Abstract
Accurately predicting ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties early in drug development is essential for selecting compounds with optimal pharmacokinetics and minimal toxicity. Existing ADMET-related benchmark sets are limited in utility due to their small dataset sizes and the lack of representation of compounds used in drug discovery projects. These shortcomings hinder their application in model building for drug discovery. To address this issue, we propose a multi-agent data mining system based on Large Language Models that effectively identifies experimental conditions within 14,401 bioassays. This approach facilitates merging entries from different sources, culminating in the creation of PharmaBench. Additionally, we have developed a data processing workflow to integrate data from various sources, resulting in 156,618 raw entries. Through this workflow, we constructed PharmaBench, a comprehensive benchmark set for ADMET properties, which comprises eleven ADMET datasets and 52,482 entries. This benchmark set is designed to serve as an open-source dataset for the development of AI models relevant to drug discovery projects.
Subject terms: Computational chemistry, Cheminformatics
Background & Summary
Optimization of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties plays a pivotal role in drug discovery. These pharmacokinetic properties directly influence a drug’s efficacy, safety, and ultimately clinical success. Early assessment and optimization of ADMET properties are essential for mitigating the risk of late-stage failures and for the successful development of new therapeutic agents1.
The development of computational approaches provides a fast and cost-effective means for drug discovery, allowing researchers to focus on candidates with better ADMET potential and reduce labor-intensive and time-consuming wet-lab experiments2–4. One of the key factors contributing to the success of computational approaches in drug discovery is the decent volume of compound-related biomedical data5. The number of bioassays is increasing each year, and many of their screening results are publicly accessible in databases such as ChEMBL6, PubChem7, and BindingDB8 etc.
Manual curation of ADMET data based on public data sources has been reported and some of them have been widely used as benchmark datasets for model evaluation. Wu et al.9, who constructed a large-scale benchmark for molecular machine learning named MoleculeNet, included 17 datasets and more than 700,000 compounds covering categories of physical chemistry and physiology related to ADMET experiments. Huang et al.10 published the Therapeutics Data Commons, which includes 28 ADMET-related datasets with over 100,000 entries by integrating multiple curated datasets from previous work. For specific ADMET experiment, Meng et al.11 present B3DB, which includes 1,058 compounds containing log BB values and 7,807 compounds with classification labels for the blood-brain barrier as one of the distribution properties. Meng et al.12 collected seven aqueous solubility datasets and presented a dataset curation workflow to establish solubility datasets as one of the physicochemical properties.
However, serious concerns about these benchmark datasets still exist. Firstly, most of these benchmarks include only a small fraction of the publicly available bioassay data. For instance, the ESOL dataset13 within MoleculeNet provides water solubility data for 1,128 compounds, while the PubChem7 database contains more than 14,000 relevant entries. Secondly, the entries in these benchmarks differ substantially from those in the industrial drug discovery pipeline. For example, the mean molecular weight of compounds in the ESOL dataset is only 203.9 Dalton, whereas compounds typically within the drug discovery projects have molecular weights ranging from 300 to 800 Dalton14.
These limitations of compiled open-source benchmark datasets are primarily due to the high complexity of data annotation for biological and chemical experimental records. Frequently, experimental results for identical compounds can vary significantly under different conditions, even within the same type of experiment15. For example, aqueous solubility can be influenced by various factors, such as different types of buffers, pH level, and experimental procedure. Thus, the same compound might be annotated with different solubility values depending on those experimental conditions16. This sort of variability poses a big challenge in the fusion of experimental results.
Recently developed Large Language Models (LLMs) like ChatGPT17, PubMedBERT18, and BioBERT19 represent a novel approach of effectively extracting data from a large body of text, therefore a potential method for addressing data curation challenges. Some of these LLMs demonstrate state-of-the-art performance through one-shot or few-shot learning as a form of multi-task learning17,20,21. Compared to supervised methods or models requiring thousands of data for fine-tuning, this approach allows us to develop condition extraction models more efficiently with only a few examples.
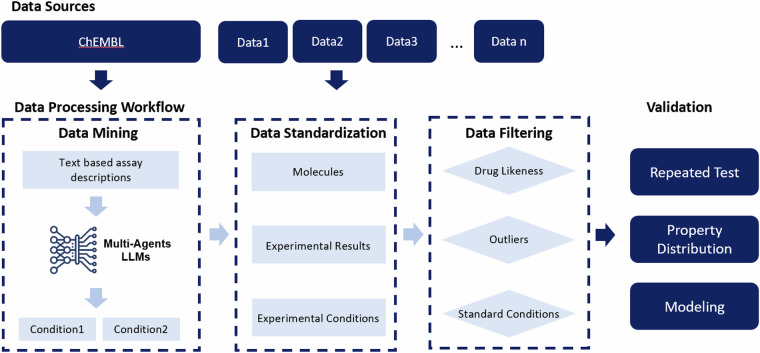
In the current study, we leveraged these LLMs as a core engine to extract experimental conditions from assay descriptions within biomedical databases, and an automated data processing framework was established for processing them for facilitating compilation of ADMET benchmark datasets as shown in Fig. 1. We implemented the pipeline to process bioassay data from the ChEMBL database and extract the experimental conditions missing from the table descriptions. These data, along with some other public datasets, were standardized and filtered to create PharmaBench22.
Fig. 1.
Data processing workflow for building PharmaBench: From left to right, the multi-agent LLM system extracts experimental conditions from the ChEMBL database, combines other data sources and standardizes the data, filters various data types, and validates them through repeated tests, property distribution, and AI modeling.
Eventually, PharmaBench22, a data package including eleven ADMET properties, was curated and provided to cheminformatics community serving as a benchmark set for ADMET predictive model evaluation. These properties are recognized as key factors in real-world drug development efforts, and both the size and diversity of the data are significantly greater than those of previous datasets. We also included multiple validation steps to confirm the data quality, molecular properties, and modeling capabilities of PharmaBench22.
Methods
The Methods section provides a detailed overview of the data processing workflow used in constructing PharmaBench22, as depicted in Fig. 1. The Data Collection subsection outlines the data sources employed to build PharmaBench22. It includes a comprehensive description of the multi-agent LLM system for extracting experimental conditions from assay descriptions, detailed in the Data Mining subsection. Following the identification of experimental conditions in the Data Mining stage, we merge experimental results from various sources and standardize and filter the data based on drug-likeness, experimental values, and conditions, as summarized in the Data Standardization and Filtering section. Finally, we post-process the datasets by removing duplicate test results and dividing the dataset based on Random and Scaffold splitting methods for AI modeling purposes.
We establish a final benchmark set that comprises experimental results in consistent units and under standardized experimental conditions. In addition, the data processing workflow described in the Methods section can eliminate inconsistent or even contradictory experimental results for the same compounds, enabling other researchers to effectively construct datasets from public data sources. For code reproduction, all data processing tasks were conducted within a Python 3.12.2 virtual environment, established using Conda on an OSX-64 platform. This environment included pandas 2.2.1, NumPy 1.26.4, Matplotlib 3.8.3, rdkit 2023.9.5, scikit-learn 1.4.1.post1, scipy 1.12.0, seaborn 0.13.2, and openai 1.12.0. A detailed description of the environment requirements can be found on GitHub at https://github.com/mindrank-ai/PharmaBench.
Data collection
Our data primarily originated from the ChEMBL database, a manually curated collection of SAR (Structure-Activity Relationship) and related physicochemical property data, largely sourced from peer-reviewed journal articles. The data type within the ChEMBL database typically includes experimental value, chemical structure, assay description, type of experiment, and certain experimental conditions. Table 1 summarizes the original entries we collected, along with the number of bioassays of the ChEMBL database used for PharmaBench22. We analysis through 97,609 raw entries based on 14,401 different bioassays in PharmaBench22.
Table 1.
