Abstract
Despite much information as to the structure and function of the general transcription factors, little is known about the regulation of their expression. Transcription of the Saccharomyces cerevisiae SUA7 (TFIIB) gene results in the formation of two discrete transcripts. It was originally reported that the two transcripts were derived from two promoters separated by ~80 bp. We have found that the two transcripts are instead derived from a common promoter and differ at the 3′-end by ~115 bp. The longer of the two transcripts has an unusually long 3′-untranslated region. We have analyzed the levels of these transcripts under different cell growth conditions and find that the relative amounts of the two transcripts vary. Approximately equal amounts of each transcript are observed during exponential growth, but stresses and growth limiting conditions lead to a decrease in the relative amount of the larger transcript. These results suggest that the expression of the SUA7 gene may be controlled by regulation of 3′-end formation or mRNA stability. One of the general transcription factors, then, may be subject to regulation by a general response of the mRNA processing machinery.
INTRODUCTION
The SUA7 gene of Saccharomyces cerevisiae encodes TFIIB, one of the general transcription factors required for expression of all protein encoding genes (1). This essential transcription factor has two primary roles in the assembly of the initiation complex. First, it is required for the association of the RNA polymerase with the promoter bound transcription factor TFIID by direct contacts with both the TATA binding protein (TBP) and the RNA polymerase (2,3). In addition, clear genetic and biochemical evidence exists that TFIIB is one determinant of start site selection in yeast (4–7).
As a critical point in initiation complex assembly, TFIIB has been suggested to be a transcription-limiting step in assembly that can be increased by interaction with activator proteins (8,9). Consistent with this, protein–protein interactions have been detected for TFIIB with a number of activators (10,11) and activation defective mutants of TFIIB have been isolated (12). Addition of TFIIB has been observed to overcome the effect of activator squelching in vitro (13) and in vivo (11). In addition, in vitro mechanistic experiments have suggested that TFIIB addition to the TFIID–TATA complex is a limiting step that can be overcome by the effect of activator proteins (14,15). Although the idea that TFIIB levels play a role in promoter activity and activation is not without controversy (16–18), these other experiments suggest that the concentration of TFIIB in the cell could be important to promoter activity and raise the question of what cellular conditions might affect its intracellular concentration.
Despite the potential importance of cellular levels of the general transcription factors, little is known about how their expression might be controlled. The promoters from the human (19), mouse (20), Acanthamoebae (21), Drosophila (22) and yeast (23) TBP genes have been examined. Although these studies identified sequences required for expression in vitro and/or in vivo, they did not address the question of whether cellular conditions affect the expression of the gene. Studies with yeast have found that the levels of both TBP and some of the TATA-associated factors decrease dramatically in post exponential phases of growth (24). Both TFIIB and TBP have been shown to be regulated during some phases of mammalian development (25,26), suggesting that, at least under some conditions, increased amounts of the general transcription factors are required. To address the question of what conditions might normally affect transcription factor expression, we began an analysis of the SUA7 gene in yeast.
Analysis of in vivo RNA showed that the SUA7 gene was encoded in two discrete transcripts that differed by about 100 bases in size (5). Start sites for these two transcripts were mapped to two discrete points by primer extension. The more proximal of the two start sites contains a nearly consensus TATA sequence (TATAAAT) at the appropriate distance from the start site. The more distal of the two start sites does not contain a reasonable TATA sequence and is therefore a ‘TATA-less’ promoter. The presence or absence of a TATA box has been shown to impact the interaction of the general transcription factors and RNA polymerase with the promoter and can lead to differences in regulation for the promoter (reviewed in 27,28). In two well-characterized examples (29–31) different types of core promoters contribute to the expression of the genes examined. In both of these cases, evidence exists for differential usage of the two types of core promoters, TATA-containing and TATA-less, under different conditions.
To address the question of transcription factor regulation, we examined the effect of different cell growth conditions on the levels of each of the two transcripts for the SUA7 gene. Such an analysis would let us address the possible significance of the two transcripts and of the two promoters proposed to direct their synthesis.
MATERIALS AND METHODS
Yeast strains and plasmids
The haploid strain 23-3a (MATa bar1 leu2 ura3-52 his4 trp1) was obtained from L. Marsh (Albert Einstein College of Medicine, New York, NY). The diploid strain GLM100 was derived from a mating between XLM44-3-27 (L. Marsh) and EG124 (I. Herskowitz, University of California, San Francisco, CA) and has the genotype a/α TRP1/trp1 LEU2/leu2 ura3-52/ura3-52 lys2/LYS2 HIS4/his4 ade2-100/ADE2 CAN1/can1 (G.L.Matika and B.C.Hoopes, unpublished). The plasmid pBlsc-SUA7 was used for probe synthesis. It was constructed by isolating the ClaI–BamHI restriction endonuclease generated fragment from pDW5462 (5) that contains SUA7 DNA from –789 to +972 relative to the start point of translation. This DNA fragment was ligated into the ClaI and BamHI sites of pBluescript KS+ (Stratagene, Inc.). Standard procedures were used for recombinant DNA techniques (32), media preparation and the growth of yeast strains (33,34).
Materials
Saturated acid phenol for RNA work (Ambion, Inc.), Nuclease S1, Klenow enzyme, T7 RNA polymerase, ribonucleotides, RQ1 DNaseI (Promega Corporation), deoxyribonucleotides (Boehringer Mannheim), Taq polymerase (Boehringer Mannheim or Sigma Corporation), MSI nylon membrane, formamide, urea, agarose, 40% acrylamide:bis acrylamide (19:1) (Fisher), diethylpyrocarbonate (Aldrich Chemical Corporation, Sigma Corporation), [α-32P]dATP, [γ-32P]ATP and [α-32P]CTP (New England Nuclear/Dupont), SequenaseTM sequencing kit (US Biochemicals) were purchased from the indicated sources.
