Abstract
The Microarray Explorer (MAExplorer) is a versatile Java-based data mining bioinformatic tool for analyzing quantitative cDNA expression profiles across multiple microarray platforms and DNA labeling systems. It may be run as either a stand-alone application or as a Web browser applet over the Internet. With this program it is possible to (i) analyze the expression of individual genes, (ii) analyze the expression of gene families and clusters, (iii) compare expression patterns and (iv) directly access other genomic databases for clones of interest. Data may be downloaded as required from a Web server or in the case of the stand-alone version, reside on the user’s computer. Analyses are performed in real-time and may be viewed and directly manipulated in images, reports, scatter plots, histograms, expression profile plots and cluster analyses plots. A key feature is the clone data filter for constraining a working set of clones to those passing a variety of user-specified logical and statistical tests. Reports may be generated with hypertext Web access to UniGene, GenBank and other Internet databases for sets of clones found to be of interest. Users may save their explorations on the Web server or local computer and later recall or share them with other scientists in this groupware Web environment. The emphasis on direct manipulation of clones and sets of clones in graphics and tables provides a high level of interaction with the data, making it easier for investigators to test ideas when looking for patterns. We have used the MAExplorer to profile gene expression patterns of 1500 duplicated genes isolated from mouse mammary tissue. We have identified genes that are preferentially expressed during pregnancy and during lactation. One gene we identified, carbonic anhydrase III, is highly expressed in mammary tissue from virgin and pregnant mice and in gene knock-out mice with underdeveloped mammary epithelium. Other genes, which include those encoding milk proteins, are preferentially expressed during lactation. MAExplorer may be accessed at http://www.lecb.ncifcrf.gov.MAExplorer.
INTRODUCTION
Traditionally, researchers have studied the regulation of biochemical and genetic pathways, as well as developmental programs, on a gene-by-gene basis. The development of large-scale gene expression profiling holds promise for the discovery of new genes and biochemical pathways involved in the development and regulation of mammalian cells. We created mammary enriched cDNA microarrays and analyzed them with a new program we developed, the Microarray Explorer (MAExplorer).
The MAExplorer data mining tool helps support efforts of the research community in analyzing the expression patterns of individual genes, gene families and gene clusters from microarray data. Data mining is the uncovering of relevant patterns of interest in data from a particular problem domain (1–3). Typically this involves using various database, data mining, statistical and direct-manipulation user interface techniques to identify similar expression patterns (4).
There is a range of approaches for how data mining of microarray data is performed over the Internet. However, all of these approaches assume rapid access to underlying databases and the ability to transform data from one presentation mode to another so that differences might be easily observed. One extreme is the server-centric model, which assumes that all data search and analysis is performed on a back-end server and graphic or tabular results from the server are sent back to the researcher over the Internet. This model has the advantage of keeping the data up to date, but the disadvantage of performing all computations and graphics generation on the back-end server. The other extreme is the client-centric model. Here all of the data being analyzed is copied to a user’s computer and computationally expensive analyses are done there. This allows more effective data mining with many alternate views and avoiding excessive Internet delays. A good intersection of the server-centric and client-centric methods is to distribute the computation and data to the systems where they can be handled most effectively. It is now possible to distribute some of the data and computations to the desktop because of the following three advances in computer technology. (i) Java enables computation in a Web browser or stand-alone program. (ii) Personal computers have enormous power and memory approaching that of the Cray supercomputers of the previous decade. (iii) High-speed Internet connections are readily available. If high-speed direct manipulation methodology is to be made available on the Internet for microarray data mining, then it must be brought to the user’s desktop browser rather than residing solely on the back-end server.
Using Java, MAExplorer brings the required data to the user’s computer, allowing real-time, interactive manipulation of the data. This interactive data mining emphasizes direct graphical and tabular manipulation of the data using human visualization as part of the analysis. The computational tools in MAExplorer were specifically designed for microarray data mining (in conjunction with other biological Internet databases) with several goals in mind. The first goal was to provide the tools to analyze gene expression patterns during different physiological conditions that may be easily changed during the analysis. The second goal was to allow the identification of subsets of genes with similar expression patterns based on logical and statistical tests selected by the investigator. The third goal was to be able to analyze cDNA arrays from different sources, allowing MAExplorer to be applied to data from a variety of laboratories. The fourth goal was to be able to perform the same analysis on different computational platforms using the same software. Cross-platform analysis requires either the use of a Web-server based system or the use of Java. Java was chosen because it allows greater portability as well as having decreased bandwidth requirements. Finally, the fifth goal was expedite data sharing between investigators by automatically downloading data from Web servers, but then having the ability to analyze and re-analyze the data locally.