Summary of data sources for PharmaBench, from left to right: the broad ADMET category, property name, number of ChEMBL entries and bioassays, number of other entries, and a summary of the sources with references.
| Category | Property Name | ChEMBL Entries (Bioassays Number) | Other Entries | Sources Summary |
|---|---|---|---|---|
| Physochemical | LogD | 25,332 (2,261) | 4,132 | AstraZeneca43 |
| Water Solubility | 17,141 (1,849) | 15,692 | Delaney (ESOL)13, Cuietal32, Boobier44, Wang45 | |
| Absorption | Blood-Brain Barrier (BBB) | 14,107 (1,881) | 11,427 | B3DB46, martin33, adenot47 |
| Distribution | Plasma Protein Binding (PPB) | 3,381 (1,296) | — | — |
| Metabolism | CYP 2C9 | 14,775 (3,651) | — | — |
| CYP 2D6 | ||||
| CYP 3A4 | ||||
| Clearance | Human liver microsomes clearance (HLMC) | 16,822 (2,194) | — | — |
| Rat liver microsomes clearance (RLMC) | ||||
| Mouse liver microsomes clearance (MLMC) | ||||
| Toxicity | AMES | 6,051 (1,269) | 27,758 | Xu48, EFSA49, ECVAM49, Hansen50 |
| Total | 97,609 (14,401) | 59,009 |
These entries from different bioassays in the ChEMBL database were analyzed through our Data Mining workflow to extract the experimental conditions. This is mainly because most of the experimental conditions recorded in ChEMBL are not explicitly specified. For instance, for solubility experiments, entries in the ChEMBL database do not include explicit data columns such as buffer type, pH condition, and experimental procedure, which are critical factors influencing experimental results. Although these conditions can be found in the assay descriptions, they cannot be directly used as a filter to distinguish experiments due to their unstructured nature. Manual mining work would be labor-intensive, which necessitates an automatic data processing framework to identify important experimental conditions from the description texts.
Thus, our multi-agent LLM system uses the entries from the ChEMBL database as the original sources and identifies various conditions for different ADMET experiments as summarised in Table 2. Additionally, we have augmented our datasets with some public datasets that have associated assay descriptions as illustrated in Fig. 1. Table 1 presents a summary of the 59,009 entries we have compiled from various public datasets, along with a delineation of their respective sources.
Table 2.
Table of Experimental Conditions and Filters Across Datasets.
| Property Name | Experimental Conditions | Filter |
|---|---|---|
| LogD | pH, Analytical Method, Solvent System, Equilibration Technique, Incubation Time, Shaking Condition | pH = 7.4, Analytical Method = HPLC, Solvent System = octanol-water, Incubation Time < 24 hours, Shaking Condition = shake flask |
| Water Solubility | pH Level, Solvent/System Composition, Time Period, Measurement Technique, Temperature Range | 7.6 > = pH Level >= 7, Solvent/System Composition = Water, 24 hr > Time Period > 1 hr, Measurement Technique = HPLC, Temperature Range < = 50 degree |
| BBB | Cell Line Models, Temperature Conditions, Permeability Assays, pH Levels, Concentration and Dosing Parameters | Cell Line Models = BBB, Permeability Assays! = effective permeability, pH Levels = 7.4 |
| PPB | Species/Origin of Plasma or Serum, Concentration of Tested Compound, Duration of Incubation, Analytical Detection Method, Equilibrium Dialysis for Protein Binding Assessment | Species/Origin of Plasma or Serum = Human, Concentration of Tested Compound < 1 g, Duration of Incubation < = 24 hr |
| CYP | Enzyme Source, Incubation Time, Temperature Range, pH Level, Substrate Concentration, Inhibitor Concentration, Cofactors, Detection Method, Protein Expression System, CYP sources | Enzyme Source = CYP3A4,CYP2D6,CYP2C9, Incubation Time > 1 mins, CYP sources = Human |
| LMC | Compound Concentration, Incubation Time, Presence of NADPH/NADP, Enzyme Source, Temperature Range, Analytical Technique, Species, Route of Administration, Type of Microsomes, Protein Amount or Microsomal Protein Concentration | Enzyme Source = liver microsomes, Incubation Time < 24 hrs, Species = human, mouse, rat, Route of Administration = None |
| AMES | Compound Concentration, Incubation Time, Presence of NADPH/NADP, Enzyme Source, Temperature Range, Analytical Technique, Species, Route of Administration, Type of Microsomes, Protein Amount or Microsomal Protein Concentration | For the Ames test, we didn’t filter out entries based on conditions because all positives are important for the Ames results. |
Overall, we have used more than 150,000 entries from public data sources to construct PharmaBench22, and the data mining process has analyzed 14,401 different bioassays.
Data mining
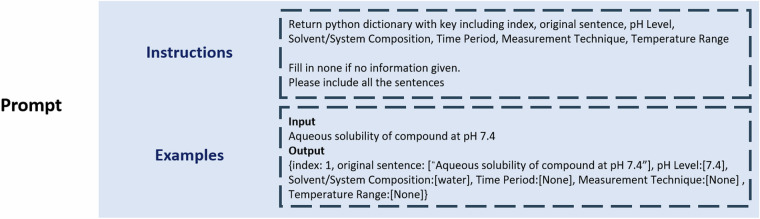
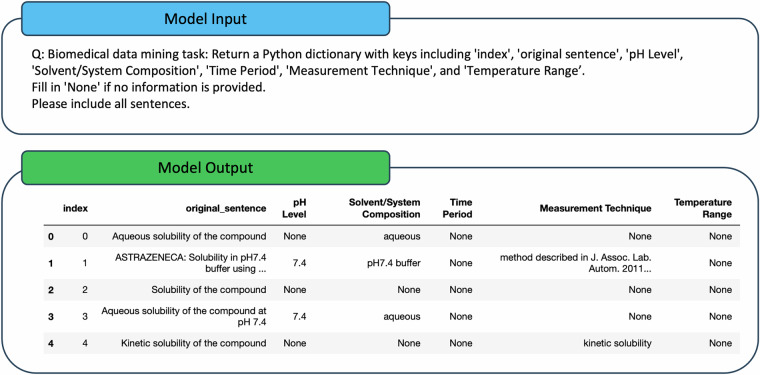
GPT-417, a model created by OpenAI, was utilized as the core LLM for the data-mining task. Based on previous research, to obtain optimized results from GPT-4, a prompt with clear instructions and examples is required for every specific task17,23–25. As shown in Fig. 2, the prompt for our data-mining process includes both instructions and examples. The instructions summarize the experimental conditions as the data mining goal and specify the requirements for the output formats. The examples, on the other hand, provide few-shot learning examples for the LLM. This prompt engineering is an important process for improving the results of GPT-417.
Fig. 2.
Sample Prompt for LLM Interaction: Illustration of a Typical User Query Input, Including Instructions and Example Parts.
However, constructing prompts for various tasks requires domain knowledge of the ADMET experiments, and creating examples for these data mining tasks remains labor-intensive. We wish to explore whether the LLMs can automatically identify key experimental conditions from different types of experiments, generate examples, and complete the complex data mining process with minimal human effort.
As a result, a multi-agent LLM data mining system was proposed in this study to extract experimental conditions from the descriptions of various bioassays26–28. An agent is a module or entity that utilizes the LLM to perform specific tasks, such as understanding, generating, or processing natural language texts28. Instead of using a single LLM-powered agent, a multi-agent system was proposed to customize LLMs into various agents, each with different capability, to automatically complete the complex data mining process, as shown in Fig. 326.
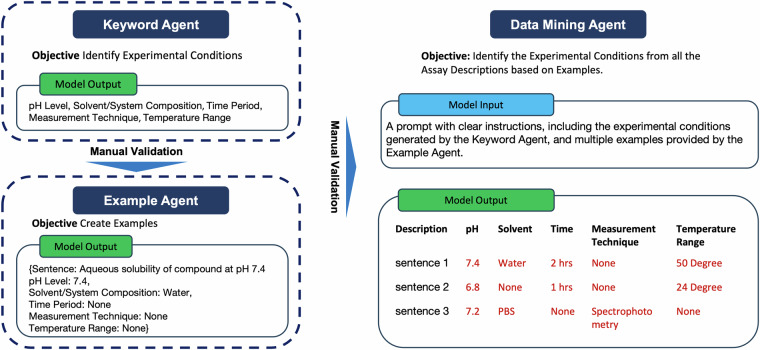
Fig. 3.
Overview of the Multi-Agent LLM Data Mining Workflow. This figure presents a summary of the multi-agent LLM data mining workflow, which includes three key components: the Keyword Agent, responsible for identifying experimental conditions; the Example Agent, tasked with generating examples; and the Data Mining Agent, designed to extract experimental conditions from assay descriptions.
The multi-agent system consists of three agents, namely keyword extraction agent (KEA), example forming agent (EFA), and data mining agent (DMA), as illustrated in Fig. 3. The KEA will pick out and summarize the key experimental conditions for ADMET experiments. The EFA will then generate examples based on these experimental results summarized by the KEA. We will manually validate the outcomes of the KEA and EFA to ensure their quality. Finally, the DMA will mine through all the assay descriptions and identify all the experimental conditions within these texts. The following sections will introduce these three agents in more detail.