Synthetic DNA
Oligonucleotides were purchased from Midland, Inc. or Integrated DNA Technologies and were used without further purification. The sequences of the oligonucleotides were: SUA7-1 (5′-ATCTATGCTCTCCCTAG-3′), Ro-RNAP (5′-CAAAACTAAAGAGTTTGG-3′), 3′ primer (5′-GACGAGCTCGGATCCTGCAGT18VN-3′), 3′ CLMP (5′-GACGAGCTCGGATCCTGCAGT6-3′), SUA7-3′ (5′-ATAACTTACCGGGCGTTG-3′), SUA7-5′ (5′-TATCACCGGATAACAAGAC-3′), RNA-L (5′-ATCTATGCTCTCCCTAG-3′). The SUA7-1 primer is the same primer used for primer extension in (5). The 3′ primer and 3′ CLMP sequences are the same sequences as those described by Russo et al. (35). The 3′ primer is used as a degenerate primer for cDNA synthesis from all mRNA. The 3′ CLMP primer can then be used as an amplification primer for the cDNA synthesized with the 3′ primer.
RNA isolation
Cells were isolated from 10 ml portions of cultures by centrifugation, rinsed with 1 ml of water, then reisolated by centrifugation and stored at –70°C until use. Total RNA was prepared from frozen cells by the glass bead–phenol method described (34). The final RNA pellet was resuspended in 50 µl of water treated with diethylpyrocarbonate and the concentration determined by UV spectrophotometry. All RNA samples had a 260:280 ratio of greater than 1.7.
Northern blots
A modification of the procedure described by Rose et al. (33) was used. Either a formaldehyde–agarose gel or a 1× MOPS non-denaturing agarose gel was used for separation of total RNA. Equal amounts of each RNA sample were ethanol precipitated in the presence of sodium acetate. Samples were resuspended by vigorous pipetting in denaturing sample buffer containing formamide and formaldehyde (33) and were heated to 60°C for 10 min before loading on the gel. After electrophoresis, RNA was transferred to a nylon membrane by upward capillary transfer (32) or by using a Turbo Blotter (Schleicher and Schuell) without alkaline denaturation. Nylon membranes were analyzed for ribosomal RNA bands by staining with methylene blue in 0.3 M sodium acetate as described by Herrin and Schmidt (36). Background was reduced by floating the membrane in boiling water until it cooled to room temperature (33), then the residual methylene blue was removed by incubation with shaking in 0.2× SSC, 1% SDS (36). Pre-hybridization and hybridization were carried out as described by Rose et al. (33) except that dextran sulfate was omitted and hybridization was performed at 50°C. The probe used was an anti-sense RNA containing SUA7 sequences from +972 to +618 synthesized from DraI digested pBlsc-SUA7 with T7 RNA polymerase under conditions recommended by the manufacturer. Quantitation of transcript levels was performed on a Storm 820 Phosphorimager (Molecular Dynamics) using the Image Quant software provided by the manufacturer.
S1 nuclease analysis
The probe used for S1 nuclease analysis was a single stranded DNA containing SUA7 sequence from +24 to –456 generated by extension of the primer SUA7-1 end-labeled with [γ-32P]ATP using single stranded pBlsc-SUA7 DNA as a template and Klenow enzyme as described in (34). After primer extension, the products were digested with BstEII to generate the 3′-end at –456. The labeled probe was purified from the template DNA by separation on a 5% polyacrylamide–7 M urea gel and the labeled product detected by autoradiography. The band was excised and soaked overnight in 0.5 M ammonium acetate, 1 mM EDTA, then the eluted DNA was purified after addition of 10 µg carrier tRNA by phenol extraction and ethanol precipitation and was resuspended in 40 µl TE. Reactions contained 20–30 µg of total RNA and 2 µl labeled probe, and reactions were performed as previously descibed (32) with 1000 U/ml Nuclease S1. After S1 digestion and ethanol precipitation, samples were resuspended in 6 µl water and 4 µl sequencing stop mix was added. Samples were separated by electrophoresis on a 6% polyacrylamide (19:1)–7 M urea sequencing gel. DNA sequence used to determine the size of protected products was generated using a Sequenase kit and conditions recommended by the manufacturer with double stranded pBlsc-SUA7 denatured with alkali as a template and the SUA7-1 oligonucleotide as a primer.
RT–PCR reactions
RNA used for RT–PCR using two SUA7-specific primers was first treated with DNase I to remove residual chromosomal DNA present in the isolated RNA. The isolated RNA was brought to 1 mM dithiothreitol, 10 mM MgCl2 and 20 mM Tris–HCl pH. 7.5 before the addition of 20 U placental RNase inhibitor and 1 U RQ1 DNase I in a total reaction volume of 50 µl. After incubation for 15 min at 37°C, 25 µl of DNase I stop mix (1.5 M sodium acetate, 50 mM EDTA) was added and the RNA was purified by phenol–chloroform extraction and ethanol precipitation. Total RNA or RNA treated with DNase I (2 µg) was reverse transcribed with AMV reverse transcriptase (10 U/reaction) using buffer and conditions recommended by the manufacturer. The final 20 µl reaction containing cDNA was diluted to 100 µl after heat inactivation of the reverse transcriptase and was used without further purification. PCR reactions (25 µl) contained 2.5–5 µl of the appropriate cDNA, 1 U Taq polymerase, 125 µM of all four deoxynucleoside triphosphates, 1.5 mM MgCl2, and 15 pmol of each primer in the buffer recommended by the manufacturer. Reactions were subjected to a 3 min incubation at 94°C to denature DNA followed by 35 cycles of 94°C (30 s), 50°C (30 s), 72°C (30 s).