Many of the data mining concepts used in MAExplorer are derived from our earlier exploratory analysis systems with 2-D electrophoretic protein gels, GELLAB-II (5–7), Flicker (8–10) and WebGel (11). These systems work with sets of corresponding protein spots across sets of 2-D polyacrylamide electrophoretic gels, while MAExplorer works with microarray cDNA spots across sets of hybridization probes. It would be useful to illustrate here many of the displays and options available through MAExplorer, but because space is limited, please refer to the MAExplorer on-line reference manual (http://www.lecb.ncifcrf.gov/MAExplorer/hmaeHelp.html) which describes in detail the requirements, philosophy and approaches and methods for data mining of cDNA microarrays.
Direct manipulation (12,13) on a variety of graphic and tabular displays makes it easier for the user to discover and save sets of interesting clones. Such interaction by selecting data lets users identify outliers or members of particular sets of clones. They can quickly understand what the abstract graphics data represent in terms of the particular cluster clones or genes involved. The expression profile of a clone is the plot of its normalized intensity as a function of sample number. Scrollable lists of expression profile plots may be generated allowing multiple clones to be viewed simultaneously. There are several plotting, histogram, clustering methods and clustering visualization tools available. Clones in the database may be reduced to a more interesting subset using the data filters. The intersection of clone sets passing each active test is then used as the final set of clones. Sets may be saved as well as used in set-theoretic computations. Individual tests include coefficient of variation, ratio and intensity histograms and ranges, t-test and various clustering methods, clone subsets, etc. and are described in detail in the reference manual.
MATERIALS AND METHODS
Overview of Microarray Explorer
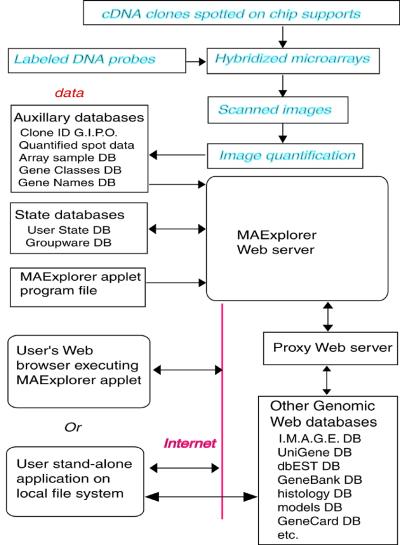
An overview is presented in Figure 1 showing where MAExplorer fits into the data analysis process after arrays have been hybridized and the images quantified. If it is used as a stand-alone application, then data can reside either on the Web database server or locally on the user’s computer. When used as a Web-browser applet, the data must reside on the Web server, but additional capabilities are made available with support from the back-end Web database server. This server design includes several distinct functions. These include hosting of log-in-protected microarray quantitative and auxiliary data required for support basic MAExplorer operations. The public database does not require a log-in while the collaborator subset of the database does. Another support function is the use of a Web proxy server to access other genomic Web sites for the Java applet.
Figure 1.
Overview of MAExplorer exploratory data analysis system. Initial data preparation steps are performed prior to analysis by MAExplorer and are indicated by cyan italics at the top of the figure (see Materials and Methods).
The primary data (Fig. 1) consists of quantified microarray image spot data as well as corresponding qualitative clone ID, gene-in-plate-order (GIPO), gene name, hypertext base-references and related information. After the microarrays are hybridized, they are scanned and spots quantified using image spot quantification programs saved for each sample in a tab-delimited file. Typical microarray image quantification software includes Research Genetics’ Pathways™, Molecular Dynamics ImageQuant™, etc. These auxiliary databases and the MAExplorer Java applet are copied to the Web server or local file system (in the case of the stand-alone version) where they are then available to be downloaded by users. When a user invokes a Web page containing the Java applet, it first downloads the applet that then downloads auxiliary databases including a configuration file that describes the array data. It then downloads the subset of quantified microarray spot data files requested for the set of hybridization probe samples being investigated. Additional probes may be downloaded at any time. When the user selects an operation that requires access to Web databases not residing on the MAExplorer Web server, implicit Java security restrictions prevent the applet from going directly to these other Web servers. Instead, it requests the MAExplorer proxy server request the data from the foreign Web server and then returns it back to the user’s Web browser. When used as a stand-alone application, data may be cached or saved on the local computer for local off-line use and direct access to other Internet genomic databases may be made without using a proxy server.
MAExplorer facilitates microarray data mining through four key features. First, it permits the organization of the experiments and associated data by defining features such as developmental stage, strain and tissue source. This allows the investigator to design multiple mining experiments to compare gene expression patterns between different probe hybridizations of the same microarray or to compare a single probe within one microarray. Second, MAExplorer allows the investigator to explore, compare, record analyses and share these analyses with other investigators. Third, it uses modern techniques of graphical direct manipulation (requiring the real-time response of Java) with statistical, clustering and spreadsheet techniques. Fourth, the MAExplorer connects to other public Internet genomic databases.