Keyword extraction agent
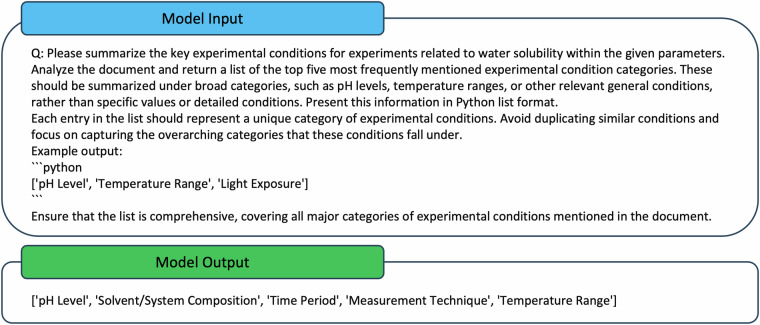
The KEA is designed to summarize key experimental conditions from various ADMET experiments. A prompt, as illustrated in Fig. 4, along with texts from 50 randomly selected assay descriptions, was created as the model input for the KEA. This prompt instructs GPT-4 to summarize the experimental conditions from selected assay descriptions of bioassays in ChEMBL. The model’s task is to identify and summarize the top five most frequently mentioned experimental conditions. For more complex experiments, such as microsome clearance and CYP inhibition, the model was asked to summarize the top ten conditions. GPT-4 is required to generalize these conditions rather than just listing specific conditions and duplicating or listing similar conditions should be avoided. An example of a Python list is provided to KEA to illustrate the desired output format for GPT-4. This process will leverage GPT-4’s internal knowledge to generate a list of significant experimental conditions. An example of the input and output for the KEA is shown in Fig. 4.
Fig. 4.
Sample Prompt for the Keyword Extraction Agent.
The experimental conditions summarized by the KEA are listed in the ‘Experimental Condition’ column of Table 2. Domain experts were invited to confirm if these conditions are key conditions for ADMET experiments. These experimental conditions are then used as the primary data mining goal for the DMA to extract from each assay description.
Example forming agent
The EFA focuses on generating examples from assay description texts. The prompt for this agent includes clear instructions incorporating the key experimental conditions summarized by the Keyword Agent, along with forty assay descriptions for analysis purposes. The Example Agent returns a Python dictionary containing the index, original sentences, and key experimental conditions as the keys. It will return ‘None’ if no information is provided within the sentences.
For each ADMET experiment, forty examples will be generated through these automatic pipelines. Manual examination is conducted on the examples to eliminate errors and confirm the format. This fast labeling process generates few-shot learning examples for DMA, which avoids intensive human labeling. The example input and output for this agent is shown in Fig. 5.
Fig. 5.
Sample Prompt for the Example Forming Agent.
Data mining agent
The DMA aims to complete the mining task for all assay descriptions from the ChEMBL database. As shown in Fig. 2, the prompt for this agent includes instructions containing the experimental conditions summarized by the KEA and forty examples generated by the EFA. As shown in Fig. 5, the prompt defines the data mining task, identifying experimental conditions and outputting them in the desired format. These examples provide few-shot learning data for the DMA to learn how to standardize the output format and improve the overall output quality.
GPT-4 has a limit on the number of tokens to be processed in a single request17. Thus, we divided the assay descriptions with a batch size of twenty to mitigate the risk of overloading the model. This batching technique allows for a more accurate and reliable analysis, especially when dealing with complex assay descriptions.
The DMA will return a Python Dictionary for every batch input. A routine was written to convert the Python Dictionary from a Markdown file into a Pandas DataFrame, which is then stored. Eventually, the Data Mining Agent goes through all the assay descriptions in the raw data and stores the output of every batch. The experimental conditions mined based on this multi-agent system are then merged back into the original file for data standardization and filtering.
Overall, this multi-agent system mines through 14,401 assay descriptions to identify the experimental conditions for seven different ADMET experiments. It largely minimizes human effort to extract structured experimental conditions which will be then used in the following data standardization and filtering procedures.
Data standardization
The data obtained from different sources exhibit significant variability in the format of structure, data type, name of experimental condition, and the unit and range of experimental value. For standardizing the data, we design a data standardization workflow to clean the data obtained from the previously described data mining step and it includes standardization of structure format, experimental condition, and experimental value.
Structure Standardization: A standard pipeline using RDKit29 is used to convert compound SMILES into canonical SMILES. This pipeline includes checking validity, stripping salts, and removing molecules containing metal atoms.
Standardization of experiment condition: Experiment conditions from various sources are standardized into a unified format. For conditions being numerical values, such as pH, temperature, and compound concentration, they are converted into floating numbers. String values, such as buffer type, CYP type, cell strain type, etc., are standardized using the same naming format. For binary variables, such as the addition of S9 in an Ames experiment, a boolean value of ‘True’ or ‘False’ is used. The experimental conditions across different sources are standardized using a consistent naming strategy, thereby facilitating the data filtering section.
Standardization of experiment value: A similar standardization procedure is also carried out on experimental readouts. For regression tasks, the experimental results, which may be in varying units, are converted to a consistent unit. In some cases, log transformation is applied to experimental results to reduce data range. For classification tasks, thresholds are defined to assign class labels on datasets.
Data filtering
A data filtering process aims to filter out entries with abnormal molecules and irregular experimental results, to construct the final benchmark set that contains experimental results in consistent units and experimental conditions.
Molecule Filter: Molecules containing metal atoms are removed. In addition, amino acids, peptides, or antibodies are removed.
Filter of experiment value: For filtering experiment results, entries containing results outside the normal data range, e.g. negative values for half-life data, are removed. Additionally, upper and lower limits for experiment results are set. Outliers and abnormal distributions in the regression values are manually validated and eliminated if they cannot be self-explained.
Filter of experiment condition: The extreme experiment conditions are eliminated while preserving the rest of the entries. For experiment conditions that contain a few ‘None’ values, we typically only retain entries within a specific range of result value, as indicated in the ‘Filter’ column of Table 2, and remove the entries that fall outside of this range or contain ‘None’. For instance, we only preserve the pH value equal to 7.4 for the LogD experiments and remove the rest. We exclude experiment conditions of which the majority is a ‘None’ entry, as they do not provide useful filters due to the predominance of unknown information. The details for the Experimental Condition Filter can be found in Table 2.
Data preparation for AI modeling
After the above data processing workflow, a series of ADMET datasets were constructed from various bioassays. The count of entities is summarized in Table 3. However, multiple experimental results for the same compounds occur under the same conditions within the datasets after the processing workflow.
Table 3.
Summary of Datasets in PharmaBench: ‘Property Name’ refers to the name of the dataset.
| Category | Property Name | Entries After Data Processing | Final Entries for AI modeling | Unit | Mission Type |
|---|---|---|---|---|---|
| Physochemical | LogD | 14,141 | 13,068 | — | regression |
| Water Solubility | 14,818 | 11,701 | log10nM | regression | |
| Absorption | BBB | 12,486 | 8,301 | — | classification |
| Distribution | PPB | 1,310 | 1,262 | % | regression |
| Metabolism | CYP 2C9 | 4,507 | 999 | Log10uM | regression |
| CYP 2D6 | 1214 | Log10uM | regression | ||
| CYP 3A4 | 1980 | Log10uM | regression | ||
| Clearance | HLMC | 5,252 | 2286 | Log10(mL.min-1.g-1) | regression |
| RLMC | 1129 | Log10(mL.min-1.g-1) | regression | ||
| MLMC | 1403 | Log10(mL.min-1.g-1) | regression | ||
| Toxicity | AMES | 24,780 | 9,139 | — | classification |
| Total | 77,294 | 52,482 |
‘Entries After Data Processing’ indicates the number of entries remaining after the data processing workflow. ‘Final Entries for AI Modeling’ denotes the number of entries used in the final AI modeling process. ‘Unit’ specifies the measurement unit for regression tasks. ‘Mission Type’ encompasses two categories: regression and classification.
Thus, we employed various strategies to unify these repeated results in the final datasets. For regression tasks, for compounds with repeated data, the mean value was taken as the unified value. There are two classification datasets in our benchmark set. For the BBB experiment, we eliminate all compounds with contradictory results, while for AMES, we label the compounds as positive if at least one positive result occurs in these experiments. This approach is primarily due to the fact that AMES is a toxicity-related experiment, which requires the model to be highly sensitive to positive results30.
Additionally, we divided the datasets for each property into training and test sets with a ratio of 0.8:0.2 respectively, utilizing both random and scaffold splitting methods. Random splitting involves distributing the compounds arbitrarily across the training and test sets, whereas scaffold splitting is designed to create sets with distinct structural features by allocating compounds that share the same core scaffold exclusively to either the training or the test set10. This approach ensures that the test set is structurally different from the training compounds, making it more challenging for models to predict.
