Summary
Imbalanced dietary intake, such as a high-fat diet (HFD) during pregnancy, has been associated with adverse offspring outcomes. Metabolic stress from imbalanced food intake alters the function of epigenetic regulators, resulting in abnormal transcriptional outputs in embryos to cause congenital disorders. We report herein that maternal HFD exposure causes metabolic changes in pregnant mice and non-compaction cardiomyopathy (NCC) in E15.5 embryos, accompanied by decreased 5-hydroxymethylcytosine (5hmC) levels and altered chromatin accessibility in embryonic heart tissues. Remarkably, maternal vitamin C supplementation mitigates these detrimental effects, likely by restoring iron, a cofactor for Tet enzymes, in a reduced state. Using a genetic approach, we further demonstrated that the cardioprotective benefits of vitamin C under HFD conditions are attributable to enhanced Tet activity. Our results highlight an interaction between maternal diet, specifically HFD or vitamin C, and epigenetic modifications during early heart development, emphasizing the importance of balanced maternal nutrition for healthy embryonic development.
Subject areas: Pregnancy, Epigenetics, Developmental biology, Model organism, Diet
Graphical abstract

Highlights
-
•
Maternal high-fat diet causes developmental defects in embryonic heart
-
•
High-fat diet alters the function of Tet enzymes and reshapes chromatin accessibility
-
•
Vitamin C boosts TET activity and provides a protective effect on the embryonic heart
Pregnancy; Epigenetics; Developmental biology; Model organism; Diet
Introduction
Balanced nutrition exposure during pregnancy is crucial for promoting normal embryonic development. Maternal environmental exposure, such as a high-fat diet(HFD), toxin, and pollution, can have negative consequences on the offspring.1,2,3 For instance, maternal HFD exposure has been found to cause metabolic abnormalities and neurodevelopmental aberrations in offspring.1,3,4,5 Mechanistically, this has been linked to epigenetic dysregulation, including abnormal DNA methylation changes in the offspring, which in turn could impact transcriptional outputs to influence early development.
The crosstalk between metabolic pathways and epigenetic regulators has been reported in various biological systems.6,7 Nutrients, such as fatty acids and vitamins, are utilized by the metabolic machinery to produce metabolites that are essential for supporting the proper function of epigenetic regulators. In parallel, epigenetic machinery can in turn modulate the transcriptional network essential for metabolic homeostasis. For example, the catalytic activity of the Ten-eleven Translocation (TET) family of dioxygenases, which are pivotal in the stepwise oxidation of methylated DNA toward demethylation, is tightly governed by nutritional levels and metabolic availability.8,9,10,11 Essential cofactors for TET enzymatic activity include 2-oxoglutarate (2-OG) and Fe2+. Vitamin C can augment the activity of TET enzymes by keeping iron in its reduced state.9,10,12,13 Furthermore, lipid assimilation and metabolism are also intimately linked to TET function,13,14,15 as TET enzymes have been shown to regulate fatty acid oxidation in liver tissues by controlling the transcription of peroxisome proliferator activated receptor alpha (PPARα), a key transcription factor that regulates lipid metabolism.16 These findings lend strong support to the intimate crosstalk between HFD and TET-mediated epigenetic modulation.
Although the association between maternal HFD exposure and congenital heart disease (CHD) has been reported;17,18 the potential roles of epigenetic alterations induced by such diets in developmental anomalies are yet to be elucidated. In the current study, we evaluated the impact of maternal HFD on the expression and enzymatic function of murine Tet proteins in myocardium during embryonic development. We further investigated the beneficial effect of maternal vitamin C exposure in countering the negative impacts of HFD treatment. Our study establishes a connection between maternal HFD exposure and altered TET-mediated epigenetic regulation during heart development. From a translational standpoint, this work underscores the potential of maternal vitamin C intake as a preventative strategy to protect against abnormal cardiac development associated with maternal obesity.
Results
High-fat diet treatment alters Tet protein expression and function in dams
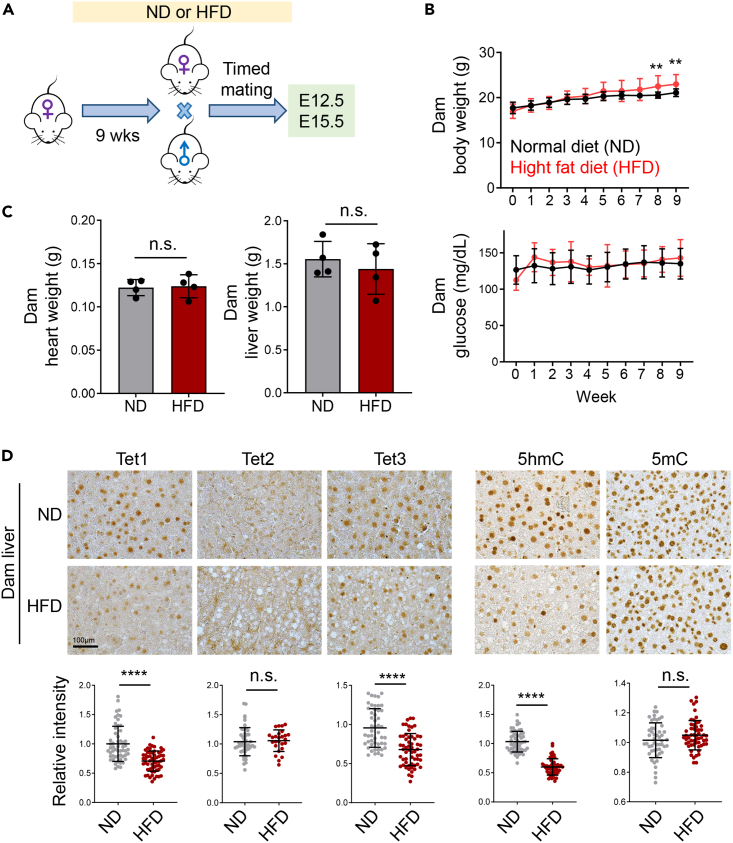
To assess the impact of maternal HFD on offspring cardiac development, female mice were administered either a normal diet (ND; 10 kcal% fat) or a HFD (60 kcal% fat) for nine weeks before mating with age-matched males on a normal diet. The embryos were then harvested for subsequent analysis at developmental stages E12.5 and E15.5 (Figure 1A). Body weight and blood glucose levels of the female mice (dams) were measured on a weekly basis during the dietary intervention (Figure 1B). For the initial seven weeks, the body weight of the dams on both diets remained comparable. However, dams subjected to the HFD exhibited a marked increase in body weight during the 8th and 9th weeks compared to the control group (Figure 1B, top). No significant differences were observed in the blood glucose levels between dams on the normal diet and those on the HFD throughout the nine weeks (Figure 1B, bottom). Additionally, no discernible differences were noted in the heart and liver sizes between dams on the normal and HFDs (Figure 1C).
Figure 1.
Maternal HFD alters Tet activities in the liver of dams
(A) Experimental timeline for normal diet (ND) or high-fat diet (HFD) feeding of timed mated female mice.
