Abstract
The NEREA (Naples Ecological REsearch for Augmented observatories) initiative aims to establish an augmented observatory in the Gulf of Naples (GoN), designed to advance the understanding of marine ecosystems through a holistic approach. Inspired by the Tara Oceans expedition and building on the scientific legacy of the MareChiara Long-Term Ecological Research (LTER-MC) site, NEREA integrates traditional physical, chemical, and biological measurements with state-of-the-art methodologies such as metabarcoding and metagenomics. Here we present the first 10 months of NEREA data, collected from April 2019 to January 2020, encompassing physico-chemical parameters, plankton biodiversity (e.g., microscopy and flow cytometry), prokaryotic and eukaryotic metabarcoding, a prokaryotic gene catalogue, and a collection of 3818 prokaryotic Metagenome-Assembled Genomes (MAGs). NEREA’s efforts produce a significant volume of multifaceted data, which enhances our understanding of marine ecosystems and promotes the development of scientific hypotheses and ideas.
Subject terms: Ecology, Ecology
Background & Summary
Oceans harbour invaluable biodiversity that plays a pivotal role in driving biogeochemical cycles and enhancing ecosystem productivity. Coastal ecosystems provide humankind with a wide array of goods and services, including food, biochemical compounds, fossil fuels, and recreational spaces1.
Coastal systems face threats from a variety of intensive human activities including agriculture, fisheries, aquaculture, shipping, urbanisation, and tourism. Addressing such increasing threats requires integrated approaches within well-established monitoring frameworks2, such as the Long-Term Ecological Research (LTER) sites, crucial for identifying significant ecological events under a changing global climate, while providing valuable insights for society management and policymakers3,4. Among LTERs, the MareChiara LTER site (LTER-MC) in the Gulf of Naples (GoN; Central Tyrrhenian Sea5,6, Western Mediterranean Sea), established in 1984, represents one of the world’s longest ecological time series focused on the study of plankton, as well as on the physico-chemical state of the water column7.
The GoN, spanning roughly 195 km of coastline, faces significant pressures from a variety of human activities. The city of Naples and its metropolitan area, with about 4 million inhabitants, see an influx of tourists during summer, further impacting both the mainland and the highly popular islands of Capri, Ischia and Procida. The region is also a hub for industrial and commercial activities, including one of the largest ports in the Mediterranean Sea, which amplifies the anthropogenic impact. Associated anthropogenic stresses, such as illegal dumping and discharges, intense maritime traffic and industrial activities are only examples of the continuous human pressures on this area8. It is within this context that the LTER-MC was established and continues to operate.
Implementing existing observation sites with data generated through innovative and diversified approaches is crucial for the development of “augmented observatories”, as stated by the G7 Turin declaration9, which encourages multidisciplinary marine biodiversity observations. This effort is essential to pursue the ‘Ocean Health Index’ (OHI) assessment10 which aims to employ scientific methods to quantitatively characterise specific biological, physical, economic, and social aspects and to guide policymakers towards the sustainable management of ocean resources. In line with this, a multidisciplinary and integrative investigation of plankton, ranging from virus to jellyfish, was carried out by the Tara Oceans project, an oceanographic expedition aimed at using cutting-edge molecular, microscopical and optical techniques to acquire multifaceted data on the wide variety of species contributing to marine plankton11. Plankton encompasses autotrophic, heterotrophic, and mixotrophic species from all kingdoms of life12, spanning six orders of magnitude in size13. These organisms play a pivotal role in driving the Earth’s biogeochemical cycles14 and account for half of the planet’s oxygen production15.
Based upon the conceptual and methodological framework developed by the Tara Oceans project and building up on the scientific legacy of the LTER-MC site, we here present NEREA (Naples Ecological REsearch for Augmented observatories)16. This initiative aims to establish an augmented observatory in the GoN to study the dynamics of marine plankton communities. The project integrates expertise from various fields, including physics, oceanography, chemistry, physiology, molecular biology, bioinformatics, data science, modelling, and theoretical ecology. This integration facilitates a synergistic approach where laboratory experiments and computational analyses complement field observations. Such observations are refined by experimental and theoretical insights, creating a feedback loop that enhances ecological research methods. The collaborative nature of NEREA produces a comprehensive dataset that advances our understanding of marine ecosystems and addresses ecological questions.
Additionally, NEREA is committed to fostering international collaborations and it is actively involved in the global-scale research program BioGeoSCAPES, aimed to improve our understanding of the microbial biogeochemistry of the oceans. NEREA is also part of the UN Decade of Ocean Sciences (UNDOS) under the Ocean Biomolecular Observing Network (OBON) Program.
Here we present the first 10 months of NEREA data, collected from April 2019 to January 2020, encompassing physico-chemical parameters, plankton biodiversity (microscopy and flow cytometry), prokaryotic and eukaryotic metabarcoding, a prokaryotic gene catalogue, and a collection of 3818 prokaryotic Metagenome-Assembled Genomes (MAGs).
Methods
Study area
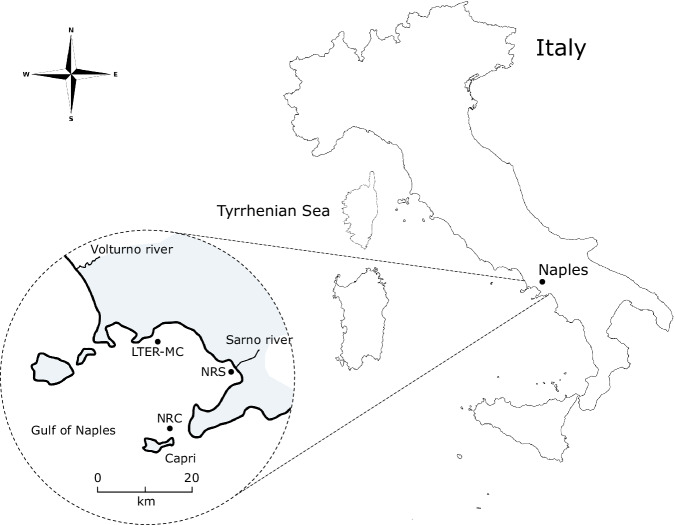
The GoN is one of the most representative Mediterranean coastal basins, with key interplay between physical drivers and ecosystem responses (e.g., 17–20). It is characterised by five persistent water masses17: Coastal Surface Water (CSW), Tyrrhenian Surface Water (TSW), Tyrrhenian Intermediate Water (TIW), Atlantic Water (formerly referred to as Modified Atlantic Water), and Levantine Intermediate Water (LIW)6. Local climatic conditions, including wind patterns and river discharges, significantly influence these waters, particularly near the shorelines (e.g., 18,19,21). NEREA covers the whole GoN area with monthly sampling at the LTER-MC site (Lat 40°48.5′ N, Long 14° 15′ E; bottom depth: ca. 75 m), located two nautical miles off downtown Naples; this site experiences alternating influences from nutrient-rich coastal waters and the nutrient-poor waters of the Mid Tyrrhenian Sea7. The LTER-MC site (DEIMS iD: https://deims.org/0b87459a-da3c-45af-a3e1-cb1508519411) belongs to the Long-Term Ecological Research national and international networks (LTER-Italy, LTER-Europe and ILTER).
In addition to sampling at LTER-MC site, NEREA includes seasonal sampling at the River Sarno mouth and plume (Lat 40°43′ N, Long 14°27′ E; bottom depth: ca. 15 m) and a biannual sampling at a deep station located at the Dohrn Canyon (Lat 40°36′ N, Long 14°08′ E; bottom depth ca. 1300 m)22 near the Island of Capri (Fig. 1). The Sarno river is highly polluted23, bringing intermittent loads of nutrients and chemical pollutants to the coast from intensive agricultural activities and leather factories24. On the other hand, the Dohrn Canyon, extending 25 km and located around 12 nautical miles off Naples, is a deep bifurcated submarine structure. It represents an active geological site contributing to the upwelling of deep waters and generating turbidity currents at the seafloor25. While primarily investigated for its geological attributes25, the canyon biodiversity remains largely unknown26. It represents a biodiversity hotspot, offering critical ecological benefits and serving as a breeding ground for various marine species along the coastline27.
Fig. 1.
