Summary
Given the resurgence of syphilis, research endeavors to improve current assays for serological diagnosis and management of this disease are a priority. A proteome-scale platform for high-throughput profiling of the humoral response to Treponema pallidum (T. pallidum) proteins during infection could identify antigens suitable to ameliorate the performance and capabilities of treponemal tests for syphilis. Additionally, because infection-induced immunity is partially protective, profiling the response to T. pallidum outer membrane proteins (OMPs) could help select vaccine candidates. Therefore, we developed a pan-proteome array (PPA) based on the Nichols and SS14 strain complete proteomes and used it to define the immunoglobulin M (IgM) and IgG humoral response to T. pallidum proteins in sera collected longitudinally from long-term infected rabbits and from rabbits that were infected, treated, and re-infected. We identified antigens that could facilitate early diagnosis and immunity to a core set of OMP that could explain protection upon reinfection.
Subject areas: Diagnostics, Immune response, Bacteriology, Proteomics, Model organism
Graphical abstract
Highlights
-
•
We constructed a panproteomic array (PPA) based on the T. pallidum proteome
-
•
The PPA was tested with sera from syphilis-infected rabbits
-
•
Only a small portion of the T. pallidum proteome was recognized
-
•
Reactive antigens might be used to improve current diagnostics and develop vaccines
Diagnostics; Immune response; Bacteriology; Proteomics; Model organism
Introduction
Syphilis is a multistage chronic sexually transmitted infection that is still a significant burden for public health despite being treatable. The World Health Organization (WHO) estimated the syphilis global prevalence and incidence to be between 18 and 36 million cases and 5.6–11 million new annual cases, respectively.1,2 Although most cases occur in low- and middle-income countries, syphilis rates have also steadily increased for two decades in high-income countries.3,4,5,6,7,8 In the US, the rate of primary and secondary syphilis in 2022 (17.7 cases per 100,000 population) represented a 743% increase compared to the 2.1 cases per 100,000 population reported in 2000.3 If untreated, syphilis might progress to affect the cardiovascular and central nervous systems, potentially leading to severe manifestations such as aortic aneurism, stroke-like syndrome, dementia, and paralysis.9 Because the syphilis agent, Treponema pallidum subsp. pallidum (T. pallidum), can cross the placenta, mother-to-child transmission of the infection during pregnancy can lead to stillbirth, perinatal death, and a plethora of other adverse pregnancy outcomes, with severe repercussions for maternal and infant health. Syphilis global epidemiology strongly supports the need for novel research endeavors to curtail the spread of this serious infection. Research efforts that aimed at improving diagnostic approaches to both reduce the temporal gap between exposure and diagnosis to minimize the consequences of T. pallidum dissemination to virtually all bodily organs from the site of infection and assess treatment efficacy, for example, are highly warranted.
Laboratory diagnosis of syphilis is mainly achieved through serology, with a testing algorithm encompassing the use of two separate and complementary tests.10 Lipoidal serologic tests, such as the venereal disease research laboratory (VDRL), detect antibodies to lipoidal molecules released by both damaged host cells and T. pallidum during active infection,11 and they are generally referred to as non-treponemal tests (NTT, albeit this is now considered a misnomer). Seroconversion, however, occurs 3–6 weeks after exposure to the pathogen. The relatively low sensitivity of these assays (74%–99%) sometimes causes failure to detect infection in patients with primary syphilis. The fluctuation of serum titer of lipoidal tests is, however, important to monitor successful response to treatment, relapse, or reinfection following a syphilis episode. An effective antibiotic therapy should induce a 4-fold decrease in lipoidal test titer within 3–12 months (depending on the infection stage at diagnosis).12 If a history of syphilis is known, a rise in lipoidal test titer generally supports reinfection or treatment failure and the need for additional treatment.
A positive lipoidal test result is verified with a treponemal test (TT) to detect antibodies to specific T. pallidum antigens, such as the T. pallidum particle agglutination assay (TPPA), the fluorescent treponemal antibody absorption (FTA-ABS) test, or the chemiluminescence and enzyme immunoassays (CIA, EIA). Treponemal tests like the TPPA and FTA-ABS are performed manually and use whole T. pallidum as the source of antigen (either intact or sonicated), whereas automated high-throughput assays like the EIA and CIA employ recombinant T. pallidum lipoproteins, known to be highly specific for T. pallidum subspecies, abundantly expressed, and immunodominant. Although TTs have excellent sensitivity and specificity, their efficacy is still limited in the diagnosis of early syphilis,10 do not discriminate between active or past infections like lipoidal tests do, and cannot be used to assess syphilis stage, even though limited research suggests that differential immune responses to specific T. pallidum antigens might occur during infection progression and in response to therapy.13
Developing and testing an antigen array encompassing the whole T. pallidum proteome could lead to the identification and recruitment of additional antigens to increase the sensitivity and overall capabilities of TTs based on recombinant proteins and perhaps provide evidence on whether a TT could allow for disease staging based on differential immunoreactivity over time or help monitor treatment response, relapse, or reinfection in previously treated patients.
Furthermore, it is established that long-term infected rabbits (>90 days) develop partially protective immunity, which leads to attenuation of disease manifestations upon reinfection,14,15 and clinical studies have suggested that patients who experienced a syphilis episode are more likely to be asymptomatic with a subsequent episode.16,17,18,19,20,21 These findings strongly suggest that an effective syphilis vaccine may be attainable if the right antigens are targeted. Because evidence exists that antibodies to T. pallidum surface-exposed outer membrane proteins (OMPs) are significant mediators of protective immunity,22,23,24 the identification of the OMP that preferentially elicit a humoral response during experimental infection could provide clues regarding candidates to include in an experimental vaccine design.
In this study, we constructed a panproteomic array (PPA) based on the annotated proteomes of the Nichols and SS14 T. pallidum isolates, which are the model laboratory strains for the two genetically distinct clades of this pathogen currently circulating worldwide.25 The PPA was tested with sera collected longitudinally from long-term Nichols- and SS14-infected animals and from animals that were infected with either Nichols or SS14, treated, and then reinfected with the homologous strain two months post-treatment. Here, we report on the portion of T. pallidum proteome recognized during experimental infection with either strain, the immunoglobulin G (IgG) and IgM antibody profiles that develop in infected rabbits, and the antibody kinetic profiles in long-term infected animals and in rabbits that were reinfected following effective treatment with benzathine penicillin G (BPG).
Results
Rabbit infection and serology
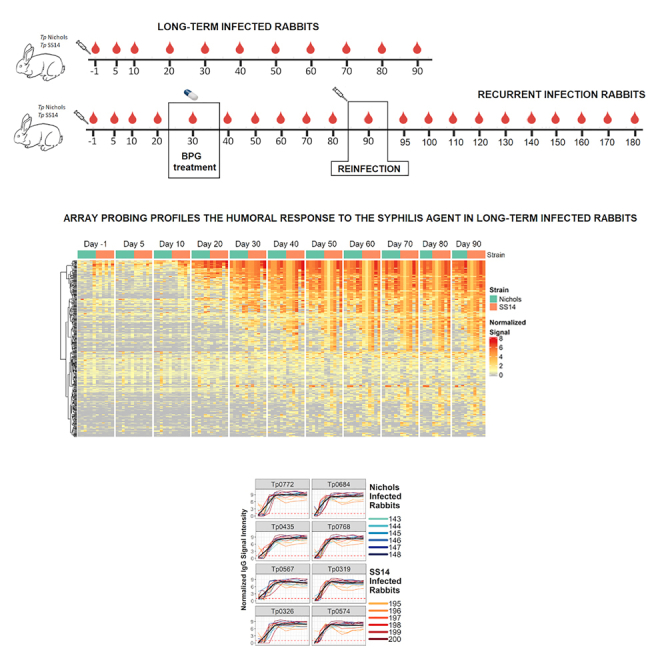
For the experimental infections, two groups of 12 animals were infected intradermally (ID) with either the Nichols strain or the SS14 strain. Each group was then divided into sub-groups of six animals. The first subgroup of Nichols-infected animals was labeled Nichols long-term (N.LT) because these animals were infected and followed longitudinally for 90 days, during which serum samples were collected at day 5 and day 10 post-infection and, subsequently, every 10 days until day 90 post-infection. The second subgroup of six Nichols-infected animals was labeled Nichols reinfection (N.RI) because animals were injected intramuscularly (IM) at day 30 post-inoculation with 200,000 units of BPG (Bicillin L-A, Pfizer, New York, NY) and, two months post-treatment (at study day 90), reinfected. From these animals, serum samples were obtained before initial inoculation, at day 5 and day 10 post-initial infection and every 10 days after for the remainder of the 180-day experiment. The same experimental design was applied to the 12 animals infected with the SS14 strains, which were identified as the SS14 long-term (S.LT) and SS14 reinfection (S.RI) groups, respectively.
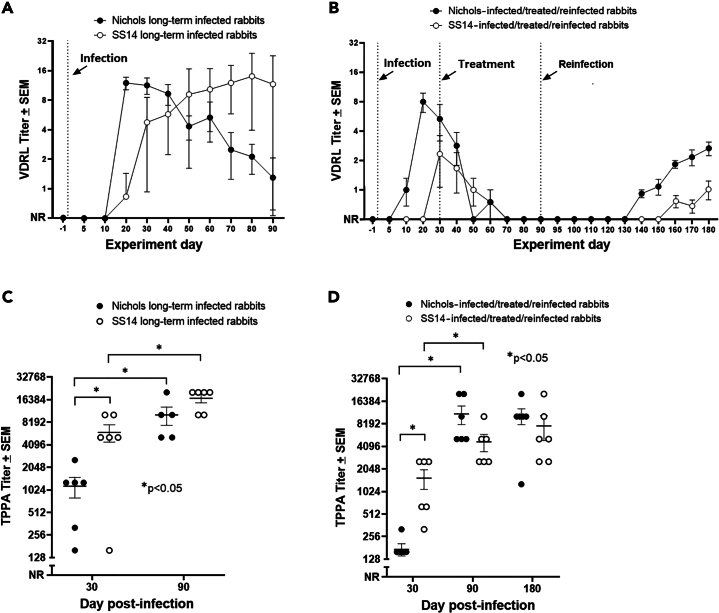
Following infection, the VDRL and TPPA tests were used post-inoculation with T. pallidum to assess the establishment and progression of the infection. Over time, all rabbits in both the long-term infected N.LT and S.LT groups became VDRL positive, even though rabbits inoculated with the Nichols strain seroconverted earlier than those inoculated with the SS14 strain (Figure 1A). More specifically, all the N.LT rabbits were VDRL positive by day 20 post-inoculation, whereas all S.LT rabbits were positive by day 40 post-inoculation (Figure 1A). The highest VDRL titer was 1:16 (for three and four animals in the N.LT and S.LT groups, respectively). By day 50 post-inoculation, titers had declined in most N.LT animals, whereas a decline began at later time points for the S.LT rabbits. These results are consistent with an active infection established in all animals. Consistently, all the rabbits in the N.RI group (Figure 1B) were VDRL positive by day 20 post-inoculation, whereas only two (33%) rabbits in the S.RI group (Figure 1B) seroconverted before treatment (day 30). All of the N.RI and S.RI VDRL-positive rabbits seroconverted to a nonreactive VDRL within a month following treatment administration. Upon animal reinfection (60 days post-BPG treatment; experiment day 90), an increase in the mean VDRL reactivity was measurable starting at day 140 for the animals in the N.RI group (∼50 days after reinoculation; Figure 1B) and day 160 for the SS14-infected animals (∼70 days after reinoculation; Figure 1B). At the end of the experiment (day 180 post-infection), all N.RI and S.RI animals had seroconverted, albeit titers remained lower in S.RI rabbits than in rabbits from the N.RI group. These results support that infection was established, cured, and established again in the RI animal groups.
Figure 1.
Serological test results for rabbits enrolled in this study
(A) VDRL titers for long-term (LT) infected rabbits inoculated with either the Nichols or SS14 strain measured over 90 days post-infection. Vertical dotted line indicates the infection event (experiment day 0).
