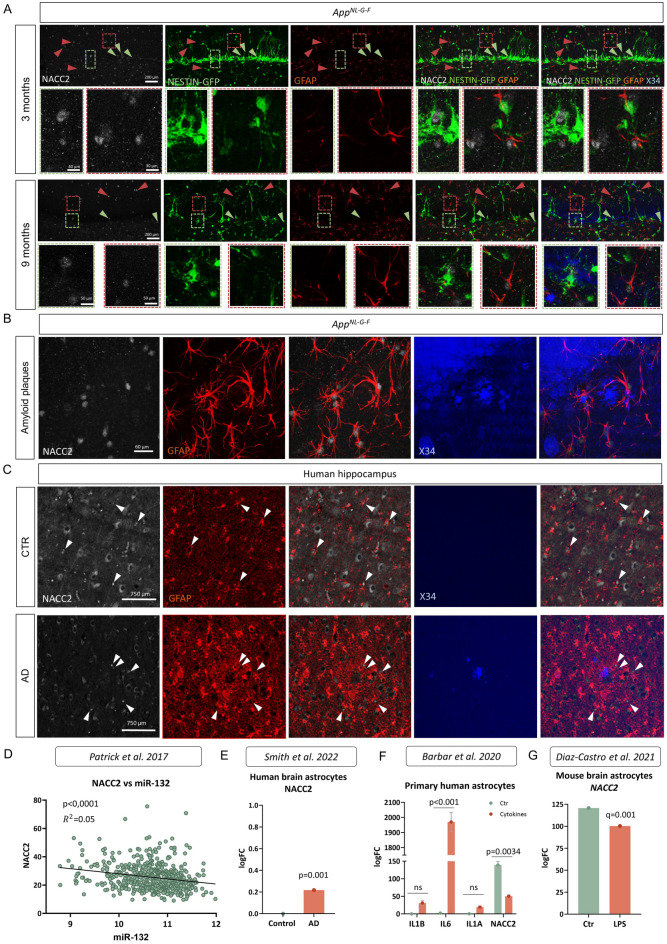
To assess whether our findings on the regulatory relationship between miR-132 and NACC2 could have any implications for AD pathology, we subsequently profiled NACC2 expression in the dentate gyrus of a knock-in amyloidosis mouse model (AppNL-G-F) (MGI:5637817, source indicated in Supplementary Table 3) crossed in-house to the nestin-GFP reporter line51. NACC2 immunostaining in young (3-month-old) and old (9-month-old) AppNL-G-F x nestin-GFP mice showed a similar expression profile to what we previously observed in wild-type nestin-GFP animals (Fig. 3G), with NACC2 immunoreactivity detected mainly in nestin-GFP+ RGLs and astrocytes, and to a lesser extent in neurons (Fig. 5A, Supplementary Fig. 2). Old AppNL-G-F x nestin-GFP mice harbored fewer GFP+ RGLs, as we previously reported12. Interestingly, while no change in total dentate gyrus Nacc2 levels between wild-type (MGI:2670020, source indicated in Supplementary Table 3) and AppNL-G-F mice was present (Supplementary Fig. 5), we observed NACC2 expression in reactive astrocytes surrounding amyloid beta (Aβ) plaques in old mice (Fig. 5B) and a (small) trend for increased numbers of NACC2+/GFP+ and NACC2+/GFAP+ cells after normalizing for total numbers of GFP+ RGLs and GFAP+ astrocytes (Supplementary Fig. 3). Next, we confirmed NACC2 expression in areas of astrogliosis around Aβ plaques also in human AD hippocampus (Fig. 5C). Correlation analysis using a previously published transcriptomic dataset revealed a significant anti-correlation between miR-132 and NACC2 levels in the brains of individuals participating in the ROSMAP longitudinal cohort study of aging, further suggesting a relationship between miR-132 and NACC2 in human brain52 (Fig. 5D).
Fig. 5.
NACC2 is expressed in the mouse and human AD brain and is linked to reactive astrogliosis and inflammation. (A) Immunolabeling of NACC2+, GFP+ and GFAP+ cells, and amyloid plaques (X34) in the dentate gyrus of AppNL-G-F x nestin-GFP mice at 3 months and 9 months of age. Scale bar, 200 μm. Green arrowheads indicate NACC2+ and GFP+ labeled cells, red arrowheads indicate NACC2 and GFAP-positive cells. Lower panels in 3- and 9-months groups are magnified inlets of the areas indicated in dashed-line boxes in the upper panels. Dashed line color corresponds to arrowhead color. (B) Immunolabeling of NACC2+ and GFAP+ cells around amyloid plaques (X34) in the dentate gyrus of 9-month-old AppNL-G-F x nestin-GFP. Scale bar, 60 μm. (C) Immunolabeling of NACC2+ and GFAP+ cells, and amyloid plaques (X34) in human healthy control and AD hippocampus. Scale bar, 500 μm. White arrows indicate NACC2+/GFAP+ labeled cells in control brain (upper panel) and around an amyloid plaque in AD brain (lower panel). (D) Re-plotting of data from Patrick et al.52 depicting an anti-correlation between miR-132 and NACC2 levels in the aging human brain. Simple linear regression analysis was used. (E) Re-plotting of data from Smith et al.53 showing significantly increased NACC2 expression in astrocytes in human AD brain. (F) Re-plotting of data from Barbar et al.54 depicting significantly decreased NACC2 expression in primary human astrocytes upon cytokine incubation. (G) Re-plotting of data from Diaz-Castro et al.55 showing significantly reduced Nacc2 expression in mouse brain astrocytes upon induction of inflammation with LPS.
Interestingly, a recently published single-nucleus RNA sequencing dataset in human AD brain also yielded a significant NACC2 upregulation53, specifically in astrocytes (Fig. 5E), suggesting that NACC2 may contribute to the neuroinflammatory response to Aβ accumulation in AD. Even though miR-132 is consistently downregulated in AD brain22–25 there are currently no data supporting a link between reduced miR-132 levels and increased NACC2 expression in Aβ-responsive astrocytes (with or without an association to the neurogenic niche). Yet, previously published data show an initial upregulation of NACC2 upon induction of inflammation in both human54 and mouse astrocytes55 (Fig. 5F,G), a reverse pattern than that observed for miR-132 under similar conditions56–58.