Geographical map of the Gulf of Naples (GoN) with NEREA sampling sites. LTER-MC: Long-Term Ecological Research MareChiara; NRC: NEREA-Capri (Dohrn Canyon); NRS: NEREA-Sarno (Sarno river).
NEREA operational units
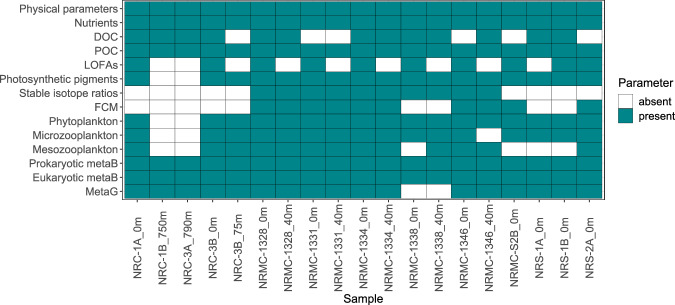
NEREA’s multidisciplinary approach leads to the production of a wide variety of data (Fig. 2), which can be listed as follows:
-
(i)
Physical parameters;
-
(ii)
Chemical data, including nutrients, particulate and dissolved organic matter, particulate lipid-derived oxylipins and stable isotope ratios;
-
(iii)
Plankton biodiversity data, including picoplankton quantification by flow cytometry; species identification and quantification by light microscopy of phyto- micro- and mesozooplankton;
-
(iv)
Plankton metabarcoding and metagenomic data.
Fig. 2.
Presence and absence matrix of analysed parameters. The matrix displays the data availability for each parameter across NEREA samples. Torquoise squares indicate the presence of data, while white squares represent the absence of data for the corresponding parameter in each sample. DOC: Dissolved Organic Carbon; POC: Particulate Organic Carbon; LOFAs: Linear Oxygenated Fatty Acids; FCM: Flow Cytometry; metaB: metabarcoding; MetaG: metagenomics.
All scientists operate in a coordinated way both in laboratory and during onboard activities, where simultaneous sampling and observations are run (Fig. 3).
Fig. 3.
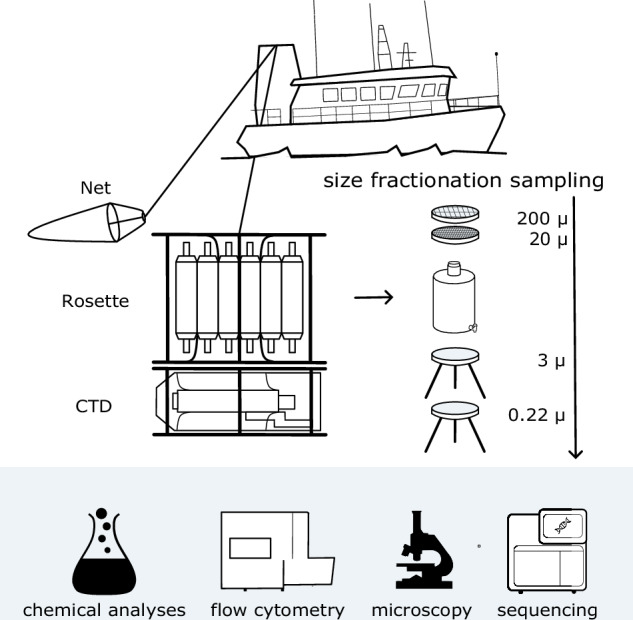
NEREA sampling strategy. A Conductivity, Temperature, and Depth (CTD) probe attached to a rosette equipped with multiple Niskin bottles collects distinct water samples. Plankton samples are also collected by deploying nets of appropriate mesh size (20 µm for phytoplankton and 200 µm for zooplankton). Part of the collected water is subjected to size fractionation, involving a series of sieves and filters with decreasing mesh and pore sizes, for the separation of plankton into distinct size categories. Samples obtained from Niskin bottles and nets are subsequently chemically analysed and processed via flow cytometry, microscopy, and DNA/RNA extraction followed by sequencing.
Physical state of the system
This task is firstly pursued onboard by the real-time production and observation of vertical profiles for multiple parameters, such as temperature (°C), salinity, density (σ), dissolved oxygen (% and mL L−1), fluorescence (µg L−1), transmittance (%), turbidity (NTU), pH and photosynthetically active radiation (PAR). The above-mentioned observations are obtained using a SeaBird Electronic SBE 911 plus v2 multi-parameter profiler. This probe acquires 24 data variables per second with an accuracy of 0.002 °C for temperature and 0.001 S/m for conductivity, and it is equipped on a rosette-type sampler with twelve Niskin bottles of 10 L each, which are employed in the collection of water samples at different depths, based on the physical structure of the water column.
The procedures described are essential for analysing the water column characteristics based on its vertical physical structure, enabling real-time planning for further water sampling. In the GoN, particularly near the coast, the water column tends to be mixed and homogeneous during winter but becomes highly stratified from late spring to autumn28. This seasonal variability leads to different environmental conditions, which in turn influence the composition of planktonic communities at various depths. Consequently, sampling strategies need to be adjusted seasonally. For example, in the context of meta-omic studies, at the coastal LTER-MC site only surface water is collected during winter, while during periods of stratification an additional sample is taken at 40 m depth to account for the different conditions and planktonic communities. In addition, a thermosalinograph is used during the transit between sampling sites, collecting continuous data for the superficial temperature, conductivity, and salinity.
Chemical state of the system
The seawater used for the chemical analyses is collected from Niskin bottles at different depths. The analyses involve the measurement of nutrients, total nitrogen (TN), total phosphorus (TP), dissolved organic carbon (DOC), particulate organic carbon (POC), particulate nitrogen (PN), particulate lipid-derived oxylipins (Linear Oxygenated Fatty Acids, LOFAs), chlorophyll a and other photosynthetic pigments.
At the LTER-MC site, nutrients, TN, TP, and DOC are collected at 0, 2, 5, 10, 20, 30, 40, 50, 60, and 70 m depths; POC and HPLC at 0, 10, 20, 40, and 60 m depths; LOFAs at 0 m depth, chlorophyll a at 0, 2, 5, 10, 20, 40, and 60 m depths. At the Dohrn Canyon station, all the chemical analyses are carried out on samples collected at 0, 10, 20, 40, 60, 80, 100 m depths. Nutrients, TN, TP, DOC and POC are analysed also at the deeper depths of 150, 200, 250, 300, 400, 600, 800 m. LOFAs are analysed at 0 m depth. At Sarno, all the chemical analyses are carried out on samples collected at 0 m.
Nutrients, TN, and TP
Determination of inorganic nutrients (NO3−, NO2−; PO4−, SiO4−) is performed on samples directly collected from the Niskin bottles into high-density polypropylene vials and frozen at −20 °C. Analyses are performed using a Flow-Sys III Systea auto-analyzer using the methods reported by29. For the determination of TN and TP, samples are taken directly from the Niskin bottle in 100 ml HDPE containers and immediately frozen at −20 °C. Analyses are performed after digestion at 120 °C for 30 minutes, following the method proposed by30.
DOC and POC
Samples for DOC and POC analysis are collected directly from the Niskin bottles into acid-washed Nalgene polycarbonate bottles. Samples for DOC are stored at −20 °C until filtration and analysis. Filtration is carried out using 0.22 µm polyethersulfone (PES) filters (Minisart®), DOC analysis is carried out by high-temperature catalytic oxidation with a Shimadzu TOC analyzer (TOC-Vcsn) following the methods described in31. For POC determination, filtrations are conducted on board using a variable volume (1–3 L) of seawater filtered on pre-combusted 25 mm GF/F filters. After filtration, filters are rinsed with deionized water and stored at −20 °C. The analyses are carried out using a Thermo Scientific FlashEA 1112 elemental analyzer (Thermo Fisher Scientific) following32 and using cyclohexanone-2,4-dinitrophenylhydrazone as standard.
