Abstract
Although the typical mitochondrial DNA (mtDNA) is portrayed as a circular molecule, a large number of organisms contain linear mitochondrial genomes classified by their telomere structure. The class of mitochondrial telomeres identified in three yeast species, Candida parapsilosis, Pichia philodendra and Candida salmanticensis, is characterized by inverted terminal repeats each consisting of several tandemly repeating units and a 5′ single-stranded extension. The molecular mechanisms of the origin, replication and maintenance of this type of mitochondrial telomere remain unknown. While studying the replication of linear mtDNA of C.parapsilosis by 2-D gel electrophoresis distinct DNA fragments composed solely of mitochondrial telomeric sequences were detected and their properties were suggestive of a circular conformation. Electron microscopic analysis of these DNAs revealed the presence of highly supertwisted circular molecules which could be relaxed by DNase I. The minicircles fell into distinct categories based on length, corresponding to n × 0.75 kb (n = 1–7). Similar results were obtained with two other yeast species (P.philodendra and C.salmanticensis) which possess analogous telomeric structure.
INTRODUCTION
Chromosomes may be circular or linear, the circular forms being more frequently associated with simpler organisms and linear chromosomes with higher eukaryotes. Cells have evolved complex terminal elements termed telomeres that protect them from mistaking chromosome ends for broken DNA, which can trigger unwanted repair and apoptotic responses. Almost all telomeres share two common features: multiple repeats varying from a few nucleotide units to large multi-kilobase retrotransposons, and a single-stranded protrusion at the terminus (for review see 1,2). Telomere specific proteins have also been identified. In cases such as the macronuclear DNA of Oxytricha the proteins cap the DNA ends (3–5) while in mammalian telomeres TRF1 and TRF2 proteins bind along the duplex repeat segments (6–8) and may catalyze the long range looping of the terminal single-stranded overhang back into the preceding duplex repeat segment to form a large loop (t-loop) (9). Such looping may provide an architectural solution for disguising the DNA end from the repair machinery. Understanding the way in which telomeres are maintained and have evolved is fundamental to studies of cancer and aging.
Telomeres are replicated during S phase by the cellular replication machinery that reproduces the genome. When the end of the telomere is encountered, the requirement for the lagging strand polymerase to initiate synthesis from an RNA primer could result in the loss of a short segment of telomeric sequences at each round of replication and, hence, progressive shortening over subsequent generations (10). These sequences are restored in most organisms by a terminal replicase termed telomerase (11, for review see 12). Telomerase thus maintains the integrity of telomeres in cells in which it is active. The relation of telomerase activity to cell imortalization and cancer has been suggested (for review see 13,14). However, the interrelationship between telomerase activity and oncogenesis is still under debate. Other mechanisms exist for telomere maintenance and may possibly reflect evolutionary independent and/or earlier mechanisms.
It has been demonstrated that telomerase-negative human tumors and tumor-derived immortal cell lines can acquire very long telomeric blocks through unknown telomere-lengthening mechanisms. The maintenance of telomeres by alternative, telomerase-independent mechanisms may be important in certain types of cancer and in therapeutic attempts, the emergence of such mechanisms may represent a potential source of resistance of tumor cells toward the telomerase inhibitors (15–20).
The study of Saccharomyces cerevisiae mutants with defects in telomerase revealed that these cells may reconstitute chromosomal ends by RAD52-dependent recombination mechanisms (21–24), and recombinational mechanisms were proposed for mouse cells lacking telomerase (25,26). Schizosaccharomyces pombe containing mutations in the gene coding for telomerase RNA maintains its telomeres either by a recombinational RAD52-dependent mechanism or by circularization of its chromosomes (27). Rare survivors of Kluyveromyces lactis with the double mutation Δter1 Δrad52 might suggest the existence of additional mechanisms (28). Also, in several organisms that include insect (Chironomus, Anopheles, Drosophila) and plant (Allium) species telomerase-independent strategies of telomere elongation naturally operate as a primary mechanism (for review see 15). Although in many cases an unequal recombinational mechanism may be operative, Drosophila melanogaster maintains the ends of its chromosomes by repeated transfer of telomere-associated retrotransposons (29,30). It is possible that these latter mechanisms may reflect some of the earliest molecular strategies used for the generation and maintenance of telomeres.
While it is often presumed that a long evolutionary distance exists between genomes arranged as circles and those that are linear, cases exist in which closely related organisms with nearly identical genomes can be found that are arranged either as a circle or as a linear molecule with canonical telomeres. Mitochondrial genomes of a variety of yeast species provide one such example. The mitochondrial genomes in many organisms are represented by linear DNAs of defined length (31). The close similarity in genetic organization and homology of the coding sequences between the linear and circular yeast mitochondrial genomes suggest their close relationship and argue against a possibility that linear genomes represent an independent evolutionary line (31).
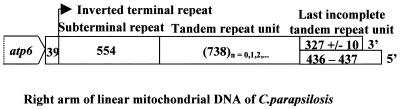
According to the phylogenetic analysis described by Barns et al. (32), three pathogenic yeast species Candida tropicalis, Candida albicans and Candida parapsilosis form a closely related cluster on the phylogenetic tree. The first two have circular mitochondrial DNA (mtDNA) while C.parapsilosis has a linear mtDNA (31) with a terminal structure (mitochondrial telomere) that consists of inverted terminal repeats themselves consisting of long (738 bp) units repeated in tandem. The telomeres end in an incomplete repeat unit and a 5′ single-stranded extension of 100–120 nt (Fig. 1) (33). The number of repetitive units varies, thus giving rise to a heterogeneous population of molecules differing in size (33). Since the shortest molecules lack a complete repeat unit they would be unable to restore the missing sequences without special mechanisms and might be preferentially lost over-repeated replication events. The exact replication mechanism is not known.