(B) Measurements of the body weight (top) and blood glucose levels (bottom) for female mice fed with ND or HFD over a nine-week period (n = 11 mice/group; mean ± SD); ∗∗p < 0.01 (two-sided unpaired Student’s t test).
(C) Weight measurement of heart and liver collected from dams fed with ND or HFD for 9 weeks (n = 4 mice/group; mean ± SD). n.s., not significant.
(D) (Top) Representative images showing immunohistochemistry (IHC) staining of the indicated Tet1-3 proteins and DNA modifications (5hmC or 5mC) in liver tissues collected from dams (n = 4–5) exposed to ND or HFD for 9 weeks. Scale bar, 100 μm. (Bottom) Quantifications of the relative intensities of the corresponding markers (n = 50–60 cells; mean ± SD); ∗∗∗∗p < 0.0001 (two-sided unpaired Student’s t test).
To further investigate the effects of a HFD on Tet protein expression and function, we performed immunohistochemistry (IHC) staining to quantify the levels of Tet proteins (Tet1-3), 5-hydroxymethylcytosine (5hmC), and 5-methylcytosine (5mC) in liver and heart tissues of dams on normal or HFD (Figures 1D and S1A). We observed a notable decrease in the levels of Tet1 and Tet3 proteins, as well as 5hmC, in the livers of female mice on the HFD. By contrast, Tet2 protein and 5mC levels remained largely unaltered between the two dietary conditions. No significant differences in Tet1, 5hmC, and 5mC levels were observed in heart tissues between the two conditions. Collectively, these results indicate that a 9-week HFD regimen seems to exert rather minor effects on the body weight and the glucose levels in pregnant mice, yet it significantly alters the Tet-mediated epigenetic machinery in the liver of the dams, hinting a potential epigenetic reprogramming due to the high-fat dietary uptake during pregnancy.
Maternal HFD exposure leads to offspring cardiac anomalies
Maternal HFD has been linked to adverse pregnancy outcomes and is associated with an increased risk of overweight and obesity in offspring.1,11,18,19 Furthermore, offspring from mothers exposed to a HFD has been found to exhibit cardiac hypertrophy at postnatal day 77.19 To further study the impact of maternal HFD on heart development in utero, we performed timed mating and assessed cardiac tissues of embryos at E12.5 and E15.5 from dams on either a normal or a HFD. We found that while the HFD did not alter the number of embryos per pregnancy (Figure S2A), it significantly increased embryo weight at E15.5 but not at E12.5 (Figure S2B). Subsequent histological analyses of E15.5 embryonic cardiac tissues revealed that a maternal HFD induced enlarged heart size and ventricular non-compaction cardiomyopathy (NCC), characterized by diminished ventricular wall thickness and enlarged trabecular areas compared to controls (Figure 2A). Additionally, maternal HFD exposure resulted in a decrease in the expression of Tet1/3 proteins (but not at the transcription level) and reduced levels of 5hmC in cardiac tissues at E15.5 but not at E12.5 (Figures 2B–2D, S2C, and S2D).
Figure 2.
Maternal HFD suppresses TET protein expression and causes ventricular non-compaction cardiomyopathy (NCC) in embryos
(A) Representative H&E-stained sections (Left) and quantification of ventricular area and ventricular wall thickness (Right) from E15.5 embryonic heart tissues following maternal ND or HFD (n = 3–6 embryos per group, mean ± SD). ∗p < 0.05 (two-sided unpaired Student’s t test). Scale bar, 200 μm (4x) or 100 μm (20x).
(B) Representative IHC images (top) and quantification (bottom) of Tet1, Tet3 and 5hmC levels in the ventricular area of E15.5 embryonic hearts from the ND and HFD groups (n = 50–60 cells/group; mean ± SD. ∗p < 0.05, ∗∗∗∗p < 0.0001 (two-sided unpaired Student’s t test). Scale bar, 100 μm.
(C) Western blot analysis of Tet1 in heart tissues collected from E15.5 embryos exposed to maternal ND or HFD (n = 2 embryos/group). Tubulin was used as a loading control.
(D) 5hmC dotblot analysis on DNA samples from heart tissues collected from E12.5 and E15.5 embryos exposed to maternal ND or HFD (n = 2 embryos/group). Methylene blue (ME-Blue) staining is used as loading control.
(E) Genomic distribution of differentially enriched ATAC-seq peaks in heart tissues collected from E15.5 embryos following maternal ND or HFD exposure.
(F) GREAT analysis of functionally annotated differentially enriched ATAC-seq peaks from cardiac tissues of E15.5 embryos under maternal ND or HFD.
(G) Genome browser views of ATAC-seq data at the Ppara and Tnnt2 loci in heart tissues collected from E15.5 embryos exposed to maternal ND (blue) or HFD (red).
(H) hMeDIP analysis of 5hmC enrichment at the promoters of Ppara and Tnnt2 in heart tissues collected from E15.5 embryos exposed to maternal ND or HFD. IgG is used as a negative control (n = 4 embryos, mean ± SD; ∗p < 0.05, ∗∗p < 0.01; two-sided unpaired Student’s t test).
(I) Real-time qPCR analysis on Ppara and Tnnt2 expression in heart tissues collected from E15.5 embryos exposed to maternal ND or HFD (n = 3–4 embryos; mean ± SD). ∗p < 0.05 (two-sided unpaired Student’s t test).
Next, we explored the molecular pathways affected by maternal HFD by conducting the assay for transposase-accessible chromatin using sequencing (ATAC-seq) on cardiac tissues from E15.5 embryos. Our systematic analysis yielded 644 genomic regions with differential enrichment, predominantly enriched at promoters and enhancers (Figure 2E; Table S1). Subsequent analysis using genomic regions enrichment of annotations tool (GREAT) indicated that maternal HFD primarily influenced genes involved in chromosome organization, actomyosin organization and muscle development in the offspring (Figure 2F). To ascertain if HFD-induced chromatin accessibility changes precipitate transcriptional modifications, we focused on Tnnt2 and Ppara, two genes that are identified as important downstream targets of Tet protein in our previous publication20 and are pivotal for myocardial development and metabolic processes.21,22 ATAC-seq and hMeDIP-qPCR analysis unveiled a notable reduction of chromatin accessibility and 5hmC levels, respectively, at the promoter of these two genes (Figures 2G and 2H). Real-time quantitative PCR (qPCR) analysis further confirmed a reduction in the expression of Tnnt2 and Ppara genes in the embryo hearts subjected to HFD compared to those on a normal diet (Figure 2I). Taken together, our findings suggest maternal HFD exposure remodels the epigenome and leads to alterations in transcriptional outputs to ultimately affect cardiac development in offspring.