LOFAs
For the analysis of particulate oxylipins a variable volume (0.75–3 L) of seawater is collected using Niskin bottles, pre-filtered onto 200 μm mesh nylon net, and then filtered through polycarbonate filters (47 mm diameter, 2 μm mesh size); filters are kept in 2 ml Eppendorf tubes and promptly frozen in liquid nitrogen. After thawing, filters are sonicated in Milli-Q water, left at room temperature for 30 min and then oxylipins extracted and quantified through Liquid Chromatography-Mass Spectrometry (LC-MS) after addition of a known amount of 16-hydroxyhexadecanoic acid as internal standard. According to the targeted-metabolomic approach used, only LOFAs deriving from hexadecatrienoic (C16), eicosapentaenoic (C20), and docosahexaenoic (C22) fatty acid precursors are quantified. Amount of LOFAs is normalised by the volume of the seawater filtered and reported as ng-LOFAs/L33.
Total chlorophyll a
The seawater for the determination of the total chlorophyll a is filtered (200 to 1080 ml) on GF/F (25 mm diameter), which are quickly frozen in liquid nitrogen. Once thawed, chlorophyll a is extracted in acetone 90% and its concentration determined according to34 using a spectrofluorometer (Shimadzu RF-5301 PC), which is daily calibrated using a chlorophyll a standard solution (from Anacystis nidulans; Sigma).
Photosynthetic pigments
For the analysis of the pigment spectrum using High Performance Liquid Chromatography (HPLC), 2-3 L of seawater are collected at each depth and filtered through GF/F filters (47 mm in diameter), which are then stored in liquid nitrogen. The pigment separations are conducted using an Agilent 1100 HPLC (Agilent technologies, United States) according to the method outlined in35 and modified by36. The HPLC system, equipped with an HP 1050 photodiode array detector and a HP 1046 A fluorescence detector, is specifically used for the measurement of chlorophyll degradation products. Calibration of the instrument is performed using external standard pigments supplied by the International Agency for 14C determination-VKI Water Quality Institute.
Carbon and nitrogen stable isotope ratios of plankton
Water samples for pico- and nanoplankton (<20 µm) are collected at a depth of 1 m using a Niskin bottle and then transferred into plastic bins. These samples are transported to the laboratory on ice, where they are pre-filtered through a 20 µm mesh. The filtered water is subsequently concentrated onto pre-combusted (4 hours at 500 °C) GF/F filters. These filters are individually frozen and later freeze-dried. Water samples for microplankton (20–200 µm) and mesozooplankton (200–2,000 µm) are collected through vertical tows from approximately 70 m depth to the surface using 20 µm and 200 µm plankton nets, respectively. Samples are transferred into plastic jars and transported to the laboratory on ice. In the lab, microplankton samples are concentrated onto 20 µm filters, and mesozooplankton samples onto 200 µm filters. These are then transferred into vials, frozen, and freeze-dried. For the determination of carbon and nitrogen stable isotopes, 1.0 ± 0.1 mg of each sample is sent to the Stable Isotope Facility at the University of California, Davis, USA. Analysis is performed using a PDZ Europa ANCA-GSL elemental analyser interfaced with a PDZ Europa 20–20 isotope ratio mass spectrometer (Sercon Ltd., Cheshire, UK). The instrumental precision is ±0.02‰ for δ13C and ± 0.03‰ for δ15N. Final δ13C and δ15N values are expressed relative to the international standards Vienna Pee Dee Belemnite (VPDB) for carbon and nitrogen. To ensure reproducibility, duplicate aliquots of 26 samples are analysed, yielding a reproducibility of ± 0.04‰ for δ13C and ± 0.06‰ for δ15N.
Plankton biodiversity - Flow Cytometry
Discrete samples for picoplankton are collected from the Niskin bottles and kept in the dark at 4 °C until analysis (less than 2 h). Subsamples of 1 ml are either run unstained for the picophytoplankton counts or fixed with a mix of 1% paraformaldehyde and 0.5% glutaraldehyde, frozen in liquid nitrogen and stored at −80 °C37. Once thawed, samples are stained with SYBRGreen I for heterotrophic bacteria38 and processed using a Becton-Dickinson FACVerse flow cytometer (BD BioSciences) equipped with a 488 nm solid-state laser and a standard filter set. Picophytoplankton (the phototrophic part) and bacteria are classified based on their scatter versus fluorescence properties and counted by the volumetric device of the instrument, after appropriate adjustments due to dilution. Gating is on red fluorescence for the picophytoplankton and green fluorescence from the SYBRGreen stain for the heterotrophic prokaryotes. One µm beads (PolySciences) are used as internal standards and for normalisation of scatter and fluorescence values (arbitrary units, a.u.). Files generated are elaborated using the FCS Express software (DeNovo Software Inc).
Plankton biodiversity - Light Microscopy
Identification and quantification by light microscopy of unicellular eukaryotic organisms, i.e., protists, is carried out following two different analytical protocols, depending on the size and abundance of the organisms that roughly correspond to phytoplankton and microzooplankton. We grouped under the “Phytoplankton” label mainly autotrophic unicellular species but also small-sized mixotrophic and heterotrophic taxa. “Microzooplankton” includes ciliates and large heterotrophic and mixotrophic dinoflagellates. Phytoplankton and microzooplankton samples are collected from the same Niskin bottles used for the meta-omic analyses. 250 ml of water in a dark glass bottle in duplicate is fixed with Lugol’s solution (1%) and stored in the dark. Phytoplankton and microzooplankton analysis are performed according to the Utermöhl method after the settling of a variable volume of sample39. Settled volumes vary in relation to the concentration of target cells ranging from 3 to 100 ml for phytoplankton and from 100 to 250 ml for microzooplankton. Counting is done over varying fractions of the sedimentation chamber, generally 2–4 transects for phytoplankton, half, or entire chamber for microzooplankton. Identification and counting are done using an inverted light microscope Zeiss Axiovert 200 (Carl Zeiss, Germany) at 400x (phytoplankton) and 200x (microzooplankton) magnifications. Specimens are identified at the lowest possible taxonomic level based on classic taxonomy books40–45 supplemented by more recent specialised papers. Lists of taxa have been updated and checked for nomenclature and synonyms as reported in the taxonomic reference websites (Algaebase http://www.algaebase.org/ and WoRMS http://www.marinespecies.org/).
In the laboratory, mesozooplankton samples are analysed after being fixed in 95% ethanol. Initially, the sample is concentrated and re-suspended in 200 ml of ethanol. Depending on the sample density, aliquots are taken using the Huntsman beaker subsampling method at ratios ranging from 1/4 to 1/3246. These aliquots are thoroughly mixed and then subsampled using a large bore graduated pipette. The aliquots are transferred into a 10 ml mini-Bogorov chamber for examination under a Leica M165C stereomicroscope. In most cases, copepods are identified at the highest taxonomic levels, i.e. species, genus and family. Among other mesozooplankton, some groups are identified at the species level (e.g., Cladocerans, Chaetognaths, Siphonophores), while other holoplankton (e.g., Amphipods, Mysids, Ostracods, Euphausiids, Pteropods, Hydromedusae) and meroplanktonic taxa (e.g., Decapods, Cirripeds, Echinoderms, Bivalves, Molluscs, Gastropods) are identified at higher taxonomic levels. All the mesozooplanktonic species abundances are reported as individuals m−3.
Plankton metabarcoding and metagenomics – sampling
The sampling strategy for the biological samples used herein, from the sampling devices to the size fraction division of organisms, is based on that used in the Tara Oceans project47,48, integrating and adapting those procedures to the coastal system studied in NEREA. The size fractions used are efficient for effectively separating the principal components of the marine plankton: 0.2–3 µm (picoplankton: e.g., archaea, heterotrophic bacteria, cyanobacteria); 3–20 µm (nanoplankton: small eukaryotic protists, e.g., diatoms, flagellates, chrysophyta, chlorophyta); 20–200 µm (microplankton: larger eukaryotic protists, e.g., diatoms; protozoa; foraminifera; tintinnids; rotifers; juvenile metazoans); 200–20000 µm (mesozooplankton: metazoans, e.g., copepods and cladocera).
At each station, 10-L tanks are filled with seawater taken directly from the Niskin bottles. From each bottle, seawater is collected using a double sieve in series, the first with a mesh of 200 µm and the second one of 20 µm. After that, using a peristaltic pump and two “tripods” (e.g., stainless steel filters holders of 142 mm diameter), the seawater is filtered in series on two polycarbonate filters (diameter 142 mm, porosity 3 µm and 0.22 µm, respectively) to divide the size fractions 3–20 µm and 0.22–3 µm. The 10 L of water collected is used to obtain two replicates (5 L for replicate) of each size class.