Figure 1.
Arrangement of the DNA elements at C.parapsilosis telomeres. The arrangement of the repeat units at both arms of the linear mitochondrial DNA of C.parapsilosis as described previously (33) is illustrated. Units are in base pairs.
In this article we present the identification and analysis of extragenomic circular molecules derived from mitochondrial telomere repeats from C.parapsilosis using electrophoretic methods and electron microscopic analysis. We show that the presence of these molecules is not limited to C.parapsilosis since we have isolated analogous structures from mitochondria of two other yeast species (Candida salmanticensis and Pichia philodendra) with the same type of mitochondrial telomere organization.
MATERIALS AND METHODS
Yeast strains and DNA
Candida parapsilosis SR23 (CBS 7157) is a laboratory strain from the collection of the Department of Biochemistry (Comenius University, Bratislava, Slovakia). The strains C.salmanticensis CBS 5121 and P.philodendra CBS 6075 are type strains of the species from Centraalbureau voor Schimmelcultures (CBS), Delft, and were kindly provided by H. Fukuhara (Institut Curie, Orsay, France).
Mitochondrial DNA fragments (738 bp EcoRI fragment representing full telomeric repeat and the 2.9 kb BglII internal fragment of mtDNA) were described previously (33). Oligonucleotides were synthesized by Genset, France. TEL51 represents the sequence of the last 51 nucleotides from the 5′ overhang of the extreme end of mtDNA of C.parapsilosis. TEL51C is complementary to TEL51.
DNA isolation and analysis
Candida parapsilosis was grown in YPD medium (1% yeast extract, 1% peptone, 2% glucose) on a rotary shaker at 30°C to mid-log phase. The cells were then harvested, resuspended in 0.1% sodium azide, 100 mM EDTA pH 8.0 and incubated for 10 min on ice. Subsequently, the cells were collected by centrifugation and total DNA was isolated as described (34). Restriction enzymes were from New England Biolabs and used according to the manufacturer’s instructions. [α-32P]dATP (3000 Ci/mmol) and [γ-32P]ATP (3000 Ci/mmol) were from Amersham Pharmacia Biotech (Piscataway, NJ). The DNA fragments used as probes in hybridization experiments were isolated from agarose gels by the GENECLEAN II Kit (Bio101, Inc., CA) according to the manufacturer’s instructions. Southern blotting, DNA labeling using random oligonucleotide primers (Gibco BRL), terminal labeling of oligonucleotides with [γ-32P]ATP, and DNA hybridization, were performed essentially as described (35).
Benzoylated-naphthoylated DEAE (BND)–cellulose chromatography
Samples of total DNA were fractionated on BND cellulose (Sigma) chromatography as described (36). Briefly, DNA samples (50–100 µg), in 300 mM NaCl were applied to a BND–cellulose column (0.5 ml) previously equilibrated with 10 mM Tris–Cl, 300 mM NaCl, 1 mM EDTA pH 8.0. Double-stranded DNA molecules were eluted with 3 ml of 10 mM Tris–Cl, 800 mM NaCl, 1 mM EDTA, pH 8.0 (salt wash) and subsequently molecules with single-stranded regions were eluted with 3 ml of 10 mM Tris–Cl, 1 M NaCl, 1.8% caffeine, 1 mM EDTA, pH 8.0 (caffeine wash). The DNA was precipitated with ethanol and taken up in 10 mM Tris–Cl, 1 mM EDTA, pH 8.0.
Neutral-neutral 2-D agarose gel electrophoresis
2-D gel electrophoresis was performed according to Brewer and Fangman (37). In the first dimension samples were separated in 0.4–0.5% agarose gels in 1× TBE (90 mM Tris–borate, 2 mM EDTA) at 0.5–0.6 V/cm for 13–17 h at room temperature. Subsequently, the gels were stained with ethidium bromide (0.3 µg/ml in 1× TBE). The entire lane was excised and embedded in a 1% agarose gel prepared in 1× TBE containing 0.3 µg/ml of ethidium bromide. Separation in the second dimension was at 5 V/cm for 3–6 h at 4°C with continual buffer circulation. After electrophoresis, the DNA was transferred onto nylon membranes and hybridized with the corresponding DNA probes as described in the text.
Preparation of mitochondria and purification of mitochondrial DNA on CsCl gradients
Crude mitochondria were prepared as previously described (33). The mitochondria were further purified by flotation (38), with the modifications described by Newman et al. (39).
Mitochondrial DNA was purified by isopycnic centrifugation in the presence of bisbenzimide as described (40) with minor modifications. Briefly, 100 µl of crude or purified mitochondria were lysed in 300 µl of the mitochondria lysis buffer (10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 25 mM EDTA, 1% sarcosyl). Proteinase K was added to a final concentration of 100 µg/ml and the lysate was incubated for 30 min at 37°C. DNA was then isolated by phenol:chloroform:isoamylalcohol (25:24:1) extraction and ethanol precipitation. The final mtDNA pellet was solubilized in 4.5 ml of TE, 4.5 g of CsCl and bisbenzimide (final concentration 40 µg/ml) were added, and the sample was centrifuged for 5 h in a T836B vertical rotor at 55 000 r.p.m. at 15°C. The mtDNA band was collected, extracted four times with isopropanol saturated with 5.9 M CsCl, diluted with 5 vol of TE and precipitated with 2 vol of 95% ethanol. The purified mtDNA was solubilized in TE. For the preparation of the probes employed in the Southern blot analysis, mtDNA was purified directly from total cell DNA preparations by centrifugation in a CsCl–bisbenzimide gradient as described (41).