(B) VDRL titers for rabbits in the reinfection (RI) groups inoculated with either the Nichols or SS14 strain and measured over 180 days post-infection. Vertical dotted lines mark the infection, treatment, and reinfection events (experiment day 0, 30, and 90, respectively). For calculating the mean ± SEM, titers were converted to log2 with nonreactive = −1.0; weakly reactive = −0.5; reactive 1:1 = 0, reactive 1:2 = 1, and so forth. Antilogs of mean ± SE of titers are shown in this figure with nonreactive set at y = 0.
(C) TPPA titers for long-term (90 days) infected rabbits inoculated with either the Nichols or SS14 strain measured at day 30 and day 90 post-infection.
(D) TPPA titers for rabbits in the RI groups inoculated with either the Nichols or SS14 strain measured at day 30 (time of treatment), day 90 (time of reinfection), and 180 post-infection. Data are represented as mean ± SEM. Asterisks identify significant differences following analysis with Student’s t test with significance set at p < 0.05.
All infected rabbits in the N.LT and S.LT groups were TPPA positive at day 30 post-inoculation and displayed significantly higher mean titers at day 90 post-infection than at day 30 (Figure 1C), which was consistent with infection progression. The mean titer was significantly higher in the S.LT group than in the N.LT group at day 30 but not significantly different between the two groups at day 90 post-infection. All the N.RI and S.RI rabbits had seroconverted by day 30 post-inoculation (Figure 1D), supporting establishment of the infection also in the three S.RI animals that did not become VDRL positive before treatment (Figure 1B). Compared to day 30, TPPA titers were significantly higher also for the N.RI and S.RI groups at the time of reinfection (day 90) but not significantly different between the two groups at day 90 post-infection. Day 180 titers were not significantly different from day 90 titers for both groups (Figure 1D). Overall, these serological data further support that inoculation with T. pallidum led to a productive infection in all animals.
Profiling of the humoral responses in long-term infected rabbits
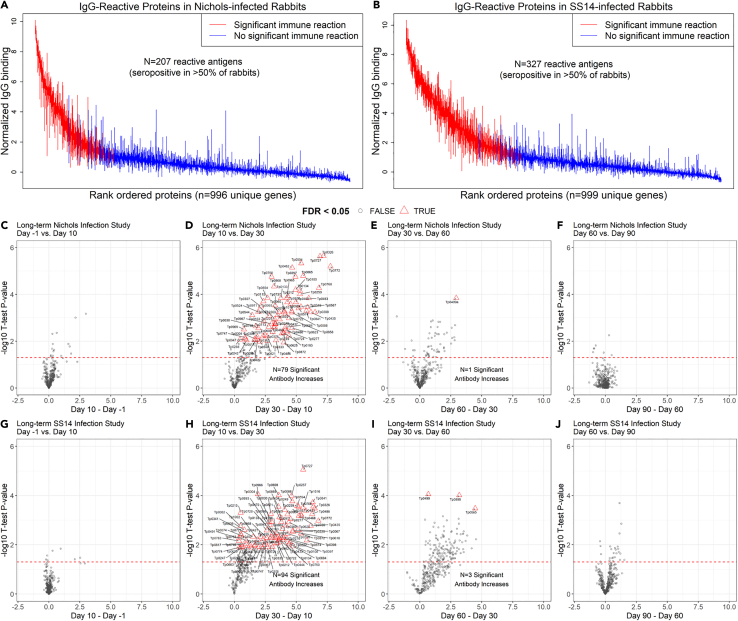
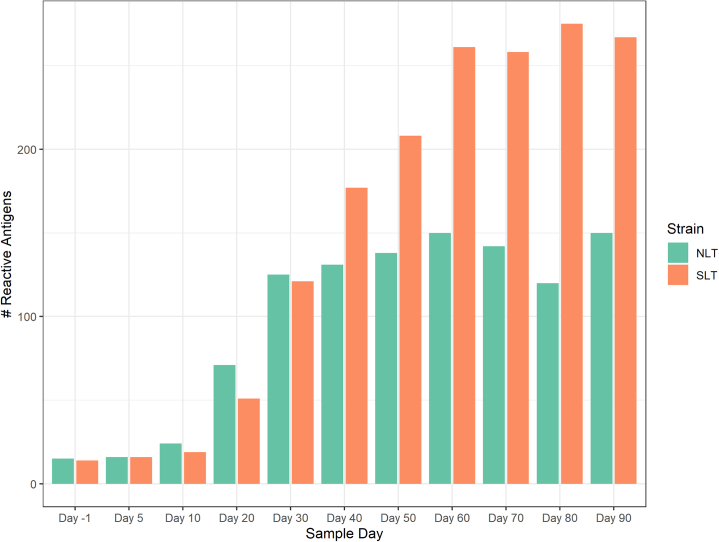
Overall, sera collected from the N.LT rabbits showed that a total of 207 antigens over 1,008 unique targets (encompassing core and Nichols-specific proteins) were significantly recognized over the course of the infection (Figure 2A). Positive targets represented approximately ∼20% of the Nichols strain proteome (as annotated in NC_021490.2). At the end of the 3-month infection period, sera from S.LT animals recognized a total of 327 antigens, corresponding to ∼31% of the SS14 proteome (as annotated in NC_021508.1; Figure 2B). In N.LT animals, there were no significant increases in specific IgG responses to T. pallidum antigens by 10 days post-infection, although there was a non-significant positive trend for a few antigens (Figure 2C). By day 30, there were 79 antibody responses that significantly increased from day 10 (Figure 2D). From day 30 to day 60, no additional antibody responses were significantly increased based on our set false discovery rate (FDR), except for Tp0409a/YajC preprotein translocase subunit. At this time point, however, a non-significant positive trend was detectable for numerous antigens (Figure 2E), represented by circles above the dotted line. The change in the number of targeted proteins observed between day 60 and day 90 post-infection was not significant (Figure 2F). Animals infected long-term with the SS14 strain (S.LT) also had no significant changes by day 10 post-infection (Figure 2G), whereas 94 antibody responses were significantly increased from day 10 to day 30 (Figure 2H) and three from day 30 to day 60 (Figure 2I), corresponding to the hypothetical proteins Tp0565 and Tp0895 and the Tp0499/MreD, a rod-shape determining protein. As for the N.LT animals, no significant changes in antibody levels were observed between day 60 and day 90 post-infection in S.LT animals (Figure 2J), aside from a positive trend for a few targets. The discrepancy between total number of reactive antigens and number of significantly increased antibodies between time points is explained by the statistical power to detect incremental increases in antibody levels between sequential 10-day intervals and variation in the numbers of reactive antigens at different time points, as opposed to reactivity by maximal responses across the study period. The increase in the number of proteins significantly targeted by IgG antibodies began between day 10 and day 20 for both N.LT and S.LT animals and continued through day 30 but were not significantly increased thereafter (Figure S1), likely due to limits in statistical power. In contrast, the numbers of reactive antigens at each time point (i.e., antigens seropositive in over half of animals at each study day) were similar for N.LT and S.LT animals until day 30, thereafter plateauing for N.LT but continuing to increase until day 60 post-infection for S.LT animals, to then reach plateau (Figure 3). The breadth of reactive antigens with increasing antibody levels was more consistent with the number of reactive antigens when time points were compared against baseline (Figure S2). The complete list of antigens recognized in both the N.LT and S.LT strains and time to a seropositive response is provided in Table S1 (IgG summary statistics).
Figure 2.
IgG reactivity and longitudinal trajectory in long-term (N.LT and S.LT) infected rabbits
(A and B) Interquartile range plots showing the maximal normalized IgG binding signal for each rabbit for each T. pallidum protein over the 90-day period of the long-term infection. Each bar represents a protein on the array, and proteins are ordered by the median signal of (A) Nichols-infected (N.LT) rabbits and (B) SS14-infected (S.LT) rabbits. Red bars represent a significantly reactive protein or proteins with seropositive responses in at least four out of six total rabbits in the group.
(C–J) Volcano plots showing the difference in normalized IgG binding between time points for each reactive protein on the array (x axis) by the inverse log10p value of paired Student’s t tests. In each plot, the horizontal red dashed line represents an unadjusted p value of 0.05. All values above the red line are below 0.05. After adjustment for the false discovery rate, antibody responses that remained statistically significant were highlighted as red triangles and labeled with the antigen annotated identifier.
(C–F) Plots showing sequential time point differences for N.LT animals.
(G–J) Plots showing sequential time point differences for S.LT animals. Data are represented as mean ± SEM.
Figure 3.
Antigens with significant immune responses at each time point in long-term (N.LT and S.LT) infected rabbits
Bar plot showing the count of T. pallidum proteins with significant reactivity or proteins with seropositive responses in at least four out of six total rabbits in each group at each given time point. The grouped orange bars represent Nichols-infected (N.LT) animals, whereas green bars represent long-term SS14-infected (S.LT) animals. Data are represented as mean ± SEM.
The robust IgG response that developed in both LT rabbit groups was accompanied by a weak IgM response. More specifically, in the N.LT- and S.LT-infected animals, respectively, a total of 13 proteins elicited a weak but significant IgM response (Table S1, IgG summary statistics; Figure S3A). The rabbits in the N.LT group recognized 11 targets, whereas in the S.LT animals only five (Figure S3A). In N.LT animals, our stepwise regression model associated significant IgM reactivity to the Tp0319-TmpC, Tp0435-TpN17, and Tp0684-MglB lipoproteins, while the remaining targets were annotated as either hypothetical proteins (Tp0707 and Tp0990) or proteins with random functions such as Tp0154 (one of the three T. pallidum pseudouridylate synthase), albeit only at two time points post-inoculation, Tp0567 (PrfB peptide chain release factor II), TP0653 (PotB ABC transporter permease), and Tp0727 (FlgE flagellar hook protein). The only putative OMP recognized was Tp0733 (an OmpW homolog) (Figure S3A). The S.LT rabbits, on the contrary, only recognized the Tp0435-TpN17 and Tp0768-TmpA lipoproteins, the Tp0772 hypothetical protein, Tp0733-OmpW, and the Tp0567-PrfB factor. In both groups, the Tp0435-Tp17 protein elicited the strongest IgM response (Figure S3A).