The size fraction 20–200 µm is obtained by rinsing the 20-µm mesh previously used, with 10 L of water resulting from the serial filtration of the fractions 3–20 µm and 0.22–3 µm, recovering in this way the material attached to the mesh. The 10 L of water in the tank containing the 20–200 µm fraction is then filtered in two rounds on two polycarbonate filters of 3 µm (5 L per duplicate). The filters collected from each of the above-mentioned size classes are finally placed into 5 ml cryovials and frozen in liquid nitrogen.
At 0 and 40 m depths, we collect the 0.22–3 µm, 3–20 µm and 20–200 µm size fractions, while we have chosen to collect only the 0.22–3 µm and >3 µm sizes in the deeper station of the Dohrn Canyon using 10 L of water for each of the two replicates due to the limited concentration of material found at 800 m depth.
The 200–2000 µm size fraction is gathered with a double WP2 plankton net, featuring a mouth area of 0.25 m² and a mesh opening of 200 µm, provided with a 500 ml plexiglass filtering cod-end. This net is vertically deployed from a depth of 50 m to the surface at LTER-MC, and both from 200 m to the surface and from 50 m to the surface at Dohrn Canyon. To calculate the volume of water that has been filtered, a mechanical flowmeter from Hydrobios is utilised. After collection, the 500 ml of the mesozooplankton sample are divided into two equal parts of 250 ml each. Both aliquots are filtered through a 200 µm mesh and then fixed in ethanol 95%. One aliquot is used for counting, while the other is used for reference library. The other 500 ml are filtered through a 200 µm mesh, treated, and stored in liquid nitrogen for meta-omics.
Nucleic acid extraction
Nucleic acid extractions are performed according to the protocol used within the Tara Oceans project49. Briefly, it is carried out by cryogenically grinding cryopreserved filters and zooplankton pellets. This process is followed by simultaneous RNA/DNA co-extraction utilising the NucleoSpin RNA and DNA Elution buffer kits from Macherey-Nagel, Düren, Germany. To disintegrate cells, the cryopreserved filters are ground as follows: first, each filter is treated with 1 ml of RA1 lysis buffer and 1% β-mercaptoethanol and then subjected to a grinding program: pre-cooling (2 min), first cycle of 10 knocks/s grinding (1 min), cooling time (1 min) and a final cycle of 10 knocks/s grinding (1 min). The powder resulting from cryogrinding is then subjected to nucleic acid extraction with the NucleoSpin RNA kits (Macherey-Nagel) and DNA Elution buffer kit (Macherey-Nagel). Cryoground powder is resuspended in 2 ml of RA1 lysis buffer mixed with 1% β-mercaptoethanol and then transferred to a NucleoSpin filter from the RNA Midi kit. This solution is centrifuged for 10 minutes at 1,500 g. Following this, an additional 1 ml of RA1 lysis buffer with 1% β-mercaptoethanol is added, and the filter is again centrifuged for 3 minutes at 1,500 g. The resulting eluate is then transferred to a new tube containing an equal volume of 70% ethanol. This mixture is loaded onto a NucleoSpin RNA Mini spin column and washed twice with a DNA washing solution. The DNA is eluted with 100 µl of DNA elution buffer three times and subsequently stored at −20 °C. Finally, the DNA is quantified using a dsDNA-specific fluorometric quantitation method (Broad Range and High Sensitivity Assays) using a Qubit fluorometer. Under such conditions, median quantities of 0.97 and 0.81 μg of DNA and RNA were obtained.
Library preparation from amplicon PCRs
Two markers are targeted for metabarcoding analyses: (i) the hypervariable region V9 of the 18S rDNA for eukaryotes (515 F: 5′ GTG YCA GCM GCC GCG GTA A 3′; 926 R: 5′ CCG YCA ATT YMT TTR AGT TT 3′)50, and (ii) the hypervariable region V4-V5 of the 16S rDNA gene for prokaryotes (1389 F: 5′ TTG TAC ACA CCG CCC 3′; 1510 R: 5′ CCT TCY GCA GGT TCA CCT AC 3′)51. The libraries are constructed using a Barcode IDentifier (BID). In particular, 12 different BIDs are incorporated into the amplification primers, enabling pooling of 6 to 12 PCR products before the library preparation. Then, from pools of PCR products tagged by different BIDs, a single library is constructed and indexed by a NextFlex DNA barcode. The libraries are prepared using 100 ng of amplicon directly end-repaired, A-tailed at 3’ end and ligated with Illumina adaptors using the NEBNext DNA Module and NextFlex DNA barcodes. After two 1x AMPure XP clean ups (only 1 for the 18S), the ligated products are amplified with Kapa Hifi HotStart NGS library Amplification kit and then purified by 1x AMPure XP. Libraries are initially quantified by Quant-it dsDNA HS on a Fluoroskan Ascent instrument (Thermo scientific). Following this, they are quantified by qPCR with the KAPA Library Quantification Kit for Illumina Libraries (Kapa Biosystems) on an MXPro instrument (Agilent Technologies). Library profiles are evaluated using a high throughput microfluidic capillary electrophoresis system (LabChip GX, Perkin Elmer, Waltham, MA). Metabarcoding libraries exhibit low diversity sequences at the beginning of the reads due to the primer sequences used for tag amplification. This low diversity can interfere with the correct cluster identification, leading to a significant loss of data output. For this reason, loading concentrations of these libraries and PhiX DNA spike-in (20% instead of 1%) are adapted to minimise the impacts on the run quality. Metabarcoding libraries are sequenced on a NovaSeq 6000 instrument (Illumina, San Diego, CA, USA) using 2 × 150 or 250 base-length read chemistry for 18SV9 or 16SV4-V5 libraries, respectively.
Library preparation for metagenomics
DNA is first fragmented to a size range of 100–1,000 bp using the E220 Covaris Focused ultrasonicator (Covaris, Woburn, MA, USA). These fragments undergo end-repair and are then adenylated at the 3′ end. NEXTflex HT Barcodes (Bioo Scientific, Austin, TX, USA) are subsequently added using products from the NEBNext DNA modules (New England Biolabs, Ipswich, MA, USA). The library construction process includes two consecutive clean-up steps using 1 × AMPure XP (Beckman Coulter, Brea, CA, USA). After these clean-ups, the ligated product is amplified through 12 PCR cycles using the Kapa Hifi Hotstart NGS library amplification kit (Kapa Biosystems, Wilmington, MA, USA), followed by 0.6x AMPure XP purification. Metagenomics libraries, similarly to metabarcoding libraries, are quantified using Quant-iT dsDNA HS on a Fluoroskan Ascent instrument (Thermo scientific) and by qPCR with the KAPA Library Quantification Kit for Illumina Libraries (Kapa Biosystems) on an MXPro instrument (Agilent Technologies). Subsequently, library profiles are assessed with a high-throughput microfluidic capillary electrophoresis system (LabChip GX, Perkin Elmer, Waltham, MA). Metagenomic libraries are sequenced by a NovaSeq 6000 instrument (Illumina, San Diego, CA, USA) by paired ends of 150 bases in length.