Maternal vitamin C intake attenuates HFD-induced cardiac developmental defects in the offspring
Vitamin C is known to augment the activity of Tet enzymes by ensuring the maintenance of iron as a cofactor in its reduced state.9,10 Its role as an antioxidant in mitigating fat accumulation is also well documented.23,24 To examine if vitamin C could have beneficial effects on dams and their offsprings, we treated maternal mice with vitamin C when they were exposed to either a normal or HFD (Figure 3A). We noted that vitamin C supplementation mitigated weight gain in dams subjected to a HFD, with negligible effects on dams consuming a normal diet (Figure 3B). A similar trend was also observed in the embryos collected from the dams exposed to the corresponding conditions (Figure 3C). This protective effect of vitamin C was further evidenced by improved glucose tolerance test (GTT) outcomes, where vitamin C administration lowered maternal glucose levels (Figure 3D). This observation is consistent with the immunohistochemical analysis of Tet, 5hmC, and 5mC levels in liver tissue collected from the dam (Figure S3A). Vitamin C treatment partially restored Tet and 5hmC levels in the liver tissues from the dams exposed to HFD. Collectively, these findings indicate that vitamin C supplementation can partially counteract the adverse metabolic perturbations induced by a HFD during pregnancy in female mice.
Figure 3.
Vitamin C ameliorates maternal HFD-induced glucose intolerance and ventricle wall thinning in embryos
(A) Experimental timeline of maternal vitamin C treatment under ND or HDF conditions.
(B) Changes in body weight of dams on ND or HFD, with and without vitamin C (ViC) treatment (n = 7–8 dams per group; mean ± SD). ∗∗∗p < 0.001 (two-sided unpaired Student’s t test).
(C) Embryos weights in maternal ND or HFD groups, with or without vitamin C (ViC) treatment (n = 17–23 embryos per group; mean ± SD; ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05; two-sided unpaired Student’s t test).
(D) Glucose tolerance test (GTT) results from dams on ND or HFD, with and vitamin C (ViC) treatment (n = 3–7 dams/group; mean ± SD). ∗∗∗p < 0.001 (two-sided unpaired Student’s t test).
(E and F) Quantification of levels of reduced (Fe2+), oxidized (Fe3+), and total iron in placenta (E, left), embryonic heart tissue (E, middle), embryonic liver (E, right), and maternal serum (F), under maternal ND or HFD conditions, with and without vitamin C treatment (n = 3–5/group; mean ± SD). ∗∗p < 0.01, ∗∗∗p < 0.001 (two-sided unpaired Student’s t test).
(G) Representative H&E images (left) and quantification (right) of ventricular wall thickness of embryonic heart tissues at E15.5 from dams on ND and HFD, with or without vitamin C treatment (n = 4–10 embryos/group; mean ± SD. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (two-sided unpaired Student’s t test). Scale bar, 100 μm.
(H) Representative IHC images (left) and quantification (right) of 5hmC levels in the trabecular area of embryonic heart tissue at E15.5 from dams on HFD, with or without vitamin C treatment (n = 35–45 cells/group; mean ± SD). ∗∗∗∗p < 0.0001 (two-sided unpaired Student’s t test). Scale bar, 40 μm.
(I) Real-time qPCR analysis of Ppara expression in heart tissues collected from E15.5 embryos exposed to maternal HFD with and without vitamin C treatment (n = 3–4 embryos; mean ± SD). ∗∗p < 0.001 (two-sided unpaired Student’s t test).
Vitamin C acts as an antioxidant, essential for sustaining iron in its reduced state, which is a crucial cofactor for dioxygenases, including TET proteins.9,10,13 We subsequently quantified the various forms of iron in the embryos, placenta, and serum from dams (Figures 3E and 3F). Notably, there was a significant decrease in reduced iron in the serum of dams subjected to a HFD compared to those on a normal diet (Figure 3F). Vitamin C supplementation restored levels of reduced iron in dams on a HFD but did not affect those on a ND (Figure 3F). However, vitamin C had a minimal impact on placental iron levels (Figure 3E). Notably, in the cardiac tissues of embryos from HFD-exposed mothers, we observed a pronounced reduction in iron content (both reduced and oxidized forms) when compared to the normal diet group (Figure 3E). Vitamin C treatment was found to elevate iron levels (both forms) in embryonic hearts under both HFD and ND conditions relative to their respective controls (Figure 3E, middle). To determine the tissue specificity of these effects, we further assessed iron levels in the liver tissues of the same embryos and observed no significant differences (Figure 3E, right). These findings suggest that maternal HFD exposure may preferentially compromise iron bioavailability in both dams and embryos, while vitamin C supplementation has the potential to counteract iron loss associated with HFD treatment.
We next moved on to investigate the effects of maternal vitamin C intake on cardiac development in offspring. We discovered that vitamin C supplementation in mothers significantly mitigated the incidence of myocardial ventricular NCC in offspring subject to a HFD, as evidenced by normalized ventricular wall thickness in E15.5 embryos (Figure 3G). Furthermore, vitamin C markedly increased 5hmC levels (Figures 3H and S3B) and Ppara gene expression (Figure 3I) in E15.5 cardiac tissues of embryos from HFD-fed dams. Collectively, these findings underscore the protective role of vitamin C on heart development in embryos derived from HFD-challenged female mice.