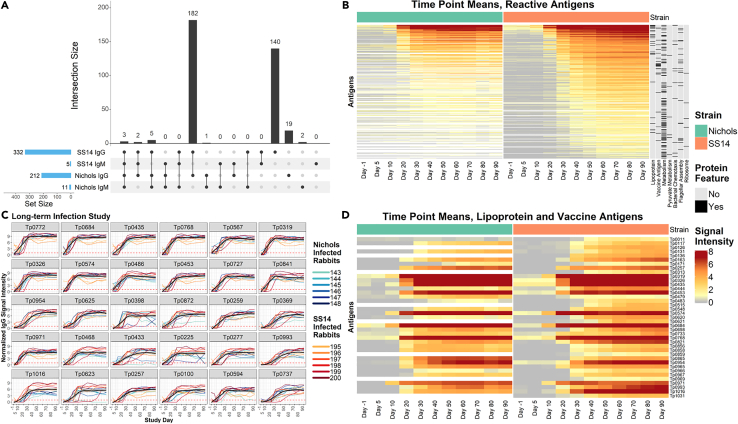
Of the 351 total targets in the array found to have elicited a significant IgG response in the long-term infected animals (regardless of the infecting strain), 182 antigens were recognized by both the Nichols- and SS14-infected rabbits, whereas 140 antigens were recognized only by sera from the SS14-infected animals, and 19 antigens elicited a response exclusively in Nichols-infected rabbits (Figure 4A; Table S1, IgG summary statistics). Stepwise regression identified seven main categories of reactive proteins/enzymes (Figure 4B). In order of significance, such categories were lipoproteins (p = 2.0−9), metabolism-associated enzymes (p = 6.8−6), flagellar assembly proteins (p = 6.2−5), enzymes specific for pyruvate metabolism (p = 2.0−4), vaccine candidates (i.e., OMPs; p = 1.3−4), ribosomal proteins (p = 2.9−2), and chemotaxis-related proteins (p = 2.1−2). Antibody kinetics shown for individual rabbits for the 30 most reactive antigens in both N.LT and S.LT groups support the early development of reactivity to these targets, and a limited rabbit-to-rabbit variability (Figure 4C; Table 1). The analysis of reactivity limited to annotated/putative T. pallidum lipoproteins and putative OMPs/vaccine candidates (Figure 4D) showed that almost the entirety of these targets was recognized by the S.LT rabbit group, although at various degrees of intensity. The Tp0684-MglB, Tp0435-TpN17, Tp0768-TmpA, Tp0319-TmpC, Tp0574-TpN47, Tp0453 (a concealed outer-membrane-associated protein),26 Tp0954 (a putative placental adhesin),27 Tp0971-TpD iron transporter, Tp0993-RlpA, and Tp1016 (a putative lipoprotein with peptidyl prolyl cis/trans isomerase function) were the most significantly recognized lipoproteins, whereas Tp0326-BamA was the most highly recognized OMP (Figure 4D). Aside from Tp0326-BamA, most OMPs currently evaluated as vaccine candidates elicited a significant but overall weak immunity. Compared to the S.LT rabbits, the Nichols-infected animals failed to recognize the Tp0126-OmpW, Tp0313-TprE, Tp0515-LptD, Tp0621-TprJ, Tp0859-FadL, Tp0969-TolC, Tp01031-TprL OMPs, and the Tp0136 putative outer membrane lipoprotein/adhesin.28 Tp0011-TprB was recognized in both strains, but the signal intensity was higher in S.LT rabbits than N.LT ones.
Figure 4.
IgG binding profiles in N.LT and S.LT rabbits
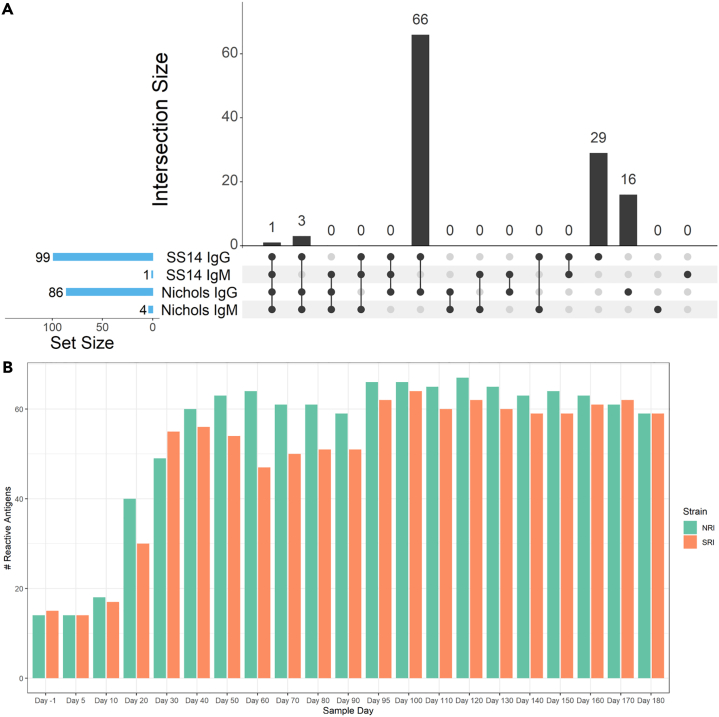
(A) UpSet plot summarizing both the set and set size of antigens recognized by IgG and IgM during experimental infection of rabbits with either the Nichols or SS14 strains of T. pallidum. The UpSet plot is equivalent to a Venn diagram, where each vertical bar represents antibody responses that “intersect” or overlap between two or more categories (i.e., antigens that are reactive in each of the categories specified by the connected dot matrix below the bar graph). The blue horizontal bars represent the total number of reactive antigens in each category.
(B) Heatmap showing the mean IgG binding signal intensity for each reactive antigen in either Nichols or SS14-strain-infected rabbits. Columns represent the means of each group of rabbits at each sequential time point, and rows represent T. pallidum proteins sorted by hierarchical clustering. Protein features that were significantly associated with antibody reactivity in stepwise regression models are shown in the black and gray columns to the right of the heatmap. Responses are grouped by Nichols- (green header) and SS14 (orange header)-strain-infected rabbits.
(C) Line plots of reactivity to the 30 most reactive antigens in animals infected long-term with the Nichols strain (teal to dark blue lines) and the SS14 strain (orange to dark red lines). In each graph, the black line represents the running mean of all samples at each time point post-infection.
(D) Heatmap of the antigens annotated as lipoproteins or OMPs/vaccine candidates (ordered by gene ID) recognized in Nichols-infected animals (left side, green header), and SS14-infected rabbits (right side, orange header) over the 90-day infection period. Data are represented as mean ± SEM.
Table 1.
Most significantly recognized T. pallidum antigens in N.LT and S.LT rabbits
| ORF #a | Putative gene function | Normalized IgG binding (Nichols-infected rabbits/SS14-infected rabbits)b | Seroprevalence (%) N.LT/S.LT rabbits |
Time to 100% seroprevalence (day) N.LT/S.LT rabbits |
|---|---|---|---|---|
| Tp0772 | Hypothetical protein/transcriptional regulator | 7.20/7.34 | 100/100 | 20/20 |
| Tp0684 | MglB/glucose-galactose binding lipoprotein | 6.89/7.09 | 100/100 | 10/20 |
| Tp0435 | TpN17 lipoprotein | 6.31/6.90 | 100/100 | 20/20 |
| Tp0768 | TmpA lipoprotein | 6.74/6.91 | 100/100 | 20/20 |
| Tp0567 | FlbB/flagellar protein | 6.37/6.34 | 100/100 | 20/20 |
| Tp0319 | TmpC/membrane lipoprotein; possible ABC transporter solute binding lipoprotein | 6.31/6.47 | 100/100 | 20/10 |
| Tp0326 | BamA/OMP assembly factor | 6.05/6.42 | 100/100 | 20/30 |
| Tp0574 | TpN47 lipoprotein | 6.01/6.03 | 100/100 | 20/10 |
| Tp0486 | Hypothetical protein | 5.54/6.78 | 100/100 | 10/20 |
| Tp0453 | Concealed OMP | 5.69/5.77 | 100/100 | 20/20 |
| Tp0727 | FlgE/flagellar hook protein | 5.68/5.83 | 100/100 | 20/20 |
| Tp0841 | DegQ/serine endoprotease | 5.36/6.06 | 100/100 | 20/20 |
| Tp0954 | Lipoprotein/putative adhesin | 4.78/5.37 | 100/100 | 30/30 |
| Tp0625 | Hypothetical protein | 4.85/5.41 | 100/100 | 30/30 |
| Tp0398 | FliE/flagellar hook-basal body complex protein | 4.29/4.88 | 100/100 | 30/50 |
| Tp0872 | FliD/flagellar filament capping protein | 4.76/5.41 | 100/100 | 30/50 |
| Tp0259 | LysM/peptidoglycan-binding domain-containing protein | 4.99/5.14 | 100/100 | 20/20 |
| Tp0369 | BamD/OMP assembly factor BamD | 3.95/4.65 | 100/100 | 30/40 |
| Tp0971 | TpD/iron transporter lipoprotein | 4.73/5.06 | 100/100 | 10/20 |
| Tp0468 | Tetratricopeptide repeat protein | 4.08/5.36 | 100/100 | 20/30 |
| Tp0433 | Hypothetical protein | 3.77/4.52 | 100/100 | 40/70 |
| Tp0225 | Leucine-rich repeat domain-containing protein | 3.62/5.65 | 100/100 | 20/20 |
| Tp0277 | Ctp peptidase | 3.84/5.25 | 100/100 | 30/30 |
| Tp0993 | RlpA/septal ring lytic transglycosylase | 3.24/4.14 | 100/100 | 30/40 |
| Tp1016 | Peptidyl-prolyl cis-trans isomerase | 2.86/5.40 | 100/100 | 30/30 |
| Tp0618 | Hypothetical protein | 3.31/4.43 | 100/100 | 30/40 |
| Tp0100 | TlpA/disulfide reductase | 3.66/4.71 | 100/100 | 30/40 |
| Tp0257 | GpD/glycerophosphodiester phosphodiesterase | 4.02/4.19 | 100/100 | 20/20 |
| Tp0594 | Hypothetical protein | 3.83/4.29 | 100/100 | 30/40 |
| Tp0737 | Periplasmic binding protein ABC transporter | 3.76/3.46 | 100/100 | 30/50 |
The line plot is selecting antigens by the means of the maxes for each rabbit. For each protein, the maximum of all time points for each rabbit is calculated, and then the mean of those maxes is assigned as the value to sort by.
Based on the annotation of the Nichols (NC_021490.2) and SS14 strain (NC_021508.1) genomes.
Mean reactivity values over 90 days post-infection.
Profiling of the humoral responses in treated/reinfected rabbits
A subset of T. pallidum antigens elicited a gradual and robust IgG response also in the two groups of RI animals (Figure 5A). More specifically, at the end of the 180-day experiment, following treatment and reinfection, a total of 86 antigens had elicited a significant response in Nichols-infected rabbits, whereas 99 targets were recognized by rabbits infected with the SS14 strain (Figure 5A). Most of the recognized targets (n = 66) were shared between the two N.RI and S.RI animals, whereas 29 and 16 proteins, respectively, were recognized only by SS14- or Nichols-infected rabbits (Figure 5A; Table S2, Complete list of antigens recognized in LT rabbits). Compared to LT animals, fewer targets (n = 4) elicited an IgM response in RI rabbits (Figures 5A and S3B). More specifically, the N.RI rabbits recognized the Tp0435-TpN17 and Tp0574-TpN47 lipoproteins, as well as the hypothetical protein Tp0707 and the Tp0727-FlgE flagellar hook protein. The S.RI rabbits, on the contrary, only recognized the Tp0707 hypothetical protein (Figure S3B).
Figure 5.
Comparison of antibody responses
(A) Overlap in IgG and IgM antibody responses between N.RI and S.RI rabbit groups. The UpSet plot is equivalent to a Venn diagram, where each vertical bar represents antibody responses that “intersect” or overlap between two or more categories, i.e., antigens that are reactive in each of the categories specified by the connected dot matrix below the bar graph. The blue horizontal bars represent the total number of reactive antigens in each category.
(B) Antigens with significant immune responses at each time point in the N.RI and S.RI rabbit groups. The bar plot shows the count of T. pallidum proteins with significant “reactivity” or proteins with seropositive responses in at least four of six rabbits at each given time point. The grouped green bars represent Nichols-infected (N.RI) animals, whereas orange bars represent long-term SS14-infected (S.RI) animals.
As seen for the LT rabbits, significant increases in the number of IgG-reactive proteins began between day 10 and day 20 for both N.RI and S.RI animals and continued throughout day 30. Following treatment (day 30 post-infection), the number of reactive antigens increased in N.RI rabbits to reach plateau at experiment day 40 but remained virtually unchanged in the S.RI rabbits. Following successful reinfection (per VDRL serology; Figure 1B), the total number of reactive proteins slightly increased in both animal groups (Figure 5B). The complete list of antigens recognized in both the N.RI and S.RI rabbits and time to a seropositive response is provided in Table S1 (IgG summary statistics).