Metabarcoding - 16S ASV generation and taxonomic assignment
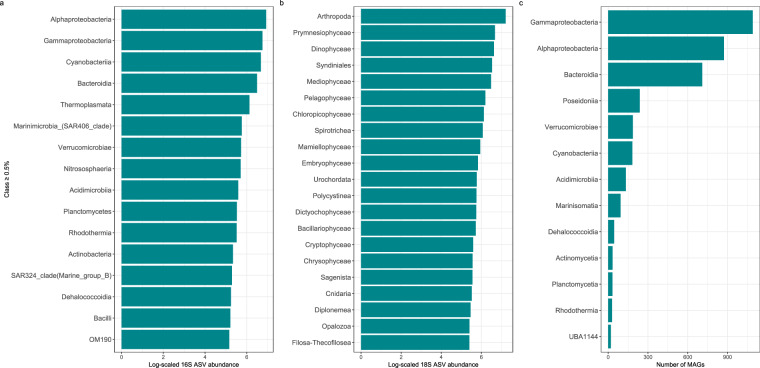
Raw 16S paired-end sequences were subjected to a data quality control step49 and subsequently imported into the QIIME2 pipeline v.2022.2.052. Leftover primers and adapter sequences were removed through cutadapt53. The Amplicon Sequence Variants (ASVs) table, which represents true biological sequences within each sample, was generated using the denoised-paired method including truncation, denoising, dereplication, merging, and chimera filtering of the DADA2 (Divisive Amplicon Denoising Algorithm 2) plugin inside QIIME254. Default parameters were used with the exception of the forward and reverse sequence length (–p-trunc-len-f and–p-trunc-len-r), that were set to 220 and 180, respectively. Processed reads that passed all these filters were used for taxonomy classification (Fig. 4a). The V4-V5 regions were extracted from the pre-formatted reference sequences and taxonomy file built on the SILVA 138 99% OTUs database55–57 and the vsearch (v.2.6.2) global alignment implemented in QIIME2 was used58.
Fig. 4.
Class-level abundance of Amplicon Sequence Variants (ASVs) and Metagenome-Assembled Genomes (MAGs). The panels show the log-scaled read counts for 16S (a) and 18S (b) ASVs and the number of prokaryotic MAGs (c) ASVs and MAGs representing more than 0.5% of the total abundance and assigned to a known class are displayed.
Metabarcoding - 18S ASV generation and taxonomic assignment
Illumina paired-end V9-18S raw reads (FASTQ format, 2 × 150 PE) were pre-processed with cutadapt53 and vsearch to remove primer sequences, trim low quality bases and unify mixed orientation reads produced in the ligation-based library preparation; the procedure was implemented in a custom bash script. Processed reads were then used to generate ASVs using the DADA2 R library54; the pipeline was adapted from the one described on the program website (https://benjjneb.github.io/dada2/tutorial.html); no further quality filtering was implemented at this stage, except for discarding all reads with ambiguities (parameter maxN = 0 of function filterAndTrim). Filtered forward (F) and reverse (R) reads were used to train the error model and then denoised by applying the trained error model to generate ASVs. Finally, F and R reads were merged and checked for chimeras, allowing no mismatches in read merging (default parameter maxMismatch = 0 of function mergePairs). ASVs were then classified with BLAST against the PR2 (v.5.01) reference database59, integrated with 1,293 environmental sequences from GoN protist strains and fungi. Highest bit scores matching with the best taxonomic resolution were then selected among the returned results (Fig. 4b).
Metagenomic data processing
Metagenomic data processing was performed as described in60. Briefly, all sequencing reads were quality filtered using BBMap (v.38.79). This step involved removing sequencing adapters, eliminating reads that matched the PhiX control genome, and discarding low-quality reads based on specific criteria (trimq = 14, maq = 20, maxns = 1, minlength = 45). For downstream analyses, either quality-controlled reads were used, or in some cases, reads were merged using bbmerge.sh with a minimum overlap of 16. All metagenomes were assembled individually with metaSPAdes (v.3.15.2)61. The assembled scaffolds were filtered to retain only those longer than 0.5 kbp. Gene prediction on these scaffolds was performed using Prodigal (v.2.6.3)62 with the parameters -c -q -m -p meta. For MAGs reconstruction, quality-controlled metagenomic reads from all samples were mapped back to the scaffolds (≥1 kbp) of each sample. The mapping was executed using BWA (v.0.7.17-r1188)63, configured to allow reads to map at secondary sites with the use of the -a flag. This step ensured that alignments were only considered if they were at least 45 bases long, had an identity of at least 97%, and provided a coverage of 80% or more of the read sequence. The alignments were then compiled into BAM files, which were processed using the jgi_summarize_bam_contig_depths script from MetaBAT2 (v.2.12.1)64. This script facilitated the calculation of within-sample and between-sample coverages for each scaffold. Finally, the scaffolds were binned using MetaBAT2, applying parameters –minContig 2000 and–maxEdges 500 to enhance the sensitivity of the binning process. Metagenomic bin quality was assessed using two methods: the ‘lineage workflow’ of CheckM (v.1.1.3)65 and Anvi’o (v.7.1)66. Bins were retained for further analysis if they met the following criteria from either tool: a completeness of at least 50% and a contamination level of 10% or less. Following the quality assessment, 1070 prokaryotic MAGs were taxonomically classified using GTDB-Tk (v.2.1.0)67 utilising default parameters against the GTDB R207 release68 (Fig. 4c). Gene prediction on these MAGs was carried out using Prodigal (v.2.6.3)62 with parameters -c -q -m -p single. Gene sequences predicted from prokaryotic MAGs of all samples and gene sequences predicted from assemblies of prokaryote-enriched samples were clustered at 95% identity, retaining the longest sequence within each cluster as the representative using CD-HIT (v.4.8.1)69. The specific parameters used for this clustering were -c 0.95 -M 0 -G 0 -aS 0.9 -g 1 -r 1 -d 0. The representative gene sequences obtained from the clustering were then aligned against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, using the DIAMOND aligner (v.2.0.15)70. The filtering criteria for these alignments included a minimum query and subject coverage of 70%, and a requirement for the bitScore to be at least 50% of the maximum expected bitScore, which references the score against itself. The 59 metagenomes were mapped to the 5,515,266 million cluster representatives using BWA (v.0.7.17-r1188)63 with the -a parameter to allow alignment at secondary sites. The resulting BAM files from these alignments were then refined to retain only the ones exhibiting a minimum percentage identity of 95% and a minimum alignment length of 45 bases. For the calculation of length-normalised gene abundance, the process began by counting inserts from the best unique alignments. In cases of ambiguously mapped inserts, fractional counts were allocated to the corresponding target genes based on their unique insert abundances. Subsequently, the total insert counts were divided by the length of each respective gene to normalise the gene abundance data accurately. Gene-length normalised read abundances were further converted into per-cell gene copy numbers, dividing them by the median abundance of 10 single-copy marker gene copies71 in each sample. For taxonomic profiling, the mOTUs database (v.3.1)71 was extended with 1070 prokaryotic MAGs forming 433 new species level clusters. Quality controlled sequencing reads of 59 metagenomes were profiled using the extended mOTUs database with default parameters.
Data Records
We assign a specific label to each NEREA sampling event. We define the station code using the letters NR for all the sites followed by MC, S and C respectively for MareChiara, Sarno and Dohrn Canyon. The sampling code at the MareChiara site follows the specific LTER-MC code campaign to unify the two samplings when conducted simultaneously.
Data described here are available in the Sample Registry “NEREA- Naples Ecological REsearch for Augmented observatories” on Zenodo (https://zenodo.org/communities/nerea/records), which contains the following projects:
Physical Oceanography: CTD and Thermosalinograph72 (https://zenodo.org/records/10986789);
Biogeochemical data73 (https://zenodo.org/records/11035857);
LOFAs data: particulate lipid-derived oxylipins74 (https://zenodo.org/records/11058615);
HPLC data75 (https://zenodo.org/records/11035381);
Carbon and nitrogen stable isotope ratios of plankton76 (https://zenodo.org/records/12743875);
Plankton biodiversity data: Flow cytometry data77 (https://zenodo.org/records/11035933);
Plankton biodiversity data: Phytoplankton78 (https://zenodo.org/records/10987241);
Plankton biodiversity data: Microzooplankton79 (https://zenodo.org/records/10987215);
Plankton biodiversity data: Mesozooplankton80 (https://zenodo.org/records/10987255).
The datasets included in the above-listed projects share a similar structure: they are all xlsx files including three specific sheets:
a “Readme”, which contains the dataset title, a brief description of the dataset and the methods used to produce it, and any additional note;
a “Coordinates” sheet, including the coordinates of all the sampling stations included in the datasets;
a “Data” sheet with the data.
Prokaryotic and eukaryotic metabarcoding Zenodo projects include xlsx files structured as described above. Moreover, they contain additional files with bioinformatic scripts and intermediates, including tsv and fasta files:
Metabarcoding data: 16S ASVs abundance table and taxonomic assignment81 (https://zenodo.org/records/12801913);
Metabarcoding data: 18S ASVs abundance table and taxonomic assignment82 (https://zenodo.org/records/12801941).