Tet proteins mediate the protective effects of vitamin C in offspring
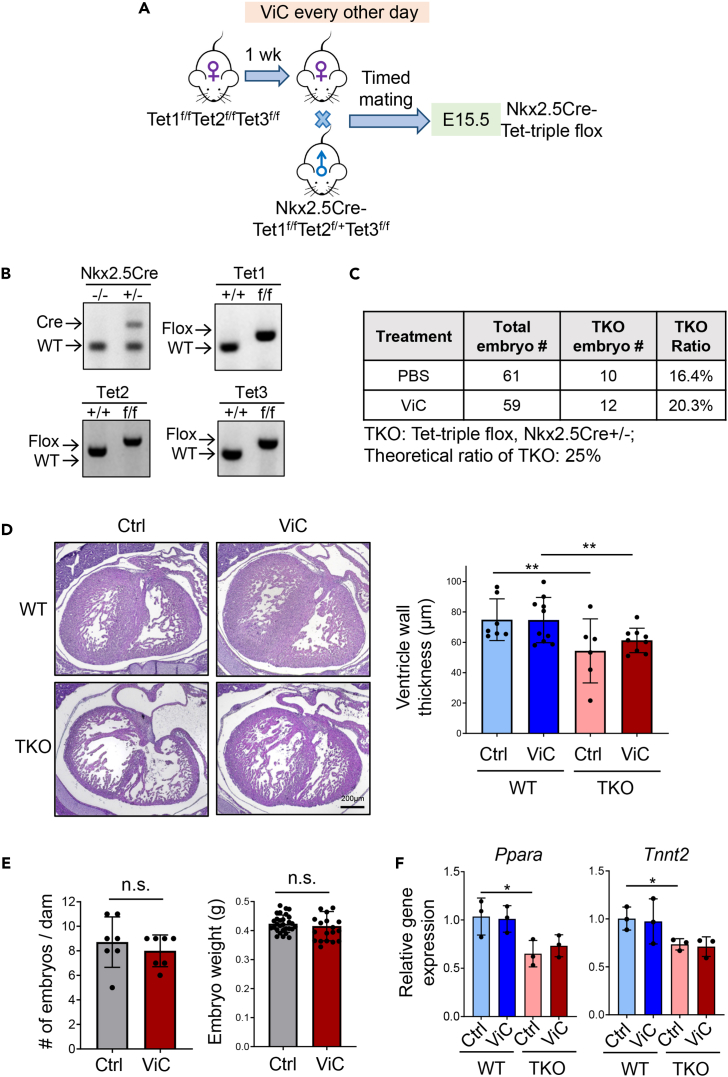
We next moved on to explore the essential role of TET enzymes in mediating the aforementioned beneficial effects of vitamin C during maternal HFD treatment. In our previous study, we generated a Nkx2.5Cre-Tet2f/fTet3f/f (Tet-DKO) mouse model to specifically deplete Tet enzymes in cardiomyocytes.20 Based on this Tet-DKO line, we further generated a Nkx2.5Cre-Tet1f/fTet2f/fTet3f/f (Tet-TKO) mouse model with a clean Tet-null background (Figures 4A and 4B). Similar to the Tet-DKO mouse model, ablation of all three Tet proteins led to embryonic lethality, with a notable decline in embryo count at E15.5 (Figure 4C). We also observed a ventricular NCC phenotype in the myocardial tissue of Tet-TKO embryos, analogous to that observed in offspring from HFD-exposed mothers (Figure 4D). Unlike the WT mice, we observed that maternal vitamin C treatment had minor effects on embryo numbers and weight compared with the untreated group for the Tet-TKO mice (Figure 4E). Furthermore, histological assessments indicated that the ameliorative effect of maternal vitamin C on the NCC phenotype was not recapitulated in Tet-TKO embryos, unlike in WT progeny subjected to maternal HFD (Figure 4D). At the mRNA level, deletion of Tet protein reduces expression of Ppara and Tnnt2, which is consistent with an earlier report,20 while vitamin C treatment failed to restore the expression of these two genes in Tet-deficient embryos (Figure 4F). Together, findings from our genetic and functional studies imply that while maternal vitamin C supplementation may modulate the enzymatic functions of various proteins,25 the cardioprotective outcomes of vitamin C treatment in the context of maternal HFD exposure are, at least in part, attributable to the enhanced activity of TET enzymes.
Figure 4.
Tet proteins contribute to vitamin C-mediated cardioprotection during development in maternal HFD-exposed embryos
(A) Schematic of the experimental setup for administering vitamin C to myocardium-specific Tet-TKO mouse models under a normal diet (ND).
(B) Genotyping results confirm cardiac specific deletion of Tet genes in embryos.
(C) Total count of Tet-TKO embryos obtained from dams treated with PBS or vitamin C.
(D) Representative H&E-stained sections of embryonic hearts (left) and quantification of ventricular wall thickness of heart tissues (right) collected from E15.5 Tet-TKO embryos, with or without vitamin C treatment (n = 6–10 embryos/group; mean ± SD. ∗∗p < 0.01 (two-sided unpaired Student’s t test). Scale bar, 200 μm.
(E) Quantification of the number and weight of embryos collected from Tet-TKO dams treated with or without vitamin C (n = 7 dams/group, n = 20–29 embryos/group; mean ± SD). n.s., no statistical significance (two-sided unpaired Student’s t test).
(F) Real-time qPCR analysis on Ppara and Tnnt2 expression in WT and Tet-TKO heart tissues collected from E15.5 embryos treated with or without vitamin C (n = 3 embryos; mean ± SD; ∗p < 0.05; two-sided unpaired Student’s t test).
Discussion
In this study, we reported a suppressive effect of a maternal HFD on both the expression and enzymatic function of TET proteins during embryonic cardiac development. Notably, embryos subjected to a maternal HFD displayed a ventricular NCC phenotype, consistent with our prior findings in embryos with cardiac-specific Tet ablation.20 These observations suggest that Tet loss-of-function might contribute to maternal HFD-induced cardiac developmental defect. Our study demonstrates that maternal HFD leads to diminished expression of Tet1 and Tet3 proteins in embryonic heart tissues without affecting their mRNA levels. This aligns with existing literature, such as a study indicating that AMPK phosphorylation enhances TET2 protein stability.26 The decrease in Tet1 and Tet3 protein levels may stem from attenuated AMPK-mediated phosphorylation upon maternal HFD exposure. Further biochemical investigation is warranted to elucidate the interplay between AMPK and Tet1/3 following maternal HFD exposure.
Our previous studies indicated that targeted ablation of Tet in cardiac tissues alters the transcription of pivotal genes involved in myocardial development, as well as genes crucial for metabolic regulation.20 In the current study, we found a marked reduction in chromatin accessibility and transcription of Ppara in embryonic heart tissues from dams subjected to a HFD. Remarkably, vitamin C supplementation was able to restore the expression of Ppara in these tissues. The gene Ppara encodes the peroxisome proliferator-activated receptor alpha (PPARA), a key transcription factor that regulates lipid metabolism.27 A growing body of evidence suggests a strong association between dysregulated lipid metabolism and cardiovascular diseases (CVDs).28,29 Inhibiting fatty-acid oxidation has been reported to benefit heart regeneration.30 Our study provides direct evidence to support that maternal exposure to HFD can attenuate TET function and reshape the chromatin accessibility to alter gene expression. In addition, PPARA is a recognized therapeutic target for various metabolic disorders, including hyperlipidemia and non-alcoholic fatty liver disease.31,32,33 PPARA agonists are under active clinical trials for NAFLD treatment. Hence, the potential for PPARA agonists to mitigate the adverse effects of maternal HFD warrants further exploration.
Our data suggested a protective role of vitamin C during heart development in embryos exposed to HFD during pregnancy. Vitamin C supplementation was observed to regulate body weight and glucose metabolism in mothers on a HFD, likely attributed to its antioxidant capacity that helps prevent excessive fat accumulation. In parallel, we observed restoration of Tet enzymatic activity in embryonic heart tissues under maternal vitamin C treatment, which is possibly due to increased iron levels at the reduced form in embryonic heart tissues. Vitamin C-induced restoration of Tet function appears to be a key mechanism in safeguarding cardiac development during gestational HFD exposure. On the other hand, a previous study suggested a suppressive effect of HFD on vitamin C levels in guinea pigs, indicating a negative regulatory mechanism between HFD and vitamin C.34
Worth noting is that previous studies reported hypertrophic effects, including increased heart weight, body weight, thicker interventricular septum, and enlarged cardiomyocytes, in the heart tissues collected from progenies under maternal metabolic dysregulation, including diabetes, obesity, or HFD exposure conditions.35,36 In our study, we observed an increase in embryo size and heart size under maternal HFD exposure conditions, which aligns with previous reports. Other hypertrophic features were not observed in our study, which might be due to variations in the treatment conditions and species. These aspects could be explored further in future investigations. Nevertheless, maternal metabolic dysregulations are strongly associated with various congenital heart diseases.18 For example, maternal diabetes leads to fetal cardiomyopathy and neonatal hypertrophic cardiomyopathy.37,38 Therefore, maternal metabolic balance is essential for supporting normal embryonic development.