Analysis of RT–PCR products
Products produced as described above were separated by agarose gel electrophoresis (2.5% Nusieve GTG agarose) and the individual products amplified from excised gel slices using Pfu DNA polymerase as per the manufacturer’s instructions (Promega Corporation). The PCR products were purified using Wizard PCR Purification system (Promega Corporation) and ligated into SmaI restricted pBluescript KS+. Ligations were transformed into chemically competent XL10 cells (Stratagene, Inc.) and colonies containing plasmids with inserts were identified by restriction digestion. Insert containing plasmids were sequenced using the Big-Dye Terminator Cycle Sequencing Kit (PE Applied Biosystems) and analyzed on an ABI prism 310 Genetic Analyzer. Analysis of sequences for 3′-end-processing signals was performed using MacVector 6.5 (Eastman Kodak Co.).
RESULTS
To begin our investigation of the regulation of expression of the TFIIB gene of S.cerevisiae, we analyzed the levels of TFIIB specific transcripts as a function of cellular growth conditions where transcriptional activity is affected. Cultures of haploid yeast were grown to exponential phase, then subjected to one of several conditions. For amino acid starvation, cells were collected by centrifugation, washed with sterile water, then cultured for 48 h in a synthetic media containing nitrogen and glucose but no amino acids or required nucleotides. The yeast strain used was auxotrophic for several amino acids (see Materials and Methods) so this induced an amino acid starvation response (37). Cells were also allowed to grow for 5 days to stationary phase in rich, glucose-containing medium. Cells in stationary phase show a general decrease in protein synthesis and specific changes in gene expression (38). To examine a well-characterized stress response, the cells were subjected to heat shock (39). The temperature of the culture was rapidly raised to 37°C and the culture incubated at this temperature for 2 h. Total RNA was isolated from portions of the cultures and equal quantities of RNA analyzed by northern analysis.
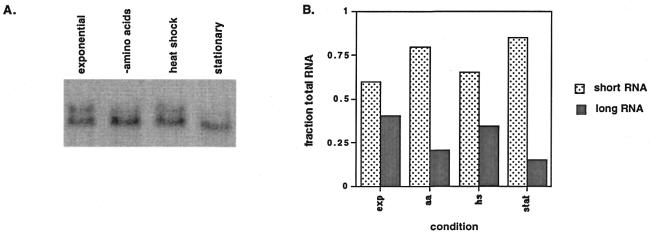
As previously reported by Pinto et al. (5), we observed two discrete SUA7-specific transcripts for cells grown under exponential conditions. RNA isolated from the cells is shown in Figure 1A. However, under the stressed conditions, the relative amount of each transcript changes. It is apparent that the different conditions lead to a decrease in the amount of the longer transcript with an increase in the amount of the shorter. Quantitation of the northern analysis is shown in Figure 1B. Although the relative amount of each transcript changes, the absolute levels of total RNA does not change dramatically (data not shown).
Figure 1.
Northern analysis of SUA7-specific transcripts as a function of cell growth conditions. Total RNA (15 µg) isolated from cells grown under different conditions was separated by denaturing agarose gel electrophoresis and transferred to a nylon membrane as described in Materials and Methods. (A) Results of phosphorimager visualization of the membrane after hybridization to an anti-sense SUA7 RNA probe. RNA was isolated from cells harvested in exponential phase, after starvation for amino acids, after a heat shock or after growth to stationary phase. (B) Quantitation of relative transcript levels for different conditions. The volume of each transcript was measured using Image Quant software, then the fraction of each transcript making up the total RNA for that condition was calculated.
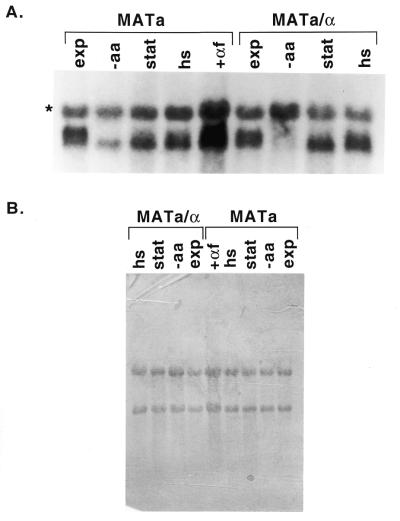
These results are also observed for diploid cells. RNA isolated from haploid and diploid cells in exponential phase is shown in Figure 2. [The third band marked with an asterisk is only observed under low stringency hybridization conditions and has a mobility consistent with it resulting from hybridization of the probe to 18S ribosomal RNA (data not shown).] We observe only subtle differences for the transcripts isolated from haploid and diploid cells; there may be more of the longer transcript relative to the shorter transcript for the haploid strain (23-3a) compared to the diploid strain (GLM100). As shown in Figure 2, arresting the cell cycle in the G1 phase by the addition of α-factor to the haploid a cells does not result in a similar shift to the shorter transcript. Although in this experiment it appears that the absolute levels of total RNA changes, this effect is not reproducible.
Figure 2.
Diploid cells show a similar effect of stress. Total RNA (10 µg) isolated from cells grown under different conditions was separated by agarose gel electrophoresis and transferred to a nylon membrane as described in Materials and Methods. (A) Results of autoradiography are shown after hybridization of the membrane to an antisense SUA7 RNA probe. Total RNA was prepared from cells isolated in exponential phase (lanes 1 and 6), after amino acid starvation (lanes 2 and 7), in stationary phase (lanes 3 and 8), after a 2 h heat shock (lanes 4 and 9), and after a 2 h treatment with α-factor (lane 5). (B) Results of methylene blue staining of the membrane before hybridization to demonstrate equal loading of RNA samples.