Preparation of mitochondrial telomeric minicircles by alkaline lysis
Aliquots (30 µl) of purified mitochondria were resuspended in 250 µl of 50 mM Tris–HCl, pH 8.0, 10 mM EDTA, 100 µg/ml RNase A and lysed by addition of 250 µl of 200 mM NaOH, 1% SDS. After 4 min incubation at room temperature, 350 µl of ice-cold 3.0 M potassium acetate (pH 5.5) was added for 10 min on ice followed by 10 min centrifugation at 10 000 g at 4°C. The supernatants were extracted with an equal volume of phenol:chloroform:isoamylalcohol (25:24:1), and the DNA precipitated with isopropanol and resuspended in 10 µl of TE. To relax supertwisted minicircles, 4 µl of DNA was incubated for 60 min at 15°C in 10 µl of 50 mM Tris–HCl, pH 7.4, 10 mM MgSO4, 0.1 mM DTT containing 0.1 ng of pancreatic DNase I (Boehringer Mannheim). The reaction was stopped by addition of EDTA to a concentration of 50 mM.
Electron microscopy (EM)
Preparation of the DNAs for EM, including the droplet variation of the Kleinschmidt method (42), direct adsorption of the DNA to carbon foils, and length analysis, was carried out as described previously (9). A Philips CM12 was used and images were scanned from film using a Nikon LS4500 film scanner and the contrast adjusted using Adobe Photoshop software. A plasmid pGEMEX-1 (Gibco BRL) was used as an internal standard.
RESULTS
Neutral-neutral 2-D agarose gel electrophoresis of C.parapsilosis DNA reveals the presence of distinct classes of telomeric fragments
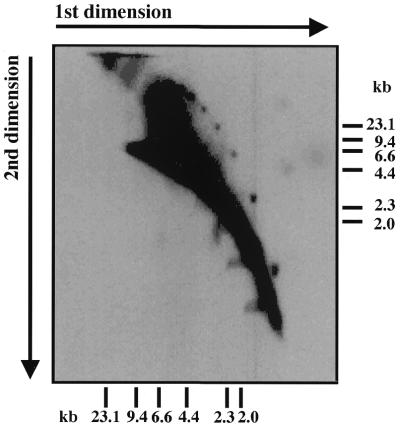
2-D gel electrophoresis was used to investigate the replication of mitochondrial telomeres of C.parapsilosis. For this purpose the preparation of total DNA was digested with BglII, which cleaves the terminal fragments to produce a telomere ladder pattern (33), followed by enrichment for molecules containing single-stranded regions using BND–cellulose chromatography. The samples were then separated by 2-D gel electrophoresis. Southern hybridization analysis with a 32P-labeled 738 bp EcoRI fragment representing the full telomere repeat unit revealed the presence of a discrete family of spots migrating apart from the main population (Fig. 2). Estimation of the size of the DNA in these spots indicated that they might represent multimers of the 0.75 kb telomere repeat unit.
Figure 2.
Neutral-neutral 2-D agarose gel electrophoresis of C.parapsilosis DNA reveals the presence of distinct classes of telomeric fragments. A BglII digest of total DNA of C.parapsilosis was separated by neutral-neutral 2-D electrophoresis (Materials and Methods) and hybridized with a radiolabeled probe containing the C.parapsilosis 738 bp telomeric repeat unit. The set of discrete spots of decreasing size are shown below to be circular multimers of the telomeric repeat.
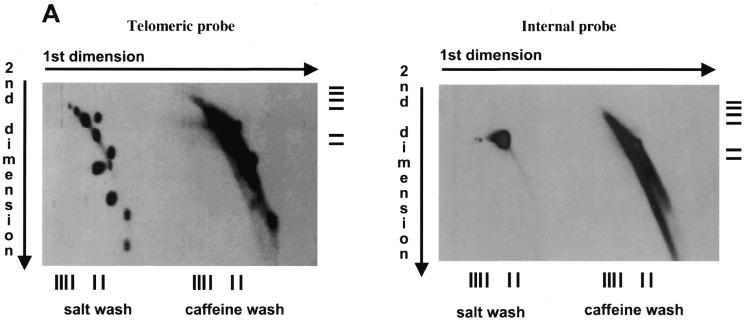
The eluates from the salt and caffeine washes from BND–cellulose chromatography were then analyzed by 2-D electrophoresis (Fig. 3A) revealing that the DNA forming the set of spots is present in the salt wash eluate. When the terminally labeled telomere-derived oligonucleotides TEL51 and its complementary counterpart TEL51C (43) were employed as probes, similar results were obtained (data not shown). Low affinity of material in the spots to BND–cellulose and the presence of both telomeric strands revealed by oligonucleotide probes argues that the spots represent different forms of double-stranded DNA molecules. Probing the Southern blot with a 2.9 kb BglII internal fragment of mtDNA showed no hybridization to the spots suggesting that they specifically contain terminal sequences.
Figure 3.
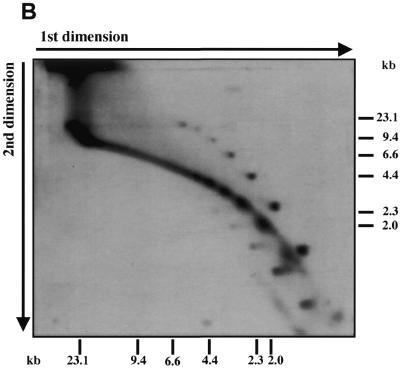
Telomeric fragments elute from BND–cellulose as duplex DNA and are present in the undigested preparations of C.parapsilosis DNA. (A) Total DNA of C.parapsilosis digested with BglII was loaded on a BND–cellulose column and eluted first with 0.8 M NaCl (salt wash) followed by 1.8% caffeine in 1 M NaCl (caffeine wash). The eluted DNA was separated by neutral-neutral 2-D electrophoresis as in Figure 1, and hybridized with a radiolabeled 738 bp telomeric tandem repeat unit (telomeric probe) or a 2.9 kb BglII fragment of mtDNA of C.parapsilosis internal to the telomere (internal probe). The bars represent DNA marker fragments of 23.1, 9.4, 6.6, 4.4, 2.3 and 2.0 kb, respectively. (B) Undigested total DNA of C.parapsilosis (not subjected to BND–cellulose chromatography) was separated by neutral-neutral 2-D electrophoresis and hybridized with a radioactively-labeled 738 bp telomeric tandem repeat unit probe.