Discussion
Whether neurogenesis persists in the adult human brain has been a longstanding debate59–61, from the initial evidence identifying dividing cells in the dentate gyrus of cancer patients62, all the way to the recent -seemingly- conflicting results of single-nucleus RNA sequencing studies in adult human hippocampus9,63–66. In animal models, boosting AHN can modify AD pathology and ameliorate cognitive performance12,13. In humans, evidence is more scarce, with some studies suggesting reduced cell numbers and altered transcriptional programs in immature-like neuronal populations9–11. Even though evidence this far primarily stems from rodent studies, the mere addition of new neurons to the hippocampal network, and the contribution of the immature cells to intercellular signaling homeostasis at the adult neurogenic niche, or the combination thereof, could potentially be beneficial in the degenerating human brain59.
While miRNA alterations in the human brain are often difficult to replicate due to a series of technical and biological limitations67–69, miR-132 has been one of the few miRNAs that is consistently reported by several independent studies to be downregulated in AD22–25. miR-132 has emerged as a master regulator of neuronal function and a putative therapeutic target in AD, where its supplementation can ameliorate several aspects of pathology, including amyloidosis, TAU hyperphosphorylation and memory deficits12,23,26–32. We recently demonstrated that miR-132 can restore AHN in AD mouse models12, while it is also involved in the transition of microglia from a more activated to a more homeostatic cell state, repressing disease-associated microglial signatures, which have been functionally linked to AD pathology21,70–78. Understanding the miR-132-regulated molecular targets that mediate its effects on AHN in the AD brain can help profile the mechanisms and outcome of a putative miR-132-based therapeutic strategy in AD.
Our meta-analysis identified NACC2, SALL1 and SOX5 as putative miR-132 targets involved in both AHN and AD. However, only Nacc2 became upregulated upon miR-132 knockdown in an RGL-specific manner, suggesting a putative role of the miR-132-NACC2 interaction at initial stages of NSC activation and differentiation. In adult mouse hippocampal NSCs, Nacc2 has been previously shown to be bound by Ascl1, a transcription factor required for the exit of NSCs from quiescence in response to activating signals in the adult hippocampus74. Indeed, NACC2 expression did no longer anticorrelate to that of miR-132 once cells enter terminal differentiation stages in a human NSC line, while it exhibited a gradual downregulation profile along the differentiation trajectory from human iPSCs to early neurons, as opposed to miR-132 levels that kept increasing. The rather unexpected inverse expression pattern of NACC2 at the terminal differentiation state of NSCs may be attributable to a burst in astrogliogenesis, reflected by profoundly increased AQP4 levels (Fig. 3C), as NACC2 levels are highest expressed in astrocytes in both mouse and human brain75,76. The functional significance of the tight regulation of NACC2 by miR-132 during early neurogenesis was confirmed in cultured human NSCs, where NACC2 deficiency reduced cell proliferation and apoptosis, while it induced differentiation of these cells towards neurons and astrocytes. The ability of miR-132 to regulate glial progenitor fate choice had previously already been shown during embryogenesis77. Of note, we previously identified a set of putative miR-132 targets in adult RGLs after increasing the levels of miR-132 in mouse brain12. Yet, in the present work, among these previously identified transcripts, only Hdac3 was upregulated following miR-132 knockdown (Supplementary Table 4). The differential, context-dependent sensitivity of miRNA targets towards miRNA suppression has been well documented79,80. In particular, only a small fraction of target genes are actually repressed by a given miRNA under physiological conditions, often resulting in largely distinct sets of altered transcripts upon overexpression or knockdown of the same miRNA.
Interestingly, miR-132 knockdown triggered pathways related to inflammatory signaling in RGLs isolated from the adult mouse hippocampus, similarly to what we previously observed upon miR-132 overexpression12. Although still a relatively unexplored field of research, recent studies show that under homeostatic conditions, murine adult NSCs harbor intrinsic inflammatory properties and also receive inflammatory signals from the niche, which need to be proactively suppressed in order to enable their proliferative and differentiation capacity36,78. miR-132 has been shown to act as anti-inflammatory regulator in a variety of cellular populations, including astrocytes, peripheral macrophages and microglia, wherein inflammatory cues initially trigger miR-132 upregulation56–58. Interestingly, NACC2 expression shows a reverse profile, becoming repressed upon treatment of mouse and human astrocytes with inflammatory cytokines and lipopolysaccharide54,55, indicating that in these conditions miR-132 may be keeping NACC2 levels below a critical threshold to control inflammation.
Our earlier work showed that in human AD brain, miR-132 levels initially increase, similarly to other homeostatic, compensatory programs, like those including signatures of synaptic plasticity22. However, as disease progresses, miR-132 becomes significantly downregulated22. Of note, as opposed to miR-132, NACC2 is upregulated in late-stage AD brain52,53,81. A re-analysis of previously published data52 confirmed a linear relationship between miR-132 levels and NACC2 levels in individuals participating in two longitudinal cohort studies of aging. Here, we demonstrate that NACC2 is expressed in reactive astrocytes surrounding amyloid plaques in AD mouse and human brain. Our work did not address whether there is a direct link between NACC2 astrocytic expression and miR-132 in AD brain. Yet, aberrant inflammatory signalling due to NACC2 de-repression upon miR-132 deficiency in an AHN-independent manner, for instance in pathology-responsive astrocytes, could potentially be of functional relevance in AD.
Whether miR-132 acts as a compensatory regulator at initial stages of AD pathology also at the adult neurogenic niche, and if this involves NACC2 regulation, are still open questions. Our work provides a first proof-of-concept that NACC2 is a novel target of miR-132 with possible implications in both AHN and AD (Fig. 6). Further work is necessary to dissect the functional impact of this interaction in specific cellular populations, related to AHN or not, along the AD trajectory.
Fig. 6.