Several design decisions (detailed in the reference manual) help us achieve the above key features that allow such easy manipulation of the microarray data. They include the use of (i) bit-sets to efficiently implement sets of clones, critical for implementing efficient data filtering, (ii) tab-delimited ASCII data for all file I/O to simplify addition of new data tables, (iii) extended classes whenever possible simplify the code, making it easier to maintain, (iv) pop-up windows for 2-D plots, histograms, expression profiles, clustergrams, reports, dialog boxes, etc. rather than sharing a single window and (v) a pop-up window registry that notifies all windows requiring updates when any changes were made to the state of the analysis.
Microarray membrane design
In order to identify differentially expressed genes in mammary tissue we have generated mammary enriched cDNA microarrays and analyzed them with the MAExplorer. As part of the Mammary Genome Anatomy Program (MGAP, http://mammary.nih.gov/mgap) ∼50 000 ESTs from mammary tissue of a virgin mouse (library NbMMG) and >10 000 mammary ESTs from a lactating mouse (library NMLMG) have been sequenced. We have identified 1500 gene sets from the NMLMG library (C57BL/6 lactating mammary gland).
Washington University sequenced these clones from the 3′-end and 1500 unique sequences were identified. These 1500 unique sequences were PCR amplified and at least 5 ng of each was spotted onto the 5 × 7 cm nylon microarray membrane by Research Genetics (http://www.resgen.com/). The membrane is laid out in two fields that are identically spotted. Each field contains eight grids (A–H) with eight columns and 24 rows. Control spots of house keeping genes are also spotted throughout the membrane for normalization and aligning the membrane in the computer analysis program, Pathways 2.01 from Research Genetics.
RNA extraction
Tissue samples were harvested from C57BL/6, Stat5a knock-out, C/EBP-β knock-out, and β-b-inhibin knock-out mice at the stages indicated. Pooled samples (at least four mice) were homogenized and RNA was extracted by the acid guanidinium thiocyanate method (14).
Microarray probe preparation and hybridization
Total RNA (5 µg) was radio-labeled with [α-33P]dCTP in a reverse transcription reaction as described in the protocol from Research Genetics. Incorporation of the radioactivity was determined by scintillation counting. Pre-hybridization was performed in 10 ml of QuikHyb with 1 µg/µl of Human Cot1 DNA and oligo (dA) for blocking. Hybridization was allowed to proceed for 16 h and the membranes were then washed according to the Research Genetics protocol.
Imaging and analysis
Microarrays were placed on wetted rectangles of Whatmann filter paper and sealed in saran wrap. These were exposed on PhosphorImager screens (Packard Bell) for 15 min to 24 h (several exposures of each membrane to achieve the highest quality exposure). The Phosphor screens were then imaged by the Cyclone Storage Phosphor System (Packard Instruments Company) and stored as data files. These data files were imported into the Pathways 2.01 software analysis package for microarrays (Research Genetics) and intensities were recorded for each spot. The intensity data was then imported into the MAExplorer for analysis.
Northern analysis and probe preparation
Total RNA from C57BL/6 mice from several stages of mammary gland development was prepared as above. Total RNA (20 µg) was separated through 1.26% formaldehyde gels and blotted onto Gene Screen nylon membranes. The membranes were cross-linked and pre-hybridized for 30 min in QuikHyb and then hybridized for at least 8 h at 65°C (with sheared salmon sperm DNA blocking). Clones were picked from the 96 well master plates provided by Research Genetics that were used in the microarray membrane spotting. Each clone was PCR amplified with M13 forward and reverse primers and gel purified using the Qiagen kit for gel extraction. Each clone was sequenced (described below) to confirm its identity. Probes were prepared with the Stratagene Kit Prime It II for random end-labeling 1 µg of a clone with [α-32P]dCTP. Probes were purified through G-50 Sephadex columns and their incorporation of radioactivity was determined by scintillation counting. Washing was done at 65°C with 2 × SSC, 0.1% SDS twice for 20 min, 1 × SSC, 0.1% SDS once for 20 min and 0.1 × SSC, 0.1% SDS once for 20 min. The membranes were sealed in plastic bags and exposed on film.
Library screening
One particular clone of interest (1382656) was found from the northern analysis and was screened out of a differential cDNA library prepared from Stat5a null mice (as compared to C57BL/6 mice). The screening process was performed as described in earlier work (15). Sequencing of the clone was performed with the vector primers and then by designing specific primers to walk along. The sequences were analyzed and aligned in a contig with the Sequencher 3.0 software. BLAST searches compared the sequence of the clone with the GenBank database.
Sequencing
Clones were isolated as described above in the probe preparation. The clones were amplified with M13 reverse primer by the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit, the reaction was cleaned through Princeton Separation’s Cetrisep columns and separated on the ABI Prism 310 sequencer. Sequences were then checked against the sequences listed for a particular clone in the GenBank EST database.