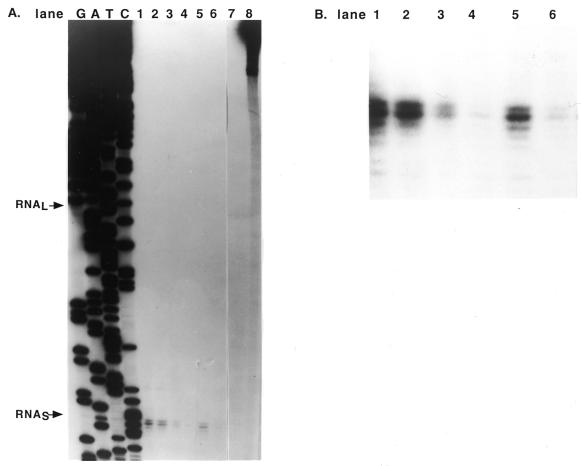
To pursue the question of the differential regulation of the two transcripts, we wished to more accurately measure the relative amounts of each transcript under different conditions. Because the two SUA7-specific transcripts are so similar in size, it is difficult to accurately quantify the relative amount of each transcript by northern analysis. We synthesized a strand specific labeled DNA probe containing SUA7 sequences from +24 to –454 relative to the start point of translation to use for S1 nuclease protection experiments. This labeled DNA probe is expected to generate protected products of 68 bp for the short RNA and 148 bp for the long RNA. RNA was isolated from cells for the growth conditions described above and the amount of each 5′-end assessed by S1 nuclease protection (see Materials and Methods). However, as shown in Figure 3A, only one protected product was observed. As demonstrated by the sequencing reactions produced from the same primer used for production of the DNA probe, this product corresponded to that expected for the start site for the short RNA reported by Pinto et al. (5). The position at which we expect to see the product corresponding to hybridization to the long RNA is marked; there is clearly no protected product at that position. A darker exposure of the autoradiogram also showed no evidence of the longer transcript (data not shown).
Figure 3.
S1 protection analysis of SUA7 transcripts. Total RNA was hybridized with a single stranded template strand DNA probe containing SUA7 DNA from +24 to –454 relative to the start of translation. The hybridized region was analyzed by S1 protection as described in Materials and Methods. (A) Autoradiogram showing the protected products after separation on a sequencing gel. The size of the protected products was determined by comparison to the sequencing ladder generated with a primer containing the same 5′-end as the single stranded DNA probe. The positions for the expected short and long RNAs are marked with arrows. RNA was isolated from cells in exponential phase (lane 1), after heat shock (lanes 2 and 5), after amino acid starvation (lanes 3 and 6) and in stationary phase (lane 4). Samples from the amino acid starved cells contained 20 µg of total RNA; the other samples contained 30 µg of total RNA. Lanes 7 and 8 contain labeled probe in the presence and absence of nuclease S1, respectively. (B) A darker exposure of the protected products from the autoradiogram shown in (A).
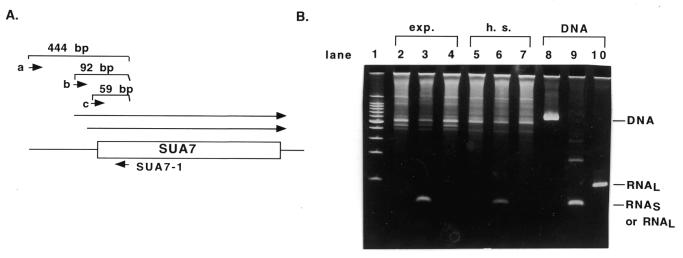
The nuclease protection results suggest that original primer extension assays might not have identified an actual transcriptional start site. We suspected that this might be the case because experiments in our laboratory using total RNA instead of poly A+ RNA yielded large amounts of primer extension product corresponding to the longer RNA, but only small amounts of the product corresponding to the shorter RNA (C.Coffeen and B.C.Hoopes, unpublished). These results were clearly inconsistent with the relative amounts of each transcript observed by northern analysis and with the published work. One possible explanation for our results would be that the shorter primer extension product corresponds to the start site also identified by nuclease protection assays and that the longer primer extension product is the result of DNA synthesis primed from a different RNA template. To test this hypothesis, we performed an RT–PCR analysis of SUA7- specific RNA. Reverse transcriptase was used for cDNA synthesis from RNA samples using the same primer used in this study and others (5) for primer extension assays. The resulting cDNA was then amplified using three different combinations of primers. One reaction contained the primer extension primer and an oligonucleotide with a 5′-end at –421 relative to the translational start site for Sua7p. These primers should only produce a product from genomic DNA that may be contained in the reaction; this region of the SUA7 chromosomal region is not expected to be found in either transcript. As shown in lanes 2, 5 and 8 of Figure 4, products are only observed from reactions containing DNA and not from the cDNA synthesized with the primer extension primer. A second set of primers contained the primer extension primer and an oligonucleotide with a 5′ at –36 relative to the translational start site for Sua7p. This set of primers should generate a 59 bp product from either transcript. As shown in lanes 3, 6 and 9 of Figure 4, this primer set generates a product of the expected size from reactions containing DNA or the synthesized cDNA. The third set of primers contained the primer extension primer and an oligonucleotide with a 5′-end at –69 relative to the translational start site for Sua7p. This set of primers would produce a product of 92 bp from the longer RNA but could not produce a product from the shorter RNA. As shown in lanes 4, 7 and 10 of Figure 4, this primer set only generated a product of appropriate size from DNA; no product was observed for the reaction containing the cDNA for either normal or heat shocked cells. These results support our proposal that the TATA-less start site identified by primer extension (5) does not direct the synthesis of the longer SUA7-specific RNA identified by northern analysis.