Data Records
We have compiled 11 ADMET datasets to form PharmaBench, which is freely available at figshare22. Table 3 includes the number of entries after the data processing workflow for each dataset and the final entries for AI modeling. The final entries consist of one experimental result for each molecule, based on the experimental condition as described in the ‘Filter’ column of Table 2. The mission type of the different datasets is also summarized in the ‘Mission Type’ column of Table 3, including regression and classification.
Overall, PharmaBench22 comprises a total of 52,482 entries. It is stored in comma separated values (CSV) format and includes a unified SMILES representation, experimental results, property names, and training labels based on both scaffold and random splitting, as summarized in Table 4. The data are also openly accessible on GitHub at https://github.com/mindrank-ai/PharmaBench, along with the processing workflow.
Table 4.
List of information in the final datasets.
| Column Name | Description | Data Type |
|---|---|---|
| Smiles_unify | Standardized SMILES representation of compounds is based on standardization methods described in the Data Standardization. | String |
| value | Experimental values for different datasets including regression values and classification values | Float |
| property | Different ADMET property name for the experiment | String |
| scaffold_train_test_label | Training labels based on scaffold splitting, where 1 represents the training data and 0 represents the testing data. | Float |
| random_train_test_label | Training labels based on random splitting, where 1 represents the training data and 0 represents the testing data. | Float |
Including the column name, the description for the column information and the data type.
The following section will introduce different datasets in more detail, including a general introduction to various ADMET properties, the units for different datasets, and the number of molecules.
LogD LogD31 measures a drug’s pH-adjusted lipophilicity, representing the ratio of its total concentration (both ionized and un-ionized) in oil and water phases. This is an important property to consider in drug discovery as it influences a compound’s bioavailability, permeability, and other pharmacokinetic properties. The unit for LogD, which stands for the logarithm of the distribution coefficient (D), is dimensionless. We introduce a regression task that includes 13,068 unique molecules for predicting LogD.
Water Solubility Water solubility32 denotes the maximum amount of a solute that can dissolve in water to form a uniform solution. In drug development, it significantly impacts drug bioavailability, since a drug requires adequate solubility for absorption into the bloodstream. The unit for the water solubility dataset is log10nM, and it includes 11,701 unique molecules for the regression prediction of these values. We filtered out the dynamic water solubility data in this dataset based on the experimental conditions.
The Blood-Brain Barrier (BBB) The BBB33 is a selective barrier that separates the blood from the central nervous system (CNS) and poses significant challenges for drug delivery to the CNS. Predicting BBB penetration is crucial for designing drugs targeting CNS diseases. We have chosen log BB = –1 as the threshold value, as this is the most widely used threshold, as discussed in the B3DB. Overall, there are 8,301 unique molecules for the BBB task.
Plasma Protein Binding (PPB): PPB34 is an important pharmacokinetic parameter that characterizes the extent to which a compound binds to proteins in the bloodstream. PPB can influence a compound’s distribution, elimination, and therapeutic efficacy. The experimental results for PPB experiments range from 0 to 1, representing the percentage of the drug in the plasma that is bound. For instance, if a drug has a PPB of 90%, it means that 90% of the drug molecules present in the plasma are attached to plasma proteins, leaving only 10% free and active. There are records of 1,262 molecules in the PPB datasets.
CYP: Cytochrome P450 (CYP)35 is the primary metabolic enzyme responsible for drug metabolism in the body. CYP enzymes catalyze the oxidation of organic substances, a process that often represents the first step in the metabolism of many drugs. Multiple CYP isoforms exist in the human body, each with unique specificity for various substrates. The unit for the CYP datasets is Log10uM, indicating the binding affinity of compounds to different CYP enzymes. There are three different CYP datasets in this benchmark, namely CYP 2C9 (999 molecules), CYP2D6 (1,214 molecules), and CYP 3A4 (1,980 molecules).
Liver Microsome Clearance (LMC): Liver Microsome Clearance36 refers to the process by which compounds are metabolized and cleared in the liver microsomal system. This in vitro assessment is crucial in drug discovery and development, as it offers an early estimation of a compound’s in vivo clearance rate and potential for drug-drug interactions. The unit for LMC is Log10(mL.min-1.g-1), indicating the clearance speed of microsomes for different drugs. We have included three different LMC datasets in this benchmark, namely human LMC (2,286 molecules), rat LMC (1,129 molecules), and mouse LMC (1,403 molecules).
AMES: The AMES test30 evaluates a compound’s mutagenic potential by assessing whether specific bacteria regain the ability to grow without histidine. It serves as a cost-effective, preliminary toxicity screening method widely used in various industries, particularly in drug development, to identify potential carcinogens. A positive AMES result indicates that the compound may have mutagenic potential, characterized by abnormal bacterial growth speed. We have included 9,139 molecules for the AMES test.
Technical Validation
Once the data collection is done, we evaluate the datasets from three aspects. Firstly, we use the repeated test results for the datasets before and after the implementation of the data processing workflow to demonstrate the improvement in data quality resulting from this workflow. Secondly, we illustrate the characteristics of PharmaBench22 by showing distributions of various molecular properties. Lastly, we trained various machine learning and deep learning models on the datasets and presented model performance on the test sets.
Repeated test for data quality assessment
A comparison for repeated test results is a methodological approach where the same experiment is conducted multiple times to verify the consistency of the results37. Limited by the scope of this work, we cannot verify each data point through wet lab experiments or review each literature to confirm the direct data quality of the dataset. Thus, we implement an indirect approach, namely repeated testing, to confirm the data quality before and after data processing. A raw dataset often contains multiple records for the same compound due to different sources and varying experimental conditions. Repeated testing compares the maximum and minimum values for the same compound under the same condition to validate the data quality.
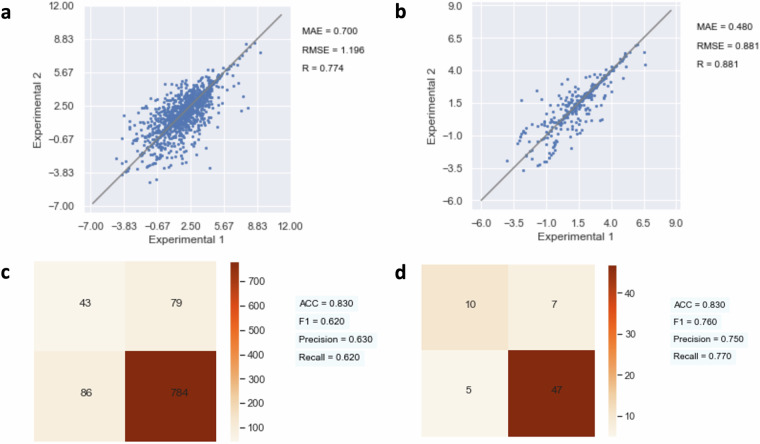
As shown in Fig. 6, the repeated test plot is used to analyze regression results, and the confusion matrix is used to analyze the classification results. If the experimental results are consistent for different data sources, the repeated test plot will exhibit higher correlation and a lower mean absolute error (MAE) for regression tests, and the confusion matrix will show higher accuracy (ACC), precision, and recall for classification tests. In contrast, low-quality data will have opposite metric scores.
Fig. 6.
Comparison of Data Quality Before and After the Data Processing Workflow Through Repeated Test Plots and Confusion Matrices (a) Repeated Test Plot for the LogD Experiment Before Data Processing. (b) Repeated Test Plot for the LogD Experiment After Data Processing. (c) Repeated Test Plot for the BBB Experiment Before Data Processing. (d) Repeated Test Plot for the BBB Experiment After Data Processing. Additional data can be found in Table 5.
We use this method to compare data quality before and after considering the experimental conditions mined through our data mining process, thereby demonstrating improvement in data quality based on our approach. The data quality before and after the data processing workflow can be compared and evaluated through the metrics mentioned above.
Specifically, we group data entries for the same molecules from the raw data to create the ‘before data processing’ plot, and we group data entries for the same molecules under identical conditions for the ‘after data processing’ plot. The maximum and minimum experimental results for each group are selected as the worst-case scenario. We have created a scatter plot for the regression tests and a confusion matrix for the classification tests, as shown in Fig. 6. The R, MAE, and RMSE for regression tasks, and ACC, F1, precision, and recall for classification tasks, have been calculated and are recorded within Tables 5 and 6.
Table 5.