In summary, findings made from the current study provide compelling evidence to support that a maternal HFD can alter the function of epigenetic regulators (such as the Tet family of dioxygenases) and reshape the chromatin accessibility, ultimately causing developmental defects in embryos, as exemplified by ventricular NCC phenotypes described herein. From a translational perspective, our study offers proof-of-concept that vitamin C can be therapeutically exploited to modulate the enzymatic activities of epigenetic regulators, thereby exerting a protective effect to safeguard proper heart development during embryogenesis.
Limitations of the study
In the present study, we demonstrated an intimate connection between maternal diet, specifically HFD or vitamin C, and epigenetic modifications during early heart development. However, some limitations need to be addressed in future studies. First, we did not consider the sex of the embryos when analyzing the data. Male and female offspring might respond differently to maternal HFD exposure. Secondly, current techniques are limited in assessing heart function during the embryonic stage. While we observed a NCC phenotype via histological analysis, the effects of maternal HFD on heart function in embryos remain unknown. Lastly, vitamin C has multiple beneficial effects on the dam and embryos beyond promoting Tet catalytic function. Future studies are needed to further clarify the interplay between maternal metabolism and epigenetic remodeling during early heart development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-TET1 (clone 5D8) | Dr. Leonhardt Heinrich | N/A |
| Rabbit polyclonal anti-TET1 | ABclonal | Cat# A1506; RRID: AB_2761941 |
| Rabbit polyclonal anti-TET2 | Abcam | Cat# Ab124297; RRID: AB_2722695 |
| Rabbit polyclonal anti-TET3 | GeneTex | Cat# GTX121453; RRID: AB_10723106 |
| Mouse monoclonal anti-5-methylcytosine (clone 33D3) | Sigma-Aldrich | Cat# MABE146; RRID: AB_10863148 |
| Rabbit polyclonal anti-5-hydroxymethylcytosine | Active Motif | Cat# 39769; RRID: AB_10013602 |
| Mouse monoclonal anti-alpha Tubulin (clone DM1A) | Santa Cruz | Cat# sc-32293; RRID: AB_628412 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling | Cat# 7074; RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling | Cat# 7076; RRID: AB_330924 |
| Rabbit IgG Isotype Control | Thermo Scientific | Cat# 31235; RRID: AB_243593 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | T5648 |
| L-Ascorbic acid | Sigma-Aldrich | A4403 |
| Rodent Diet With 60 kcal% Fat | Research Diets | D12492i |
| Rodent Diet With 10 kcal% Fat | Research Diets | D12450Ji |
| Glucose Solution | Gibco | A2494001 |
| Novolin R Insulin 100u/ml | Novolin | 183311 |
| Hematoxylin | Poly Scientific R+D Corp | S212a |
| Eosin | Poly Scientific R+D Corp | S176 |
| Antigen Unmasking Solution, Citrate-Based | Vector Laboratories | H-3300-250 |
| Xpert Protease Inhibitor Cocktail Solution (100x) | GenDEPOT | P3100 |
| Critical commercial assays | ||
| EmeraldAmp GT PCR Master Mix | TaKaRa | RR310B |
| ImmPRES HRP Horse Anti-Rabbit IgG Polymer Detection Kit, Peroxidase | Vector Laboratories | MP-7401 |
| ImmPRESS HRP Goat Anti-Mouse IgG Polymer Detection Kit, Peroxidase | Vector Laboratories | MP-7452 |
| ImmPRESS HRP Goat Anti-Rat IgG, Mouse adsorbed Polymer Detection Kit, Peroxidase | Vector Laboratories | MP-7444 |
| DAB Substrate Kit, Peroxidase (HRP) | Vector Laboratories | SK-4100 |
| NucleoSpin RNA Plus Kit | MACHEREY-NAGEL | 740984 |
| PrimeScript RT Master Mix | TaKaRa | RR036B |
| 2X Universal SYBR Green Fast qPCR Mix | ABclonal | RK21203 |
| Iron Assay Kit | Sigma-Aldrich | MAK025 |
| ATAC-Seq Kit | Active Motif | 53150 |
| Agilent High Sensitivity DNA kit | Agilent | 5067-4626 |
| NucleoSpin Tissue, Mini kit for DNA from cells and tissue | MACHEREY-NAGEL | 740952 |
| West-Q Pico Dura ECL Solution | GenDEPOT | W3653 |
| Deposited data | ||
| ATAC-seq Data | This paper | GSE252640 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 |
| Mouse: B6.129S1(SJL)-Nkx2-5tm2(cre)Rph/J | The Jackson Laboratory | 024637 |
| Oligonucleotides | ||
| Pparα qPCR primer forward | IDT | AGAGCCCCATCTGTCCTCTC |
| Pparα qPCR primer reverse | IDT | ACTGGTAGTCTGCAAAACCAAA |
| Tnnt2 qPCR primer forward | IDT | CAGAGGAGGCCAACGTAGAAG |
| Tnnt2 qPCR primer reverse | IDT | CTCCATCGGGGATCTTGGGT |
| Gapdh qPCR primer forward | IDT | TGACCTCAACTACATGGTCTACA |
| Gapdh qPCR primer reverse | IDT | CTTCCCATTCTCGGCCTTG |
| Tet1 qPCR primer forward | IDT | GAGCCTGTTCCTCGATGTGG |
| Tet1 qPCR primer reverse | IDT | CAAACCCACCTGAGGCTGTT |
| Tet2 qPCR primer forward | IDT | TGCAAAACCTGGCTACTGTC |
| Tet2 qPCR primer reverse | IDT | AACATGCAGTGACTCCTGAG |
| Tet3 qPCR primer forward | IDT | TCCGGATTGAGAAGGTCATC |
| Tet3 qPCR primer reverse | IDT | CCAGGCCAGGATCAAGATAA |
| Pparα hMeDIP qPCR primer forward | IDT | TTGGGCAGTCCCTTCACCTA |
| Pparα hMeDIP qPCR primer reverse | IDT | GATGCCCATTTAGTGCCTCG |
| Tnnt2 hMeDIP qPCR primer forward | IDT | CCAGGAAATCCATGGCCCTT |
| Tnnt2 hMeDIP qPCR primer reverse | IDT | ATGCAAGCACTCTCCACTCC |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism 9 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| BioRender | BioRender | https://www.biorender.com/ |
| Trim Galore v0.5.0 | The Babraham Institute | https://github.com/FelixKrueger/TrimGalore |
| Bowtie2 | Johns Hopkins University | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Genrich v0.5 | Dr. John M. Gaspar | https://github.com/jsh58/Genrich |
| Bedtools | Quinlan Laboratory | https://github.com/arq5x/bedtools2 |
| Deseq2 | Dr. Michael Love | https://www.bioconductor.org/packages/release/bioc/vignettes/DESeq2/inst/doc/DESeq2.html |
| GREAT | Bejerano Laboratory | http://great.stanford.edu/public/html/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yun Huang (yun.huang@tamu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The sequencing datasets were deposited into the NCBI BioProject under the accession number of GSE252640.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animal studies were approved by the Institutional Animal Care Use Committee (IACUC) of the Institute of Biosciences and Technology at Texas A&M University. Mice were housed in a temperature- and humidity-controlled room under a 12 h light/dark cycle (lights on at 06:00), with ad libitum access to food and water. Littermates of the same sex were randomly assigned to experimental groups. Most mouse strains bear a C57BL/6 genetic background unless otherwise noted. Nkx2.5-Cre (The Jackson Laboratory; Catalog No. 024637) mouse strain was obtained from The Jackson Laboratory. All strains were housed in the same room, with the same cage content. For genotyping, mouse tails were cut and boiled in 50 mM NaOH for 45 min and then neutralized in 10 mM Tris–HCl at pH 5.4. PCR was carried out using the EmeraldAmp GT PCR Master Mix (TaKaRa). For the maternal high-fat diet experiments, naïve female mice at 8–10 weeks old were fed with high-fat diet (60 kcal% fat, D12492i, Research Diets, USA) for 9 weeks before timed mating. The age-matched mice were fed normal diet (10 kcal% fat, D12450Ji, Research Diets) for 9 weeks before timed mating and used as control. Timed mating was applied and the day when a vaginal plug was found was defined as E0.5. Vitamin C (0.5g / kg) was applied every other day via intraperitoneal (i.p.) injection one week before timed mating till the embryos at E15.5 days. The number of animals used in each experiment was indicated in the corresponding figure legends.