Figure 4.
RT–PCR analysis of the 5′-end of the SUA7 transcripts. (A) Diagram of the RT–PCR assay used for detection of transcripts differing at the 5′-end. Total RNA isolated from GLM100 cells harvested in exponential phase or after heat shock was treated with DNase I as described in Materials and Methods and used for cDNA synthesis using the SUA7-1 oligonucleotide (SUA7 sequences from +8 to +24) as a primer. This cDNA was then used for PCR using the SUA7-1 primer and either primers a (SUA7-5′), b (RNAL), or c (Ro-f). (B) The products of PCR reactions are shown after separation on a 6% native polyacrylamide gel. Lane 1, a 100 bp DNA ladder; lanes 2–4, cDNA made from exponential cells; lanes 5–7, cDNA made from heat shocked cells; lanes 8–10, plasmid pDW5462. Lanes 2, 5 and 8, primer a; lanes 3, 6 and 9, primer c; lanes 4, 7 and 10, primer b.
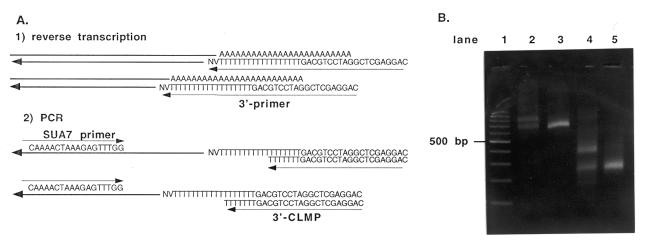
Since there are clearly two different SUA7-specific transcripts and the results described above suggest that the transcripts have a single 5′-end with a standard TATA-containing promoter, the differences in length must occur elsewhere in the transcript. Since few transcripts in yeast are spliced, the most likely explanation for differences in length for two would be two potential sites for 3′-end formation and polyadenylation. If this were true, we would expect that RT–PCR analysis of the 3′-ends of the transcript would show two discrete products. The relative amounts of these two products should vary for RNA made from cells grown under different conditions. As shown in Figure 5, RT–PCR analysis of the 3′-end of SUA7-specific transcripts confirms this hypothesis. Samples of RNA were used for cDNA synthesis with a primer designed for analysis of 3′-end formation (40). RNA isolated from exponentially grown and amino acid-starved 23-3a cells was used for cDNA synthesis with this 3′ primer. The cDNA was then amplified by PCR with an SUA7-specific primer and a primer containing the arbitrary sequence used as a ‘tag’ for the cDNA. One of the SUA7-specific primers used had a 5′-end at +593 and the other a 5′-end at +1010. As predicted by our hypothesis that the two SUA7-specific transcripts differ at the 3′-end, amplification of cDNA made from RNA isolated from 23-3a cells isolated in exponential phase showed two discrete products for the 3′-end. The two products are ~730 and 880 bases long for the primer with a 5′-end at +593 (Fig. 5B, lane 2) and 275 and 410 bp long for the primer with the 5′-end at +1010 (Fig. 5B, lane 4). For the RNA isolated from the amino acid starved cells, only a single product is obtained whose size corresponds to the smaller product observed for RNA isolated from cells grown to exponential phase (Fig. 5B, lanes 3 and 5). These results confirm our hypothesis that the two SUA7 transcripts differ at the 3′-end and not at the 5′-end as was originally reported.
Figure 5.
RT–PCR analysis of the 3′-end of the SUA7 transcripts. (A) Diagram of the RT–PCR assay used for 3′-end determination. Total RNA isolated from exponential phase and amino acid starved 23-3a cells was treated with DNase I and used for cDNA synthesis using the ‘tagged’ 3′ primer shown. SUA7 specific transcripts were then amplified by PCR using the SUA7-specific primers and the tag primer (see Materials and Methods for primer sequences). (B) Products of the RT–PCR reaction were analyzed by electrophoresis on a 2.5% Nusieve agarose gel in TAE and detected by staining with ethidium bromide. Lane 1 is a 100 bp ladder used as a size standard, lane 2 contains products from RNA made from exponential phase cells with the Ro-RNAP primer (5′-end at +594), lane 3 contains products from RNA made from amino acid starved cells with the Ro-RNAP primer, lane 4 contains exponential RNA with the SUA7-3′ primer (5′-end at +1010) and lane 5 contains RNA from starved cells with the SUA7-3′ primer.
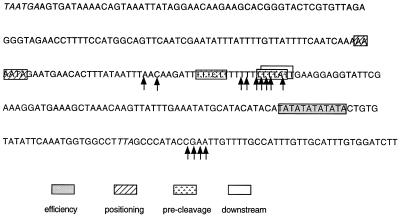
The two polyadenylation sites appear to differ in the degree to which they specify a precise endpoint for the 3′-end. To obtain information that might allow us to understand this difference, we isolated the short and long RT–PCR products and ligated them into a plasmid vector. Individual clones were then sequenced to determine the sequence at which polyadenylation had occurred. These data are summarized in Figure 6. Sequences of 13 individual clones showed the short transcript resulted from 3′-end formation occurring in a very T-rich region encompassing about 28 bases. Sequences of six clones derived from the long RT–PCR product showed 3′-end formation occurring in a cluster of bases occurring about 35 bases downstream of a TA repeat region. Potential 3′-end-processing sequences in this region were identified by searching the 3′-untranslated region (3′-UTR) for the four classes of sequences identified by Graber et al. (41). The results of this analysis are presented in Table 1 and are shown by the boxed sequences in Figure 6.
Figure 6.