Comparison of Metrics Between the Regression Datasets Before and After the Data Processing Workflow.
| ADMET Property Name | Before Data Processing Workflow | After Data Processing Workflow | ||||
|---|---|---|---|---|---|---|
| R | RMSE | MAE | R | RMSE | MAE | |
| LogD | 0.774 | 1.196 | 0.7 | 0.881 | 0.881 | 0.48 |
| Water Solubility | 0.554 | 1.02 | 0.64 | 0.788 | 0.745 | 0.305 |
| Plasma Protein Binding (PPB) | 0.875 | 13.468 | 6.699 | 0.951 | 2.61 | 2.033 |
|
CYP 2C9 CYP 2D6 CYP 3A4 |
0.428 | 0.933 | 0.66 | 0.578 | 0.915 | 0.52 |
|
Human liver microsomes clearance Rat liver microsomes clearance Mouse liver microsomes clearance |
0.627 | 0.839 | 0.59 | 0.737 | 0.676 | 0.354 |
Table 6.
Comparison of Metrics Between the Classification Datasets Before and After the Data Processing Workflow.
| ADMET Property Name | Before Data Processing Workflow | After Data Processing Workflow | ||||||
|---|---|---|---|---|---|---|---|---|
| ACC | F1 | Precision | Recall | ACC | F1 | Precision | Recall | |
| AMES | 0.87 | 0.78 | 0.77 | 0.8 | 0.92 | 0.85 | 0.85 | 0.85 |
| BBB | 0.83 | 0.62 | 0.63 | 0.62 | 0.83 | 0.76 | 0.74 | 0.78 |
Table 5 demonstrates the results of repeated tests for regression tasks within PharmaBench22, while Table 6 summarizes the classification tasks. All metrics improved following the data processing workflow, validating the quality increment. The results of repeated tests for certain experiments, such as the LogD experiment, have significantly improved data quality after the data processing workflow, reaching a level comparable to that of traditional wet lab experiments. However, the results of repeated tests for CYP and clearance experiments remain relatively low, due to the complex nature of these in vitro experiments.
Analysis of property distribution
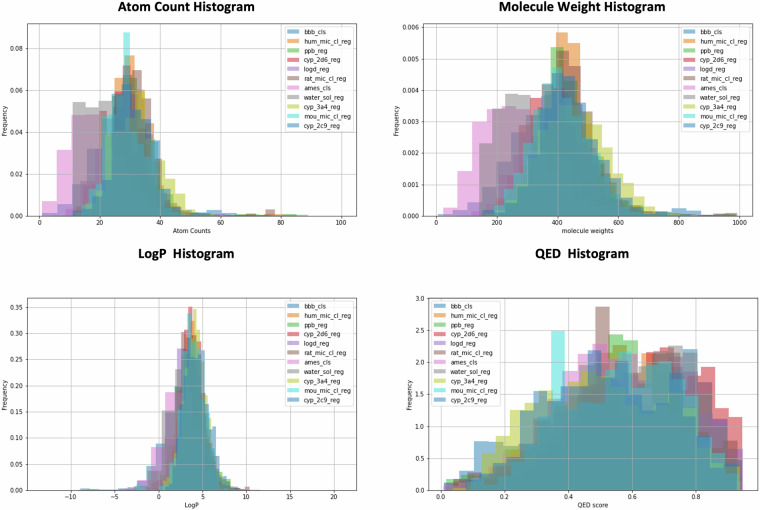
Basic physicochemical properties of the compounds, including atom counts, molecular weight, LogP, and QED, were calculated using RDKit. Histograms representing the frequency of these properties were calculated and are presented in Fig. 7 to illustrate the characteristics of the molecules within PharmaBench22,38.
Fig. 7.
Frequency Histograms for All PharmaBench Datasets: Count Distribution of Atom Numbers, Molecular Weights, LogP, and QED Scores Across Different Datasets.
This histogram demonstrates that compounds in PharmaBench22 exhibit a broad distribution. The number of non-hydrogen atoms per molecule typically ranges from 10 to 50, and molecular weights range from 200 to 600 Daltons, which are consistent with the range of drug-like small molecules39. Additionally, the LogP values of these datasets are in the range from 0 to 8, indicating a tendency towards lipophilicity, which is also well aligned with that of drug-like compounds39. QED is a metric that evaluates the potential of a compound to be developed as a successful drug, based on a multi-factorial analysis of molecular properties of marked drugs39. The QED distribution for PharmaBench22 is skewed towards 1, suggesting that many compounds in the datasets possess favorable physio-chemical properties.
Overall, the molecules within PharmaBench22 demonstrate preferable characteristics, which are similar to those in the small molecule drug discovery projects.
Deep learning and machine learning modeling
Modeling protocol
Similar to the repeated test mentioned above, we used MAE, RMSE, and Pearson correlation coefficient R to evaluate the regression results. For classification results, we utilized AUC (area under the receiver operating characteristic curve), ACC, and the F1 score (F1) for evaluation.
We selected two machine-learning approaches and seven deep-learning models for this evaluation process. The machine learning models include XGBoost40 and Random Forest (RF)41, utilizing the Extended Connectivity Fingerprints (ECFP) as descriptors for the molecules42. We selected seven deep learning models, some of them need a pre-training process, and their input is either 2D graph or 3D conformation. Detailed descriptions of these models can be found in Table 7.
Table 7.
Summary of AI Models Utilized in the Validation Process.
| Model Name | Type of Model | Whether Including Pretraining | 2D/3D |
|---|---|---|---|
| Random Forest (RF)41 | Machine Learning | Na | NA |
| XGBoost40 | Machine Learning | Na | NA |
| CMPNN51 | Graph Deep Learning | No | 2D |
| FPGNN52 | Graph Deep Learning | No | 2D |
| DHTNN53 | Transformer Deep Learning | No | 2D |
| KANO54 | Graph Deep Learning | Yes | 2D |
| MPG55 | Graph Deep Learning | Yes | 2D |
| Unimol56 | Transformer Deep Learning | Yes | 2D + 3D |
| Transformer-M57 | Transformer Deep Learning | Yes | 2D + 3D |
All models were built using default parameters, without additional fine-tuning. Although hyper-parameter optimizing strategy might improve the results, our intention is not to select the best model but rather to use these models to verify the quality of the PharmBench.
Modeling results
We present the metrics for the regression models and for the classification models, trained using both random as shown in Table 8 and scaffold splitting as shown in Table 9 datasets.
Table 8.
Summary of final results for the PharmaBench based on random split.