Method details
Glucose tolerance test (GTT)
GTT experiments were performed on dams subjected to either HFD or ND when embryos reaching E12.5 days old. Following an overnight fast, the mice received an intraperitoneal injection of glucose at a dose of 1.5 g / kg body weight the next morning. Blood glucose levels were then measured at 0, 15, 30, 60, 90 and 120 mins post-injection using the True Metrix Blood Glucose Test Meter (TRUE METRIX).
Antibodies
An anti-Tet1 antibody was kindly provided by Dr. Leonhardt Heinrich (1:100), anti-Tet2 (Abcam ab124297, 1:100), anti-Tet3 (GeneTex GTX121453, 1:200), anti-5mC (Millipore MABE146, 1:1000) and anti-5hmC (Active Motif 39769, 1: 40,000) were purchased from commercial sources for immunohistochemistry analysis. Anti-rabbit IgG, HRP-linked Antibody (Cell Signaling #7074), anti-mouse IgG, HRP-linked Antibody (Cell Signaling #7076), anti-5hmC (1:4000, Active Motif, #39769), anti-Tet1 (ABclonal A1506, 1:800) and anti-α-Tubulin (Santa Cruz Biotechnology sc-32293, 1:1000) were purchased from commercial sources for Western blot or dot-blot assays.
Histological analyses
Hematoxylin-eosin (H&E) staining was performed to study the heart structure. Briefly, all embryos were dissected in PBS and fixed overnight in 4% PFA at 4°C. The next day dehydrated them with graded ethanol (from 70% to 100%), 100% xylene and embedded in paraffin. Slides were sectioned at the thickness of 7 μm and stained with hematoxylin and eosin. Results were imaged using a Nikon Eclipse Ci microscopy. ImageJ was used for the quantification.
Immunohistochemistry staining
Slides were dewaxed for 3 min in xylene twice and then rehydrated in a graded series of ethanol (100% to 70%). The antigen was retrieved by boiling slides in Antigen Unmasking Solution, Citrate-Based (Vector Laboratories) for 15 min. For DNA modifications staining, slides were treated with 2 N HCl for 30 min to expose the epitopes and then neutralized with 100 mM Tris–HCl (pH 8.5) for 10 min. ImmPRES HRP Reagent Kit (Vector Laboratories) was used to block background and incubate antibodies against target proteins or DNA modifications. Slides were rinsed with PBS with 0.05% Tween-20 and incubated with secondary antibodies. A DAB peroxidase substrate kit (Vector Laboratories) was used for signal detection. IHC stained slides were imaged using a Nikon Eclipse Ci microscopy.
RNA extraction
Whole ventricles from embryos at E15.5 were dissected and immediately snap-frozen. The tissues were then homogenized using a Fisherbrand Bead Mill 4 Mini Homogenizer. Subsequently, RNA was extracted using the NucleoSpin RNA Plus kit (MACHEREY-NAGEL), ensuring the removal of DNA contamination.
Real-time quantitative PCR (qPCR)
Purified total RNA (100 ng to 500ng) was reverse transcribed into cDNA using the PrimeScript RT Master Mix (TaKaRa). Real-time quantitative PCR was performed with a QuantStudio3 (Thermo Fisher Scientific) instrument using a 2X Universal SYBR Green Fast qPCR Mix kit (ABclonal). A three-step cycling program was used with a 5-min denaturation at 95°C, then proceeded through 30 cycles, each consisting of a 10-sec denaturation at 95°C, a 20-second annealing at 60°C, and a 30-second extension at 72°C. The mRNA levels of the target genes were quantified relative to GAPDH as the normalization control. All the primers were obtained from Integrated DNA Technologies.
Iron measurement
Both ferrous (Fe2+) and ferric iron (Fe3+) contents were measured using the Iron Assay Kit (Sigma-Aldrich) by following the manufacturer’s instructions. OD values were measured at 593 nm and the iron concentration was calculated using standard curve. The concentrations of iron in the samples were normalized to the number or the weight of the tissue.
ATAC-seq library preparation and data analysis
ATAC-seq libray preparation was performed using the ATAC-seq kit (Active motif, 53150) in accordance with the manufacturer’s instructions. Briefly, prior to ATAC-seq, embryonic heart tissues were first rinsed with ice-old PBS. Nuclei were then extracted using a dounce homogenizer in ATAC lysis buffer. Subsequently, the homogenates were passed through a 40 μm mesh strainer to filter the samples, followed by counting of the isolated nuclei. Tagmentation, amplification, and subsequent purification of the DNA libraries were carried out by following the manufacturer's instructions. The quality of purified DNA libraries was assessed by the Agilent High Sensitivity DNA kit (Agilent Technologies). Libraries were pooled and sequenced on an Illumina HiSeq sequencer by Novogene.
Adaptor trimming of raw reads was performed by TrimGalore v0.5.0 with default parameters. High-quality reads (Q ≥ 20) were uniquely aligned to the mm10 reference genome using Bowtie2 with the ‘--very-sensitive’ option. Only uniquely mapped reads were finally extracted for the downstream analysis. The resulting alignment of each sample (with two biological replicates) was analyzed by Genrich v.0.5 (https://github.com/jsh58/Genrich) with ATAC-seq mode (option: -j, -q 0.05, -d 150). The options to remove PCR duplicates (-r) and to discard alignments to chrM (-e chrM) were applied to call chromatin accessible peak regions. Bedtools merge was used to count the reads that fell into non-overlapped peak regions, and differential chromatin accessible regions were detected using DESeq2 with normalized peak signals (fold change ≥ 2; FDR < 0.05). GREAT [39] analysis was used to perform the functional annotation of differential chromatin accessible regions.