Further analysis revealed that the DNA forming the spots is present in crude preparations of mtDNA but absent from CsCl purified samples (data not shown; 33). This suggested that it may represent extragenomic telomere fragments. To confirm this, a sample of total undigested DNA was separated by 2-D electrophoresis and hybridized with a telomeric probe as described above (Fig. 3B) revealing the separate spots again. Together, these results show that the DNAs forming the spots are physically dissociated from full-length linear mtDNA molecules and contain a sub-set of sequences, derived from telomeres. These fragments could be linear or circular.
Electron-microscopic analysis of mitochondrial telomeric minicircles of C.parapsilosis
To determine the conformation of the mitochondrial telomeric fragments observed by agarose gel electrophoresis, various preparations of mtDNA from C.parapsilosis were prepared for EM by surface spreading on denatured films of cytochrome C (Materials and Methods). Examination of the DNA revealed no indication of a family of discrete linear molecules other than full-length mtDNAs. Rather, very small circles (minicircles) were observed although they were rare (2–3 for every 100 mtDNAs). Their sizes, however, corresponded to the size of the DNAs in the spots detected by 2-D electrophoresis, suggesting that these unusual DNAs are small telomere-derived circles.
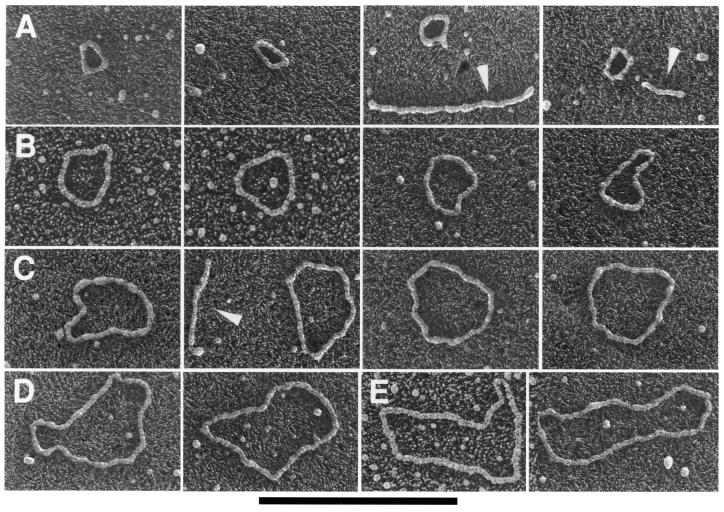
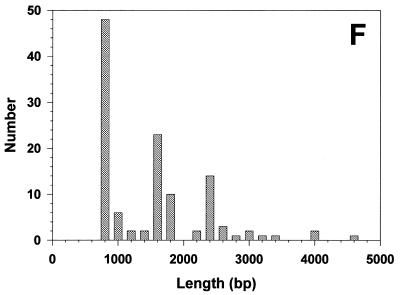
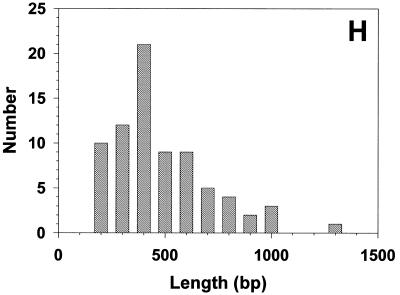
The circular nature of these DNAs suggested that they might be separated from the linear DNA by the alkaline lysis protocol used for the isolation of small bacterial plasmids (44). When this was carried out (Materials and Methods), inspection of the sample by EM revealed the presence of short, stiff linear DNAs often with small balls at the ends, and somewhat thicker than normal linear DNA (Fig. 4A and C). This appearance is diagnostic of highly supertwisted DNA. Because their highly twisted nature made measurement of the circular contour difficult, the DNA was treated with a low concentration of pancreatic DNase I in the presence of Mg2+ to nick the circles. This resulted in the appearance of a high proportion of open circular molecules (Fig. 4A–E) whose lengths (Fig. 4F) fell into several distinct classes corresponding to n × (750–800 bp) (n = 1–7). Southern blot analysis of the sample in which the 738 bp telomeric tandem repeat unit was used as a probe revealed the presence of several distinct classes of DNA molecules whose size and distribution correlates with those observed by EM (Fig. 4G).
Figure 4.