Metagenomic data are grouped in two Zenodo projects. The first project contains:
the full prokaryotic gene catalogue (tsv);
the Metagenome-Assembled Genomes (MAGs) sequences (fasta) and annotation (tsv);
the Taxonomic profiling using mOTUs (MG-based operational taxonomic units) database83 (https://zenodo.org/records/11035656).
The second project contains the single-assemblies and the genes (fasta and gff files)84 (https://zenodo.org/records/11046519).
Plankton metabarcoding and metagenomic raw reads were registered in the European Nucleotide Archive (ENA) at EMBL-EBI, under the umbrella project permanent identifier PRJEB7464985 (https://www.ebi.ac.uk/ena/browser/view/PRJEB74649). Raw sequencing data are recorded in three component projects all linked to the umbrella project:
-
(i)
Prokaryotic metabarcoding sequencing data: The 16S raw sequence data files (FastQ format) are available under the BioProject ID PRJEB7464186 (https://www.ebi.ac.uk/ena/browser/view/PRJEB74641);
-
(ii)
Eukaryotic metabarcoding sequencing data: the 18S raw sequence data files (FastQ format) are available under the project ID PRJEB7465887 (https://www.ebi.ac.uk/ena/browser/view/PRJEB74658);
-
(iii)
Metagenomics: Raw sequence data files (FastQ format) are available under the project ID PRJEB7465988 (https://www.ebi.ac.uk/ena/browser/view/PRJEB74659).
Technical Validation
The NEREA sampling methodology follows the one used during the Tara Oceans expedition32. This methodology is based on the study from the literature of size, abundance, and richness values of all the plankton organisms used to characterise each size fraction division. DNA obtained from extraction is quantified by a dsDNA-specific fluorometric method (Broad Range and High Sensitivity Assays) using a Qubit Fluorometer instrument. All metabarcoding libraries are quantified by measurement of the Qubit dsDNA HS Assay and then a size profile analysis is performed using an Agilent 2100 Bioanalyzer and through qPCR with the KAPA Library Quantification Kit for Illumina Libraries on an MXPro instrument. For metagenomics, the size profiles are visualised using an Agilent Bioanalyzer DNA High Sensitivity chip.
Acknowledgements
The NEREA activities have received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 101000858 (TechOceanS), No 862626 (EuroSea), No 101081642 (OBAMA-NEXT). NEREA is an UN Ocean Decade Action No 64.2 under the OBON Program. NEREA is also partly funded under the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4 - Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU. The Long-Term Ecological Research site MareChiara program is funded by Stazione Zoologica Anton Dohrn, Naples (Italy). L.C. acknowledges MARCO-BOLO (ID: 101082021) and AtlantECO (ID: 862923) projects. R.C. acknowledges the OBAMA-NEXT (ID: 101081642) project. L.R. has been supported by the BiOcean5D project (ID: 101059915). V. D., L.M., E.R., and D. GV. acknowledge a fellowship funded by the Stazione Zoologica Anton Dohrn (SZN) within the SZN-Open University Ph.D. program. E.R. was also supported by the project “Antitumor Drugs and Vaccines from the Sea (ADViSE)” (CUP B43D18000240007–SURF 17061BP000000011; PG/2018/0494374) funded by POR Campania FESR 2014–2020 “Technology Platform for Therapeutic Strategies against Cancer”- Action 1.1.2 and 1.2.2. The sequencing lab at Genoscope acknowledges financial support from France Génomique National infrastructure, funded as part of “Investissement d'Avenir” program managed by Agence Nationale pour la Recherche (contract ANR‐10‐INBS‐09). S.S. acknowledges funding from the Swiss National Science Foundation (205320_215395) and support from the ETH IT services for calculations that were carried out on the ETH Euler cluster. The authors thank Alexandre Epinoux, Carmen Minucci and Francesco Manfellotto for technical assistance.
Author contributions
Conceptualization: R.C., F.C., D.DA., D.I., S.L., F.M., M.Mo.; Funding: R.C., D.I.; Sampling design and data collection: U.C., Y.C., R.C., D.DA., I. DA., I.DC., V.D., R.G., D.I., P.L., F.M., L.M., M.Mo., C.M., A.Pa., I.P., E.R., L.R., D.S., F.T., AC.T., D.GV.; Data processing: M.A., A.A., G.B., C.B., A.B., Y.C., R.C., G.C., I. DA., I.DC., G.dI., R.G., M.F., K.L., F.M., M.Ma., PH.O., A.Pa., I.P., A.Pe., R.P., F.R., HJ.R., E.R., M.S., C.S., D.S., AC.T., G.Z.; Data curation: A.A., G.B., C.B., D.B., L.C., Y.C., R.C., G.C., I. DA., I.DC., G.dI., R.G., M.F., K.L., A.L., F.M., M.Ma., PH.O., A.Pa., I.P., A.Pe., R.P., F.R., HJ.R., L.R., M.S., D.S., AC.T., G.Z.; Data management: L.C., R.C., F.M., L.R., AC.T.; Visualisation: L.C., D.DA., A.L., L.R.; Writing - Original Draft: R.C., D.DA., M.Mo., L.R.; Writing - Review and Editing: G.B., D.B., L.C., Y.C., ML. C., R.C., D.DA., I. DA., I.DC., R.G., K.L., F.M., L.M., PH.O., A.Pa., I.P., F.R., V.R., HJ.R., L.R., M.S., D.S., S.S., G.Z.; Supervision: Y.C., R.C., ML.C., D.DA., A.F., D.I., F.M., M.Mo., R.P., D.S., S.S., P.W.
Code availability
Coding scripts for 16S and 18S ASVs generation and taxonomic assignment can be found at [https://zenodo.org/records/12801913] and [https://zenodo.org/records/12801941], respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lucia Campese, Luca Russo.
These authors jointly supervised this work: Raffaella Casotti, Fabio Conversano, Domenico D’Alelio, Daniele Iudicone, Francesca Margiotta, Marina Montresor.
Contributor Information
Lucia Campese, Email: lucia.campese@szn.it.
Raffaella Casotti, Email: raffaella.casotti@szn.it.
Fabio Conversano, Email: fabio.conversano@szn.it.
Domenico D’Alelio, Email: domenico.dalelio@szn.it.
Daniele Iudicone, Email: daniele.iudicone@szn.it.
Francesca Margiotta, Email: francesca.margiotta@szn.it.
Marina Montresor, Email: marina.montresor@szn.it.
References
- 1.Barbier, E. B. Marine ecosystem services. Curr. Biol.27, R507–R510 (2017). 10.1016/j.cub.2017.03.020 [DOI] [PubMed] [Google Scholar]
- 2.Mirtl, M. et al. Genesis, goals and achievements of Long-Term Ecological Research at the global scale: A critical review of ILTER and future directions. Sci. Total Environ.626, 1439–1462 (2018). 10.1016/j.scitotenv.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 3.Hughes, B. B. et al. Long-Term Studies Contribute Disproportionately to Ecology and Policy. BioScience67, 271–281 (2017). 10.1093/biosci/biw185 [DOI] [Google Scholar]
- 4.Wells, H. B. M., Dougill, A. J. & Stringer, L. C. The importance of long‐term social‐ecological research for the future of restoration ecology. Restor. Ecol.27, 929–933 (2019). 10.1111/rec.13000 [DOI] [Google Scholar]
- 5.Occhipinti-Ambrogi, A. et al. Alien species along the Italian coasts: an overview. Biol. Invasions13, 215–237 (2011). 10.1007/s10530-010-9803-y [DOI] [Google Scholar]
- 6.Iacono, R., Napolitano, E., Palma, M. & Sannino, G. The Tyrrhenian Sea Circulation: A Review of Recent Work. Sustainability13, 6371 (2021). 10.3390/su13116371 [DOI] [Google Scholar]
- 7.Zingone, A. et al. Time series and beyond: multifaceted plankton research at a marine Mediterranean LTER site. Nat. Conserv.34, 273–310 (2019). 10.3897/natureconservation.34.30789 [DOI] [Google Scholar]
- 8.Tornero, V. & Ribera d’Alcalà, M. Contamination by hazardous substances in the Gulf of Naples and nearby coastal areas: A review of sources, environmental levels and potential impacts in the MSFD perspective. Sci. Total Environ.466–467, 820–840 (2014). 10.1016/j.scitotenv.2013.06.106 [DOI] [PubMed] [Google Scholar]
- 9.Summary, E.G7 Future Seas Oceans Working Group. Progress since may 2016. Turin 2017.