Hydroxymethylated DNA immunoprecipitation (hMeDIP)
E15.5 ventricles were dissected and then homogenized using a Fisherbrand Bead Mill 4 Mini Homogenizer. Genomic DNA was extracted using the NucleoSpin Tissue, Mini kit for DNA from cells and tissue (MACHEREY-NAGEL). DNA was sonicated with Covaris (Peak Incident power 50W, Duty factor 10%, cycles per burst 200cpb, treatment time 70s) to obtain 300–500-bp fragments. Sonicated DNA was denatured in 0.4 M NaOH, 10 mM EDTA at 95°C for 10 min, then neutralized with ice-cold 2 M ammonium acetate (pH 7.0). Then the denatured DNA were immunoprecipitated with antibodies against 5hmC (Active Motif, #39769) or Rabbit IgG Isotype Control (Thermo Scientific 31235) overnight at 4°C in IP buffer (100 mM Na-Phosphate buffer pH 7.0, 1.4 M NaCl, 0.5% Triton X-100). DNA was purified and used for real-time qPCR.
Dot-blot assay
Purified genomic DNA were denatured and loaded on a nitrocellulose membrane at two-fold serial dilutions using an assembled Bio-Dot apparatus (Bio-Rad) according to the manufacturer’s instructions. The membrane was washed, air-dried and vacuum-baked at 80°C for 2 h. Then the DNA hybridized membrane was blocked with 5% Bovine Serum Albumin in TBST for 1 h at room temperature and incubated with an anti-5hmC antibody (1:4000, Active Motif, #39769) overnight at 4°C, followed by incubation with a secondary antibody (Anti-rabbit IgG, HRP-linked Antibody, Cell Signaling #7074) at room temperature for 1 hour. West-Q Pico Dura ECL Solution (GenDEPOT) was used for development, and the antigen–antibody complexes were detected using the ChemiDoc Imaging system (Bio-Rad). The membrane was washed with ddH2O briefly and then stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) to confirm the total amounts of loaded DNA samples.
Protein extraction and Western blot analysis
Tissues were rinsed with PBS twice and lysed in a radio-immunoprecipitation assay (RIPA) buffer (150mM NaCl, 1% TritonX-100, 0.1% SDS, 0.5% Sodium deoxycholate, 50mM Tris-HCl pH8.0), supplemented with Xpert Protease Inhibitor Cocktail Solution (100X) (GenDEPOT) on ice for 10 minutes. Cell debris was removed by centrifugation at 12,000 x g at 4°C for 10 minutes, collect the supernatant. The samples were loaded onto 7% SDS-PAGE gels after mixing with SDS loading buffer (100 mM Tris-HCl, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM DTT, pH 6.8) and denaturing at 100°C for 10 minutes. Proteins were transferred onto Nitrocellulose membranes (Millipore) and blocked in 5% Bovine Serum Albumin (GenDEPOT) for 1 hour at room temperature. Membranes were then incubated with the primary antibodies at 4°C overnight, followed by incubation with a secondary antibody at room temperature for 1 hour. West-Q Pico Dura ECL Solution (GenDEPOT) was used for development, and the antigen–antibody complexes were detected using the ChemiDoc Imaging system (Bio-Rad).
Quantification and statistical analysis
The statistical analysis was performed using unpaired student’s t-test using GraphPad Prism version 9.0. All tests were two-tailed and a p value of less than 0.05 was considered significant. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001. All values are represented as the mean ± SD.
Acknowledgments
This work was supported by the National Institutes of Health (R35HL166557, R01CA211773, and R01DK132286 to Y.H., R01CA232017 to Y.Z.), the Welch Foundation (BE-1913-20220331 to Y.Z.), and the Cancer Prevention and Research Institute of Texas (RP210070 to Y.Z.).
Author contributions
Y.H. and Y.Z. directed and supervised the project. Y.Y.H. performed most animal-related work, molecular characterization, and sequencing library preparation. L.R. performed all the bioinformatics analysis on high-throughput sequencing data. S.F. generated Tet-TKO mouse model, M.C. and A.S. supported histology analysis. Y.H. and Y.Z. wrote the manuscript. All the authors participated in the discussion, data interpretation and manuscript editing or discussion.
Declaration of interests
The authors declare no competing interests.
Published: July 31, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110631.
Supplemental information
References
- 1.Urbonaite G., Knyzeliene A., Bunn F.S., Smalskys A., Neniskyte U. The impact of maternal high-fat diet on offspring neurodevelopment. Front. Neurosci. 2022;16 doi: 10.3389/fnins.2022.909762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandes D.J., Spring S., Roy A.R., Qiu L.R., Yee Y., Nieman B.J., Lerch J.P., Palmert M.R. Exposure to maternal high-fat diet induces extensive changes in the brain of adult offspring. Transl. Psychiatry. 2021;11:149. doi: 10.1038/s41398-021-01274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Xiao X., Zheng J., Li M., Yu M., Ping F., Wang T., Wang X. A Maternal High-Fat Diet Induces DNA Methylation Changes That Contribute to Glucose Intolerance in Offspring. Front. Endocrinol. 2019;10:871. doi: 10.3389/fendo.2019.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keleher M.R., Zaidi R., Hicks L., Shah S., Xing X., Li D., Wang T., Cheverud J.M. A high-fat diet alters genome-wide DNA methylation and gene expression in SM/J mice. BMC Genom. 2018;19:888. doi: 10.1186/s12864-018-5327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mdaki K.S., Larsen T.D., Wachal A.L., Schimelpfenig M.D., Weaver L.J., Dooyema S.D.R., Louwagie E.J., Baack M.L. Maternal high-fat diet impairs cardiac function in offspring of diabetic pregnancy through metabolic stress and mitochondrial dysfunction. Am. J. Physiol. Heart Circ. Physiol. 2016;310:H681–H692. doi: 10.1152/ajpheart.00795.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai Z., Ramesh V., Locasale J.W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat. Rev. Genet. 2020;21:737–753. doi: 10.1038/s41576-020-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etchegaray J.P., Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol. Cell. 2016;62:695–711. doi: 10.1016/j.molcel.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martínez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A., et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z., et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- 11.Chen B., Du Y.R., Zhu H., Sun M.L., Wang C., Cheng Y., Pang H., Ding G., Gao J., Tan Y., et al. Maternal inheritance of glucose intolerance via oocyte TET3 insufficiency. Nature. 2022;605:761–766. doi: 10.1038/s41586-022-04756-4. [DOI] [PubMed] [Google Scholar]
- 12.Yin R., Mao S.Q., Zhao B., Chong Z., Yang Y., Zhao C., Zhang D., Huang H., Gao J., Li Z., et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 13.Yuan Y., Liu C., Chen X., Sun Y., Xiong M., Fan Y., Petersen R.B., Chen H., Huang K., Zheng L. Vitamin C Inhibits the Metabolic Changes Induced by Tet1 Insufficiency Under High Fat Diet Stress. Mol. Nutr. Food Res. 2021;65 doi: 10.1002/mnfr.202100417. [DOI] [PubMed] [Google Scholar]