RESULTS
We created the MAExplorer to facilitate the data mining of cDNA microarrays. The MAExplorer enables the investigator to mine data from microarrays using different computing platforms, with hybridizations from different support platforms and cDNA-labeling. We use the following notation: the sample probe cDNA is labeled (from total RNA) and then hybridized against the known cDNA targets tethered to the microarray. An alternative notation that reverses these terms is also commonly used (16). In our study of the mammary gland arrays, MAExplorer was used to analyze data from microarray hybridizations on nylon membranes with 33P-labeled probes. The MAExplorer was used to address two issues. (i) Is it possible to verify the expression pattern of hormonally regulated genes during pregnancy and lactation? (ii) Is it possible to determine the expression pattern of differentiation-specific genes in normal mammary tissue and in tissue from gene knock-out mice?
Identification of genes expressed during pregnancy and lactation
Identification of genes expressed during pregnancy and lactation was achieved through the isolation of known genes and ESTs prepared from mouse mammary tissue, the establishment of cDNA arrays with 1500 UniGene sets enriched for mammary sequences and the hybridization with probes from different developmental stages of pregnancy and lactation. We used the MAExplorer to identify genes that are preferentially expressed in mammary tissue during pregnancy and lactation.
A typical scenario for analysis (which could be used for any analysis) incorporates many methods for viewing the data. The analysis steps are listed below in the order they might be performed. Note that iteration of some of these steps may be required for data mining complex sets of conditions, especially in the setting of constraints for the ‘data filter’ (step iv) when the user focuses on subtle patterns of interest.
Select a set of hybridization probes to be analyzed.
Select query type: 2-probe (X versus Y) or N-probe expression.
Select normalization method.
Restrict search using the ‘data filter’ to subset clones by gene class, clone subset, ratio range, intensity range, coefficient of variation, t-test, expression profile, cluster membership, etc.
Review scatter and expression plots, ratio and intensity histograms.
Cluster clones by similar expression profiles and review clustergram and dendrogram plots.
Select subset(s) of clones of interest in plots or reports.
Access other Web genome databases from spreadsheets or plots.
Save clones of interest in named clone sets and perform set operations if required.
Create reports for export to Excel.
Save state of exploratory data analysis for later use or sharing.
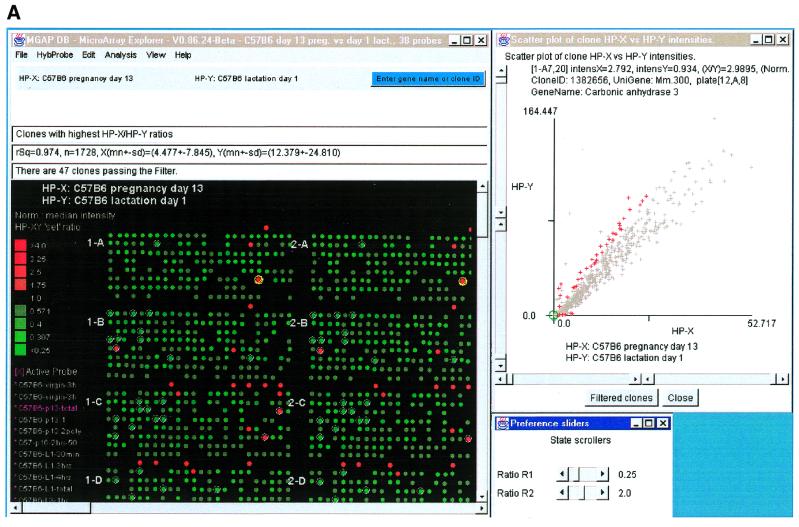
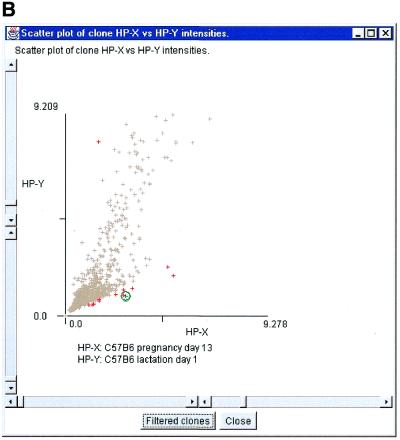
Figure 2A shows a computer screen with the primary MAExplorer window and various pop-up windows. This exploration compares probes HP-X, a set of three hybridizations of day 13 of pregnancy, and HP-Y, a set of five hybridizations from day 1 of lactation. The data was normalized by the median for each sample, and then filtered for mean ratios of pregnancy/lactation outside the range of 0.25–2.0 (set by adjusting the slider controls) and for milk protein genes (step iv). The scatter plot of this comparison is shown in Figure 2A. The scatter plot method (step v) allows the user to plot the intensity data between two probes, HP-X and HP-Y (pregnancy and lactation in this case). Scatter plots are useful for obtaining a better understanding of the outliers when comparing different hybridizations. Figure 2B shows a zoomed-in region of Figure 2A using the scatter plot scroll bars. The outliers on the scatter plots represent those genes that are expressed at higher levels during lactation. Most of the outliers in this comparison are milk protein genes, showing that milk protein genes are expressed at higher levels during lactation than during pregnancy. This was confirmed by northern blot analysis (Fig. 3).