| Property | Metrices | RF | XGBoost | CMPNN | FPGNN | DHTNN | KANO | MPG | Unimol | Trans-M |
|---|---|---|---|---|---|---|---|---|---|---|
| LogD | R | 0.573 | 0.786 | 0.86 | 0.865 | 0.801 | 0.898 | 0.888 | 0.901 | 0.901 |
| MAE | 0.866 | 0.618 | 0.528 | 0.489 | 0.613 | 0.434 | 0.464 | 0.452 | 0.431 | |
| RMSE | 1.14 | 0.868 | 0.757 | 0.715 | 0.844 | 0.631 | 0.672 | 0.671 | 0.639 | |
| Solubility | R | 0.49 | 0.629 | 0.645 | 0.706 | 0.547 | 0.703 | 0.699 | 0.723 | 0.612 |
| MAE | 0.711 | 0.592 | 0.619 | 0.536 | 0.658 | 0.54 | 0.545 | 0.516 | 0.635 | |
| RMSE | 0.871 | 0.76 | 0.801 | 0.712 | 0.838 | 0.712 | 0.72 | 0.695 | 0.812 | |
| PPB | R | 0.389 | 0.581 | 0.411 | 0.705 | 0.236 | 0.655 | 0.705 | 0.697 | 0.719 |
| MAE | 0.14 | 0.122 | 0.15 | 0.103 | 0.19 | 0.111 | 0.101 | 0.111 | 0.106 | |
| RMSE | 0.202 | 0.19 | 0.273 | 0.168 | 0.257 | 0.189 | 0.191 | 0.208 | 0.2 | |
| CYP 3A4 | R | 0.247 | 0.467 | 0.368 | 0.609 | 0.387 | 0.485 | 0.584 | 0.593 | 0.564 |
| MAE | 10.977 | 9.761 | 9.865 | 7.892 | 10.744 | 10.033 | 8.33 | 8.545 | 8.536 | |
| RMSE | 18.374 | 17.565 | 19.903 | 15.735 | 17.324 | 17.036 | 16.128 | 16.842 | 16.619 | |
| CYP 2C9 | R | 0.073 | 0.141 | 0.148 | 0.251 | 0.19 | 0.154 | 0.388 | 0.29 | 0.332 |
| MAE | 10.823 | 10.62 | 8.733 | 9.746 | 10.503 | 9.858 | 8.518 | 8.567 | 8.882 | |
| RMSE | 17.08 | 16.959 | 17.849 | 16.419 | 16.864 | 16.916 | 16.788 | 17.9 | 16.174 | |
| CYP 2D6 | R | 0.213 | 0.232 | 0.328 | 0.477 | 0.278 | 0.498 | 0.527 | 0.47 | 0.463 |
| MAE | 12.129 | 13.097 | 10.802 | 10.999 | 13.251 | 10.697 | 10.196 | 10.227 | 10.927 | |
| RMSE | 19.826 | 20.685 | 21.612 | 19.948 | 20.467 | 19.385 | 20.012 | 20.695 | 20.664 | |
| HLMC | R | 0.462 | 0.652 | 0.223 | 0.712 | 0.497 | 0.684 | 0.738 | 0.538 | 0.705 |
| MAE | 0.521 | 0.427 | 0.707 | 0.419 | 0.576 | 0.465 | 0.41 | 0.604 | 0.442 | |
| RMSE | 0.697 | 0.605 | 1.008 | 0.628 | 0.789 | 0.668 | 0.599 | 0.862 | 0.631 | |
| RLMC | R | 0.492 | 0.675 | 0.363 | 0.704 | 0.564 | 0.738 | 0.753 | 0.683 | 0.721 |
| MAE | 0.511 | 0.395 | 0.589 | 0.378 | 0.494 | 0.376 | 0.357 | 0.426 | 0.381 | |
| RMSE | 0.692 | 0.557 | 0.867 | 0.52 | 0.683 | 0.514 | 0.495 | 0.609 | 0.543 | |
| MLMC | R | 0.621 | 0.766 | 0.349 | 0.811 | 0.6 | 0.785 | 0.8 | 0.738 | 0.772 |
| MAE | 0.628 | 0.447 | 0.765 | 0.409 | 0.629 | 0.45 | 0.414 | 0.521 | 0.428 | |
| RMSE | 0.862 | 0.62 | 1.106 | 0.567 | 0.863 | 0.633 | 0.569 | 0.747 | 0.59 | |
| BBB | ACC | 0.796 | 0.81 | 0.842 | 0.852 | 0.842 | 0.867 | 0.865 | 0.861 | 0.847 |
| AUC | 0.698 | 0.726 | 0.901 | 0.93 | 0.903 | 0.934 | 0.925 | 0.918 | 0.922 | |
| F1 | 0.867 | 0.873 | 0.814 | 0.886 | 0.882 | 0.907 | 0.849 | 0.842 | 0.829 | |
| AMES | ACC | 0.726 | 0.791 | 0.78 | 0.79 | 0.751 | 0.795 | 0.807 | 0.788 | 0.803 |
| AUC | 0.727 | 0.791 | 0.847 | 0.87 | 0.83 | 0.869 | 0.879 | 0.857 | 0.88 | |
| F1 | 0.715 | 0.788 | 0.78 | 0.782 | 0.756 | 0.8 | 0.807 | 0.787 | 0.802 |
Table 9.
Summary of final results for the PharmaBench based on scaffold split.
| Property | Metrices | RF | XGBoost | CMPNN | FPGNN | DHTNN | KANO | MPG | Unimol | Trans-M |
|---|---|---|---|---|---|---|---|---|---|---|
| LogD | R | 0.527 | 0.688 | 0.854 | 0.815 | 0.784 | 0.859 | 0.862 | 0.875 | 0.867 |
| MAE | 0.934 | 0.776 | 0.548 | 0.604 | 0.681 | 0.528 | 0.526 | 0.517 | 0.504 | |
| RMSE | 1.249 | 1.071 | 0.807 | 0.838 | 0.912 | 0.766 | 0.758 | 0.745 | 0.737 | |
| Solubility | R | 0.337 | 0.485 | 0.463 | 0.625 | 0.535 | 0.615 | 0.627 | 0.674 | 0.522 |
| MAE | 0.734 | 0.654 | 0.663 | 0.573 | 0.634 | 0.58 | 0.581 | 0.53 | 0.626 | |
| RMSE | 0.918 | 0.832 | 0.858 | 0.747 | 0.828 | 0.772 | 0.758 | 0.707 | 0.834 | |
| PPB | R | 0.292 | 0.489 | 0.514 | 0.543 | 0.197 | 0.581 | 0.718 | 0.733 | 0.668 |
| MAE | 0.142 | 0.122 | 0.13 | 0.114 | 0.173 | 0.106 | 0.099 | 0.095 | 0.097 | |
| RMSE | 0.204 | 0.186 | 0.236 | 0.179 | 0.235 | 0.185 | 0.17 | 0.179 | 0.172 | |
| CYP 3A4 | R | 0.101 | 0.154 | 0.327 | 0.368 | 0.144 | 0.483 | 0.457 | 0.425 | 0.317 |
| MAE | 10.572 | 10.619 | 8.114 | 9.925 | 10.43 | 8.425 | 8.201 | 8.532 | 8.955 | |
| RMSE | 16.54 | 16.123 | 16.701 | 15.606 | 16.156 | 15.307 | 14.376 | 15.895 | 15.867 | |
| CYP 2C9 | R | 0.05 | 0.061 | 0.203 | 0.32 | −0.118 | 0.147 | 0.307 | 0.318 | 0.229 |
| MAE | 11.911 | 11.201 | 9.684 | 10.028 | 10.58 | 11.64 | 9.504 | 9.524 | 10.194 | |
| RMSE | 18.471 | 17.582 | 18.377 | 16.933 | 17.449 | 17.35 | 17.417 | 17.774 | 18.08 | |
| CYP 2D6 | R | 0.293 | 0.197 | 0.201 | 0.301 | 0.23 | 0.37 | 0.482 | 0.269 | 0.464 |
| MAE | 11.728 | 12.024 | 10.453 | 11.173 | 12.199 | 10.881 | 10.13 | 10.283 | 9.961 | |
| RMSE | 18.041 | 17.819 | 19.156 | 17.611 | 17.89 | 17.622 | 17.527 | 18.071 | 17.677 | |
| HLMC | R | 0.469 | 0.628 | 0.345 | 0.665 | 0.609 | 0.722 | 0.751 | 0.722 | 0.733 |
| MAE | 0.592 | 0.459 | 0.63 | 0.444 | 0.519 | 0.395 | 0.375 | 0.426 | 0.392 | |
| RMSE | 0.813 | 0.647 | 0.921 | 0.604 | 0.729 | 0.554 | 0.541 | 0.613 | 0.567 | |
| RLMC | R | 0.337 | 0.564 | 0.194 | 0.642 | 0.44 | 0.621 | 0.731 | 0.645 | 0.664 |
| MAE | 0.698 | 0.557 | 0.696 | 0.496 | 0.694 | 0.522 | 0.433 | 0.483 | 0.463 | |
| RMSE | 0.958 | 0.819 | 0.939 | 0.716 | 0.915 | 0.762 | 0.685 | 0.651 | 0.677 | |
| MLMC | R | 0.488 | 0.672 | 0.399 | 0.732 | 0.61 | 0.729 | 0.749 | 0.727 | 0.753 |
| MAE | 0.73 | 0.579 | 0.808 | 0.568 | 0.665 | 0.549 | 0.499 | 0.568 | 0.518 | |
| RMSE | 0.987 | 0.844 | 1.13 | 0.774 | 0.926 | 0.767 | 0.723 | 0.824 | 0.744 | |
| BBB | ACC | 0.825 | 0.837 | 0.84 | 0.851 | 0.85 | 0.846 | 0.858 | 0.844 | 0.871 |
| AUC | 0.731 | 0.75 | 0.887 | 0.923 | 0.909 | 0.915 | 0.923 | 0.92 | 0.935 | |
| F1 | 0.885 | 0.892 | 0.809 | 0.887 | 0.895 | 0.888 | 0.833 | 0.819 | 0.846 | |
| AMES | ACC | 0.762 | 0.769 | 0.793 | 0.771 | 0.765 | 0.743 | 0.798 | 0.818 | 0.799 |
| AUC | 0.761 | 0.768 | 0.858 | 0.858 | 0.844 | 0.865 | 0.869 | 0.878 | 0.869 | |
| F1 | 0.776 | 0.783 | 0.793 | 0.786 | 0.767 | 0.78 | 0.798 | 0.818 | 0.799 |
For the datasets associated with regression tasks, the prediction results achieve desirable metrics for LogD, water solubility, BBB, and microsomal clearance, exhibiting relatively high R values and low MAE and RMSE. However, the prediction results for the CYP remain relatively low, which indicates that further improvements in data quality and modeling approaches are required for these datasets. The metrics for the classification tasks are all relatively high, which indicates that the models can effectively predict the classification results for the BBB and AMES datasets.