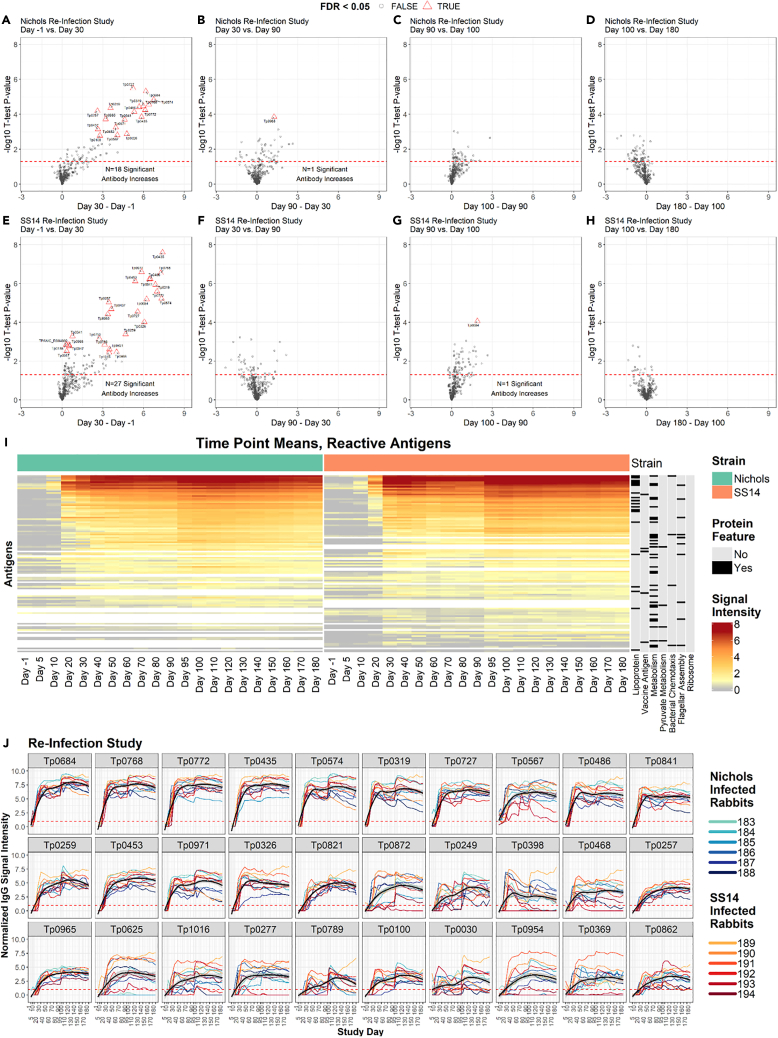
Volcano plots comparing seroreactivity to T. pallidum proteins in animals prior to infection and in sera collected at day 30 post-infection (time of treatment) with the Nichols strain showed that significant reactivity had developed to 18 antigens (Figure 6A; Table S1, IgG summary statistics). The same analysis using later time point sera supported that the increment in the number of significantly recognized antigens was virtually nonexistent (Figures 6B–6D), although a positive trend was seen for numerous proteins (circles above the dotted line). Unlike the animals in the LT groups, rabbits in the S.RI group developed reactivity to a similar number of antigens over the course of the experiment when compared to the N.RI rabbits. Volcano plots comparing the reactivity between different time points in the S.RI animals showed that reactivity to most antigens (n = 27) developed before treatment (Figure 6E) and was not significantly increased by reinfection (Figures 6F–6H).
Figure 6.
IgG reactivity profiles of rabbits that were infected, treated, and then reinfected
(A–H) Volcano plots show the difference in normalized IgG binding between time points for each reactive protein on the array (x axis) by the inverse log10p value of paired Student’s t tests. The horizontal red dashed lines represent an unadjusted P-value of 0.05, and all values above the red line are below 0.05. After adjustment for the false discovery rate, antibody responses that remained statistically significant were highlighted as red triangles and labeled with the antigen ID. (A–D) are sequential time point differences for Nichols-infected (N.RI) animals, and (E–H) are for SS14-infected (S.RI) animals.
(I) Heatmap showing the mean IgG binding signal intensity for each of the 115 antigens that were reactive in either N.RI or S.RI rabbits. Columns represent the means of each group of rabbits at each sequential time point, and rows represent T. pallidum proteins sorted by hierarchical clustering. Protein features that were significantly associated with antibody reactivity in stepwise regression models are shown in the black and gray columns to the right of the heatmap. Responses are grouped by Nichols- (green header) and SS14 (orange header)-strain-infected rabbits.
(J) Line plots showing the longitudinal trajectories of the 30 most reactive antigens by seroprevalence and normalized intensity. Each individual rabbit’s normalized IgG binding signal intensity is plotted as a colored line. Nichols-infected rabbits are teal to dark blue lines, and SS14-infected rabbits are orange to dark red lines. In each graph, the black line represents the running mean of all samples at each time point post-infection. Values are reported in Table S1 (IgG summary statistics).
The heatmap of reactive antigens (Figure 6J) graphically shows the effect of BPG treatment (day 30) and reinfection (day 90) in RI animals. Reactivity to several antigens declined over the two months that followed treatment, to then increase again starting at day 95, five days after animal reinoculation. Overall, a total of 115 proteins were collectively recognized in these two rabbit groups (Figure 5A; Table S2, Complete list of antigens recognized in LT rabbits). Antibody kinetics shown for individual rabbits for the 30 antigens in both the N.RI and S.RI that were most significantly recognized in both rabbit groups (Figure 6J) showed that a decline in reactivity (black line, representing the running mean of the values form each rabbit from each time point) occurred for several antigens when individual reactivities were averaged but was never found to be significant at experiment day 90 (at the time of reinfection) compared to experiment day 30, at the time of treatment. Analysis of the kinetic for other antigens, also considering the number of rabbits that seroconverted over the observation period, did not support that any of the reactive targets could be effectively used to monitor treatment response in a fashion comparable to lipoidal serological tests. Table S1 (IgG summary statistics) provides reactivity values for each antigen recognized in both the N.RI and the S.RI rabbits.
Antibody response to strain-specific targets
Several targets, reported in Tables 2, S1, S2, and S3 (IgG summary statistic and Complete list of antigens recognized in LT and RI rabbits, respectively), were labeled as strain-specific proteins due to their partial sequence identity between the Nichols and SS14 strains or because differently annotated by in silico pipelines, like in the case of Tp0040 and Tp0134c. Of the 13 strain-specific targets that elicited a significant IgG response in any of the rabbit groups analyzed in our study (Table 2), there were two putative OMPs, represented by the putative porins Tp0131 (the TprD/TprD2 antigens in Nichols and SS14, respectively) and Tp0548 (a FadL homolog, whose encoding sequence is widely used for T. pallidum strain typing).29 Immunity to both Tp0131 variants was detected in both N.LT and S.LT rabbits, although not in the RI animals, likely due to early treatment of the animals and the low level of expression of OMPs in this pathogen.30,31 Consistent with the fact that the TprD/TprD2 variants differ significantly within their central region, that sequence identity exists also with other Tpr paralogs of the syphilis agents (e.g., TprF and TprI),32 and that our array does not have a peptide corresponding uniquely to the TprD2 targeted, however, it is unclear whether the detected immunity is truly specific to Tp0131. The SS14-Tp0548 variant was only recognized in S.LT-infected rabbits, whereas reactivity to the Nichols variant was detected in both N.LT and S.LT animals, supporting that there is enough antigenic similarity for antibody cross-reactivity. The two variants of the Tp0136 putative surface fibronectin-binding protein28 were recognized by both the S.LT and S.RI rabbit groups, but not by any Nichols-infected animals, suggesting that this target might be in general more expressed in the SS14 strain than in Nichols. Overall, none of the SS14- or Nichols-specific antigens exhibit a pattern of recognition fully strain specific.
Table 2.
Reactivity to strain-specific antigens
| Target variant SS14 (S) Nichols (N) |
Gene coordinatesa | Significantly reactive in N.LT vs. S.LT | Significantly reactive in N.RI vs. S.RI | Annotated function | Percent identity (%) |
|---|---|---|---|---|---|
| Tp0006 (N)b Tp0006 (S) |
7014.7181 7014.8261 |
X X ✓ ✓ |
X X ✓ X |
Hypothetical protein/probable lipoprotein | 75.4% |
| Tp0027 (N) Tp0027 (S) |
34211.35434 34209.35432 |
Not represented in the arrayc X X |
Not represented in the arrayc X X |
HlyC/CorC family transporter | 99.7% |
| Tp0040 (N) Tp0040 (S) |
47597.49390 46945.49395 |
✓ X ✓ X |
✓ X X X |
Methyl-accepting chemotaxis protein | 73.4% |
| Tp0126c (N) Tp0126c (S) |
149381.149866 149381.149866 |
X X X X |
X X X X |
Hypothetical protein | 88.8% |
| Tp0127 (N) Tp0127 (S) |
149809.150477 149860.150240 |
✓ ✓ X ✓ |
X X X X |
Hypothetical protein | 51.12% |
| Tp0131 (N) Tp0131 (S) |
154160.152370 154110.152314 |
✓ ✓ ✓ ✓ |
X X X X |
Putative OMP/porin transporter; TprD/D2 | 79.0% |
| Tp0134c (N) Tp0134c (S)d |
Not annotatede 156884.157591 |
N/A X ✓ |
N/A X X |
hypothetical protein | N/A |
| Tp0136 (N) Tp0136 (S) |
157747.157819 157797.157869 |
X ✓ X ✓ |
X ✓ X ✓ |
Putative surface lipoprotein/adhesin | 91.9% |
| Tp0318 (N) Tp0318 (S) |
Not annotatede 335991.335812 |
N/A X X |
N/A X X |
Hypothetical protein | N/A |
| Tp0461 (N) Tp0461 (S) |
492057.492416 492090.492506 |
X X X X |
X X X X |
Helix-turn-helix transcriptional regulator | 81.5% |
| Tp0470 (N) Tp0470 (S) |
499845.498736 499709.498768 |
✓ ✓ ✓ ✓ |
✓ ✓ X X |
TPR domain protein | 84.8% |
| Tp0479 (N) Tp0479 (S) |
511161.510487 510890.510351 |
✓ ✓ ✓ ✓ |
X X X X |
Putative membrane protein | 79.9% |
| Tp0548 (N) Tp0548 (S) |
593270.594574 593141.594457 |
✓ ✓ X ✓ |
X X X X |
Putative OMP; FadL homolog | 94.5% |
As annotated in GenBank: NC_021490.2 and Genbank: NC_021508.1.
SS14 Tp0006 sequence is encompassed by Tp0006 and Tp0007 ORFs in the Nichols strain. Neither Nichols’ annotated Tp0006 nor Tp0007 is represented in the array.
No amplification of the target gene.
Also annotated in the T. pallidum DAL-1 strain.33
Not annotated in the Nichols strain by any pipeline.
Discussion
Advancements in genomics, transcriptomics, and proteomics approaches have recently led to a significant increase in the number of T. pallidum genome sequences25 and to the definition of a hierarchy of highly to poorly expressed genes at both the mRNA and protein level in treponemes grown in vitro or recovered from experimentally infected rabbits.31,34,35,36 Comparative analyses of the 514 assembled T. pallidum genomes sequenced to date from all continents, for example, showed that currently circulating strains fall within either the Nichols or the SS14 clades of this pathogen.25 Such evidence led to including SS14- and Nichols-specific targets in our PPA and the core proteome shared by both strains. Furthermore, the recent resequencing of both the Nichols and SS14 laboratory isolates allowed the use of updated annotations to define the boundaries of the individual open reading frames (ORFs) cloned into the plasmid library for IVTT.37,38 The choice of a PPA containing the complete T. pallidum proteome was also supported by gene expression studies showing detectable messages for virtually all annotated genes and by the results of mass-spectrometry-based studies collectively demonstrating that 90% of the T. pallidum annotated proteome is translated. Not all T. pallidum antigens are however represented in the array.31,34,35,36 Not represented targets are reported in Table S4 (Failed cloning). The TprK/Tp0897 putative OMP was purposely omitted due to its intra-strain hypervariability.39,40 Although a recombinant TprK fragment corresponding to the protein-less variable amino-terminus (aa 29/AQV-274/ALA; carrying two predicted invariant conserved surface-exposed loops) was spotted on the arrays as a control, and this target was significantly recognized in rabbits infected with either strain (Table S1, IgG summary statistics), we did not perform a formal analysis of reactivity, given that no data could be collected on the response to the missing portion of the protein. Ongoing work using a phage immunoprecipitation/sequencing approach (PhIP-Seq) employing a phage library of TprK variable and conserved sequences based on deep sequencing of the tprK gene will define which TprK epitopes elicited a significant humoral immunity in these infected animals.