Polyadenylation sites for the SUA7 gene. Shown is the DNA sequence of the 3′-UTR of the SUA7 gene as deposited in GenBank as part of the sequencing of Chromsome XVI. The stop codons for the SUA7 and SRP54 genes are indicated in italics. The sites for polyadenylation determined by sequencing of individual plasmids is denoted by the arrows. Potential efficiency, positioning, pre-cleavage and downstream elements are shown as boxed sequences (41).
Table 1. Distribution of 3′-end processing signals in the 3′-UTR for SUA7.
| Efficiency | Positioning | Pre-cleavage | Downstream | |
|---|---|---|---|---|
| Optimal sequence | TATATA | AAAATA | TTTTAT | TTTTCT |
| Distance found | –80 to –30 | –35 to –20 | –20 to +5 | +5 to +30 |
| RNAS | none | AAAATA | TTTTATTTTTTC | TTTATT |
| RNAL | TATATA | None | none | none |
The optimal sequences are those given by Graber et al. (41). The sequences found for SUA7 are shown in boxes in Figure 6.
DISCUSSION
Our examination of the effect of growth conditions on the transcript levels of the SUA7 gene has led to two main conclusions. First, we have demonstrated that the synthesis of the two SUA7 transcripts is directed by a single promoter and that the transcripts differ in their 3′-UTR by about 115 bases. Secondly, we have shown that the relative amounts of the long and short transcripts vary as a function of cellular growth conditions and stresses.
The original observation that the two SUA7 transcripts differed at the 5′-end was prompted by a correspondence between the difference in size of the two transcripts observed on a northern blot and the size of two prominent primer extension products (5). Our primer extension assays showed a much greater amount of the longer than the shorter product despite the fact that northern analysis showed roughly equal amounts of the two transcripts. This suggested to us that one of the two primer extension products might be an experimental artifact. Primer extension assays are prone to several types of artifacts, including premature terminations and extensions of complementary hairpins to produce double stranded products (32,34). Although the latter reaction would result in a longer product, we favor the idea that the longer primer extension product may actually result from hybridization of the primer used for synthesis to a different, more abundant RNA, than that which encodes SUA7. We suggest this because we observe products even larger in size than the prominent longer product, and because the amount of this product observed is not affected by conditions (such as amino acid starvation) that reduce the level of SUA7 transcription (data not shown).
Nuclease protection assays support the idea that the more proximal (TATA-containing) start site is used and that the distal start site is not. There are, of course, potential artifacts associated with the use of nuclease protection assays as well (32,34), but we have supported the presence of a single promoter with RT–PCR assays. We see evidence for only the single 5′-end but for two 3′-ends separated by the expected distance. In contrast to the primer extension experiments, the relative amount of the different 3′-ends observed is responsive to cellular growth conditions in a manner predicted by the northern analysis.
The existence of two different 3′-ends for SUA7 is interesting in light of the fact that this region contains two closely spaced convergently transcribed genes. SUA7 is transcribed convergently with SRP54 (SRH1) (5), a gene encoding a subunit of the signal recognition particle (42). There are only 275 bp between the translational stop codons for the two genes. As shown in Figure 6, this means that the longer RNA clearly contains a portion of the SRP54 coding sequence. Depending on where the 3′-end for the SRP54 gene is, both transcripts could in fact overlap with the SRP54 transcript. This raises the question of how the effects we observe for the two SUA7 transcripts may affect the expression of SRP54. There are several other pairs of genes for which this question has already been addressed (43,44). These studies have suggested that convergent transcription and antisense RNA production does not affect the regulation of gene pairs. Therefore, although this is an issue that might be pursued, the effects of stress conditions that we see on SUA7 3′-end formation probably do not affect the expression or function of the essential SRP54 gene.
Both of the SUA7 transcripts contain the entire coding sequence for TFIIB and the longer transcript contains an additional ∼115 bases of 3′-UTR. Sequences within the 3′-UTR have been shown to affect mRNA stability, mRNA localization and translation (reviewed in 45–47). Any of these characteristics could differ for the two transcripts, affecting the extent to which they contribute to TFIIB protein levels within the cell under different conditions. The longer of the two RNAs is interesting in that its 3′-UTR is quite long compared to the yeast transcripts that have been analyzed so far. Analysis of the size distribution seen for yeast 3′-UTRs shows a narrow distribution peaking at 100 bases (41). It has been proposed that the short length of yeast 3′-UTRs is a consequence of the action of the nonsense-mediated mRNA decay pathway generally involved in mRNA surveillance (48). The question of how (or whether) the longer of the two SUA7 transcripts escapes nonsense-mediated mRNA decay is an interesting one.
Although we do not know what functional differences may exist for the two SUA7 transcripts, we do know that the longer of the two transcripts is more sensitive to changes in growth conditions. The amount of the longer transcript decreases when cells are starved for amino acids or other nutrients, when they are grown to stationary phase or when cells are subjected to a 2 h heat shock. We have purposely examined these phenomena under conditions where cells have been subjected to the altered condition for an extended period of time rather than looking at very transient changes. Most of these situations have been shown to result in transcriptional and post-transcriptional effects on gene expression (39,49,50). It is interesting that in the case of the diploid strain under amino acid starvation conditions, the cells are actually only being starved for uracil. However, since limitation for purines is linked to the general amino acid control response (51), starvation for pyrimidines may actually result in a similar response.