Schematic representation of the crosstalk between miR-132 and NACC2 along the neurogenic trajectory and in AD. Under physiological conditions, increasing miR-132 levels regulate early neurogenic decisions in NSCs involving effects on cell proliferation, apoptosis, neuronal differentiation and inflammatory signaling, by keeping NACC2 levels under control. In AD, miR-132 deficiency may lead to the de-repression of NACC2 and thereby contribute to cellular phenotypes in NSCs and astrocytes related to a response to amyloidosis.
Methods
Meta-analysis
A systematic search was performed in the PubMed database to collect all articles encompassing RNA sequencing (RNAseq) and proteomics studies related to AD, AHN or miR-132. Eight independent searches were conducted between 2021/01/25 to 2021/02/26 (Fig. 1B). All 1877 hits were collected and saved in Zotero (v5.0.95.1) and Microsoft Excel for further selection. The collected publications were elaborately screened based on inclusion and exclusion criteria defined prior to the search (Fig. 1B). This approach yielded a final list of 32 datasets from 28 different articles (Supplementary Table 1).
Genes were considered to be up- or downregulated when the fold-change (FC) was > 1.20 or < 0.80 (< − 1.20 for the dataset in Hansen et al. 2016) or the logFC was > 0.25 or < − 0.25. Only those genes with appropriate (log)FC values and an adjusted p-value (or false discovery rate (FDR)/q-value) below 0.05 were overlaid with the in silico predicted miR-132 targets. Overlapping genes were considered putative miR-132 targets relevant to AD pathology or miR-132 levels. The following exceptions were made: For the Mostafavi et al.82 dataset, genes were considered if they were positively correlated to neurofibrillary tangles and Aβ (> 1.00) and negatively to cognition score (< − 1.00). For the Bossers et al. (2010) dataset, genes were considered as miR-132 targets if they showed an anti-correlated expression pattern to miR-132 at all Braak stages as indicated by Lau et al.22,83. To determine if identified genes were relevant to AHN, targets were compared to genes enriched in early neurogenesis-associated cell types, as identified by Hochgerner et al.33. Overlapping genes were considered potential miR-132 targets relevant to AHN as well as AD and/or miR-132 levels (Supplementary Table 2).
Target prediction
In silico miR-132 target prediction was performed using three different miRNA target prediction tools: Targetscan (v7.2)84, DIANA MicroT-CDS (v5.0; Threshold at 0.7)85,86 and miRDB (v6.0)87,88. Targetscan predictions were set to include poorly conserved binding partners. Targets that were predicted by all three prediction tools were incorporated into the final selection. This approach aimed to ensure the inclusion of the most robust targets, while minimizing the risk of false positives. All predictions were done separately for mouse and human miR-132 (mmu-mir-132 and hsa-mir-132, respectively) targets.
Animals
Mice were bred according to standard laboratory approaches and housed under standard 12 h light–dark conditions. Food and water were provided ad libitum. Wild-type mice were, depending on the experiment, C57BL/6JRj (MGI:2670020, source indicated in Supplementary Table 3) and nestin-GFP (MGI:5523870, source indicated in Supplementary Table 3), while the AD mouse models used in the study included AppNL-G-F animals and a crossbred line between nestin-GFP mice and AppNL-G-F (MGI:5637817, source indicated in Supplementary Table 3). To visualize neural stem cells and neuronal progenitor cells in vivo and to isolate cells from the adult dentate gyrus for single-cell sequencing, we utilized nestin-GFP mice.
All animal experiments conducted in this study received approval from the ethical committees of KU Leuven (LA1210596), and the Royal Dutch Academy of Arts and Sciences (AVD80100202010904) and adhered to the European guidelines for the care and use of laboratory animals (Council Directive 86/6009/EEC). Additionally, the study is reported in accordance with the ARRIVE guidelines.
Human hippocampus
Postmortem hippocampal tissue from AD and healthy control donors was obtained from the Netherlands Brain Bank. The medical ethics committee of the VU Medical Center in Amsterdam, The Netherlands, has approved the procedure for brain donation to the Netherlands Brain Bank (NBB) and the use of clinical and pathological information for research, under approval number 2009/148. In accordance with national ethics guidelines, all donors had previously provided informed consent for brain autopsy and the utilization of their material and clinical data for research purposes. The certification of AD pathology was conducted by a qualified neuropathologist (Supplementary Table 5).
Intracerebroventricular (i.c.v.) injections
I.c.v. injections were performed as previously described89 using the following stereotactic coordinates: AP-0.1 mm, ML-1.0 mm, and DV-3.0 mm (from the skull). For miR-132 knockdown, 2-month-old nestin-GFP mice were infused with 2 ul of miR-132 antagomir [locked nucleic acid (LNA)-, cholesterol-modified oligonucleotide] (Exiqon, QIAGEN, Denmark) at 0.33 nmol/ml in artificial cerebrospinal fluid (aCSF) (Harvard Apparatus, USA). Control mice received a scrambled LNA oligonucleotide in aCSF. Analysis of antagomiR-132-injected animals was performed at 3 months of age. Randomization of injectates was employed for all injection sessions, and animals were randomly allocated to each treatment.
Adult dentate gyrus dissociation for fluorescence activated cell sorting (FACS) of nestin-GFP+ cells
Dentate gyri (each sample consisted of 1 full dentate gyrus; right and left hemisphere) were first dissected and minced in Hibernate A Low Fluorescence (BrainBits, IL, USA) on ice. Tissue dissection was carried out in 20U/ml papain and 50U/ml DNaseI (Worthington, NJ, USA) for 20 min at 37 °C. The enzymatic digestion was stopped by incubation in 10 mg/ml ovomucoid solution (Worthington, NJ, USA), cells were dissociated by trituration, filtered through a 70 mm nylon mesh, span down and eventually resuspended in ice-cold Hibernate A. Debris and myelin removal was performed using the adult brain dissociation kit (Miltenyi Biotec, Netherlands) according to the manufacturer’s instructions. Finally, cells were resuspended in ice-cold Hibernate A and dead cells were stained with propidium iodide (ThermoFisher Scientific, Belgium). GFP+ and GFP- cells were sorted in a FACS Aria III (BD Biosciences, CA, USA) in RNase-free tubes (in bulk) or in 96-well plates (in single) for downstream analysis.