This study attempted to close a significant knowledge gap in syphilis research by analyzing the longitudinal development of antibodies to each T. pallidum protein during infection and how acquired immunity changes in response to treatment. This goal was also pursued by McKevitt et al.41 who admirably produced an 882-protein array based on the Nichols genome sequenced in 1998 by expressing each single antigen in Escherichia coli.42 Upon testing this early array with sera from long-term (86 days) Nichols-infected rabbits, however, only 106 immunoreactive antigens were identified, whereas our study identified 213 reactive proteins in N.LT rabbits. Aside from the lower number of targets on McKevitt’s array41 compared to our PPA, this discrepancy might partially be attributable to false-negative results due—as acknowledged by the authors—to poorly expressed target proteins in a system that was still reliant on E. coli for protein expression rather than on IVTT. The authors also alluded to PCR-generated errors in their plasmid clones that might have altered the immunogenicity of target epitopes to explain why T. pallidum antigens known to be immunogenic during infection failed to be detected in their study.41 Regardless, a comparison between the 30 most significantly reactive antigens in both studies (41; Table 1) showed very similar results, with 20 shared targets and only 5 proteins (Tp0872/flagellar filament capping protein, Tp0259/LysM peptidoglycan binding protein, Tp0594/hypothetical protein, and Tp0737/periplasmic binding protein), recognized by our PPA but not by the earlier array, and six targets (Tp0453/concealed OMP, Tp0272/FlgE flagellar hook, Tp0841/DegQ endoprotease, Tp0369/BamD, Tp0468/tetratricopeptide domain protein, and Tp0433/hypothetical protein), recognized by both arrays, but associated to higher reactivity in our study. In our study, no differences were seen when signals generated by IVTT proteins and control recombinant proteins expressed in E. coli were seen (Figure S4).
Of the three targets currently used in TTs based on recombinant proteins, both the 47 kDa lipoprotein/Tp0574 (TpN47) and the 17 kDa lipoprotein/Tp0435 (TpN17) were among the most highly recognized antigens by all N.LT and S.LT rabbits, also in agreement with the McKevitt study.41 However, the 15 kDa lipoprotein/Tp0171 (TpN15) was not highly immunoreactive in infected animals in either study. Here, the N.LT and S.LT rabbit sera became significantly positive to TpN15 between day 40 and day 50 post-inoculation (Table S1, IgG summary statistics), which does not fully support the use of this antigen for early diagnosis, despite being highly expressed, remarkably conserved across T. pallidum strains and subspecies, and sequence specific for the agents of human treponematoses.43 Several clinical studies, however, support the validity of TpN15 as a serodiagnostic target,44,45,46,47,48,49,50,51,52 and this discrepancy could be due to differential immunity that develops in the rabbit compared to the natural human host. We also identified several antigens worth investigating as diagnostic candidates in addition to or in substitution of immunodominant lipoproteins to increase the sensitivity of TTs (Table 1). Among those, the hypothetical transcriptional regulator, Tp0772, elicited a fast and significantly high response in enrolled animals. Significant reactivity to this target in human sera was reported by Brinkman et al.,53 albeit using a limited number of specimens. The Tp0768/TmpA lipoprotein could be another suitable target. In a previous study, using 120 patient sera and a minimal protein array also containing Tp0768, we showed a significant correlation in reactivity between Tp0768 and both the TpN17 and TpN47 lipoproteins.54 However, reactivity to TmpA was generally lower than both TpN17 and TpN47, and the spread of values was greater.54 The possible use of Tp0768 in TTs is also supported by the results of studies published by Brinkman et al.53 and by the van Embden group that analyzed ∼70 sera from patients at different stages of syphilis over two studies.55,56 The possible use of Tp0277, Tp0319, Tp0326, Tp0453, Tp0684, and Tp0768 was also supported by Runina et al.57 and by work from other investigators as well.58
A complementary approach to the use of protein array was used by McGill et al.35 who opted for characterizing the humoral response to T. pallidum antigen after separating the pathogen’s proteome using 2D-gel electrophoresis coupled with immunoblotting using sera from three rabbits infected 84 days post-IT infection and patients. Of the 88 unique T. pallidum polypeptides found to be most highly expressed with this proteomic approach, 30 were recognized by infected rabbit serum with different degrees of reactivity. Of these antigens, however, 13 were not found to be reactive in our study, which suggests the possibility that a different inoculation route might lead to differential gene expression and development of immunity to specific antigens.
The near absence of an IgM response in all sera from the N.LT, N.RI, S.LT, and S.RI animals (Figure S3) was surprising, as we expected IgM reactivity to at least partially overlap that of the IgG-positive targets. The lack of IgM signal cannot be attributed to the lack of protein target spotted on the array or to the inability of IgM to bind the target, as positive control features in the array were reactive but likely to the low level of IgM in the serum samples. Equally intriguing was the difference in the number of targets recognized in S.LT animals (n = 327) compared to the N.LT rabbits (n = 207) (Figure 2). Given that both groups of animals received the same inoculum size, in lack of a better explanation, this result could be related to the different growth rate of the two strains we used. Nichols’ generation time is, in fact, 41.5 h, whereas SS14 needs 56.6 to divide.59 This apparently minor difference causes remarkable phenotypic changes in disease manifestations in infected animals. For example, in rabbits infected ID with the same inoculum size (106 cells of either the Nichols or the SS14 strain), dermal lesion progression from erythema to ulceration and then healing requires virtually half the time in Nichols-infected animals. Likewise, in animals inoculated IT with 107 cells/testis of Nichols treponemes, orchitis (assessed by palpation) manifests in approximately 10 days, whereas ∼20 days are needed in rabbits infected IT with the same inoculum of SS14 cells. Whether this difference in generation time is due to the genetic diversity existing between the two strains or the adaptation process that the Nichols strain likely experienced during its continual propagation in rabbits since 1912, one could hypothesize that faster growth translates into an accelerated immune recognition and pathogen clearance, leading to a limited array of antigens recognized before most pathogen cells are eliminated. This hypothesis could also be consistent with the work by McKevitt et al.41 discussed above, where a treponemal inoculum 67 times higher than ours (4 x 108 Nichols cells/rabbit) was entirely administered IT to their rabbits, potentially leading to fewer antigens being recognized in their Nichols-infected rabbits than ours due to a fast host response induced by an abnormally large inoculum. Although a comparison between the results of NTTs and TTs in both studies could have corroborated our hypothesis, McKevitt et al. did not report such data.41
Another notable discrepancy was the much lower numbers of reactive antigens identified in the N.RI and S.RI animals (Figures 3 and 5B). To discard technical factors, QC reprobes were performed on the N.LT and S.LT higher reacting samples, as well as manual inspection of microarray scanner files. However, such analyses confirmed that the primary difference between the cohorts is due to heterogeneity in the six rabbits in each group, with the N.RI and S.RI groups having more “low responder” rabbits than the N.LT and S.LT groups. This would also explain the lower number of significant increases to day 30 in the N.RI and S.RI groups despite similar differential effect sizes (Figures 2D and 2H vs Figures 6A and 6E). The treatment of the groups was identical up to day 30, so the discrepancies observed may be attributable to random chance, and we posit that further studies with more animals would result in a convergence of reactive antigen counts.
New cases of syphilis occur primarily in areas characterized by poor access to health care due to low socioeconomic status. The availability of an effective vaccine would greatly help in reducing disease incidence in adults and newborns and dependence on antibiotics to avoid infection, particularly when post-exposure prophylaxis is considered.60,61,62 Early work on syphilis immunology in the rabbit model established that rabbits infected with T. pallidum for at least 3 months develop immunity to symptomatic reinfection, albeit they get reinfected if inoculated again.63,64 Defining the humoral nature of this “chancre immunity” might identify potential candidates to actively pursue as components of a syphilis vaccine. As T. pallidum clearance from infected lesions is believed to occur mainly through phagocytosis of opsonized treponemes,65 surface-exposed antigens of the syphilis spirochete are the most likely vaccine candidates and several are currently pursued to induce protective immunity. Our results suggest that antibodies to the Tp0326/BamA OMP, a protein known to be associated with biogenesis of the outer membrane,66 might play a role in protective immunity, as also suggested in recent work by Ferguson et al.67 The comparison of the immunity generated to OMPs in both N.LT and S.LT animals, however, also suggested that there might be a core number of OMPs sufficient to induce partial protection, as relatively fewer of these putative surface-exposed antigens were recognized in N.LT rabbits compared to S.LT ones (Figure 4D). Reported immunization/challenge data for some of the antigens that appear to be dispensable seem to support this hypothesis. Rabbit immunization with Tp0126/OmpW, for example, did not induce protection following infectious challenge,68 and neither did immunization with the putative surface-exposed lipoprotein Tp0136,69 even though recent work suggests a functional role for the immunity to this antigen in the inhibition of treponemal dissemination,70 consistent with its role of fibronectin-binding adhesin of T. pallidum. With regard to the Tp0859/FadL homolog, the presence of a homopolymeric G repeat within the ORFs71 suggests that this gene might undergo translational phase variation, supporting a non-essential biological role for this putative porin transporter, likely due to the presence of four additional members in this family of paralogous proteins in the T. pallidum proteome.
In summary, we have adapted a robust high-throughput proteome-synthesis process to two strains of the syphilis agent and have provided proof of concept that the resulting proteomic array can define antigen-specific antibody profiles from infected laboratory subjects. The application of this technology, both at the pre-clinical and clinical level, will contribute to improving our understanding of syphilis pathogenesis and immunology and to accelerating the discovery of potential vaccine candidates and antigens for diagnostics applications. Ongoing studies in the laboratory are evaluating the humoral response to T. pallidum antigens using 217 sera longitudinally collected pre- and post-treatment from 120 patients with syphilis enrolled between 2019 and 2021 in five sexual health clinics in Peru. Such data will allow us to further investigate whether treponemal antigens could be used to perform disease staging and monitor response to treatment in patients with different HIV status history of infection. For our ongoing syphilis in pregnancy study (SIPS), samples from two additional cohorts of pregnant subjects with syphilis will be tested to assess whether a correlation exists between immunity to specific T. pallidum antigens and pregnancy outcomes.