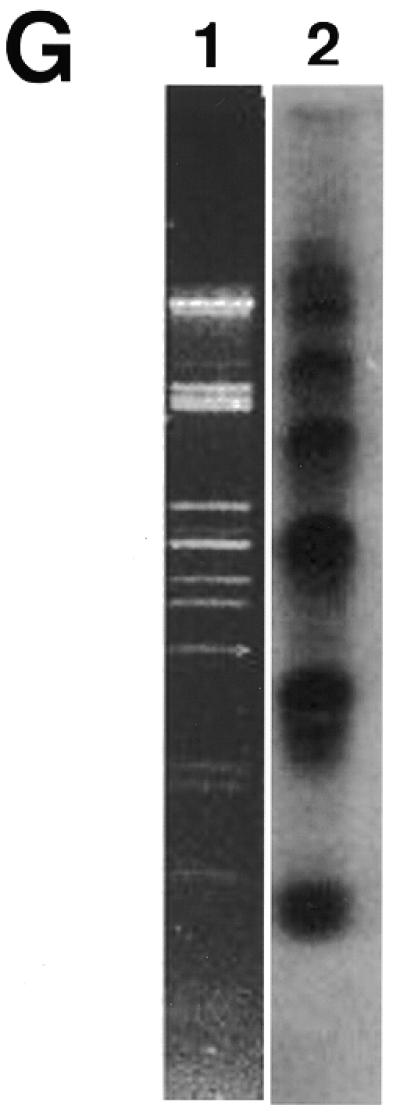
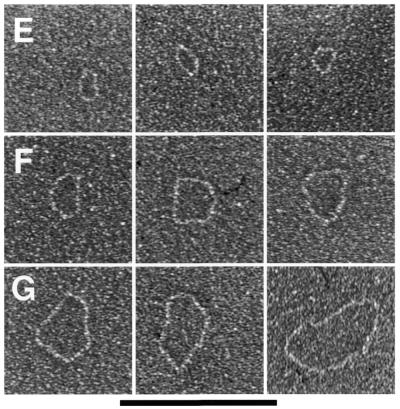
Mitochondrial telomeric minicircles of C.parapsilosis isolated by alkaline lysis of purified mitochondria. Mitochondrial telomeric minicircles of C.parapsilosis were isolated from purified mitochondria and relaxed with DNase I as described in Materials and Methods. Aliquots were prepared for EM by spreading on a denatured surface film of cytochrome C protein and rotary shadowcast with platinum-paladium (Materials and Methods). Circles with contour length classes of: 0.75 kb (A), 1.5 kb (B), 2.25 kb (C), 3.0 kb (D) and 3.75 kb (E), respectively, are shown. Shown in a reverse contrast. Bar represents 0.5 µm. (F) A histogram of circle lengths measured from micrographs as in A–E. (G) An aliquot of the purified minicircles was subjected to a Southern blot analysis using a 738 bp telomeric tandem repeat unit as a probe (C, lane 2). λDNA digested with PstI (C, lane 1).
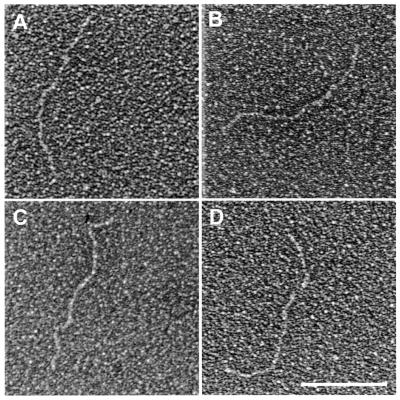
To obtain additional evidence that the minicircles observed by EM are composed of telomeric sequences, the minicircles isolated from mitochondria of C.parapsilosis were treated with EcoRI, which has a single recognition site within the telomeric tandem repeat unit (33). If the minicircles are composed of telomeric sequences, the restriction enzyme should produce a population of linear fragments whose lengths represent the size of the tandem repeat unit (738 bp in C.parapsilosis). Indeed, EM analysis of the EcoRI-treated sample revealed the presence of exclusively linear fragments whose lengths (796 ± 28 bp, n = 11) corresponded to the size of a single telomeric tandem repeat unit of C.parapsilosis. This was confirmed by gel analysis. Samples of C.parapsilosis minicircles were subjected to the Southern blot analysis, where the 738 bp telomeric tandem repeat unit was used as a DNA probe. When digested with EcoRI, the circles were linearized to fragments with an expected length (∼750 bp) (Fig. 5).
Figure 5.

Treatment of the minicircles with EcoRI produces linear fragments of ∼750 bp visaulized by EM (A–D) and Southern blot hybridization with the telomeric EcoRI fragment (E). Shown in reverse contrast. Scale bar equals 0.1 µm.
The mitochondrial telomeric minicircles are not restricted to mitochondria of C.parapsilosis
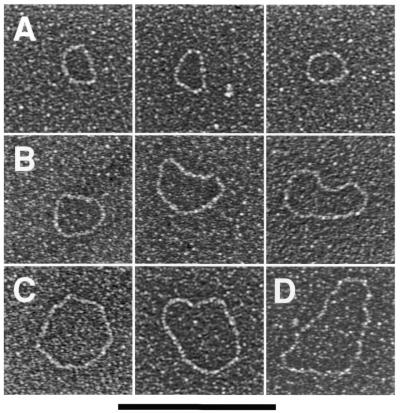
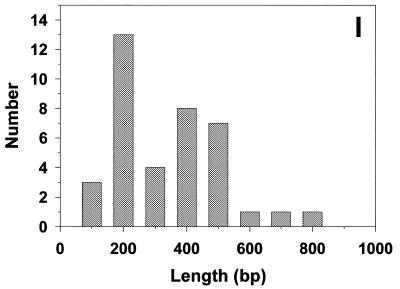
To demonstrate that these telomeric minicircles are not restricted to mitochondria of C.parapsilosis, but may be a general feature of yeast possessing this type of linear mitochondrial genomes, circular DNA molecules were isolated from mitochondria of P.philodendra and C.salmanticensis using the above approach. It had been previously shown that the telomeric tandem repeats consist of 286 bp (P.philodendra) and ∼110 bp (C.salmanticensis) in these two species (33). Alkaline lysis of purified mitochondria of P.philodendra yielded a population of circular DNA molecules whose size ranged between ∼200 and 1300 bp (Fig. 6A–D, H). The absence of distinct classes of minicircles is probably the result of the very small size of the repetitive unit thus leading to smearing of the distributions due to EM measurement errors. The telomeric tandem repeats in C.salmanticensis are almost three times smaller than those of P.philodendra and alkaline lysis of purified mitochondria of C.salmanticensis yielded a population of circular molecules where the size of the smallest circle observed was ∼100 bp (Fig. 6E–G, I).