Smart-seq2 processing and single-cell library preparation
For FACS isolation of single cells, four miR-132 antagomir-injected and four control-injected nestin-GFP male mice were used, and 96 cells were initially plated-sorted per mouse. FACS-sorted cells were processed using the Smart-seq2 protocol90. Briefly, RNA was reverse-transcribed using biotinylated oligo-dT primers (IDT DNA, Belgium) and biotinylated template-switching (TSO) oligonucleotides (Exiqon, QIAGEN, Denmark) and subsequently cDNA was pre-amplified using the KAPA HiFi Hot Start DNA Polymerase (KAPA Biosystems, Roche Diagnostics, Belgium) and biotinylated ISPCR primers (IDT DNA, Belgium). cDNA was purified using AMPURE XP Agencourt beads (Beckman Coulter, France), concentration was calculated with the Quantifluor dsDNA kit (Promega, Netherlands) and adjusted on an Echo 525 Liquid Handler (Labcyte, CA, USA). 100 pg cDNA per cell were used for library preparation with Nextera XT DNA Library Preparation Kit (Illumina, CA, USA). Finally, following purification on magnetic beads the libraries were multiplexed and sequenced using the Illumina NextSeq500 platform (Illumina, CA, USA).
Single-cell data processing
To confirm quality of the raw reads, a FastQC analysis (Version 0.11.5, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was performed. Reads were aligned to the mouse mm10 genome (Ensembl Version 88) using STAR (Version 2.5.2)91 with default options. A count table was generated from the alignments using Subread/FeatureCounts 1.5.192.
The count matrix was analyzed with Seurat 2.3.193, using the following workflow: Cells with less than 200 genes or more than 6000 genes (likely doublets) were removed, as well as cells whose mitochondrial gene expression accounted for more than 20% of the total gene expression of that cell. Genes found in less than 0.5% of cells were also excluded. Following this filtering, 723 of the original 768 cells remained. Data were log-normalized and scaled by a factor of 10,000. The normalized count data subsequently regressed on the number of reads. Principal component analysis (PCA) was performed using the top 2000 highly variable genes. The first 15 PCAs were used to identify clusters (using default parameters). Data was visualized using Uniform Manifold Approximation and Projection (UMAP). Cluster markers were determined by a differential expression analysis of each cluster versus all other clusters combined (Supplementary Table 6). Literature markers (Table S5 from Walgrave et al.12) were used for cell type annotation. Gene differential expression between experimental (miR-132 knockdown) and control conditions (within each cell population) was performed (Supplementary Table 4). Differential expression (for analysis of cluster markers and for analysis between controls and experimental conditions) was performed as follows: p-values were calculated using a Wilcoxon rank-sum test (using Seurat’s “FindMarkers” function) and corrected for multiple testing using a Bonferroni correction. Log fold changes were calculated using the natural logarithm. Genes with adjusted p-values of < 0.05 were considered significantly differentially expressed. Sequencing data reported in this study have been deposited and are publicly available at NCBI Gene Expression Omnibus (GEO Accession Number: GSE268144).
Human cell lines
ReNcell™ VM Human Neural Progenitor Cells (Millipore; RRID: CVCL_E921), an immortalized human neural stem cell and progenitor cell line, were cultured according to the manufacturer’s instructions in laminin-coated flasks (1–2 mg/ml; Sigma-Aldrich; Greiner Bio-one) at 37 °C under 95% O2 and 5% CO2. 20 ng/mL epidermal growth factor (EGF) and fibroblast growth factor 2 (FGF-2; both PeproTech) were supplemented to prevent differentiation. Undifferentiated ReNcells were passaged using 1 × Accutase (Sigma-Aldrich) every 2–3 days at > 70% confluency. To induce differentiation, growth factors were removed from the culture medium. ReNcells were collected in 1 ml TRIzol reagent (Invitrogen) and stored at − 80 °C or fixed with 4% PFA in PBS on coverslips for immunohistochemistry, depending on the purpose of the experiment.
Human induced pluripotent stem cells (iPSCs; iPSC EPITHELIAL-1, Sigma-Aldrich) were cultured in mTeSRTM1 (Stemcell Technologies) in (hESC-qualified) Matrigel-coated plates (Corning). iPSCs were differentiated into neurons according to a previously established protocol37. Briefly, cells were cultured in Neural Maintenance Medium (Fisher Scientific), supplemented with 10 µM SB431542 (Sigma-Aldrich) and 1 µM LDN-193189 (Miltenyi Biotech) at 37 °C under 95% O2 and 5% CO2 for ten days in order to induce differentiation into NPCs. Cells were passaged using 0.5 µM EDTA (Sigma-Aldrich) at > 70% confluency. Further maturation of NPCs into neurons was induced by culturing the cells in Neural Maintenance Medium supplemented with 20 ng/mL brain-derived neurotrophic factor (BDNF; Stemcell Technologies), 20 ng/mL glial cell line-derived neurotrophic factor (GDNF; Stemcell Technologies), 200 µM cAMP (Stemcell Technologies) and 200 µM ascorbic acid (Sigma-Aldrich) on 0.01% poly-L-ornithine (PLO; Sigma-Aldrich) and laminin coated coverslips at 37 °C under 95% O2 and 5% CO2.
Immunofluorescence in mouse brain sections and cultured cells
For brain immunofluorescence, 50 µm-thick vibratome-prepared coronal tissue sections were initially permeabilized in 1% (v/v) Triton X-100, followed by 2 h blocking in 1% (v/v) Triton X-100, 10% (v/v) normal goat serum in PBS at room temperature (RT). Primary antibody incubation was performed in 0.3% (v/v) Triton X-100, 3% (v/v) normal goat serum overnight at 4 °C in a humidified chamber, and incubation in secondary antibody for 2 h at room temperature. To label Aβ plaques, sections were incubated for 20 min at RT in 20 μM X-34 (Sigma-Aldrich) dissolved in 60% PBS/40% ethanol (pH 10). Finally, sections were mounted in Mowiol. All the antibodies used for immunolabeling of brain tissue are listed in Supplementary Table 7.