Limitations of the study
The rabbit reinfection experiment helped address a limitation of TTs, namely the continued reactivity of modern conventional TTs after successful treatment that makes them unsuitable for monitoring treatment response, relapse, or reinfection in previously treated subjects. In this context, the identification of novel T. pallidum antigens, whose antibodies are rapidly eliminated from the host circulation, could lead to tests that could complement the NTTs, which would be helpful in the case of serological non-responders who do not achieve the 4-fold dilution in NTT titer necessary to declare serological cure or serofast patients who do not seroconvert even if the proper reduction in NTT titer was achieved.12 In our study, although reactivity to several antigens underwent a reduction over time, we could not find a specific target recognized by most infected animals that exhibited a significant reduction in reactivity between day 30 post-infection, at the time of treatment, and day 90, right before reinfection. This could also be due to the relatively short time elapsed between treatment of these animals and reinfection, and the outcome of this experiment cannot exclude the possibility that treponemal antigens could have a role in assessing successful treatment. In a previous study by Haynes et al.54 using patient samples and a minimal proteomic array carrying only 16 antigens, we identified an association with RPR titer drop post-treatment and a reduction in the signal to Tp0679/TmpB, as previously reported in a similar study by Schouls et al.68 Although the result reported here using experimental sera did not confirm the earlier report, our ongoing analysis of clinical samples would shed further light on the possible use of TmpB or other antigens for the monitoring of treatment efficacy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Syphilis-infected rabbit sera | Giacani laboratory | N/A. Sera were produced as part of the experimental design |
| DyLight650-conjugated goat anti-rabbit IgG | Bethyl Laboratories, Montgomery, TX | cat# A120-210A; RRID:AB_10631422 |
| DyLight550-conjugated goat anti-rabbit IgM | Abcam Inc., Cambridge, UK | cat# ab98455; RRID:AB_10674442 |
| Bacterial and virus strains | ||
| Treponema pallidum subspecies pallidum Nichols strain | Giacani laboratory | Originally provided by Dr. James Miller, UCLA. (GenBank: NC_021490.2) |
| Treponema pallidum subspecies pallidum SS14 strain | Giacani Laboratory | Originally provided by Dr. Sandra A Larsen, CDC (GenBank: NC_021508.1) |
| Chemicals, peptides, and recombinant proteins | ||
| T. pallidum recombinant proteins | Antigen Discovery Inc. | N/A. Proteins were produced as part of the experimental design |
| Critical commercial assays | ||
| VDRL | BD, Franklin Lanes, NJ | 88085886-07 |
| TPPA | Fujirebio Diagnostics, Inc., Malvern, PA | 1626 |
| Experimental models: Organisms/strains | ||
| New Zealand White Rabbit | Western Oregon Rabbit Co | Bioproject PRJNA274594 |
| Recombinant DNA | ||
| Plasmid DNA to express T. pallidum proteins in vitro | Antigen Discovery Inc. | N/A. Plasmids were created as part of the experimental design |
Resource availability
Lead contact
Further information and requests for resources and reagents can be directed to the lead contact, Dr. Lorenzo Giacani (giacal@uw.edu).
Materials availability
Requests to attain rabbit sera generated during this experiment, recombinant plasmids to express T. pallidum proteins, and the proteomic array might be fulfilled based on existing policies at the University of Washington and Antigen Discovery Inc.
Data and code availability
-
•
Data are deposited at on Mendeley Data and available at Mendeley Data: https://data.mendeley.com/preview/vw72m8jkwg?a=9458c05a-b54f-4a37-ad00-10b11958288f. Deposited data include antiboby reactivities detected in analyzed serum samples from animals.
-
•
Code: not applicable.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and study participant details
Animals and animal care
New Zealand White (NZW) rabbits were used for treponemal propagation and experimental infections to obtain serum samples over time. Adult male NZW rabbits were purchased from Western Oregon Rabbit Company (Philomath, OR) and housed at the Animal Use and Care Facility (ARCF) at the UW. After a period of acclimation, blood was collected from the central ear artery to perform both a lipoidal and treponemal serological test and rule out past or current infection with Treponema paraluiscuniculi, the known agent of rabbit syphilis. More specifically, heat-inactivated sera from these rabbits were tested individually using the VDRL test (BD, Franklin Lanes, NJ, cat# 88085886-07) and the SERODIA TPPA assay (Fujirebio Diagnostics, Inc., Malvern, PA, cat#1626) according to the provided protocols. Only VDRL- and TPPA-negative rabbits were enrolled in this study. Animal care was provided in accordance with the Guide for the Care and Use of Laboratory Animals, and all experimental procedures were conducted under protocol #4243-01 (PI: Lorenzo Giacani), approved by the University of Washington (UW) IACUC. All animal procedures were conducted in compliance with the ARRIVE guidelines. Rabbit euthanasia was performed consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Association.
T. pallidum strain propagation and DNA extraction
The T. pallidum Nichols and SS14 strains were propagated through intratesticular (IT) inoculation in NZW rabbits as previously described72 to obtain treponemal cells as source of DNA to construct the PPA and, subsequently, to produce the inoculum for all experimental infections. Briefly, 107 treponemes were injected in each testis. Once peak orchitis was reached (about 10 days post-IT infection with Nichols treponemes and 20-25 days for the SS14 strain), animals were euthanized, and testes were removed and minced in 10 mL of sterile saline. The resulting suspension was spun for 10 min at 1,000 x g in a 5430 Eppendorf (Hauppauge, NY) centrifuge at room temperature to remove cellular debris. The treponemes in the supernatant were enumerated using dark-field microscopy (DFM) and diluted to the appropriate concentration to inoculate naïve rabbits IT for further propagation or ID for the actual study.
Specimens
For the experimental infections, 24 rabbits divided into two groups of twelve animals were used. One group was infected with the Nichols strain, and the other with the SS14 strain. Each animal was infected ID on six sites on their shaved backs with 106 treponemes per site. The ID route was preferred to any other (IT or intravenously) because upon ID infection, animals develop primary lesions that morphologically and histologically recapitulate those of primary syphilis patients. Each group was then divided into sub-groups of six animals. The first subgroup of Nichols-infected animals was labeled Nichols long-term (N.LT) because these animals were infected and followed longitudinally for 90 days, during which serum samples were collected at day 5 and day 10 post-infection and, subsequently, every ten days until day 90 post-infection, when the animals were bled from the central ear artery one last time and then euthanized. Pre-infection sera were obtained at the time the animals arrived at the vivarium. The second subgroup of six Nichols-infected animals was labeled Nichols re-infection (N.RI) because animals were treated at day 30 post-inoculation with 200,000 units of BPG (Bicillin L-A, Pfizer, New York, NY. NDC 60793-700-10) administered intramuscularly (IM) in a single injection, and equivalent w/w to 4.8 million units for humans. Two months post-treatment (at study day 90), the animals were re-infected as described above with the homologous Nichols strain. Re-infected animals were followed longitudinally for three additional months. From these animals, serum samples were obtained before initial inoculation, at day 5 and day 10 post-initial infection and every ten days after for the remainder of the 180-day experiment. On day 90 after re-infection (at study day 180), the animals were bled and euthanized. The same experimental design described for the N.LT and N.RI rabbits was applied to the 12 animals infected with the SS14 strains, which were identified as the SS14 long-term (S.LT; n = 6 animals), and SS14 re-infection (S.RI; n = 6 animals) groups, respectively. All 24 rabbits enrolled in this study were shaved regularly in order not to hinder dermal lesion development. Collected sera were heat-inactivated at 56°C for 30 min and aliquoted. One aliquot was used for syphilis serology, while the remaining ones were frozen at -80°C to be used later to probe the PPA.
Method details
DNA extraction
Treponemes harvested for DNA extraction were spun for 10 min at 15,000 rpm at 4°C using an Eppendorf 5425 refrigerated centrifuge. Pellets were resuspended in 400 μl 1X Lysis Buffer (10mM Tris pH 8.0, 0.1M EDTA pH 8.0, 0.5% SDS) and frozen at -80°C until extraction using the QIAamp DNA Mini Kit (Qiagen Inc., Chatsworth, CA, cat#51104) according to the provided protocol.
VDRL and TPPA serology
Both VDRL and TPPA tests were performed according to the provided protocols. When sera tested positive by VDRL, serum titration was also performed. In addition, TPPA-reactive sera collected at day 30 and day 90 (from both LT and RI animals), and at day-180 (RI animals only, as the LT rabbits were euthanized at day 90 post-infection) were also titrated.
The technologist was blinded to the treatment status and group of the animals from which the samples were collected. Mean log2 titers ± SE were calculated for each group at each time point: nonreactive = −1.0; weakly reactive = −0.5; reactive at a titer of 1:1= 0; reactive at a titer of 1:2 = 1, and so on. Antilogs of mean ± SE of titers were plotted with nonreactive set at y = 0. Following testing, serum samples were stored at -80°C until use. Throughout the study, only three serum samples could not be collected, all from the N.LT group (from two rabbits at day 80 post-inoculation and from one rabbit at day 90 post-infection). Statistical analysis was performed using Student’s t-test with significance set at p<0.05.
Array construction
A core proteome was identified using the genome sequences of the Nichols (NC_021490.2) and SS14 (NC_021508.1) strains as annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PAGAP) software (https://www.ncbi.nlm.nih.gov/refseq/annotation_prok/). Pan-proteome arrays were fabricated containing 1,040 full-length or fragmented recombinant proteins representing a total of 1,009 genes. Of these, 986 were genes encompassing the core proteome shared by the two strains, 10 were genes encoding for Nichols-specific proteins and 13 were specific for the SS14 strain. Each open reading frame (ORF) sequence was amplified by PCR, inserted into the vector pXT7 by recombination in E. coli to establish a library of partial or complete coding DNA sequences. Proteins were expressed using a coupled E. coli cell-free in vitro transcription and translation (IVTT) system (Rapid Translation System, Biotechrabbit, Berlin, Germany, cat# BR1400201) and spotted onto nitrocellulose-coated glass AVID slides (Grace Bio-Labs Inc., Bend, OR) using an Omni Grid Accent robotic microarray printer (Digilabs Inc. Hopkinton, MA). Each expressed protein included a 5’ poly-histidine (His) epitope and 3’ hemagglutinin (HA) epitope. Pan-proteome array chip printing and protein expression were quality checked by probing random slides with anti-His and anti-HA monoclonal antibodies fluorescently labeled and quantifying spot signals using a microarray scanner. As positive control antigens, six recombinant T. pallidum proteins previously expressed to construct a minimal proteomic array54 or other experimental purposes were spotted onto the array at two different concentrations. Those controls included the known immunodominant lipoproteins Tp0435/TpN17, Tp0574/TpN47, and Tp0768/TmpA, known to be conserved in both strains, the putative OMPs Tp0117/TprC and Tp0131/TprD2, and a relatively conserved amino-terminal fragment of the Tp0897/TprK antigen, also known to be targeted by the host response during infection based on previous studies.73 Except for TprK, all proteins were printed on the array as full-length molecules based on their available annotation. Antigens that could not be represented in the array are listed in Table S4 (Failed cloning).
Array probing
Serum samples were diluted (1:100) and incubated on the PPA chips overnight at 4°C on a rocker. Bound IgG was detected with DyLight650-conjugated goat anti-rabbit IgG (Bethyl Laboratories, Montgomery, TX, cat#A120-210A), and IgM was detected with DyLight550-conjugated goat anti-rabbit IgM (Abcam Inc., Cambridge, UK; cat# ab98455). Washed and dried PPA chips were scanned, and the spot and background signal intensities (SI) were exported into R package for statistical analysis. Sera collected from the LT and the RI animal groups were probed in separate batches.
Quantification and statistical analysis
Spot SIs were adjusted for local background by subtraction, and values were floored to 1. Next, the data were normalized by dividing the protein spot values by the median of IVTT control spots (IVTT expression reactions with no T. pallidum ORFs), and values were log transformed using the base-2 logarithm. Thus, normalized data represented the log2 signal-to-noise ratio, where a value of 0 represents specific antibody SI equal to the background, 1.0 represents twice the background, 2.0 represents 4-fold over background, and so forth. T. pallidum protein responses were classified as seropositive for SI of at least 1.0, or twice the background. Maximal SIs across all timepoints for each rabbit were calculated for each protein. A protein was classified as “reactive” if over half of the rabbits responded to the protein, i.e., median max SI > 1. Individual antibody responses or mean antibody responses by study day were visualized using the ComplexHeatmap package.74 The distributions of reactive antigen responses were visualized using error bar plots of all proteins. Overlap in reactivity to individual antigens between groups was visualized using the UpSetR package, which recasts the information of a complex Venn diagram as bar plots.75 The association of protein physiochemical and functional features with antibody reactivity was analyzed using logistic regression within the safeBinaryRegression package76 with separation = 'find' to identify terms separating the observations when maximum likelihood estimate is found not to exist, followed by performing stepwise regression on the previous model using the stepAIC function set to both forward and backward stepwise regression. Only protein features present in at least 5 antigens and did not return singularity errors were included. Differences in antibody levels between sequential time points were assessed using paired t-tests. Analysis was stratified by Nichols and SS14 strain infection groups, and the LT and RI experiments were analyzed separately. Differences between time points were visualized using volcano plots, and longitudinal trajectories of specific antibody responses were visualized using line plots with the ggplot2 package.77 Only reactive antigens were included in differential analysis, and p-values were adjusted for the false discovery rate using the method described by Benjamini and Hochberg.78
Acknowledgments
The authors are grateful to Angela Yee (Antigen Discovery Inc.) for critical review of the manuscript. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, writing of the report, or in the decision to submit the paper for publication. This war was supported by the NIH SBIR-R43AI149804 grant to J.J.C.