Figure 6.
Mitochondrial telomeric minicircles of P.philodendra and C.salmanticensis isolated by alkaline lysis of purified mitochondria. Mitochondrial telomeric minicircles of P.philodendra (A–D) and C.salmanticensis (E–G) were isolated from purified mitochondria (Materials and Methods). Due to the very small size of some circles, the samples were directly adsorbed to thin carbon foils in the absence of denatured protein, and rotary shadowcast with tungsten (Materials and Methods). Shown in reverse contrast. Bar represents 0.1 µm. The P.philodendra examples have contour lengths 300 bp (A), 600 bp (B), 0.9 kb (C) and 1200 bp (D), respectively. The C.salmanticensis examples have contour lengths of 200 bp (E), 400 bp (F) and 600 bp (G), respectively. (H) Histogram of circle lengths measured from micrographs as in (A)–(D). (I) A histogram of circle lengths measured from micrographs as in (E)–(G).
DISCUSSION
The structural organization of the telomeres of C.parapsilosis mtDNA resembles that of eukaryotic nuclear telomeres (33), and thus similar mechanisms for their replication and maintenance may be utilized. The exact strategy employed by these linear genophores to maintain their telomeres is, however, not known. Previous studies of the replication of linear mtDNA of C.parapsilosis by 2-D gel electrophoresis by J.Nosek, L.Tomaska and A.Pastorakova (unpublished observations) had led to the observation of a family of discrete DNA spots on 2-D gels which were suspected to consist of telomeric DNA. In this study, further 2-D gel and EM analysis has been carried out to characterize these DNAs and show that they consist of a family of telomeric DNA minicircles which consist of integral multimers of 738 bp tandem telomeric repeat unit.
Although no causal relationships between the linear mtDNA and the formation of circular forms have been directly demonstrated, the characteristics of minicircles summarized below strongly suggest that such relationships may exist. First, they hybridize only with the probes derived from the telomeric tandem repeat unit. Secondly, their size as measured by agarose gel electrophoresis and EM corresponds to integral multiples of the unit repeat. Thirdly, digestion of the minicircles with a restriction enzyme containing a single recognition site in the unit repeat released only linear monomer DNAs. Fourthly, mitochondria of C.salmanticensis that possess the same organization of mitochondrial telomeres were shown here to contain minicircles whose sizes corresponded to integral numbers of their unit repeats. In addition, the minicircles were also observed in mitochondria of P.philodendra. The absence of distinct classes of minicircles is probably the result of the very small size of the repetitive unit thus leading to smearing of the distributions due to EM measurement errors. Although it is possible that the circles in this case may arise from a more complicated process, analogous organization of mitochondrial telomeres of P.philodendra, C.parapsilosis and C.salmanticensis suggest that the existence of extragenomic telomeric minicircles in mitochondria is a general feature of the yeast species with type 2 linear mitochondrial genomes.
In view of the data presented by Nosek et al. (33) on the sequence organization of type 2 yeast linear mitochondrial genomes, the presence of small circular molecules in the mtDNA preparations from C.parapsilosis and subsequently in C.salmanticensis and P.philodendra was unexpected. In the original report, repeated searches for such circular molecules were negative. However, all the mtDNAs employed in those studies were purified by CsCl gradient isopycnic centrifugations. As shown here, a high proportion of the minicircles are covalently closed and thus most likely banded in a different position in the gradients from the linear mtDNA. Southern blot analysis of various mtDNA preparations demonstrated that following the CsCl gradient separation, the preparations lacked detectable minicircles. On the contrary, mtDNAs that were not subjected to purification by isopycnic centrifugation contained a substantial amount of minicircles (data not shown).
The presence of minicircles in mitochondria has been reported before. In trypanosomal protozoa, networks of minicircles are localized in kinetoplasts (45). In plants, the main circular mtDNA contains repetitive sequences that are recombined out to generate a population of several types of small subgenomic circular molecules (46,47). A special class of mitochondrial minicircles is represented by senDNA generated during the senescence of several filamentous fungal species (48). Additional examples of minicircular mtDNA are found in the mesozoan animal Dicyema (49). The gallery of minicircular DNA molecules found in organelles also includes several circular mitochondrial plasmids identified in plants (50) and fungi (51). Another example of organellar minicircles is represented by single-gene containing circular DNA molecules found in chloroplasts of dinoflagellates (52). Finally, it was shown recently in ρ– cells of S.cerevisiae, that activation of the general amino acid response pathway results in increased intramolecular recombination between tandemly repeated sequences of ρ– mtDNA to produce small, circular oligomers that are packaged into individual nucleoids (53).
The way in which mitochondrial telomeric circles are generated is unknown. Unequal inter- or intra-strand exchanges between paralogous sequence arrays is the most widely accepted mechanism involved in the dynamics and maintenance of tandem repeats (54). The telomeric regions of linear mtDNA in several Tetrahymena species comprise direct tandem arrays of 31–53 bp repeats (55–58). Recombination-dependent mechanisms were proposed to play a role in the replication of mitochondrial telomeres of Tetrahymena (57). As the organization of the terminal parts of mtDNA of Tetrahymena resembles those identified in the yeasts investigated in this study, it may be that the mitochondrial telomeric minicircles are generated by recombination between telomeric tandem repeats.