For immunofluorescent labeling of cells cultured on coverslips, samples were initially permeabilized and blocked as described before. Primary and secondary antibody incubation was carried out in modified blocking solution for 4 °C overnight and 2 h at RT, respectively. Finally, sections were incubated in DAPI (1:5000, Sigma-Aldrich) and mounted in Mowiol. All the antibodies used for immunolabeling of cultured cells are listed in Supplementary Table 7.
miR-132 antisense oligonucleotide treatment of human iPSC-derived neurons and ReNcells
iPSC-derived neurons (D47) were treated with a synthetic miR-132 antisense oligonucleotide (hsa-miR-132-3p Inhibitor, Qiagen) or corresponding scramble control oligonucleotide at 1500 nM and collected in Qiazol after 48 h of treatment for RNA extraction. Undifferentiated ReNcells (D0) were treated with the same oligonucleotides at 250 nM at D0 and cultured in medium without EGF and FGF until collection. For differentiated ReNcells, cells were cultured in medium without EGF and FGF for a week and then treated with the same oligonucleotides at 1000 nM at D7. For both ReNcell experiments, half of the medium was replaced 24 h after the treatment. The medium was completely renewed after 48 h. After 1 week of treatment, the cells were collected in Qiazol for RNA extraction.
ReNcell transfections for NACC2 knockdown
ReNcells were cultured according to standard protocols in Neural Maintenance Medium, with growth factors (EGF and FGF-2) and without penicillin–streptomycin, following the manufacturer’s guidelines. Transfection was performed when cell confluency reached approximately 50–60%. Cells were transfected with either ON-TARGET plus NACC2 siRNA SMART pool or ON-TARGET plus Non-targeting pool (Horizon). Transfections were carried out using DharmaFECT 1 Transfection Reagent (1:100 dilution, Horizon) and Opti-MEM™ Reduced Serum Medium (Gibco). Transfections were performed with a final siRNA concentration of 25 nM. The culture medium was replaced after 24 h, and transfected ReNcells were collected in Trizol two days post-transfection.
RNA extraction, reverse transcription and semi-quantitative real-time PCR
RNA extraction from mouse dentate gyrus and ReNcells was performed using the miRVana Paris Kit (Life Technologies) following the manufacturer’s guidelines. Tissue samples were snap-frozen and stored at -80 °C, while cells were harvested in 1 ml TRIzol (Invitrogen) and stored at -80 °C prior to isolation. Briefly, the tissue samples were homogenized in disruption buffer including phosphatase inhibitors and the cells were homogenized in TRIzol, followed by a 5-min incubation in chloroform at RT. After centrifugation for 1 min at 12,000 g, the aqueous phase was collected, and 1.25 volumes of 100% ethanol were added. Subsequently, the samples were loaded onto miRVana spin columns and processed according to the manufacturer’s protocol. The RNA was ultimately eluted in 30 µl of Elution Solution, and its concentration was determined using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc.). For the iPSCs and iPSC-derived neurons, RNA was extracted using the miRNeasy Micro Kit (Qiagen) following manufacturer’s instructions and eluted in 14 µl Nuclease Free Water.
Reverse transcription of miRNAs was performed using 100 ng of RNA and the miRCURY LNA RT Kit (QIAGEN). In the case of mRNAs, 200 ng of total RNA was used to synthesize cDNA using Superscript II reverse transcriptase (Thermo Fisher Scientific).
For miRNAs, real-time semi-quantitative PCR was performed using the Sybr Green mastermix (QIAGEN) along with miRCURY LNA primers (QIAGEN). Normalization was achieved by calculating the mean expression of RNU6 snRNA (QIAGEN) and RNU5G (QIAGEN) for the mouse samples, while RNU6 and RNU5G (QIAGEN) or SNORD48 (QIAGEN) were used for the cells. Analysis of protein-coding transcripts involved the SYBR™ PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific), with GAPDH, UBC and PSMB4 as housekeeping genes. Primer sequences for mRNA-derived cDNA are available in Supplementary Table 8. Ct values were determined using the second derivative method, and subsequently fold changes were calculated using the 2−ΔΔCT method.
Immunostaining of human hippocampal sections
For human hippocampus, coronal paraffin sections of 6 µm-thickness were initially dewaxed with xylene and rehydrated in a series of ethanol solutions of decreasing concentration (100% to 50%). Antigen retrieval was performed in boiling sodium citrate 10 mM, pH 6, followed by permeabilization of the tissue in 1% (v/v) Triton X-100, and by 1 h blocking in 5% BSA in 1% (v/v) Triton X-100 at RT. Primary antibody and secondary antibody incubation were performed in 1% BSA in 1% (v/v) Triton X-100 overnight at 4 °C and 1 h at RT, respectively. To stain for Aβ plaques, sections were incubated for 20 min at RT in 20 μM X-34 (Sigma-Aldrich) dissolved in 60% PBS 40% ethanol (pH 10). Finally, sections were mounted in Mowiol. All the antibodies used for immunolabeling of brain tissue are listed in Supplementary Table 7.
Image acquisition and processing
For immunolabelled mouse brain sections, images (z-stacks) were acquired using a DMI6000 CS Confocal microscope (Leica). The FIJI software was employed for image processing and quantification. For multi-channel confocal images, distinct channels were overlaid using the ‘Merge Channel’ function in FIJI.
Quantification and statistical analysis
Statistical analysis was performed as indicated in the figure legends. Significance was set at (adjusted) p-value threshold of 0.05. Graphs show the mean and standard deviation.
For quantification of NACC2+, GFP+ and GFAP+ cells in the mouse vibratome sections, three different mice per condition and three different sections of the dentate gyrus per mouse were used. Cells were manually counted per section in the subgranular zone (the distance below the granular cell layer and into the subgranular zone that was considered for cell counting was calculated as one third of the breadth of the granular cell layer in each image) with FIJI and average values per mouse were calculated by averaging the values of all sections of the same animal. The normalized NACC2+/GFAP+ and NACC2+/GFP+ values were calculated by dividing the number of double positive cells by the total number of GFP+ or GFAP+ cells.