The changes we have observed in transcript ratios probably reflect a change in polyadenylation site choice. There are other examples of genes for which a change in 3′-end formation occurs so that the proximal signal is used at the expense of the distal one. The CBP1 gene, required for efficient mitochondrial function, has two 3′-ends, one within the coding region of the gene (52). The ratio between the two transcripts changes when cells are grown on glycerol as opposed to glucose, and these changes are thought to be the result of a switch in 3′-end choice (53). This effect is not specific for CBP1, because it is seen for other unrelated genes (54). It is also possible that the two SUA7 transcripts could differ in stability due to the extra sequence present in the longer RNA, which could contain a regulated destabilizing region. Although most destabilizing regions are thought to act constitutively (47), a destabilizing region within the coding region of the c-fos RNA has been proposed to be responsible for rapid degradation of the transcript only upon serum stimulation in mammals (55) and transcripts encoding subunits of the yeast succinate dehydrogenase complex in yeast appear to be regulated at the level of mRNA stability by the presence or absence of glucose in the media (56).
The extent to which the two discrete transcripts for the SUA7 gene contribute to Sua7p (TFIIB) protein levels is an important and unanswered question. Experiments that have examined the levels of some of the general transcription factors in yeast have concluded that although TBP protein levels decreased dramatically in cells past exponential phase, the amount of the Toa1p (TFIIA) and Sua7p (TFIIB) proteins remained relatively constant (24). These results are interesting in light of our observation that the relative levels of the two transcripts that encode TFIIB change under different growth conditions. It could be that this change in the relative amounts of the two transcripts is necessary to allow Sua7p protein levels to stay relatively constant under growth and translation-limited conditions.
The general transcription factor TFIIB serves a pivotal role in transcription, serving as the protein link between a promoter-bound TFIID and the RNA polymerase holoenzyme. Our studies on the two transcripts that encode TFIIB in the yeast S.cerevisiae have shown that their levels are sensitive to cellular growth conditions. Determining the differences in function for the two SUA7 transcripts will be an essential first step toward an understanding of how cellular stress and growth controls impact the expression of the general transcription factors.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the contribution of Christin Coffeen to early stages of this work and the technical help of Kathleen Baier. This work was supported by grants MCB-9317062 and MCB-9809917 from the National Science Foundation. Automated sequencing was supported by grant DBI-9970204 from the National Science Foundation.
REFERENCES
- 1.Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev., 62, 465–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratowski S. and Zhou,H. (1993) Functional domains of transcription factor TFIIB. Proc. Natl Acad. Sci. USA, 90, 5633–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha I., Roberts,S., Maldonado,E., Sun,X., Kim,L.-U., Green,M. and Reinberg,D. (1993) Multiple functional domains of transcription factor IIB: Distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev., 7, 1021–1032. [DOI] [PubMed] [Google Scholar]
- 4.Pinto I., Wu,W.-H., Na,J.G. and Hampsey,M. (1994) Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem., 269, 30569–30573. [PubMed] [Google Scholar]
- 5.Pinto I., Ware,D.E. and Hampsey,M. (1992) The yeast Sua7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site initiation in vivo. Cell, 68, 977–988. [DOI] [PubMed] [Google Scholar]
- 6.Na J.G. and Hampsey,M. (1993) The Kluyveromyces gene encoding the general transcription factor IIB: structural analysis and expression in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3413–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y., Flanagan,P.M., Tschochner,H. and Kornberg,R.D. (1994) RNA polymerase II initiation factor interactions and transcription start site selection. Science, 263, 805–807. [DOI] [PubMed] [Google Scholar]
- 8.Hawley D. (1991) Transcription activation: enter TFIIB. Trends Biochem. Sci., 16, 317–318. [DOI] [PubMed] [Google Scholar]
- 9.Orphanides G., Lagrange,T. and Reinberg,D. (1996) The general transcription factors of RNA polymerase II. Genes Dev., 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- 10.Lin Y.-S., Ha,I., Maldonado,E., Reinberg,D. and Green,M.R. (1991) Binding of general transcription factor TFIIB to an acidic activating region. Nature, 353, 569–571. [DOI] [PubMed] [Google Scholar]
- 11.Colgan J., Ashali,H. and Manley,J.L. (1995) A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol. Cell. Biol., 15, 2311–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W.H. and Hampsey,M. (1999) An activation-specific role for transcription factor TFIIB in vivo. Proc. Natl Acad. Sci. USA, 96, 2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X. and Berk,A.J. (1995) Reversal of in vitro p53 squelching by both TFIIB and TFIID. Mol. Cell. Biol., 15, 6474–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundseth R. and Hansen,U. (1992) Activation of RNA polymerase II transcription by the specific DNA-binding protein LSF. Increased rate of binding of the basal promoter factor TFIIB. J. Biol. Chem., 267, 7845–7855. [PubMed] [Google Scholar]
- 15.Lin Y.-S. and Green,M.R. (1991) Mechanism of action of an acidic transcriptional activator in vitro. Cell, 64, 971–981. [DOI] [PubMed] [Google Scholar]
- 16.Lee M. and Struhl,K. (1997) A severely defective TATA-binding protein-TFIIB interaction does not preclude transcriptional activation in vivo. Mol. Cell. Biol., 17, 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou S. and Struhl,K. (1997) Transcriptional activation by TFIIB mutants that are severely impaired in interaction with promoter DNA and acidic activation domains. Mol. Cell. Biol., 17, 6794–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colgan J., Wampler,S. and Manley,J.L. (1993) Interaction between a transcriptional activator and transcription factor IIB in vivo. Nature, 362, 549–553. [DOI] [PubMed] [Google Scholar]
- 19.Foulds C.E. and Hawley,D.K. (1997) Analysis of the human TATA binding protein promoter and identification of an Ets site critical for activity. Nucleic Acids Res., 25, 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohbayashi T., Schmidt,E.E., Makino,Y., Kishimoto,T., Nabeshima,Y., Muramatsu,M. and Tamura,T. (1996) Promoter structure of the mouse TATA-binding protein (TBP) gene. Biochem. Biophys. Res. Commun., 225, 275–280. [DOI] [PubMed] [Google Scholar]
- 21.Wong J.M., Liu,F. and Bateman,E. (1992) Isolation of genomic DNA encoding transcription factor TFIID from Acanthamoeba castellanii: characterization of the promoter. Nucleic Acids Res., 20, 4817–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho N., Oh,Y., Hwang,S.Y., Han,D., Park,S.P., Yoon,J., Han,K. and Baek,K. (1998) Promoter analysis of the Drosophila genes encoding TFIIB and TATA box-binding protein. Mol. Cell., 8, 770–776. [PubMed] [Google Scholar]
- 23.Schroeder S.C., Wang,C.K. and Weil,P.A. (1994) Identification of the cis-acting DNA sequence elements regulating the transcription of the Saccharomyces cerevisiae gene encoding TBP, the TATA box binding protein. J. Biol. Chem., 269, 28335–28346. [PubMed] [Google Scholar]
- 24.Walker S.S., Shen,W.-C., Reese,J.C., Apone,L.M. and Green,M.R. (1997) Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell, 90, 607–614. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt E.E. and Schibler,U. (1995) High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development, 21, 2373–2383. [DOI] [PubMed] [Google Scholar]
- 26.Veenstra G.J.C., Destree,O.H.J. and Wolffe,A.P. (1999) Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol. Cell. Biol., 19, 7972–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smale S.T. (1994) In Conaway,R.C. and Conaway,J.W. (eds), Transcription: Mechanisms and Regulation. Raven Press, New York, NY, pp. 63–81.