- 14.Byun S., Lee C.H., Jeong H., Kim H., Kwon H.M., Park S., Myung K., An J., Ko M. Loss of adipose TET proteins enhances beta-adrenergic responses and protects against obesity by epigenetic regulation of beta3-AR expression. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2205626119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J., Song J.H., Park J.H., Chung M.Y., Lee S.H., Jeon S.B., Park S.H., Hwang J.T., Choi H.K. Dnmt1/Tet2-mediated changes in Cmip methylation regulate the development of nonalcoholic fatty liver disease by controlling the Gbp2-Ppargamma-CD36 axis. Exp. Mol. Med. 2023;55:143–157. doi: 10.1038/s12276-022-00919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., Zhang Y., Zhuo Q., Tseng Y., Wang J., Ma Y., Zhang J., Liu J. TET1 promotes fatty acid oxidation and inhibits NAFLD progression by hydroxymethylation of PPARalpha promoter. Nutr. Metab. 2020;17:46. doi: 10.1186/s12986-020-00466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerf M.E. High Fat Programming and Cardiovascular Disease. Medicina. 2018;54 doi: 10.3390/medicina54050086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins R.T., 2nd, Yang W., Carmichael S.L., Bolin E.H., Nembhard W.N., Shaw G.M., National Birth Defects Prevention Study Maternal dietary fat intake and the risk of congenital heart defects in offspring. Pediatr. Res. 2020;88:804–809. doi: 10.1038/s41390-020-0813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddeek B., Mauduit C., Chehade H., Blin G., Liand M., Chindamo M., Benahmed M., Simeoni U. Long-term impact of maternal high-fat diet on offspring cardiac health: role of micro-RNA biogenesis. Cell Death Dis. 2019;5:71. doi: 10.1038/s41420-019-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang S., Li J., Xiao Y., Lee M., Guo L., Han W., Li T., Hill M.C., Hong T., Mo W., et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 2019;10:4297. doi: 10.1038/s41467-019-12325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hershberger R.E., Pinto J.R., Parks S.B., Kushner J.D., Li D., Ludwigsen S., Cowan J., Morales A., Parvatiyar M.S., Potter J.D. Clinical and functional characterization of TNNT2 mutations identified in patients with dilated cardiomyopathy. Circ. Cardiovasc. Genet. 2009;2:306–313. doi: 10.1161/CIRCGENETICS.108.846733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montaigne D., Butruille L., Staels B. PPAR control of metabolism and cardiovascular functions. Nat. Rev. Cardiol. 2021;18:809–823. doi: 10.1038/s41569-021-00569-6. [DOI] [PubMed] [Google Scholar]
- 23.Campión J., Milagro F.I., Fernandez D., Martinez J.A. Vitamin C supplementation influences body fat mass and steroidogenesis-related genes when fed a high-fat diet. Int. J. Vitam. Nutr. Res. 2008;78:87–95. doi: 10.1024/0300-9831.78.2.87. [DOI] [PubMed] [Google Scholar]
- 24.Jeon S., Lee J., Shin Y., Yoon M. Ascorbic acid reduces insulin resistance and pancreatic steatosis by regulating adipocyte hypertrophy in obese ovariectomized mice. Can. J. Physiol. Pharmacol. 2023;101:294–303. doi: 10.1139/cjpp-2022-0339. [DOI] [PubMed] [Google Scholar]
- 25.Zhitkovich A. Nuclear and Cytoplasmic Functions of Vitamin C. Chem. Res. Toxicol. 2020;33:2515–2526. doi: 10.1021/acs.chemrestox.0c00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu D., Hu D., Chen H., Shi G., Fetahu I.S., Wu F., Rabidou K., Fang R., Tan L., Xu S., et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature. 2018;559:637–641. doi: 10.1038/s41586-018-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga T., Czimmerer Z., Nagy L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta. 2011;1812:1007–1022. doi: 10.1016/j.bbadis.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldberg I.J., Trent C.M., Schulze P.C. Lipid metabolism and toxicity in the heart. Cell Metabol. 2012;15:805–812. doi: 10.1016/j.cmet.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan A., Xie G., Ding X., Wang Y., Guo L. Effects of Lipid Overload on Heart in Metabolic Diseases. Horm. Metab. Res. 2021;53:771–778. doi: 10.1055/a-1693-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X., Wu F., Günther S., Looso M., Kuenne C., Zhang T., Wiesnet M., Klatt S., Zukunft S., Fleming I., et al. Inhibition of fatty acid oxidation enables heart regeneration in adult mice. Nature. 2023;622:619–626. doi: 10.1038/s41586-023-06585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staels B., Dallongeville J., Auwerx J., Schoonjans K., Leitersdorf E., Fruchart J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation. 1998;98:2088–2093. doi: 10.1161/01.cir.98.19.2088. [DOI] [PubMed] [Google Scholar]
- 32.Rotman Y., Sanyal A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66:180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 33.Khera A.V., Millar J.S., Ruotolo G., Wang M.D., Rader D.J. Potent peroxisome proliferator-activated receptor-alpha agonist treatment increases cholesterol efflux capacity in humans with the metabolic syndrome. Eur. Heart J. 2015;36:3020–3022. doi: 10.1093/eurheartj/ehv291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frikke-Schmidt H., Tveden-Nyborg P., Birck M.M., Lykkesfeldt J. High dietary fat and cholesterol exacerbates chronic vitamin C deficiency in guinea pigs. Br. J. Nutr. 2011;105:54–61. doi: 10.1017/S0007114510003077. [DOI] [PubMed] [Google Scholar]
- 35.Vaughan O.R., Rosario F.J., Chan J., Cox L.A., Ferchaud-Roucher V., Zemski-Berry K.A., Reusch J.E.B., Keller A.C., Powell T.L., Jansson T. Maternal obesity causes fetal cardiac hypertrophy and alters adult offspring myocardial metabolism in mice. J. Physiol. 2022;600:3169–3191. doi: 10.1113/JP282462. [DOI] [PubMed] [Google Scholar]
- 36.Fan X., Turdi S., Ford S.P., Hua Y., Nijland M.J., Zhu M., Nathanielsz P.W., Ren J. Influence of gestational overfeeding on cardiac morphometry and hypertrophic protein markers in fetal sheep. J. Nutr. Biochem. 2011;22:30–37. doi: 10.1016/j.jnutbio.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Biltagi M., El Razaky O., El Amrousy D. Cardiac changes in infants of diabetic mothers. World J. Diabetes. 2021;12:1233–1247. doi: 10.4239/wjd.v12.i8.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F., Han S., Fang L., Lin X. A fetal rat model of ventricular noncompaction caused by intrauterine hyperglycemia. Cardiovasc. Pathol. 2024;69 doi: 10.1016/j.carpath.2023.107601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The sequencing datasets were deposited into the NCBI BioProject under the accession number of GSE252640.
-
•
This study does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.