Supplementary Information
Acknowledgements
We thank Bart De Strooper for providing the resources for the animal experiments and the single-cell transcriptomics; Véronique Hendrickx and Jonas Verwaeren for animal husbandry; Katrien Horré, An Snellinx, Elke Vanden Eynde and Tom Jaspers, the VIB Nucleomics Core of Leuven and the VIB-KU Leuven FACS Core for providing technical assistance and resources; the AppNL-G-F mice were kindly provided by Takaomi Saido (RIKEN Brain Science Institute, Japan), and the nestin-GFP mice were from Grigori Enikolopov (Stony Brook University, New York). This work was supported by funding from Health-Holland (LSHM20095), Alzheimer Nederland (WE.03-2020-04), and the Friends Foundation of the Netherlands Institute of Neuroscience (Stichting Vrienden van het Herseninstituut), which was made possible by Stichting Aan Boord and Stichting Burgland. The Dutch national e-infrastructure with the support of the Dutch Research Council (Nederlandse Organisatie voor Wetenschappelijk Onderzoek, NWO) and the SURF Cooperative (EINF-1405 and EINF-4880) was used. We thank the brain donors and their families for their commitment to the Netherlands Brain Bank donor program.
Abbreviations
- AD
Alzheimer’s disease
- Aβ
Beta amyloid
- AHN
Adult hippocampal neurogenesis
- miRNA
MicroRNA
- mRNA
Messenger RNA
- miR-132
MicroRNA-132
- RGLs
Radial glia-like cells
- NSCs
Neural stem cells
- OPCs
Oligodendrocyte precursor cells
- MFOL
Myelin forming oligodendrocytes
Author contributions
A.P., S.S., O.G. and E.S. contributed to the conception and design of the work; A.P., S.S., O.G., G.T., A.H., F.D.B., J.V.H., L.Z., N.T., K.C., M.F. and E.S. contributed to the acquisition, analysis, and interpretation of data; A.P., S.S., G.T. and E.S. have drafted the manuscript and all authors have revised it.
Data availability
Single-cell RNA-seq data have been deposited and are publicly available at NCBI Gene Expression Omnibus (GEO: GSE268144). Any additional information required to re-analyze the data reported in this paper is available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Amber Penning and Sarah Snoeck.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72096-6.
References
- 1.De Strooper, B. & Karran, E. The cellular phase of Alzheimer’s disease. Cell164, 603–615 (2016). 10.1016/j.cell.2015.12.056 [DOI] [PubMed] [Google Scholar]
- 2.Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science297, 353–356 (2002). 10.1126/science.1072994 [DOI] [PubMed] [Google Scholar]
- 3.Karran, E. & De Strooper, B. The amyloid cascade hypothesis: Are we poised for success or failure?. J. Neurochem.139(Suppl), 237–252 (2016). 10.1111/jnc.13632 [DOI] [PubMed] [Google Scholar]
- 4.Walgrave, H., Zhou, L., De Strooper, B. & Salta, E. The promise of microRNA-based therapies in Alzheimer’s disease: Challenges and perspectives. Mol. Neurodegener.16, 1–16 (2021). 10.1186/s13024-021-00496-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efthymiou, A. G. & Goate, A. M. Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener.12, 1–12 (2017). 10.1186/s13024-017-0184-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet.51, 414–430 (2019). 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sierksma, A., Escott-Price, V. & De Strooper, B. Translating genetic risk of Alzheimer’s disease into mechanistic insight and drug targets. Science370, 61–66 (2020). 10.1126/science.abb8575 [DOI] [PubMed] [Google Scholar]
- 8.Peng, Y. et al. Current and future therapeutic strategies for Alzheimer’s disease: An overview of drug development bottlenecks. Front. Aging Neurosci.15, 1206572 (2023). 10.3389/fnagi.2023.1206572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou, Y. et al. Molecular landscapes of human hippocampal immature neurons across lifespan. Nature607, 527–533 (2022). 10.1038/s41586-022-04912-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, Y. et al. Reduced neurogenesis in human hippocampus with Alzheimer’s disease. Brain Pathol.10.1111/BPA.13225 (2023). 10.1111/BPA.13225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreno-Jiménez, E. P. et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med.25, 554–560 (2019). 10.1038/s41591-019-0375-9 [DOI] [PubMed] [Google Scholar]
- 12.Walgrave, H. et al. Restoring miR-132 expression rescues adult hippocampal neurogenesis and memory deficits in Alzheimer’s disease. Cell Stem Cell28, 1805-1821.e8 (2021). 10.1016/j.stem.2021.05.001 [DOI] [PubMed] [Google Scholar]
- 13.Choi, S. H. et al. Combined adult neurogenesis and BDNF mimic exercise effects on cognition in an Alzheimer’s mouse model. Science361, eaan8821 (2018). 10.1126/science.aan8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell136, 215–233 (2009). 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonas, S. & Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet.16, 421–433 (2015). 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 16.Bak, M. et al. MicroRNA expression in the adult mouse central nervous system. RNA14, 432–444 (2008). 10.1261/rna.783108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landgraf, P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell129, 1401–1414 (2007). 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olsen, L., Klausen, M., Helboe, L., Nielsen, F. C. & Werge, T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS One4, e7225 (2009). 10.1371/journal.pone.0007225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barca-Mayo, O. & De Pietri Tonelli, D. Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci.71, 2975–2995 (2014). 10.1007/s00018-014-1576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krichevsky, A. M., King, K. S., Donahue, C. P., Khrapko, K. & Kosik, K. S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA9, 1274–1281 (2003). 10.1261/rna.5980303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walgrave, H. et al. microRNA-132 regulates gene expression programs involved in microglial homeostasis. iScience26, 106829 (2023). 10.1016/j.isci.2023.106829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, P. et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med.5, 1613–1634 (2013). 10.1002/emmm.201201974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salta, E. & De Strooper, B. microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer’s disease. FASEB J.31, 424–433 (2017). 10.1096/fj.201601308 [DOI] [PubMed] [Google Scholar]
- 24.Herrera-Espejo, S., Santos-Zorrozua, B., Álvarez-González, P., Lopez-Lopez, E. & Garcia-Orad, Á. A systematic review of MicroRNA expression as biomarker of late-onset Alzheimer’s disease. Mol. Neurobiol.56, 8376–8391 (2019). 10.1007/s12035-019-01676-9 [DOI] [PubMed] [Google Scholar]