Figure 2.
Screen view of part of a MAExplorer data mining session. (A) Screen showing the main MAExplorer window, scatter-plot and ratio-range threshold sliders for selecting clones of interest. The data for each probe was normalized by the median of spot intensity (for each hybridized probe) as described in the reference manual. The numeric values and plot scales reflect this normalization method. The MAExplorer user interface is similar to a Windows PC application with a set of pull-down menus which invoke operations. These menus are fully documented in the on-line reference manual. The currently selected hybridization probe array is displayed as a microarray pseudo image. In the examples used in this paper, we started MAExplorer on a database ‘C57BL/6 pregnancy day 13 versus lactation day 1’. The ordered list of 38 probes includes three C57BL/6 pregnancy day 13 (HP-X ‘set’), five C57BL/6 lactation day 1 samples (HP-Y ‘set’), as well as other stages of mammary development from normal and knock-out mice. The current gene class of clones had been set to the lactation genes of one of the authors (31). This example shows data filtered genes with white circle’s in the array image. Clicking on a spot assigns it as the current clone with data being reported in the top most message area. The names of the averaged HP-X and HP-Y samples are listed above that area and in the scatter plot. CA-III was selected as the current clone. In this database, all clones are in the left (field F1 of the array image) and are duplicated in the right (field F2). Therefore selected clones are indicated twice. In general, clicking on a spot in the array image, a point in scatter or clustergram plots, or a cell in a spreadsheet report assigns it as the clone, labels it, and accesses Web genomic databases. CA-III is indicated by a yellow (green) circle in the array image (scatter plot). Red (gray) ‘+’ indicates clones passing (failing) the data filter. (B) Zoomed region of scatter plot showing CA-III set as the current clone (green circle).
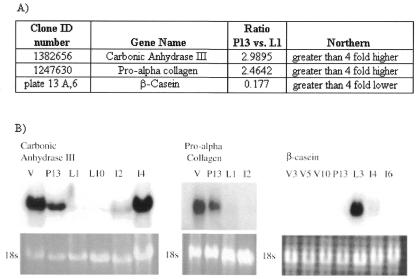
Figure 3.
Expression of mRNAs encoding CA-III, pro α-collagen and β-casein in mouse mammary tissue by microarray and northern blot analyses. (A) Table of averaged clone ratios calculated in MAExplorer for each clone (comparing C57BL/6 pregnancy day 13 mammary glands to C57BL/6 lactation day 1 mammary glands). (B) Northern blot confirmation of microarray data for each clone. Total RNA for the following stages was used in the northern: V, virgin; P13, pregnancy day 13; L1, lactation day 1; L10, lactation day 10; I2, involution day 2; I4, involution day 4; I6, involution day 6. 18S ribosomal bands are shown as loading controls.
Using the same hybridization probes as the above experiment, we restricted the search using ‘data filters’ for known genes and ESTs. The most highly expressed genes during pregnancy as compared to lactation were carbonic anhydrase-III (CA-III) and pro α-collagen type 2. Their ratios are shown in Figure 3 along with northern blot confirmation of this differential expression pattern. We also identified genes encoding α-casein and β-casein to be expressed preferentially during lactation. The ratio for β-casein is also shown in Figure 3 along with confirming northern analysis of β-casein RNA expression, showing increased expression during lactation, as expected from previous work.
Identification of genes differentially expressed in the absence of normal signaling
We have studied genetic pathways of mammary development in mice from which genes have been deleted (17). Little is known about which genetic pathways are altered in mammary tissue from these mice. We have used mammary tissue that was deficient in the transcription factor Stat5a, a key regulator of mammary gland development (18). Using the MAExplorer we have now identified several ESTs that were differentially expressed in Stat5a-null mice as well as several known genes. Specifically, CA-III was expressed at higher levels in Stat5a-null mice as compared to control mice and milk protein genes were expressed at lower levels (data not shown).
DISCUSSION
The MAExplorer is a Java-based interactive data mining application with an emphasis on direct graphical and tabular manipulation of the data. It helps users organize and analyze expression for identifying genes differentially expressed across various physiological conditions. We used MAExplorer to analyze gene expression patterns during different physiological conditions. We identified clusters of genes with similar expression patterns (milk protein genes are highly expressed during lactation and CA-III and pro α-collagen are highly expressed in virgin and pregnant mammary glands).