Author contributions
Conceptualization: J.J.C. and L.G.; data curation: J.J.C., E.R., A.O., J.V.P., C.H., A.A.T., A.D.S., and L.G.; formal analysis: J.J.C. and L.G.; funding acquisition: J.J.C. and L.G.; investigation: J.J.C., E.R., A.O., J.V.P., C.H., A.A.T., A.D.S., A.P., A.M.H., and L.G.; methodology: J.J.C., A.O., J.V.P., C.H., A.A.T., A.D.S., A.M.H., and L.G.; project administration: J.J.C. and L.G.; resources: J.J.C., C.H., A.A.T., A.D.S., E.R., A.M.H., and L.G.; supervision: L.G. and J.J.C.; validation: J.J.C.; visualization: J.J.C., L.G., and E.R.; writing—original draft: L.G. and J.J.C.; writing—review and editing: J.J.C., E.R., A.O., J.V.P., C.H., A.A.T., A.D.S., A.P., A.M.H., and L.G.
Declaration of interests
The authors have no competing interests to disclose.
Published: July 31, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110618.
Supplemental information
Complete list of antigens recognized all rabbits and time to a seropositive response.
References
- 1.WHO . World Health Organization; 2011. Prevalence and Incidence of Selected Sexually Transmitted Infections Chlamydia trachomatis, Neisseria Gonorrhoeae, Syphilis and Trichomonas vaginalis: Methods and Results Used by WHO to Generate 2005 Estimates. [Google Scholar]
- 2.Gerbase A.C., Rowley J.T., Mertens T.E. Global epidemiology of sexually transmitted diseases. Lancet. 1998;351:S2–S4. doi: 10.1016/s0140-6736(98)90001-0. [DOI] [PubMed] [Google Scholar]
- 3.CDC . US Department of Health and Human Services: Centers for Disease Control and Prevention; 2024. 2022 Sexually Transmitted Disease Surveillance. [Google Scholar]
- 4.Savage E.J., Marsh K., Duffell S., Ison C.A., Zaman A., Hughes G. Rapid increase in gonorrhoea and syphilis diagnoses in England in 2011. Euro Surveill. 2012;17 [PubMed] [Google Scholar]
- 5.Savage E.J., Hughes G., Ison C., Lowndes C.M., European Surveillance of Sexually Transmitted Infections network Syphilis and gonorrhoea in men who have sex with men: a European overview. Euro Surveill. 2009;14 doi: 10.2807/ese.14.47.19417-en. [DOI] [PubMed] [Google Scholar]
- 6.Simms I., Fenton K.A., Ashton M., Turner K.M.E., Crawley-Boevey E.E., Gorton R., Thomas D.R., Lynch A., Winter A., Fisher M.J., et al. The re-emergence of syphilis in the United Kingdom: the new epidemic phases. Sex. Transm. Dis. 2005;32:220–226. doi: 10.1097/01.olq.0000149848.03733.c1. [DOI] [PubMed] [Google Scholar]
- 7.Tucker J.D., Cohen M.S. China's syphilis epidemic: epidemiology, proximate determinants of spread, and control responses. Curr. Opin. Infect. Dis. 2011;24:50–55. doi: 10.1097/QCO.0b013e32834204bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin F., Prestage G.P., Kippax S.C., Pell C.M., Donovan B.J., Kaldor J.M., Grulich A.E., Australian-Thai HIV Vaccine Consortium Epidemic syphilis among homosexually active men in Sydney. Med. J. Aust. 2005;183:179–183. doi: 10.5694/j.1326-5377.2005.tb06989.x. [DOI] [PubMed] [Google Scholar]
- 9.LaFond R.E., Lukehart S.A. Biological basis for syphilis. Clin. Microbiol. Rev. 2006;19:29–49. doi: 10.1128/CMR.19.1.29-49.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen S.A. 9th ed. American Public Health Association; 1999. Manual of Tests for Syphilis. [Google Scholar]
- 11.Matthews H.M., Yang T.K., Jenkin H.M. Unique lipid composition of Treponema pallidum (Nichols virulent strain) Infect. Immun. 1979;24:713–719. doi: 10.1128/iai.24.3.713-719.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghanem K.G., Hook E.W., 3rd The Terms "Serofast" and "Serological Nonresponse" in the Modern Syphilis Era. Sex. Transm. Dis. 2021;48:451–452. doi: 10.1097/OLQ.0000000000001387. [DOI] [PubMed] [Google Scholar]
- 13.Castro R.R., Prieto E.S., Santo I., Azevedo J., Exposto F.L. Evaluation of the passive particle agglutination test in the serodiagnosis and follow-up of syphilis. Am. J. Clin. Pathol. 2001;116:581–585. doi: 10.1309/9TCQ-B1TA-38MV-R6UM. [DOI] [PubMed] [Google Scholar]
- 14.Bishop N.H., Miller J.N. Humoral immunity in experimental syphilis. II. The relationship of neutralizing factors in immune serum to acquired resistance. J. Immunol. 1976;117:197–207. [PubMed] [Google Scholar]
- 15.Bishop N.H., Miller J.N. Humoral immunity in experimental syphilis. I. The demonstration of resistance conferred by passive immunization. J. Immunol. 1976;117:191–196. [PubMed] [Google Scholar]
- 16.Marra C.M., Maxwell C.L., Sahi S.K., Tantalo L.C., Dunaway S.B., Lukehart S.A. Previous Syphilis Alters the Course of Subsequent Episodes of Syphilis. Clin. Infect. Dis. 2022;74:e1–e5. doi: 10.1093/cid/ciab287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenyon C., Osbak K.K., Apers L. Repeat Syphilis Is More Likely to Be Asymptomatic in HIV-Infected Individuals: A Retrospective Cohort Analysis With Important Implications for Screening. Open Forum Infect. Dis. 2018;5 doi: 10.1093/ofid/ofy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang R., Read R., Krentz H.B., Peng M., Ramazani S., Vu Q., Gill M.J. A retrospective study of the clinical features of new syphilis infections in an HIV-positive cohort in Alberta, Canada. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2018-021544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenyon C. Increases in Asymptomatic Early Syphilis May Reflect Increases in Repeated Episodes of Syphilis and Not Enhanced Screening. Clin. Infect. Dis. 2018;66:811–812. doi: 10.1093/cid/cix904. [DOI] [PubMed] [Google Scholar]
- 20.Kenyon C., Tsoumanis A., Osbak K., Van Esbroeck M., Florence E., Crucitti T., Kestens L. Repeat syphilis has a different immune response compared with initial syphilis: an analysis of biomarker kinetics in two cohorts. Sex. Transm. Infect. 2018;94:180–186. doi: 10.1136/sextrans-2017-053312. [DOI] [PubMed] [Google Scholar]
- 21.Sadowy E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid. 2018;99:89–98. doi: 10.1016/j.plasmid.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Baker-Zander S.A., Lukehart S.A. Macrophage-mediated killing of opsonized Treponema pallidum. J. Infect. Dis. 1992;165:69–74. doi: 10.1093/infdis/165.1.69. [DOI] [PubMed] [Google Scholar]
- 23.Lukehart S.A., Shaffer J.M., Baker-Zander S.A. A subpopulation of Treponema pallidum is resistant to phagocytosis: possible mechanism of persistence. J. Infect. Dis. 1992;166:1449–1453. doi: 10.1093/infdis/166.6.1449. [DOI] [PubMed] [Google Scholar]
- 24.Baker-Zander S.A., Shaffer J.M., Lukehart S.A. Characterization of the serum requirement for macrophage-mediated killing of Treponema pallidum ssp. pallidum: relationship to the development of opsonizing antibodies. FEMS Immunol. Med. Microbiol. 1993;6:273–279. doi: 10.1111/j.1574-695X.1993.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman N.A.P., Lin M.J., Xie H., Shrestha L., Nguyen T., Huang M.L., Haynes A.M., Romeis E., Wang Q.Q., Zhang R.L., et al. Treponema pallidum genome sequencing from six continents reveals variability in vaccine candidate genes and dominance of Nichols clade strains in Madagascar. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazlett K.R.O., Cox D.L., Decaffmeyer M., Bennett M.P., Desrosiers D.C., La Vake C.J., La Vake M.E., Bourell K.W., Robinson E.J., Brasseur R., Radolf J.D. TP0453, a concealed outer membrane protein of Treponema pallidum, enhances membrane permeability. J. Bacteriol. 2005;187:6499–6508. doi: 10.1128/JB.187.18.6499-6508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Primus S., Rocha S.C., Giacani L., Parveen N. Identification and Functional Assessment of the First Placental Adhesin of Treponema pallidum That May Play Critical Role in Congenital Syphilis. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.621654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke W., Molini B.J., Lukehart S.A., Giacani L. Treponema pallidum subsp. pallidum TP0136 protein is heterogeneous among isolates and binds cellular and plasma fibronectin via its NH2-terminal end. PLoS Neglected Trop. Dis. 2015;9 doi: 10.1371/journal.pntd.0003662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marra C.M., Sahi S.K., Tantalo L.C., Godornes C., Reid T., Behets F., Rompalo A., Klausner J.D., Yin Y.P., Mulcahy F., et al. Enhanced molecular typing of Treponema pallidum: geographical distribution of strain types and association with neurosyphilis. J. Infect. Dis. 2010;202:1380–1388. doi: 10.1086/656533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacani L., Molini B., Godornes C., Barrett L., Van Voorhis W., Centurion-Lara A., Lukehart S.A. Quantitative analysis of tpr gene expression in Treponema pallidum isolates: Differences among isolates and correlation with T-cell responsiveness in experimental syphilis. Infect. Immun. 2007;75:104–112. doi: 10.1128/IAI.01124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Lay B.D., Cameron T.A., De Lay N.R., Norris S.J., Edmondson D.G. Comparison of transcriptional profiles of Treponema pallidum during experimental infection of rabbits and in vitro culture: Highly similar, yet different. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centurion-Lara A., Giacani L., Godornes C., Molini B.J., Brinck Reid T., Lukehart S.A. Fine Analysis of Genetic Diversity of the tpr Gene Family among Treponemal Species, Subspecies and Strains. PLoS Neglected Trop. Dis. 2013;7 doi: 10.1371/journal.pntd.0002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houston S., Lithgow K.V., Osbak K.K., Kenyon C.R., Cameron C.E. Functional insights from proteome-wide structural modeling of Treponema pallidum subspecies pallidum, the causative agent of syphilis. BMC Struct. Biol. 2018;18:7. doi: 10.1186/s12900-018-0086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGill M.A., Edmondson D.G., Carroll J.A., Cook R.G., Orkiszewski R.S., Norris S.J. Characterization and serologic analysis of the Treponema pallidum proteome. Infect. Immun. 2010;78:2631–2643. doi: 10.1128/IAI.00173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houston S., Gomez A., Geppert A., Eshghi A., Smith D.S., Waugh S., Hardie D.B., Goodlett D.R., Cameron C.E. Deep proteome coverage advances knowledge of Treponema pallidum protein expression profiles during infection. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-45219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrosova H., Pospisilova P., Strouhal M., Čejková D., Zobaníková M., Mikalová L., Sodergren E., Weinstock G.M., Šmajs D. Resequencing of Treponema pallidum ssp. pallidum strains Nichols and SS14: correction of sequencing errors resulted in increased separation of syphilis treponeme subclusters. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romeis E, Lieberman NAP, Molini B,Tantalo, L.C., Chung, B., Phung, Q., Avendaño, C., Vorobieva, A., Greninger, A.L. Giacani, L. Treponema pallidum subsp. pallidum with an Artificially Impaired TprK Antigenic Variation System is Attenuated in the Rabbit Model of Syphilis. PLoS Pathog.,19(3), p.e1011259 [DOI] [PMC free article] [PubMed]