In regards to the splitting method, the prediction results of random splitting are better than scaffold splitting for the majority of tasks. This is understandable since the prediction performance for the majority of models is usually worse for compounds with new scaffolds. In addition, deep learning approaches significantly outperform the machine learning approaches in regression tasks. The performance gap between the deep learning and machine learning models widens for datasets with a large amount of data, such as LogD and water solubility, but narrows for smaller datasets, such as mouse microsomal clearance. This indicates that conventional machine learning approach can adapt to small datasets and has less capability to model large amounts of data compared with deep learning model. In contrast, the performance of the machine learning approach for classification tasks witnesses a significant increase. The metrics for XGBoost models for AMEs and BBB datasets surpass some deep learning approaches, indicating that machine learning approaches are more suitable for classification tasks.
Among deep learning approaches, the model with pretraining demonstrates the best performance in both regression and classification tasks for the majority of datasets. This indicates that the pretraining process can be useful for improving model performance for ADMET properties predictions. Moreover, there is no significant performance difference between graph-based and transformer-based approaches, or between 2D and 3D feature-based methods.
More research and modeling work are encouraged to utilize this benchmark set in the future. For instance, investigating approaches to improve model capabilities in predicting molecules with novel scaffolds would be valuable. The use of transfer learning and pre-training approaches is also recommended for the analysis with these datasets. Additionally, applying explainable AI techniques could provide valuable insights into the key pharmacological factors influencing ADMET properties.
Usage Notes
There are eleven ADMET datasets within PharmaBench22. Standardized SMILES representations of compounds were provided for modeling the compounds, and the experimental values are provided as the prediction targets. Users may use the labels within the scaffold_train_test_label and random_train_test_label as the train-test labels for fair comparison.
Acknowledgements
This study was funded by MindRank AI Ltd. H. C. acknowledged funding from the Guangzhou Basic and Applied Basic Research Project (202201011795) and the Pearl River Recruitment Program of Talents (No. 2021CX020227). G. Y. was supported in part by the ERC IMI (101005122), H2020 (952172), MRC (MC/PC/21013), the Royal Society (IEC\ NSFC\211235), the NVIDIA Academic Hardware Grant Program, the SABER project supported by Boehringer Ingelheim Ltd, Wellcome Leap’s Dynamic Resilience, and the UKRI Future Leaders Fellowship (MR/V023799/1).å
Author contributions
Z.N., X.X. and W.W. conceived the presented idea. X.X., W.W., Y.J. and Q.C. developed the theory and the data mining system. Q.C., W.J., G.J.Y., L.K. and J.X. collected the data. Z.N., X.X., W.W., Q.C., Y.J. and W.J. developed the data processing workflow. X.X., W.W., Q.C., Y.J., W.J., M.W. and G.J.Y. developed the technical validation for the data qualities. H. C. and G. Y. supervised the findings of this work. All authors discussed the results and contributed to the final manuscript.
Code availability
The codes used in this study have been deposited at https://github.com/mindrank-ai/PharmaBench. All the calculations were done with Python 3.12.2 under a virtual environment created with Anaconda on osx-64.
Competing interests
Z.N., X.X., W.W., Q.C., Y.J., W.J., M.W., G.J.Y., L.K. and X.J. are the employees of MindRank AI ltd.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhangming Niu, Xianglu Xiao, Wenfan Wu.
These authors jointly supervised this work: Guang Yang, Hongming Chen.
Contributor Information
Guang Yang, Email: g.yang@imperial.ac.uk.
Hongming Chen, Email: chen_hongming@gzlab.ac.cn.
References
- 1.Davis, A. M. & Riley, R. J. Predictive admet studies, the challenges and the opportunities. Current Opinion in Chemical Biology8, 378–386, 10.1016/j.cbpa.2004.06.005 (2004). 10.1016/j.cbpa.2004.06.005 [DOI] [PubMed] [Google Scholar]
- 2.Chen, H., Engkvist, O., Wang, Y., Olivecrona, M. & Blaschke, T. The rise of deep learning in drug discovery. Drug Discovery Today23, 1241–1250, 10.1016/j.drudis.2018.01.039 (2018). 10.1016/j.drudis.2018.01.039 [DOI] [PubMed] [Google Scholar]
- 3.Ferreira, L. L. & Andricopulo, A. D. Admet modeling approaches in drug discovery. Drug Discovery Today24, 1157–1165, 10.1016/j.drudis.2019.03.015 (2019). 10.1016/j.drudis.2019.03.015 [DOI] [PubMed] [Google Scholar]
- 4.Wang, Y. et al. In silico adme/t modelling for rational drug design. Quarterly Reviews of Biophysics48, 488–515, 10.1017/s0033583515000190 (2015). 10.1017/s0033583515000190 [DOI] [PubMed] [Google Scholar]
- 5.Sun, J. et al. Excape-db: an integrated large scale dataset facilitating big data analysis in chemogenomics. Journal of Cheminformatics9, 10.1186/s13321-017-0203-5 (2017). [DOI] [PMC free article] [PubMed]
- 6.Bento, A. P. et al. The chembl bioactivity database: an update. Nucleic Acids Research42, D1083–D1090, 10.1093/nar/gkt1031 (2013). 10.1093/nar/gkt1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, S. et al. Pubchem substance and compound databases. Nucleic Acids Research44, D1202–D1213, 10.1093/nar/gkv951 (2015). 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilson, M. K. et al. Bindingdb in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Research44, D1045–D1053, 10.1093/nar/gkv1072 (2015). 10.1093/nar/gkv1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu, Z. et al. Moleculenet: a benchmark for molecular machine learning. Chemical Science9, 513–530, 10.1039/C7SC02664A (2018). 10.1039/C7SC02664A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, K. et al. Therapeutics data commons: Machine learning datasets and tasks for drug discovery and development. arXiv (Cornell University)10.48550/arxiv.2102.09548 (2021).
- 11.Meng, F., Xi, Y., Huang, J. & Ayers, P. W. A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. Scientific Data8, 289, 10.1038/s41597-021-01069-5 (2021). 10.1038/s41597-021-01069-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng, J. et al. Boosting the predictive performance with aqueous solubility dataset curation. Scientific Data9, 10.1038/s41597-022-01154-3 (2022). [DOI] [PMC free article] [PubMed]
- 13.Delaney, J. S. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. ACS Publications10.1021/ci034243x.s001 (2019). [DOI] [PubMed]
- 14.Pollastri, M. P. Overview on the rule of five. Current Protocols in Pharmacology49, 10.1002/0471141755.ph0912s49 (2010). [DOI] [PubMed]
- 15.Sheridan, R. P. et al. Experimental error, kurtosis, activity cliffs, and methodology: What limits the predictivity of qsar models? Journal of Chemical Information and Modeling10.1021/acs.jcim.9b01067 (2020). [DOI] [PubMed]
- 16.Butler, J. N. Ionic equilibrium: solubility and pH calculations (Wiley, 1998).
- 17.OpenAI. Gpt-4 technical report. arXiv (Cornell University)10.48550/arxiv.2303.08774 (2023).
- 18.Gu, Y. et al. Domain-specific language model pretraining for biomedical natural language processing. ACM Transactions on Computing for Healthcare3, 1–23, 10.1145/3458754 (2022). 10.1145/3458754 [DOI] [Google Scholar]
- 19.Lee, J. et al. Biobert: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics36, 10.1093/bioinformatics/btz682 (2019). [DOI] [PMC free article] [PubMed]
- 20.Anil, R. et al. Palm 2 technical report, 10.48550/arXiv.2305.10403 (2023).
- 21.Mazurowski, M. A. et al. Segment anything model for medical image analysis: An experimental study. Medical Image Analysis89, 102918, 10.1016/j.media.2023.102918 (2023). 10.1016/j.media.2023.102918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao, X. et al. Pharmabench: Enhancing admet benchmarks with large language models. figshare10.6084/m9.figshare.25559469.v1 (2024). 10.6084/m9.figshare.25559469.v1 [DOI] [PMC free article] [PubMed]
- 23.Brown, T. et al. Language models are few-shot learners. arXiv (Cornell University)10.48550/arxiv.2005.14165 (2020).
- 24.Sahoo, P. et al. A systematic survey of prompt engineering in large language models: Techniques and applications. arXiv (Cornell University)10.48550/arxiv.2402.07927 (2024).