In addition, we were able to analyze cDNA arrays from different sources (discussed in the reference manual). MAExplorer may be used with a variety of array geometries and spot labeling methods and may be used for both membranes and glass slides with radio-labels such as 33P or fluorescent cDNA-labeling such as Cy3/Cy5. A configuration database or file defines the labeling method, number of duplicate spot fields, and geometry of the array being analyzed. Because it is Java-based, analyses may be performed on different computational platforms (discussed in the reference manual).
There is a divergence of where computation is performed in data mining Web-microarray databases. Some systems are more server-centric such as arrayDB (4) and the NCI/CIT mAdb MicroArray Database System (http://nciarray.nci.nih.gov/) where data is analyzed on the server and visualized in the browser. Other systems are more client-centric like P-SCAN (19); ScanAlyze, Cluster and TreeView programs (20); and GeneSpring (http://www.sigenetics.com/). These analysis systems run as stand-alone software on the user’s system where the primary array data resides. There is a movement between these two extremes towards both computation and data residing on both the server and on the client computers. MAExplorer falls in between these extremes, tending more toward the client-centric, but taking advantage of some server-centric features such as downloading the most recent data. There will always be a need to apply new methods for analyzing data. Ermolaeva et al. (4) suggests that a data mining facility should be flexible enough to accommodate new statistical data mining tools as they become available. One way is to add additional or use existing functionality on the back-end server, processing both to our own and other genomic servers on the Internet. Alternatively, we envision extending the MAExplorer toolbox using Java plug-ins. This method has the advantage of keeping the core MAExplorer computer memory footprint small while not inhibiting the use of new analytic methods. Plug-ins could be dynamically loaded and then communicate with the core data structures of MAExplorer to get and return data.
The MAExplorer is normally used either as a stand-alone program on a user’s computer or with a user’s Web browser over the Internet. When as a stand-alone program, the data files may be copied either from a microarray database server or from local quantified array files. The stand-alone version may be downloaded and installed on an investigator’s computer where it may be run as either a standard application or an applet (using a Web browser). It could also be installed on their own Web server to publish their data for other investigators to explore using MAExplorer. The application version lets users cache array information and sample probe study data from a Web database. This saves having to be connected in later sessions when analyzing that data. This also allows users to merge quantified array data files from their computer into a data mining session, thereby enabling a comparison of their data against data from other Web databases. In addition, users can save their user-state of an exploration on their own computer making it easier to restart a session at a later time. States may also be shared with collaborators who access the same set of samples to share results.
Analyzing a database involves dividing the samples from the database into subsets which will be compared (call them X and Y). Two developmental stages of mouse mammary gland development are presented in this paper, pregnancy versus lactation. If replicate samples are available (in this case three hybridizations of pregnancy day 13 and five hybridizations of lactation day 1), then the mean values for the X and Y sets of samples can be used. Another way of dividing the samples is by progression such as a time-study, drug-dose response, etc. We refer to this latter ordered list of samples as expression profiles (EP) of N-conditions. Because all of the data for a set of sample probes is always available in a session, users can easily switch between these interpretations.
Data from different array samples must be normalized when they are compared. Such normalization compensates for different sample concentration, hybridization times, etc. Data may be self-normalizing using double labeling methods such as Cy3/Cy5 ratio data (20) or scaled in various ways if single labels such as 33P are used. A number of methods are available including median and log median, or z-score and log z-score, etc. for computing expression ratios or z-score differences (21) and are described in detail in the reference manual.
A major goal of large-scale analysis is to find sets of clones that form natural groupings or clusters using either agglomerative or divisive methods. Commonly used clustering methods include hierarchical (20,22–25), k-means clustering (25) and self-organizing maps (SOM) (26). Hierarchical clustering divides genes into a strict divisive hierarchy of nested subsets. On the other hand, SOMs allow the imposition of partial structure by specifying an initial number of N clusters that converge on the optimal clusters whereas the k-means method may not converge as well. The faster k-means clustering method requires the specification of the number of clusters, N, but does not allow the specification of their initial positions in the k-dimensional space.
MAExplorer has a number of clustering methods including finding clones similar to a specific clone, ‘NPN-clustering’ similar to k-means and hierarchical clustering. The NPN-method lets the user set a ‘seed’ clone that will appear in the first cluster. All of these methods use normalized clone EP intensity data. Cluster distances are computed using either the Euclidean distance or correlation coefficient between EPs of pairs of clones. By directly varying distance thresholds or the number of clusters desired, performed with slider controls, users can see which genes migrate to different clusters as the parameters change. A list of cluster-means EP plots may be generated that displays the mean, standard deviation, coefficient of variation (CV) of the distances of all of the clones in the N clusters to their cluster centers. If there is a high CV, then that cluster should probably be split by increasing the number of clusters with the N clusters slider. These methods are described in the reference manual.