- 28.Enami K.H., Navarre,W.W. and Smale,S.T. (1995) Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol. Cell. Biol., 15, 5906–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corbin V. and Maniatis,T. (1990) The role of specific enhancer-promoter interactions in the Drosophila Adh promoter switch. Genes Dev., 3, 2191–2201. [DOI] [PubMed] [Google Scholar]
- 30.Hansen S.K. and Tjian,R. (1995) TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell, 82, 565–575. [DOI] [PubMed] [Google Scholar]
- 31.Struhl K. (1986) Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol. Cell Biol., 6, 3847–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Rose M., Winston,F. and Hieter,P. (eds) (1990) Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratories, Cold Spring Harbor, NY.
- 34.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1997) Current Protocols in Molecular Biology, edited by Janssen,K. John Wiley & Sons, New York, NY.
- 35.Russo P., Li,W.-Z., Guo,Z. and Sherman,F. (1993) Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7836–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herrin D.L. and Schmidt,G.W. (1988) Rapid, reversible staining of northern blots prior to hybridization. Biotechniques, 6, 196–200. [PubMed] [Google Scholar]
- 37.Warner J.R. and Gorenstein,C. (1978) Yeast has a true stringent response. Nature, 275, 338–339. [DOI] [PubMed] [Google Scholar]
- 38.Werner-Washburne M., Braun,E.L., Crawford,M.E. and Peck,V.M. (1996) Stationary phase in Saccharomyces cerevisiae. Mol. Microbiol., 19, 1159–1166. [DOI] [PubMed] [Google Scholar]
- 39.Morimoto R., Tissieres,A. and Georgopoulos,C. (1994) The Biology of Heat Shock Proteins and Molecular Chaperones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40.Guo A. and Sherman,F. (1995) 3′-end-forming signals of yeast mRNA. Mol. Cell. Biol., 15, 5983–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graber J.H., Cantor,C.R., Mohr,S.C. and Smith,T.F. (1999) Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res., 27, 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amaya Y., Nakano,A., Ito,K. and Mori,M. (1990) Isolation of a yeast gene, SRH1, that encodes a homologue of the 54K subunit of mammalian signal recognition particle. J. Biochem., 107, 457–463. [DOI] [PubMed] [Google Scholar]
- 43.Peterson J.A. and Myers,A.M. (1993) Functional analysis of mRNA 3′-end formation signals in the convergent and overlapping transcription units of the S. cerevisiae genes RHO1 and MRP2. Nucleic Acids Res., 21, 5500–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malavasic M.J. and Elder,R.T. (1990) Complementary transcripts from two genes necessary for normal meiosis in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 10, 2809–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahle E. and Keller,W. (1992) The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu. Rev. Biochem., 61, 419–440. [DOI] [PubMed] [Google Scholar]
- 46.Decker C.J. and Parker,R. (1995) Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol., 7, 386–392. [DOI] [PubMed] [Google Scholar]
- 47.Jacobson A. and Peltz,S.W. (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem., 65, 693–739. [DOI] [PubMed] [Google Scholar]
- 48.Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: Kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner-Washburne M., Braun,E., Johnston,G.C. and Singer,R.A. (1993) Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev., 57, 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yost H.J., Peterson,R.B. and Lindquist,S. (1990) RNA metabolism: strategies for regulation in heat shock response. Trends Genet., 6, 223–227. [DOI] [PubMed] [Google Scholar]
- 51.Rolfes R.J. and Hinnebusch,A.G. (1993) Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol., 13, 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayer S.A. and Dieckmann,C.L. (1991) Yeast CBP1 mRNA 3′-end formation is regulated during the induction of mitochondrial function. Mol. Cell. Biol., 11, 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sparks K.A., Mayer,S.A. and Dieckmann,C.L. (1997) Premature 3′-end formation of CBP1 mRNA results in the downregulation of cytochrome b mRNA during the induction of respiration in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 4199–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sparks K.A. and Dieckmann,C.L. (1998) Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res., 26, 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shyu A.-B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- 56.Cereghino G.P., Atencio,D.P., Saghbini,M., Beiner,J. and ScheffleI.E. (1995) Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol. Biol. Cell, 6, 1125–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]