- 25.Pichler, S. et al. The miRNome of Alzheimer’s disease: Consistent downregulation of the miR-132/212 cluster. Neurobiol. Aging50(167), e1-167.e10 (2017). [DOI] [PubMed] [Google Scholar]
- 26.El Fatimy, R. et al. MicroRNA-132 provides neuroprotection for tauopathies via multiple signaling pathways. Acta Neuropathol.136, 537–555 (2018). 10.1007/s00401-018-1880-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen, K. F. et al. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn. Mem.23, 61–71 (2016). 10.1101/lm.039578.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez-Rapp, J. et al. Memory formation and retention are affected in adult miR-132/212 knockout mice. Behav. Brain Res.287, 15–26 (2015). 10.1016/j.bbr.2015.03.032 [DOI] [PubMed] [Google Scholar]
- 29.Salta, E., Sierksma, A., Vanden Eynden, E. & De Strooper, B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med.8, 1005–1018 (2016). 10.15252/emmm.201606520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, P. Y. et al. miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum. Mol. Genet.24, 6721–6735 (2015). 10.1093/hmg/ddv377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei, Z. et al. Environmental enrichment prevents Aβ oligomer-induced synaptic dysfunction through mirna-132 and hdac3 signaling pathways. Neurobiol. Dis.134, 104617 (2020). 10.1016/j.nbd.2019.104617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, H. K. A. et al. De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer’s disease. Hum. Mol. Genet.22, 3077–3092 (2013). 10.1093/hmg/ddt164 [DOI] [PubMed] [Google Scholar]
- 33.Hochgerner, H., Zeisel, A., Lönnerberg, P. & Linnarsson, S. Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci.21, 290–299 (2018). 10.1038/s41593-017-0056-2 [DOI] [PubMed] [Google Scholar]
- 34.Mignone, J. L., Kukekov, V., Chiang, A. S., Steindler, D. & Enikolopov, G. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol.469, 311–324 (2004). 10.1002/cne.10964 [DOI] [PubMed] [Google Scholar]
- 35.Artegiani, B. et al. A single-cell RNA sequencing study reveals cellular and molecular dynamics of the hippocampal neurogenic niche. Cell Rep.21, 3271–3284 (2017). 10.1016/j.celrep.2017.11.050 [DOI] [PubMed] [Google Scholar]
- 36.Shariq, M. et al. Adult neural stem cells have latent inflammatory potential that is kept suppressed by Tcf4 to facilitate adult neurogenesis. Sci. Adv.7, eabf5606 (2021). 10.1126/sciadv.abf5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, Y., Kirwan, P. & Livesey, F. J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc.7, 1836–1846 (2012). 10.1038/nprot.2012.116 [DOI] [PubMed] [Google Scholar]
- 38.Sousa, A. M. M. et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science358, 1027–1032 (2017). 10.1126/science.aan3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Close, J. L. et al. Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron93, 1035-1048.e5 (2017). 10.1016/j.neuron.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Leemput, J. et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron83, 51–68 (2014). 10.1016/j.neuron.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 41.Darmanis, S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA112, 7285–7290 (2015). 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, M. et al. Integrative transcriptome network analysis of iPSC-derived neurons from schizophrenia and schizoaffective disorder patients with 22q11.2 deletion. BMC Syst. Biol.10, 1–20 (2016). 10.1186/s12918-016-0366-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsankov, A. M. et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol.33, 1182–1192 (2015). 10.1038/nbt.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sloan, S. A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron95, 779-790.e6 (2017). 10.1016/j.neuron.2017.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song, Y. et al. Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Mol. Cell67, 148-161.e5 (2017). 10.1016/j.molcel.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tekin, H. et al. Effects of 3D culturing conditions on the transcriptomic profile of stem-cell-derived neurons. Nat. Biomed. Eng.2, 540–554 (2018). 10.1038/s41551-018-0219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volpato, V. et al. Reproducibility of molecular phenotypes after long-term differentiation to human iPSC-derived neurons: A multi-site omics study. Stem Cell Rep.11, 897–911 (2018). 10.1016/j.stemcr.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xuan, C. et al. RBB, a novel transcription repressor, represses the transcription of HDM2 oncogene. Oncogene32, 3711–3721 (2013). 10.1038/onc.2012.386 [DOI] [PubMed] [Google Scholar]
- 49.Sierra, A. et al. Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell7, 483–495 (2010). 10.1016/j.stem.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Porter, A. G. & Jänicke, R. U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ.6, 99–104 (1999). 10.1038/sj.cdd.4400476 [DOI] [PubMed] [Google Scholar]
- 51.Saito, T. et al. Single App knock-in mouse models of Alzheimer’s disease. Nat. Neurosci.17, 661–663 (2014). 10.1038/nn.3697 [DOI] [PubMed] [Google Scholar]
- 52.Patrick, E. et al. Dissecting the role of non-coding RNAs in the accumulation of amyloid and tau neuropathologies in Alzheimer’s disease. Mol. Neurodegener.12, 1–13 (2017). 10.1186/s13024-017-0191-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, A. M. et al. Diverse human astrocyte and microglial transcriptional responses to Alzheimer’s pathology. Acta Neuropathol.143, 75–91 (2022). 10.1007/s00401-021-02372-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbar, L. et al. CD49f is a novel marker of functional and reactive human iPSC-derived astrocytes. Neuron107, 436-453.e12 (2020). 10.1016/j.neuron.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diaz-Castro, B., Bernstein, A. M., Coppola, G., Sofroniew, M. V. & Khakh, B. S. Molecular and functional properties of cortical astrocytes during peripherally induced neuroinflammation. Cell Rep.36, 109508 (2021). 10.1016/j.celrep.2021.109508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korotkov, A. et al. microRNA-132 is overexpressed in glia in temporal lobe epilepsy and reduces the expression of pro-epileptogenic factors in human cultured astrocytes. Glia68, 60–75 (2020). 10.1002/glia.23700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu, F. et al. miR-132 inhibits lipopolysaccharide-induced inflammation in alveolar macrophages by the cholinergic anti-inflammatory pathway. Exp. Lung Res.41, 261–269 (2015). 10.3109/01902148.2015.1004206 [DOI] [PubMed] [Google Scholar]