It is important for users to be able to condense their results into a report that they can take away for further data analysis. Clone and array reports may be presented either as dynamic spreadsheets with hyperlinks to external genomic databases or as tab-delimited reports that may be cut and pasted to export data directly into Excel or other applications. Dynamic reports give the user the option of branching off into one of the Internet Web genome databases to follow up ideas on roles of particular genes without leaving the context of the array explorations. These include genomic information from NCBI and NCI/CIT gene, clone or EST databases, and sample histology and model databases if they exist.
Identification of differentially expressed genes
Microarray tools have been used successfully in the identification of genetic pathways induced by physiological stimuli in simple biological systems such as yeast cultures (27,28), fibroblasts and lymphoid cells (29,30). We produced mammary-enriched cDNA microarrays and used them to profile gene expression throughout normal mammary gland development. Unlike other commercially available arrays that are focused largely on known genes and known pathways, the mammary cDNA microarray contains ∼50% ESTs without known function, and ∼200 clones specific to the mammary gland. Thus, this array should be of value in the identification of unknown genes that contribute to the physiology of mammary tissue throughout development. Mammary tissue consists of many different cell types, including secretory alveolar epithelium, ductal epithelium, myoepithelial cells, adipose cells, fibroblasts, endothelial cells, red blood cells and infiltrating lymphoid cells. Furthermore, the relative proportion of these cell types changes as the mammary gland progresses from a state of immaturity to puberty, to pregnancy, to lactation and to involution. These variables present challenges for studying gene expression with microarrays. Based on a comparison of microarray expression data from genes encoding β-casein, pro α-collagen and CA-III, with corresponding northern blot data, we have shown that microarrays are suitable to profile gene expression in mammary tissue.
In summary, MAExplorer is an easy to use tool for data mining cDNA microarray data using techniques of direct manipulation, data filtering, statistical tests and clustering. Results are presented in various graphic and tabular formats. Tables may be exported to Excel or be used to access Web genomic databases. Because it is written in Java, it runs on multiple platforms, in Web browsers, or alternatively may be easily installed as a stand-alone application. It is being made freely available to the microarray research community for use with their own arrays as well as for use with data from commercial arrays. Future enhancements will include new statistical and direct-manipulation techniques, plug-ins, data-caching, additional support for microarray Web database servers and simplified configuration for other arrays. We will be announcing these changes and new versions on our Web site.
Availability
The MAExplorer home page Web site http://www.lecb.ncifcrf.gov/MAExplorer provides full documentation, tutorials, Web browser demonstration applets and the downloadable stand-alone version. The latter stand-alone version can run on Windows 95/98/NT, Macintosh or Unix systems (Sun Solaris, Linux, etc). The Mammary Genome Anatomy Program has public microarray, histology and mouse-model databases accessible through http://mammary.nih.gov/.
Acknowledgments
ACKNOWLEDGEMENTS
We thank John Powell (CIT/NIH), Richard Simon (NCI/NIH), Greg Alvord (SAIC/FCRDC), Troy Moore (Research Genetics), the NIH Breast Cancer Think Tank (an internal organization responsible for key issues in breast cancer), Kevin Becker and Chris Cheadle (NIA), Mark Vawter (NIDA), Bob Stephens (ABCC/FCRDC), Peter Munson (CIT/NIH), Ulrike Wagner (LGP-NIDDK/NIH) and others for useful discussions and suggestions which have helped improve the MAExplorer’s capabilities and usability. Thanks also to Ellen Burchill and Tom Schneider for useful suggestions for improving this manuscript. The LECB (NCI) designed and implemented the MAExplorer in collaboration with the LGP (NIDDK).
REFERENCES
- 1.Tukey J. (1977) Exploratory Data Analysis. Addison-Wesley Pub. Co., Reading, MA, pp. 1–688.
- 2.Tufte E. (1997) Visual Explanations. Images and Quantities, Evidence and Narrative. Graphics Press, Cheshire, CT, pp. 1–156.
- 3.Cleveland W.S. (1985) The Elements of Graphing Data. Wadsworth Press, Monterey, CA, pp. 1–323.
- 4.Ermolaeva O., Rastogi,M., Pruitt,K.D., Schuler,G.D., Bittner,M.L., Chen,Y., Simon,R., Meltzer,P., Trent,J.M. and Boguski,M.S. (1998) Nature Genet., 20, 19–23. [DOI] [PubMed] [Google Scholar]
- 5.Lipkin L.E. and Lemkin,P.F. (1980) Clin. Chem., 26, 1403–1412. [PubMed] [Google Scholar]
- 6.Lemkin P.F. and Lester,E.P. (1989) Electrophoresis, 10, 122–140. [DOI] [PubMed] [Google Scholar]
- 7.Lemkin P.F. (1995) In Pickover,C. (ed.), The Visual Display of Biological Information, World Scientific Pub., River Edge, NJ, pp. 43–59.