The mechanism by which minicircles arise may be similar to that involved in the generation of extrachromosomal ribosomal DNA (rDNA) circles (ERC, 3 µm DNA) from the cluster of 100–200 tandemly repeated rDNA units on chromosome XII of S.cerevisiae (59). rDNA replication is halted in one direction at a replication fork block (60) by a product of the FOB1 gene, thus causing a DNA double-strand break within the rDNA (61). This break can be repaired by homologous recombination, which results in the formation of extrachromosomal rDNA circles (ERC, 3 µm DNA) (62–64). Deletion of RAD52, a gene required for homologous recombination, results in the absence of ERCs, thereby implicating a recombinational repair process in their formation (65). Intrachromatid recombination between telomeric repeats was suggested to be involved in the process of telomeric rapid deletion (TRD) in S.cerevisiae (66). Louis and Haber (67) demonstrated the existence of circles of S.cerevisiae subtelomeric tandem Y′ elements presumably circularized via the telomeric repeats at their ends. In addition, gene conversion mediated by the RAD52 pathway allows telomere lengthening in rare survivors of the telomerase-deficient strains (22–24). It is likely that similar mechanisms are involved in the telomerase-independent regulation of telomere size in yeast.
It is not known if the mitochondrial telomeric minicircles represent independent mitochondrial replicons. The relatively low proportion of minicircles in the mitochondria excluded our efforts to visualize replication intermediates by EM. Although the question of autonomous replication of minicircles needs to be addressed experimentally, a role for minicircles in the maintenance of mitochondrial telomeres may be envisaged. Homologous recombinantion of telomere arrays of mtDNA molecules may be involved in the processes of mitochondrial telomere dynamics. For example, non-reciprocal recombination between telomere repeat units may lead to the excision of telomeric minicircles and thus to the shortening of mtDNA molecules. More importantly, the incision of extragenomic telomeric circles containing a complete repeat unit (or an array of repeat units) into the existing mitochondrial telomere would extend the telomere length and thus prevent the subsequent loss of the molecule from the organellar pool of mtDNAs. Thus, an active role for telomeric minicircles in these yeasts may parallel alternative (telomerase independent) mechanisms of telomere maintenance. If so, further studies on linear mitochondrial genomes may shed light on how telomeres in general have evolved and are maintained.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank the members of the Griffith laboratory for discussions and L. Kovac (Department of Biochemistry, Comenius University, Bratislava, Slovakia) and H. Fukuhara (Institut Curie, Orsay, France) for helpful comments and continuous support. This work was supported by grants to J.D.G. from the National Institutes of Health (GM-31819, CA-70343) and to L.T. and J.N. from the Slovak Grant Agency (Grants 1/6168/99, 1/7248/20) and International Research Grant Program of Universidad de Complutense, Madrid, Spain. L.T. and J.N. were partially supported by the Howard Hughes Medical Institute (Grant 75195-547301).
REFERENCES
- 1.Blackburn E.H. and Greider,C.W. (1995) Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Greider C.W. (1996) Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 3.Gottschling D.E. and Zakian,V.A. (1986) Cell, 47, 195–205. [DOI] [PubMed] [Google Scholar]
- 4.Gray J.T., Celander,D.W., Price,C.M. and Cech,T.R. (1991) Cell, 67, 807–814. [DOI] [PubMed] [Google Scholar]
- 5.Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- 6.Chong L., van Steensel,B., Broccoli,D., Erdjument-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) Science, 270, 1663–1667. [DOI] [PubMed] [Google Scholar]
- 7.Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- 8.Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- 9.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 10.Watson J.D. (1972) Nature New Biology, 239, 197–201. [DOI] [PubMed] [Google Scholar]
- 11.Greider C.W. and Blackburn,E.H. (1987) Cell, 51, 887–898. [DOI] [PubMed] [Google Scholar]
- 12.Bryan T.M. and Cech,T.R. (1999) Curr. Opin. Cell Biol., 11, 318–324. [DOI] [PubMed] [Google Scholar]
- 13.Greider C.W. (1999) Trends Genet., 15, 109–112. [DOI] [PubMed] [Google Scholar]
- 14.Holt S.E. and Shay,J.W. (1999) J. Cell Physiol., 180, 10–18. [DOI] [PubMed] [Google Scholar]
- 15.Biessmann H. and Mason,J.M. (1997) Chromosoma, 106, 63–69. [DOI] [PubMed] [Google Scholar]
- 16.Bryan T.M., Englezou,A., Gupta,J., Bacchetti,S. and Reddel,R.R. (1995) EMBO J., 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raymond E., Sun,D., Chen,S.F., Windle,B. and Von Hoff,D.D. (1996) Curr. Opin. Biotechnol., 7, 583–591. [DOI] [PubMed] [Google Scholar]
- 18.Bryan T.M., Englezou,A., Dalla-Pozza,L., Dunham,M.A. and Reddel,R.R. (1997) Nature Med., 3, 1271–1274. [DOI] [PubMed] [Google Scholar]
- 19.Colgin L.M. and Reddel,R.R. (1999) Curr. Opin. Genet. Dev., 9, 97–103. [DOI] [PubMed] [Google Scholar]
