Significance
Mitotic chromosome condensation is an essential process to transmit replicated chromosomes into two daughter cells during cell division. To study the underlying physical principles of this process, we focused on depletion attraction/macromolecular crowding, which is a force that attracts large structures in crowded cell environments. Using special light microscopy, which can image the molecular density of cellular environments, we found that crowding around chromosomes increases during cell division. In vitro, higher concentrations of macromolecules condense chromatin and make it stiffer and more solid-like. Our results suggest that the rise in depletion attraction renders chromosomes more rigid, ensuring accurate chromosome transmission during cell division.
Keywords: OI-DIC, mitotic chromosome condensation, depletion attraction, chromatin, liquid droplets
Abstract
Genomic information must be faithfully transmitted into two daughter cells during mitosis. To ensure the transmission process, interphase chromatin is further condensed into mitotic chromosomes. Although protein factors like condensins and topoisomerase IIα are involved in the assembly of mitotic chromosomes, the physical bases of the condensation process remain unclear. Depletion attraction/macromolecular crowding, an effective attractive force that arises between large structures in crowded environments around chromosomes, may contribute to the condensation process. To approach this issue, we investigated the “chromosome milieu” during mitosis of living human cells using an orientation-independent-differential interference contrast module combined with a confocal laser scanning microscope, which is capable of precisely mapping optical path differences and estimating molecular densities. We found that the molecular density surrounding chromosomes increased with the progression from prophase to anaphase, concurring with chromosome condensation. However, the molecular density went down in telophase, when chromosome decondensation began. Changes in the molecular density around chromosomes by hypotonic or hypertonic treatment consistently altered the condensation levels of chromosomes. In vitro, native chromatin was converted into liquid droplets of chromatin in the presence of cations and a macromolecular crowder. Additional crowder made the chromatin droplets stiffer and more solid-like. These results suggest that a transient rise in depletion attraction, likely triggered by the relocation of macromolecules (proteins, RNAs, and others) via nuclear envelope breakdown and by a subsequent decrease in cell volumes, contributes to mitotic chromosome condensation, shedding light on a different aspect of the condensation mechanism in living human cells.
Negatively charged genomic DNA wraps around basic core histone proteins to form nucleosomes. The string of nucleosomes, together with other nonhistone proteins and RNAs, are somewhat irregularly organized in the cell as chromatin (1, 2). During interphase in higher eukaryotic cells, chromatin forms condensed domains as their functional units (3–8). With cell cycle progression, genomic DNA is duplicated by DNA replication and faithfully transmitted into the two daughter cells during mitosis (9, 10). To ensure the transmission process, interphase chromatin is further condensed into mitotic chromosomes. While several players involved in the condensation process, including condensins and topoisomerase IIα, have been identified and extensively investigated (11–16), the physical bases of the condensation process remain unclear (17, 18).
In addition to these protein factors, two kinds of physical forces governed by the chromosome milieu may contribute to the condensation process. First, free divalent cations such as Mg2+ condense chromatin or chromosomes in vitro (19–26) or by computer modeling (27). These cations have long been considered important for nucleosome–nucleosome interactions because the nucleosomes have a net negative charge and are stretched like “beads on a string” by an electrostatic repulsion force in the absence of cations. Indeed, a transient increase in free Mg2+ is observed during mitosis, contributing to chromosome condensation (28).