- 8.Lemkin P.F. (1997) Electrophoresis, 18, 461–470. [DOI] [PubMed] [Google Scholar]
- 9.Lemkin P.F. (1997) Electrophoresis, 18, 2759–2773. [DOI] [PubMed] [Google Scholar]
- 10.Lemkin P.F. (1999) Mol. Biotechnol., 12, 159–172. [DOI] [PubMed] [Google Scholar]
- 11.Lemkin P.F., Myrick,J.M., Lakshmanan,Y., Shue,M.J., Patrick,J.L., Hornbeck,P.V., Thornwall,G.C. and Partin,A.W. (1999) Electrophoresis, 20, 3492–3507. [DOI] [PubMed] [Google Scholar]
- 12.Schneiderman B. (1997) Designing the Human Interface, 3rd edn. Addison-Wesley Pub. Co., NY, pp. 1–638. [Google Scholar]
- 13.Beardsley T. (1999) Sci. Am., March, 35–36. [Google Scholar]
- 14.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 15.Stegalkina S.S., Guerrero,A., Walton,K.D., Liu,X., Robinson,G.W. and Hennighausen,L. (1999) J. Virol., 73, 8669–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phimister B. (1999) Nature Genet., supplement, 1. [Google Scholar]
- 17.Hennighausen L. and Robinson,G. (1998) Genes Dev., 12, 449–455. [DOI] [PubMed] [Google Scholar]
- 18.Liu X., Robinson,G.W., Wagner,K.U., Garrett,L., Wynshaw-Boris,A. and Hennighausen,L. (1997) Genes Dev., 11, 179–186. [DOI] [PubMed] [Google Scholar]
- 19.Carlisle A.J., Prabhu,V.V., Elkahloun,A., Hudson,J., Trent,J.M., Linehan,W.M., Williams,E.D., Emmert-Buck,M.R., Liotta,L.A., Munson,P.J. and Krizman,D.B. (2000) Mol. Carcinog., 28, 12–22. [PubMed] [Google Scholar]
- 20.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vawter M.P., Barrett,T., Cheadle,C., Sokolov,B.P., Wood,W.H.,III, Donovan,D.M., Webster,M., Freed,W.J. and Becker,K.G. (2000) Brain Res. Bull., in press. [DOI] [PubMed] [Google Scholar]
- 22.DeRisi J., Penland,L., Brown,P.O., Bittner,M.L., Meltzer,P.S., Ray,M., Chen,Y., Su,Y.A. and Trent,J.M. (1996) Genetics, 14, 457–460. [DOI] [PubMed] [Google Scholar]
- 23.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyler,V.R., Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Mol. Biol., 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White K.P., Rifkin,S.A., Hurban,P. and Hogness,D.S. (1999) Science, 286, 2179–2184. [DOI] [PubMed] [Google Scholar]
- 25.Sneath P.H.A. and Sokol,R.R. (1973) Numerical Taxonomy. W.H. Freeman Co., NY, pp. 188–308.
- 26.Tamayo P., Slonim,D., Mesirov,J., Zhu,Q., Kitareewan,S., Dmitrovsky,E, Lander E.S. and Golub,T.R. (1999) Proc. Natl Acad. Sci. USA, 96, 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalon D., Smith,S.J. and Brown,P.O. (1996) Genome Res. 6, 639–645. [DOI] [PubMed] [Google Scholar]
- 28.Wodicka L., Dong,H., Mittmann,M., Ho,M.H. and Lockhart,D.J. (1997) Nat. Biotechnol., 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- 29.Ross D.T., Scherf,U., Eisen,M.B., Perou,C.M., Rees,C., Spellman,P., Iyer,V., Jeffrey,S.S., Van De Rijn,M., Waltham,M., Pergamenschikov,A., Lee,J.C., Lashkari,D., Shalon,D., Myers,T.G., Weinstein,J.N., Botstein,D. and Brown,P.O. (2000) Nature Genet., 24, 227–235. [DOI] [PubMed] [Google Scholar]
- 30.Scherf U., Ross,D.T., Waltham,M., Smith,L.H., Lee,J.K., Tanabe,L., Kohn,K.W., Reinhold,W.C., Myers,T.G., Andrews,D.T., Scudiero,D.A., Eisen,M.B., Sausville,E.A., Pommier,Y., Botstein,D., Brown,P.O. and Weinstein,J.N. (2000) Nature Genet., 24, 236–244. [DOI] [PubMed] [Google Scholar]
- 31.Hennighausen L.G. and Sippel,A.E. (1982) Eur. J. Biochem., 125, 131–141. [DOI] [PubMed] [Google Scholar]