- 20.Reddel R.R., Bryan,T.M. and Murnane,J.P. (1997) Biochemistry (Moscow), 62, 1254–1262. [PubMed] [Google Scholar]
- 21.Wang S.-S. and Zakian,V.A. (1990) Nature, 345, 456–458. [DOI] [PubMed] [Google Scholar]
- 22.Lundblad V. and Blackburn,E.H. (1993) Cell, 73, 347–360. [DOI] [PubMed] [Google Scholar]
- 23.Le S., Moore,J.K., Haber,J.E. and Greider,C.W. (1999) Genetics, 152, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teng S.C. and Zakian,V.A. (1999) Mol. Cell. Biol., 19, 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blasco M.A., Lee,H.-W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 26.Lansdorp P.M. (1997) J. Cell. Biol., 139, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T.M., Cooper,J.P. and Cech,T.R. (1998) Science, 282, 493–496. [DOI] [PubMed] [Google Scholar]
- 28.McEachern M.J. and Blackburn,E.H. (1996) Genes Dev., 10, 1822–1834. [DOI] [PubMed] [Google Scholar]
- 29.Levis R.W., Ganesan,R., Houtchens,K., Tolar,L.A. and Sheen,F.M. (1993) Cell, 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 30.Pardue M.L., Danilevskaya,O.N., Lowenhaupt,K., Slot,F.K. and Traverse,L. (1996) Trends Genet., 12, 48–52. [DOI] [PubMed] [Google Scholar]
- 31.Nosek J., Tomaska,L., Fukuhara,H., Suyama,Y. and Kovac,L. (1998) Trends Genet., 14, 184–188. [DOI] [PubMed] [Google Scholar]
- 32.Barns S.M., Lane,D.L., Sogin,M.L., Bibeau,C. and Weisburg,W.G. (1991) J. Bacteriol., 173, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosek J., Dinouël,N., Kovac,L. and Fukuhara,H. (1995) Mol. Gen. Genet., 247, 61–72. [DOI] [PubMed] [Google Scholar]
- 34.Philippsen P., Stotz,A. and Scherf,C. (1991) Methods Enzymol., 194, 169–182. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Dijkwel P.A., Vaughn,J.P. and Hamlin,J.L. (1991) Mol. Cell. Biol., 11, 3850–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brewer B.J. and Fangman,W.L. (1987) Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- 38.Lambowitz A.M. (1979) Methods Enzymol., 59, 421–433. [DOI] [PubMed] [Google Scholar]
- 39.Newman S.M., Zelenaya-Troitskaya,O., Perlman,P.S. and Butow,R.A. (1996) Nucleic Acids Res., 24, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragnini A. and Fukuhara,H. (1989) Nucleic Acids Res., 17, 6927–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gargouri A. (1989) Curr. Genet., 15, 235–237. [Google Scholar]
- 42.Kleinschmidt A.K. and Zahn,R.K. (1959) Z. Naturforsch. B, 14, 770–779. [Google Scholar]
- 43.Tomaska L., Nosek,J. and Fukuhara,H. (1997) J. Biol. Chem., 272, 3049–3056. [DOI] [PubMed] [Google Scholar]
- 44.Birnboim H.C. and Doly,J. (1979) Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapiro T.A. and Englund,P.T. (1995) Annu. Rev. Microbiol., 49, 117–143. [DOI] [PubMed] [Google Scholar]
- 46.Palmer J.D. and Shields,C.R. (1984) Nature, 307, 437–440. [Google Scholar]
- 47.Fauron C., Casper,M., Gao,Y. and Moore,B. (1995) Trends Genet., 11, 228–235. [DOI] [PubMed] [Google Scholar]
- 48.Griffiths A.J.F. (1992) Annu. Rev. Genet., 26, 351–372. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe K.I., Bessho,Y., Kawasaki,M. and Hori,H. (1999) J. Mol. Biol., 286, 645–650. [DOI] [PubMed] [Google Scholar]
- 50.Backert S., Kunnimalaiyaan,M., Börner,T. and Nielsen,B.L. (1998) J. Mol. Biol., 284, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 51.Griffiths A.J. (1995) Microbiol. Rev., 59, 673–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang A., Green,B.R. and Cavalier-Smith,T. (1999) Nature, 400, 155–159. [DOI] [PubMed] [Google Scholar]
- 53.MacAlpine D.M., Perlman,P.S. and Butow,R.A. (2000) EMBO J., 19, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McAllister B.F. and Werren,J.H. (1999) J. Mol. Evol., 48, 469–481. [DOI] [PubMed] [Google Scholar]
- 55.Goldbach R.W., Arnberg,A.C., van Bruggen,E.F.J., Defize,J. and Borst,P. (1977) Biochim. Biophys. Acta, 477, 37–50. [DOI] [PubMed] [Google Scholar]
- 56.Middleton P.G. and Jones,I.G. (1987) Nucleic Acids Res., 15, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morin G.B. and Cech,T.R. (1988) Cell, 52, 367–374. [DOI] [PubMed] [Google Scholar]
- 58.Morin G.B. and Cech,T.R. (1988) Nucleic Acids Res., 16, 327–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Philippsen P., Thomas,M., Kramer,R.A. and Davis,R.W. (1978) J. Mol. Biol., 123, 387–404. [DOI] [PubMed] [Google Scholar]
- 60.Brewer B.J. and Fangman,W.L. (1988) Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi T. and Horiuchi,T. (1996) Genes Cells, 1, 465–474. [DOI] [PubMed] [Google Scholar]
- 62.Gangloff S., Zou,H. and Rothstein,R. (1996) EMBO J., 15, 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 63.Defossez P.A., Prusty,R., Kaeberlein,M., Lin,S.J., Ferrigno,P., Silver,P.A., Keil,R.L. and Guarente,L. (1999) Mol. Cell, 3, 447–455. [DOI] [PubMed] [Google Scholar]
- 64.Johnson F.B., Sinclair,D.A. and Guarente,L. (1999) Cell, 96, 291–302. [DOI] [PubMed] [Google Scholar]
- 65.Park P.U., Defossez,P.A. and Guarente,L. (1999) Mol. Cell. Biol., 19, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li B. and Lustig,A.J. (1996) Genes Dev., 10, 1310–1326. [DOI] [PubMed] [Google Scholar]
- 67.Louis E.J. and Haber,J.E. (1990) Genetics, 134, 547–559. [DOI] [PMC free article] [PubMed] [Google Scholar]