- 58.Gong, X., Huang, M. & Chen, L. Mechanism of miR-132–3p promoting neuroinflammation and dopaminergic neurodegeneration in Parkinson’s Disease. eNeuro10.1523/ENEURO.0393-21.2021 (2022). 10.1523/ENEURO.0393-21.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salta, E. et al. Adult hippocampal neurogenesis in Alzheimer’s disease: A roadmap to clinical relevance. Cell Stem Cell30, 120–136 (2023). 10.1016/j.stem.2023.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kempermann, G. et al. Human adult neurogenesis: Evidence and remaining questions. Cell Stem Cell23, 25–30 (2018). 10.1016/j.stem.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paredes, M. F. et al. Does adult neurogenesis persist in the human hippocampus?. Cell Stem Cell23, 780 (2018). 10.1016/j.stem.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eriksson, P. S. et al. Neurogenesis in the adult human hippocampus. Nat. Med.4, 1313–1317 (1998). 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- 63.Xu, C. et al. Automatic cell-type harmonization and integration across human cell atlas datasets. Cell186, 5876-5891.e20 (2023). 10.1016/j.cell.2023.11.026 [DOI] [PubMed] [Google Scholar]
- 64.Tosoni, G. et al. Mapping human adult hippocampal neurogenesis with single-cell transcriptomics: Reconciling controversy or fueling the debate?. Neuron111, 1714-1731.e3 (2023). 10.1016/j.neuron.2023.03.010 [DOI] [PubMed] [Google Scholar]
- 65.Franjic, D. et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron110, 452-469.e14 (2022). 10.1016/j.neuron.2021.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, W. et al. Transcriptome dynamics of hippocampal neurogenesis in macaques across the lifespan and aged humans. Cell Res.32, 729–743 (2022). 10.1038/s41422-022-00678-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linsen, S. E., Tops, B. B. & Cuppen, E. miRNAs: Small changes, widespread effects. Cell Res.1812(18), 1157–1159 (2008). 10.1038/cr.2008.311 [DOI] [PubMed] [Google Scholar]
- 68.Benesova, S., Kubista, M. & Valihrach, L. Small RNA-sequencing: Approaches and considerations for miRNA analysis. Diagnostics11, 964 (2021). 10.3390/diagnostics11060964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Brien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol.9, 402 (2018). 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell169, 1276-1290.e17 (2017). 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 71.Sala Frigerio, C. et al. The major risk factors for Alzheimer’s disease: Age, sex, and genes modulate the microglia response to Aβ plaques. Cell Rep.27, 1293–1306 (2019). 10.1016/j.celrep.2019.03.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Silvin, A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity55, 1448-1465.e6 (2022). 10.1016/j.immuni.2022.07.004 [DOI] [PubMed] [Google Scholar]
- 73.Martins-Ferreira, R. et al. The human microglia atlas (HuMicA) unravels changes in homeostatic and disease-associated microglia subsets across neurodegenerative conditions. bioRxiv10.1101/2023.08.01.550767 (2023). 10.1101/2023.08.01.550767 [DOI] [Google Scholar]
- 74.Andersen, J. et al. A transcriptional mechanism integrating inputs from extracellular signals to activate hippocampal stem cells. Neuron83, 1085–1097 (2014). 10.1016/j.neuron.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci.34, 11929–11947 (2014). 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron89, 37 (2016). 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salta, E. et al. A self-organizing miR-132/Ctbp2 circuit regulates bimodal notch signals and glial progenitor fate choice during spinal cord maturation. Dev. Cell30, 423–436 (2014). 10.1016/j.devcel.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 78.Kalamakis, G. et al. Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell176, 1407-1419.e14 (2019). 10.1016/j.cell.2019.01.040 [DOI] [PubMed] [Google Scholar]
- 79.Jin, H. Y. et al. Differential sensitivity of target genes to translational repression by miR-17~92. PLoS Genet.13, e1006623 (2017). 10.1371/journal.pgen.1006623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amin, N. D. et al. A hidden threshold in motor neuron gene networks revealed by modulation of miR-218 dose. Neuron109, 3252-3267.e6 (2021). 10.1016/j.neuron.2021.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hill, M. A. & Gammie, S. C. Alzheimer’s disease large-scale gene expression portrait identifies exercise as the top theoretical treatment. Sci. Rep.12, 17189 (2022). 10.1038/s41598-022-22179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mostafavi, S. et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat. Neurosci.21, 811–819 (2018). 10.1038/s41593-018-0154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bossers, K. et al. Concerted changes in transcripts in the prefrontal cortex precede neuropathology in Alzheimer’s disease. Brain133, 3699–3723 (2010). 10.1093/brain/awq258 [DOI] [PubMed] [Google Scholar]
- 84.Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife4, e05005 (2015). 10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reczko, M., Maragkakis, M., Alexiou, P., Grosse, I. & Hatzigeorgiou, A. G. Functional microRNA targets in protein coding sequences. Bioinformatics28, 771–776 (2012). 10.1093/bioinformatics/bts043 [DOI] [PubMed] [Google Scholar]
- 86.Paraskevopoulou, M. D. et al. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res.41, 169–173 (2013). 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen, Y. & Wang, X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res.48, D127–D131 (2020). 10.1093/nar/gkz757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu, W. & Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol.20, 1–10 (2019). 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jimenez-Mateos, E. M. et al. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol.179, 2519 (2011). 10.1016/j.ajpath.2011.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods10, 1096–1100 (2013). 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- 91.Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao, Y., Smyth, G. K. & Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 93.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420 (2018). 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq data have been deposited and are publicly available at NCBI Gene Expression Omnibus (GEO: GSE268144). Any additional information required to re-analyze the data reported in this paper is available from the corresponding author upon request.