- 38.Lin M.J., Haynes A.M., Addetia A., Lieberman N.A.P., Phung Q., Xie H., Nguyen T.V., Molini B.J., Lukehart S.A., Giacani L., Greninger A.L. Longitudinal TprK profiling of in vivo and in vitro-propagated Treponema pallidum subsp. pallidum reveals accumulation of antigenic variants in absence of immune pressure. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Addetia A., Tantalo L.C., Lin M.J., Xie H., Huang M.L., Marra C.M., Greninger A.L. Comparative genomics and full-length TprK profiling of Treponema pallidum subsp. pallidum reinfection. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0007921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKevitt M., Brinkman M.B., McLoughlin M., Perez C., Howell J.K., Weinstock G.M., Norris S.J., Palzkill T. Genome scale identification of Treponema pallidum antigens. Infect. Immun. 2005;73:4445–4450. doi: 10.1128/IAI.73.7.4445-4450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraser C.M., Norris S.J., Weinstock G.M., White O., Sutton G.G., Dodson R., Gwinn M., Hickey E.K., Clayton R., Ketchum K.A., et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 42.Centurion-Lara A., Arroll T., Castillo R., Shaffer J.M., Castro C., Van Voorhis W.C., Lukehart S.A. Conservation of the 15-kilodalton lipoprotein among Treponema pallidum subspecies and strains and other pathogenic treponemes: genetic and antigenic analyses. Infect. Immun. 1997;65:1440–1444. doi: 10.1128/iai.65.4.1440-1444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato N.S., Hirata M.H., Hirata R.D., Zerbini L.C., Silveira E.P., de Melo C.S., Ueda M. Analysis of Treponema pallidum recombinant antigens for diagnosis of syphilis by western blotting technique. Rev. Inst. Med. Trop. Sao Paulo. 1999;41:115–118. doi: 10.1590/s0036-46651999000200009. [DOI] [PubMed] [Google Scholar]
- 44.Meyer M.P., Eddy T., Baughn R.E. Analysis of western blotting (immunoblotting) technique in diagnosis of congenital syphilis. J. Clin. Microbiol. 1994;32:629–633. doi: 10.1128/jcm.32.3.629-633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun A.H., Mao Y.F., Hu Y., Sun Q., Yan J. Sensitive and specific ELISA coated by TpN15-TpN17-TpN47 fusion protein for detection of antibodies to Treponema pallidum. Clin. Chem. Lab. Med. 2009;47:321–326. doi: 10.1515/CCLM.2009.071. [DOI] [PubMed] [Google Scholar]
- 46.Wang L.N., Li J.M. Evaluation of immunoglobulin M and G Western blot and ELISA for screening antibodies to Treponema pallidum in blood donors. Sex. Transm. Dis. 2009;36:413–416. doi: 10.1097/OLQ.0b013e31819db594. [DOI] [PubMed] [Google Scholar]
- 47.de Lemos E.A., Belém Z.R., Santos A., Ferreira A.W. Characterization of the Western blotting IgG reactivity patterns in the clinical phases of acquired syphilis. Diagn. Microbiol. Infect. Dis. 2007;58:177–183. doi: 10.1016/j.diagmicrobio.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 48.Sambri V., Marangoni A., Simone M.A., D'Antuono A., Negosanti M., Cevenini R. Evaluation of recomWell Treponema, a novel recombinant antigen-based enzyme-linked immunosorbent assay for the diagnosis of syphilis. Clin. Microbiol. Infect. 2001;7:200–205. doi: 10.1046/j.1469-0691.2001.00232.x. [DOI] [PubMed] [Google Scholar]
- 49.Backhouse J.L., Nesteroff S.I. Treponema pallidum western blot: comparison with the FTA-ABS test as a confirmatory test for syphilis. Diagn. Microbiol. Infect. Dis. 2001;39:9–14. doi: 10.1016/s0732-8893(00)00213-3. [DOI] [PubMed] [Google Scholar]
- 50.Young H., Moyes A., de Ste Croix I., McMillan A. A new recombinant antigen latex agglutination test (Syphilis Fast) for the rapid serological diagnosis of syphilis. Int. J. STD AIDS. 1998;9:196–200. doi: 10.1258/0956462981922034. [DOI] [PubMed] [Google Scholar]
- 51.Young H., Moyes A., Seagar L., McMillan A. Novel recombinant-antigen enzyme immunoassay for serological diagnosis of syphilis. J. Clin. Microbiol. 1998;36:913–917. doi: 10.1128/jcm.36.4.913-917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkman M.B., McKevitt M., McLoughlin M., Perez C., Howell J., Weinstock G.M., Norris S.J., Palzkill T. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J. Clin. Microbiol. 2006;44:888–891. doi: 10.1128/JCM.44.3.888-891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haynes A.M., Konda K.A., Romeis E., Siebert J., Vargas S.K., Reyes Diaz M., Phan A., Caceres C.F., Giacani L., Klausner J.D. Evaluation of a minimal array of Treponema pallidum antigens as biomarkers for syphilis diagnosis, infection staging, and response to treatment. Microbiol. Spectr. 2024;12 doi: 10.1128/spectrum.03466-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ijsselmuiden O.E., Schouls L.M., Stolz E., Aelbers G.N., Agterberg C.M., Top J., van Embden J.D. Sensitivity and specificity of an enzyme-linked immunosorbent assay using the recombinant DNA-derived Treponema pallidum protein TmpA for serodiagnosis of syphilis and the potential use of TmpA for assessing the effect of antibiotic therapy. J. Clin. Microbiol. 1989;27:152–157. doi: 10.1128/jcm.27.1.152-157.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schouls L.M., Ijsselmuiden O.E., Weel J., van Embden J.D. Overproduction and purification of Treponema pallidum recombinant-DNA-derived proteins TmpA and TmpB and their potential use in serodiagnosis of syphilis. Infect. Immun. 1989;57:2612–2623. doi: 10.1128/iai.57.9.2612-2623.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Runina A.V., Katunin G.L., Filippova M.A., Zatevalov A.M., Kubanov A.A., Deryabin D.G. Immunochip for Syphilis Serodiagnostics with the Use of Extended Array of Treponema pallidum Recombinant Antigens. Bull. Exp. Biol. Med. 2018;165:767–771. doi: 10.1007/s10517-018-4261-0. [DOI] [PubMed] [Google Scholar]
- 57.Kubanov A., Runina A., Deryabin D. Novel Treponema pallidum Recombinant Antigens for Syphilis Diagnostics: Current Status and Future Prospects. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/1436080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edmondson D.G., DeLay B.D., Kowis L.E., Norris S.J. Parameters Affecting Continuous In Vitro Culture of Treponema pallidum Strains. mBio. 2021;12 doi: 10.1128/mBio.03536-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran N.K., Goldstein N.D., Welles S.L. Countering the rise of syphilis: A role for doxycycline post-exposure prophylaxis? Int. J. STD AIDS. 2022;33:18–30. doi: 10.1177/09564624211042444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grant J.S., Stafylis C., Celum C., Grennan T., Haire B., Kaldor J., Luetkemeyer A.F., Saunders J.M., Molina J.M., Klausner J.D. Doxycycline Prophylaxis for Bacterial Sexually Transmitted Infections. Clin. Infect. Dis. 2020;70:1247–1253. doi: 10.1093/cid/ciz866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Molina J.M., Charreau I., Chidiac C., Pialoux G., Cua E., Delaugerre C., Capitant C., Rojas-Castro D., Fonsart J., Bercot B., et al. Post-exposure prophylaxis with doxycycline to prevent sexually transmitted infections in men who have sex with men: an open-label randomised substudy of the ANRS IPERGAY trial. Lancet Infect. Dis. 2018;18:308–317. doi: 10.1016/S1473-3099(17)30725-9. [DOI] [PubMed] [Google Scholar]
- 62.Turner T.B., Hollander D.H. World Health Organization; 1957. Biology of the Treponematoses. [PubMed] [Google Scholar]
- 63.Chesney A.M., Schipper G.J. The effect of the method of inoculation on the course of experimental syphilis in the rabbit. American J. Syphilis. 1950;34:18–24. [PubMed] [Google Scholar]
- 64.Lukehart S.A., Miller J.N. Demonstration of the in vitro phagocytosis of Treponema pallidum by rabbit peritoneal macrophages. J. Immunol. 1978;121:2014–2024. [PubMed] [Google Scholar]
- 65.Desrosiers D.C., Anand A., Luthra A., Dunham-Ems S.M., LeDoyt M., Cummings M.A.D., Eshghi A., Cameron C.E., Cruz A.R., Salazar J.C., et al. TP0326, a Treponema pallidum beta-barrel assembly machinery A (BamA) orthologue and rare outer membrane protein. Mol. Microbiol. 2011;80:1496–1515. doi: 10.1111/j.1365-2958.2011.07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferguson M.R., Delgado K.N., McBride S., Orbe I.C., La Vake C.J., Caimano M.J., Mendez Q., Moraes T.F., Schryvers A.B., Moody M.A., et al. Use of Epivolve phage display to generate a monoclonal antibody with opsonic activity directed against a subdominant epitope on extracellular loop 4 of Treponema pallidum BamA (TP0326) Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1222267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haynes A.M., Godornes C., Ke W., Giacani L. Evaluation of the Protective Ability of the Treponema pallidum subsp. pallidum Tp0126 OmpW Homolog in the Rabbit Model of Syphilis. Infect. Immun. 2019;87 doi: 10.1128/IAI.00323-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brinkman M.B., McGill M.A., Pettersson J., Rogers A., Matejková P., Smajs D., Weinstock G.M., Norris S.J., Palzkill T. A novel Treponema pallidum antigen, TP0136, is an outer membrane protein that binds human fibronectin. Infect. Immun. 2008;76:1848–1857. doi: 10.1128/IAI.01424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu M., Xie Y., Zheng K., Luo H., Tan M., Zhao F., Zeng T., Wu Y. Two Potential Syphilis Vaccine Candidates Inhibit Dissemination of Treponema pallidum. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.759474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Giacani L., Lukehart S., Centurion-Lara A. Length of guanosine homopolymeric repeats modulates promoter activity of Subfamily II tpr genes of Treponema pallidum ssp. pallidum. FEMS Immunol. Med. Microbiol. 2007;51:289–301. doi: 10.1111/j.1574-695X.2007.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zobaniková M., Mikolka P., Cejkova D., Mikolka P., Čejková D., Pospíšilová P., Chen L., Strouhal M., Qin X., Weinstock G.M., Šmajs D. Complete genome sequence of Treponema pallidum strain DAL-1. Stand Genomic Sci. 2012;7:12–21. doi: 10.4056/sigs.2615838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lukehart S.A., Marra C.M. Isolation and laboratory maintenance of Treponema pallidum. Curr. Protoc. Microbiol. 2007;Chapter 12:Unit 12A.1–Unit 12A.1.8. doi: 10.1002/9780471729259.mc12a01s7. [DOI] [PubMed] [Google Scholar]
- 73.Molini B., Fernandez M.C., Godornes C., Vorobieva A., Lukehart S.A., Giacani L. B-Cell Epitope Mapping of TprC and TprD Variants of Treponema pallidum Subspecies Informs Vaccine Development for Human Treponematoses. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.862491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 75.Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konis K. University of Oxford; 2007. Linear Programming Algorithms for Detecting Separated Data in Binary Logistic Regression Models. [Google Scholar]
- 77.Ito K., Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst. Pharmacol. 2013;2 doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc. Ser. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of antigens recognized all rabbits and time to a seropositive response.
Data Availability Statement
-
•
Data are deposited at on Mendeley Data and available at Mendeley Data: https://data.mendeley.com/preview/vw72m8jkwg?a=9458c05a-b54f-4a37-ad00-10b11958288f. Deposited data include antiboby reactivities detected in analyzed serum samples from animals.
-
•
Code: not applicable.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.