- 10.Halpern, B. S. et al. An index to assess the health and benefits of the global ocean. Nature488, 615–620 (2012). 10.1038/nature11397 [DOI] [PubMed] [Google Scholar]
- 11.Karsenti, E. et al. A Holistic Approach to Marine Eco-Systems Biology. PLOS Biol.9, e1001177 (2011). 10.1371/journal.pbio.1001177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruggiero, M. A. et al. A Higher Level Classification of All Living Organisms. PLOS ONE10, e0119248 (2015). 10.1371/journal.pone.0119248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce, D. G., Frank, K. T. & Leggett, W. C. From mice to elephants: overturning the ‘one size fits all’ paradigm in marine plankton food chains. Ecol. Lett.18, 504–515 (2015). 10.1111/ele.12434 [DOI] [PubMed] [Google Scholar]
- 14.Falkowski, P. G., Fenchel, T. & Delong, E. F. The microbial engines that drive Earth’s biogeochemical cycles. Science320, 1034–1039 (2008). 10.1126/science.1153213 [DOI] [PubMed] [Google Scholar]
- 15.Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science281, 237–240 (1998). 10.1126/science.281.5374.237 [DOI] [PubMed] [Google Scholar]
- 16.Fanelli, E. et al. Towards Naples Ecological REsearch for Augmented Observatories (NEREA): The NEREA-Fix Module, a Stand-Alone Platform for Long-Term Deep-Sea Ecosystem Monitoring. Sensors20, 2911 (2020). 10.3390/s20102911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cianelli, D. et al. Dynamics of a very special Mediterranean coastal area: the Gulf of Naples. Mediterr. Ecosyst. Dyn. Conserv. 129–150 (2012).
- 18.Cianelli, D. et al. Inshore/offshore water exchange in the Gulf of Naples. J. Mar. Syst.145, 37–52 (2015). 10.1016/j.jmarsys.2015.01.002 [DOI] [Google Scholar]
- 19.Cianelli, D. et al. Disentangling physical and biological drivers of phytoplankton dynamics in a coastal system. Sci. Rep.7, 15868 (2017). 10.1038/s41598-017-15880-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Capua, I. et al. From Phenotypes to Genotypes and Back: Toward an Integrated Evaluation of Biodiversity in Calanoid Copepods. Front. Mar. Sci. 9, 833089 (2022).
- 21.Carrada, G. C. et al. Variability in the Hydrographic and Biological Features of the Gulf of Naples. Mar. Ecol.1, 105–120 (1980). 10.1111/j.1439-0485.1980.tb00213.x [DOI] [Google Scholar]
- 22.Milia, A. The Dohrn canyon: a response to the eustatic fall and tectonic uplift of the outer shelf along the eastern Tyrrhenian Sea margin, Italy. Geo-Mar. Lett.20, 101–108 (2000). 10.1007/s003670000044 [DOI] [Google Scholar]
- 23.De Pippo, T., Donadio, C., Guida, M. & Petrosino, C. The Case of Sarno River (Southern Italy). Effects of geomorphology on the environmental impacts (8 pp). Environ. Sci. Pollut. Res.13, 184–191 (2006). 10.1065/espr2005.08.287 [DOI] [PubMed] [Google Scholar]
- 24.Thiele, S., Richter, M., Balestra, C., Glöckner, F. O. & Casotti, R. Taxonomic and functional diversity of a coastal planktonic bacterial community in a river-influenced marine area. Mar. Genomics32, 61–69 (2017). 10.1016/j.margen.2016.12.003 [DOI] [PubMed] [Google Scholar]
- 25.Aiello, G., Iorio, M., Molisso, F. & Sacchi, M. Integrated Morpho-Bathymetric, Seismic-Stratigraphic, and Sedimentological Data on the Dohrn Canyon (Naples Bay, Southern Tyrrhenian Sea): Relationships with Volcanism and Tectonics. Geosciences10, 319 (2020). 10.3390/geosciences10080319 [DOI] [Google Scholar]
- 26.Taviani, M., Angeletti, L., Cardone, F., Montagna, P. & Danovaro, R. A unique and threatened deep water coral-bivalve biotope new to the Mediterranean Sea offshore the Naples megalopolis. Sci. Rep.9, 3411 (2019). 10.1038/s41598-019-39655-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciannelli, L. et al. Ichthyoplankton assemblages and physical characteristics of two submarine canyons in the south central Tyrrhenian Sea. Fish. Oceanogr.31, 480–496 (2022). 10.1111/fog.12596 [DOI] [Google Scholar]
- 28.Ribera d’Alcalà, M. et al. Seasonal patterns in plankton communities in a pluriannual time series at a coastal Mediterranean site (Gulf of Naples): an attempt to discern recurrences and trends. Sci. Mar.68, 65–83 (2004). 10.3989/scimar.2004.68s165 [DOI] [Google Scholar]
- 29.Grasshoff, K., Kremlingl, K. & Ehrhardt, M. Methods of seawater analysis. 347–395 (1983).
- 30.Valderrama, J. C. The simultaneous analysis of total nitrogen and total phosphorus in natural waters. Mar. Chem.10, 109–122 (1981). 10.1016/0304-4203(81)90027-X [DOI] [Google Scholar]
- 31.Sugimura, Y. & Suzuki, Y. A high-temperature catalytic oxidation method for the determination of non-volatile dissolved organic carbon in seawater by direct injection of a liquid sample. Mar. Chem.24, 105–131 (1988).
- 32.Hedges, J. I. & Stern, J. H. Carbon and nitrogen determinations of carbonate-containing solids1. Limnol. Oceanogr.29, 657–663 (1984). 10.4319/lo.1984.29.3.0657 [DOI] [Google Scholar]
- 33.Russo, E. et al. Density-dependent oxylipin production in natural diatom communities: possible implications for plankton dynamics. ISME J.14, 164–177 (2020). 10.1038/s41396-019-0518-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm-Hansen, O., Lorenzen, C. J., Holmes, R. W. & Strickland, J. D. H. Fluorometric Determination of Chlorophyll. ICES J. Mar. Sci.30, 3–15 (1965). 10.1093/icesjms/30.1.3 [DOI] [Google Scholar]
- 35.Vidussi, F., Claustre, H., Bustillosguzman, J., Cailliau, C. & Marty, J. Determination of chlorophylls and carotenoids of marine phytoplankton: Separation of chlorophyll a from divinyl-chlorophyll a and zeaxanthin from lutein. J. Plankton Res.18, 2377–2382 (1996). 10.1093/plankt/18.12.2377 [DOI] [Google Scholar]
- 36.Brunet, C. & Mangoni, O. Determinazione quali-quantitativa dei pigmenti fitoplanctonici mediante HPLC. vol. 56 (Roma, 2010).
- 37.Marie, D., Partensky, F., Vaulot, D. & Brussaard, C. Enumeration of Phytoplankton, Bacteria, and Viruses in Marine Samples. Curr. Protoc. Cytom.10, 11.11.1–11.11.15 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Balestra, C., Alonso-Sáez, L., Gasol, J. M. & Casotti, R. Group-specific effects on coastal bacterioplankton of polyunsaturated aldehydes produced by diatoms. Aquat. Microb. Ecol.63, 123–131 (2011). 10.3354/ame01486 [DOI] [Google Scholar]
- 39.Elder, L. & Elbrachter, M. The Utermöhl method for quantitative phytoplankton analysis. Microscopic and molecular methods for quantitative phytoplankton analysis. in Microscopic and Molecular Methods for Quantitative Phytoplankton Analysis. UNESCO 13–20 (2010).
- 40.Cupp, E. E. Marine Plankton Diatoms of the West Coast of North America. vol. 5 (University of California, Berkeley, California and London, England, 1943).