- 25.Chen, Q. et al. An extensive benchmark study on biomedical text generation and mining with chatgpt. Bioinformatics3910.1093/bioinformatics/btad557 (2023). [DOI] [PMC free article] [PubMed]
- 26.Guo, T. et al. Large language model based multi-agents: A survey of progress and challenges, 10.48550/arXiv.2402.01680 (2024).
- 27.Zhang, B. et al. Controlling large language model-based agents for large-scale decision-making: An actor-critic approach. arXiv (Cornell University)10.48550/arxiv.2311.13884 (2023).
- 28.Xi, Z. et al. The rise and potential of large language model based agents: A survey, 10.48550/arXiv.2309.07864 (2023).
- 29.Landrum, G. A. Rdkit: Open-source cheminformatics. release 2014.03.1. zenodo10.5281/zenodo.10398 (2014). 10.5281/zenodo.10398 [DOI]
- 30.Ames, B. N., Lee, F. D. & Durston, W. E. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proceedings of the National Academy of Sciences70, 782–786, 10.1073/pnas.70.3.782 (1973). 10.1073/pnas.70.3.782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsopelas, F., Giaginis, C. & Tsantili-Kakoulidou, A. Lipophilicity and biomimetic properties to support drug discovery. Expert Opinion on Drug Discovery12, 885–896, 10.1080/17460441.2017.1344210 (2017). 10.1080/17460441.2017.1344210 [DOI] [PubMed] [Google Scholar]
- 32.Cui, Q. et al. Data_Sheet_1_Improved Prediction of Aqueous Solubility of Novel Compounds by Going Deeper With Deep Learning.ZIP. Frontiers10.3389/fonc.2020.00121.s001 (2020). 10.3389/fonc.2020.00121.s001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martins, I. F., Teixeira, A. L., Pinheiro, L. & Falcao, A. O. A bayesian approach to in Silico blood-brain barrier penetration modeling. ACS Publications10.1021/ci300124c (2016). 10.1021/ci300124c [DOI] [PubMed] [Google Scholar]
- 34.Bohnert, T. & Gan, L.-S. Plasma protein binding: From discovery to development. Journal of Pharmaceutical Sciences102, 2953–2994, 10.1002/jps.23614 (2013). 10.1002/jps.23614 [DOI] [PubMed] [Google Scholar]
- 35.Martignoni, M., Groothuis, G. M. M. & de Kanter, R. Species differences between mouse, rat, dog, monkey and human cyp-mediated drug metabolism, inhibition and induction. Expert Opinion on Drug Metabolism & Toxicology2, 875–894, 10.1517/17425255.2.6.875 (2006). 10.1517/17425255.2.6.875 [DOI] [PubMed] [Google Scholar]
- 36.Brian Houston, J. & Carlile, D. J. Prediction of hepatic clearance from microsomes, hepatocytes, and liver slices. Drug Metabolism Reviews29, 891–922, 10.3109/03602539709002237 (1997). 10.3109/03602539709002237 [DOI] [PubMed] [Google Scholar]
- 37.Lord, S. J., Velle, K. B., Mullins, R. D. & Fritz-Laylin, L. K. Superplots: Communicating reproducibility and variability in cell biology. Journal of Cell Biology219, e202001064, 10.1083/jcb.202001064 (2020). 10.1083/jcb.202001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karami, T. K., Hailu, S., Feng, S., Graham, R. & Gukasyan, H. J. Eyes on lipinski’s rule of five: A new “rule of thumb” for physicochemical design space of ophthalmic drugs. Journal of Ocular Pharmacology and Therapeutics38, 43–55, 10.1089/jop.2021.0069 (2022). 10.1089/jop.2021.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nature chemistry4, 90–98, 10.1038/nchem.1243 (2012). 10.1038/nchem.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen, T. & Guestrin, C. Xgboost: A scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘16 785–794, 10.1145/2939672.2939785 (2016).
- 41.Breiman, L. Random forests. Machine Learning45, 5–32, 10.1023/A:1010933404324 (2001). 10.1023/A:1010933404324 [DOI] [Google Scholar]
- 42.Rogers, D. & Hahn, M. Extended-connectivity fingerprints. Journal of Chemical Information and Modeling50, 742–754, 10.1021/ci100050t (2010). 10.1021/ci100050t [DOI] [PubMed] [Google Scholar]
- 43.Wenlock, M. & Tomkinson, N. Experimental in vitro dmpk and physicochemical data on a set of publicly disclosed compounds. ChEMBL10.6019/CHEMBL3301361.
- 44.Boobier, S. et al. Machine learning with physicochemical relationships: solubility prediction in organic solvents and water. Nat Commun11, 5753, 10.1038/s41467-020-19594-z (2020). 10.1038/s41467-020-19594-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J., Hou, T. & Xu, X. Aqueous solubility prediction based on weighted atom type counts and solvent accessible surface areas. ACS Publications10.1021/ci800406y.s005 (2016). 10.1021/ci800406y.s005 [DOI] [PubMed] [Google Scholar]
- 46.Meng, F., Yang, X., Huang, J. & Ayers, P. W. B3db: A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. figshare10.6084/m9.figshare.15634230.v3 (2021). 10.6084/m9.figshare.15634230.v3 [DOI] [PMC free article] [PubMed]
- 47.Adenot, M. & Lahana, R. Blood-brain barrier permeation models: Discriminating between potential cns and non-cns drugs including p-glycoprotein substrates. ACS Publications10.1021/ci034205d.s001 (2019). 10.1021/ci034205d.s001 [DOI] [PubMed] [Google Scholar]
- 48.Xu, C. et al. In silico prediction of chemical ames mutagenicity. ACS Publications10.1021/ci300400a (2016).
- 49.Dimitrov, S. D. et al. Qsar toolbox – workflow and major functionalities. SAR and QSAR in Environmental Research27, 203–219, 10.1080/1062936X.2015.1136680 (2016). 10.1080/1062936X.2015.1136680 [DOI] [PubMed] [Google Scholar]
- 50.Hansen, K. et al. Benchmark data set for in silico prediction of ames mutagenicity. ACS Publications10.1021/ci900161g (2016). [DOI] [PubMed]
- 51.Song, Y. et al. Communicative representation learning on attributed molecular graphs. Griffith Research Online (Griffith University, Queensland, Australia)10.24963/ijcai.2020/392 (2020).
- 52.Cai, H., Zhang, H., Zhao, D., Wu, J. & Wang, L. Fp-gnn: a versatile deep learning architecture for enhanced molecular property prediction. Briefings in Bioinformatics2310.1093/bib/bbac408 (2022). [DOI] [PubMed]
- 53.Song, Y., Chen, J., Wang, W., Chen, G. & Ma, Z. Double-head transformer neural network for molecular property prediction. Journal of Cheminformatics1510.1186/s13321-023-00700-4 (2023). [DOI] [PMC free article] [PubMed]
- 54.Yin, F. et al. Knowledge graph-enhanced molecular contrastive learning with functional prompt. Nature Machine Intelligence5, 542–553, 10.1038/s42256-023-00654-0 (2023). 10.1038/s42256-023-00654-0 [DOI] [Google Scholar]
- 55.Li, P. et al. An effective self-supervised framework for learning expressive molecular global representations to drug discovery. Briefings in Bioinformatics2210.1093/bib/bbab109 (2021). [DOI] [PubMed]
- 56.Zhou, G. et al. Uni-mol: A universal 3d molecular representation learning framework. chemrxiv.org10.26434/chemrxiv-2022-jjm0j (2022). 10.26434/chemrxiv-2022-jjm0j [DOI] [Google Scholar]
- 57.Luo, S. et al. One transformer can understand both 2d & 3d molecular data. arXiv (Cornell University)10.48550/arxiv.2210.01765 (2022). 10.48550/arxiv.2210.01765 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Xiao, X. et al. Pharmabench: Enhancing admet benchmarks with large language models. figshare10.6084/m9.figshare.25559469.v1 (2024). 10.6084/m9.figshare.25559469.v1 [DOI] [PMC free article] [PubMed]
- Landrum, G. A. Rdkit: Open-source cheminformatics. release 2014.03.1. zenodo10.5281/zenodo.10398 (2014). 10.5281/zenodo.10398 [DOI]
- Meng, F., Yang, X., Huang, J. & Ayers, P. W. B3db: A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. figshare10.6084/m9.figshare.15634230.v3 (2021). 10.6084/m9.figshare.15634230.v3 [DOI] [PMC free article] [PubMed]
Data Availability Statement
The codes used in this study have been deposited at https://github.com/mindrank-ai/PharmaBench. All the calculations were done with Python 3.12.2 under a virtual environment created with Anaconda on osx-64.