- 41.Monks Wood Experimental Station. Provisional Atlas of the Marine Dinoflagellates of the British Isles. (Monks Wood Experimental Station, Huntingdon, 1981).
- 42.Rampi, L. & Zattera, A. Chiave per la determinazione dei Tintinnidi mediterranei. (ENEA, Roma, 1982).
- 43.Tomas, C. R. Identifying Marine Phytoplankton. (San Diego, 1997).
- 44.Lee, A. J. & Folkard, A. R. Factors Affecting Turbidity in the Southern North Sea. ICES J. Mar. Sci.32, 291–302 (1969). 10.1093/icesjms/32.3.291 [DOI] [Google Scholar]
- 45.Strüder-Kypke, M., Dr. Agatha, S., Warwick, J. & Montagnes, D. The user-friendly key to coastal planktonic ciliates. (2002).
- 46.van Guelpen, L., Markle, D. F. & Duggan, D. J. An evaluation of accuracy, precision, and speed of several zooplankton subsampling techniques. ICES J. Mar. Sci.40, 226–236 (1982). 10.1093/icesjms/40.3.226 [DOI] [Google Scholar]
- 47.Pesant, S. et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data2, 150023 (2015). 10.1038/sdata.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorsky, G. et al. Expanding Tara Oceans Protocols for Underway, Ecosystemic Sampling of the Ocean-Atmosphere Interface During Tara Pacific Expedition (2016–2018). Front. Mar. Sci. 6, (2019).
- 49.Belser, C. et al. Integrative omics framework for characterization of coral reef ecosystems from the Tara Pacific expedition. Sci. Data10, 326 (2023). 10.1038/s41597-023-02204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W. & Huse, S. M. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLOS ONE4, e6372 (2009). 10.1371/journal.pone.0006372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol.18, 1403–1414 (2016). 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- 52.Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 852–857 (2019). 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal17, 10–12 (2011). 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 54.Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583 (2016). 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596 (2013). 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yilmaz, P. et al. The SILVA and ‘All-species Living Tree Project (LTP)’ taxonomic frameworks. Nucleic Acids Res.42, D643–648 (2014). 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robeson, M. S. et al. RESCRIPt: Reproducible sequence taxonomy reference database management. PLOS Comput. Biol.17, e1009581 (2021). 10.1371/journal.pcbi.1009581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ4, e2584 (2016). 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guillou, L. et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res.41, D597–D604 (2013). 10.1093/nar/gks1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paoli, L. et al. Biosynthetic potential of the global ocean microbiome. Nature607, 111–118 (2022). 10.1038/s41586-022-04862-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res.27, 824–834 (2017). 10.1101/gr.213959.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics11, 119 (2010). 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics25, 1754–1760 (2009). 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ7, e7359 (2019). 10.7717/peerj.7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res.25, 1043–1055 (2015). 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eren, A. M. et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ3, e1319 (2015). 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaumeil, P.-A., Mussig, A. J., Hugenholtz, P. & Parks, D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics36, 1925–1927 (2020). 10.1093/bioinformatics/btz848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parks, D. H. et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol.36, 996–1004 (2018). 10.1038/nbt.4229 [DOI] [PubMed] [Google Scholar]
- 69.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods18, 366–368 (2021). 10.1038/s41592-021-01101-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruscheweyh, H.-J. et al. Cultivation-independent genomes greatly expand taxonomic-profiling capabilities of mOTUs across various environments. Microbiome10, 212 (2022). 10.1186/s40168-022-01410-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gallia, R., Passarelli, A. & Conversano, F. Physical-oceanographic data (CTD and thermosalinograph) of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10986789 (2024). 10.5281/zenodo.10986789 [DOI]
- 73.Margiotta, F. et al. Biogeochemical data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035857 (2024). 10.5281/zenodo.11035857 [DOI]
- 74.Carotenuto, Y. & d’Ippolito, G. LOFAs data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11058615 (2024). 10.5281/zenodo.11058615 [DOI]
- 75.Saggiomo, M. & Buondonno, A. HPLC data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035381 (2024). 10.5281/zenodo.11035381 [DOI]
- 76.D’Ambra, I. Stable isotope ratios data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12743875 (2024). 10.5281/zenodo.12743875 [DOI]
- 77.Casotti, R., Balestra, C. & Trano, A. C. Flow cytometry data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035933 (2024). 10.5281/zenodo.11035933 [DOI]
- 78.Sarno, D. & Percopo, I. Phytoplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987241 (2024). 10.5281/zenodo.10987241 [DOI]
- 79.Percopo, I., Maselli, M. & Sarno, D. Microzooplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987215 (2024). 10.5281/zenodo.10987215 [DOI]
- 80.Di Capua, I. Mesozooplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987255 (2024). 10.5281/zenodo.10987255 [DOI]
- 81.Trano, A. C., Casotti, R., Raffini, F. & Piredda, R. 16S Amplicon sequence variants (ASVs) data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12801913 (2024). 10.5281/zenodo.12801913 [DOI]
- 82.Montresor, M., Zampicinini, G., Piredda, R. & Di Capua, I. 18S Amplicon sequence variants (ASVs) data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12801941 (2024). 10.5281/zenodo.12801941 [DOI]
- 83.Campese, L. & Ruscheweyh, H.-J. Prokaryotic gene catalog, prokaryotic Metagenome-Assembled Genomes (MAGs) and taxonomic profiling of metagenomic data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035656 (2024). 10.5281/zenodo.11035656 [DOI]
- 84.Campese, L. & Ruscheweyh, H.-J. Assemblies of metagenomic data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11046519 (2024). 10.5281/zenodo.11046519 [DOI]
- 85.ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74649 (2024).
- 86.ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74641 (2024).
- 87.ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74658 (2024).
- 88.ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74659 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gallia, R., Passarelli, A. & Conversano, F. Physical-oceanographic data (CTD and thermosalinograph) of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10986789 (2024). 10.5281/zenodo.10986789 [DOI]
- Margiotta, F. et al. Biogeochemical data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035857 (2024). 10.5281/zenodo.11035857 [DOI]
- Carotenuto, Y. & d’Ippolito, G. LOFAs data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11058615 (2024). 10.5281/zenodo.11058615 [DOI]
- Saggiomo, M. & Buondonno, A. HPLC data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035381 (2024). 10.5281/zenodo.11035381 [DOI]
- D’Ambra, I. Stable isotope ratios data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12743875 (2024). 10.5281/zenodo.12743875 [DOI]
- Casotti, R., Balestra, C. & Trano, A. C. Flow cytometry data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035933 (2024). 10.5281/zenodo.11035933 [DOI]
- Sarno, D. & Percopo, I. Phytoplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987241 (2024). 10.5281/zenodo.10987241 [DOI]
- Percopo, I., Maselli, M. & Sarno, D. Microzooplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987215 (2024). 10.5281/zenodo.10987215 [DOI]
- Di Capua, I. Mesozooplankton data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.10987255 (2024). 10.5281/zenodo.10987255 [DOI]
- Trano, A. C., Casotti, R., Raffini, F. & Piredda, R. 16S Amplicon sequence variants (ASVs) data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12801913 (2024). 10.5281/zenodo.12801913 [DOI]
- Montresor, M., Zampicinini, G., Piredda, R. & Di Capua, I. 18S Amplicon sequence variants (ASVs) data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.12801941 (2024). 10.5281/zenodo.12801941 [DOI]
- Campese, L. & Ruscheweyh, H.-J. Prokaryotic gene catalog, prokaryotic Metagenome-Assembled Genomes (MAGs) and taxonomic profiling of metagenomic data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11035656 (2024). 10.5281/zenodo.11035656 [DOI]
- Campese, L. & Ruscheweyh, H.-J. Assemblies of metagenomic data of NEREA Augmented Observatory. Zenodo10.5281/zenodo.11046519 (2024). 10.5281/zenodo.11046519 [DOI]
- ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74649 (2024).
- ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74641 (2024).
- ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74658 (2024).
- ENA European Nucleotide Archive. https://identifiers.org/ena.embl:PRJEB74659 (2024).
Data Availability Statement
Coding scripts for 16S and 18S ASVs generation and taxonomic assignment can be found at [https://zenodo.org/records/12801913] and [https://zenodo.org/records/12801941], respectively.