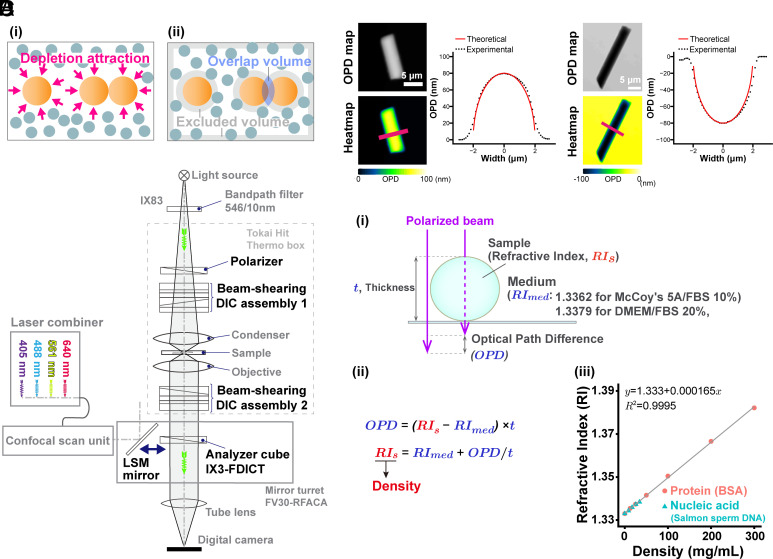
The other possible physical force is the depletion attraction/macromolecular crowding effect (29–32). This force may also be critical because the cellular environment is highly crowded with macromolecules such as proteins, RNA, DNA, and others resulting in a molecular density >100 mg/mL. The principle of the depletion attraction/macromolecular crowding effect is simple (Fig. 1A and SI Appendix, Fig. S1A). Large cellular complexes, like chromatin, are bombarded from all sides by many soluble macromolecules in a crowded environment. When two large complexes come into contact, the macromolecules exert a force equivalent to their osmotic pressure on opposite sides of the two large complexes to keep them together (Fig. 1A and SI Appendix, Fig. S1A) (29–31). This state is entropically favored because there are more accessible regions after their contact (Fig. 1A and SI Appendix, Fig. S1A). Indeed, purified nuclei (33) and chromosomes (34) condensed in vitro with ~10% (w/v) of polyethylene glycol (PEG) or polyvinyl alcohol. Such high concentrations of a macromolecular “crowder” induced folding of long synthetic chromatin fibers in vitro (35). When ~10% of bovine serum albumin (BSA) was used as a crowder, condensates of the synthetic chromatin fibers were observed in vitro (36). However, it remains unclear whether the depletion attraction/macromolecular crowding effect is really involved in mitotic chromosome condensation in the cell, especially in higher eukaryotic cells.
Fig. 1.
Schematics of the depletion attraction/macromolecular crowding effect, OI-DIC microscopy, and density quantification. (A, i) Many small spheres (blue-gray) representing soluble macromolecules bombard three large spheres (orange, representing chromatin) from all sides (arrows). When two large spheres come into contact (Right), the small ones exert a force equivalent to their osmotic pressure on opposite sides of the two large ones to keep them together (depletion attraction/macromolecular crowding effect). (ii) The shaded regions in this alternative view show regions inaccessible to the centers of mass of the small spheres (excluded volumes). When two large spheres contact each other, their excluded volumes overlap and increase the volume accessible to the small spheres. Large sphere aggregation is then favored by an increase in the entropy of the system. (B) Optical schematic of the confocal laser scanning microscope Olympus FV3000, equipped with the OI-DIC module. Details of the microcopy system are described under SI Appendix, Materials and Methods. (C) Validation of density imaging by OI-DIC microscopy using known glass rods and mineral oils. The RI of the glass rods was 1.56, and those of the oils were 1.54 (Left) and 1.58 (Right). The theoretical and experimental values are equivalent, ensuring the accuracy of our RI quantification. (Scale bar, 5 μm.) (D, i) A procedure for estimating the RI of sample (depicted as a sphere). Our OI-DIC microscopy can computationally quantify optical path differences (OPDs) at each spatial point. (ii) The formula to calculate RI of a sample. (iii) The calibration curve of RI versus the density of standard solutions for protein or nucleic acid. RI = 1.333 + 1.65 × 10−4 × C (RI, the refractive index; C, the concentration of the proteins or nucleic acids [mg/mL]).
To observe the depletion attraction/macromolecular crowding effect on chromosome condensation, it is essential to measure molecular density in the chromosome milieu of living cells. We gain insight into molecular density by using differential interference contrast (DIC) microscopy (37–40). DIC images are produced by the interference of two laterally displaced light beams passing through a sample (e.g., live cells), capturing information about the optical path length (OPL) in the sample to reveal otherwise invisible features (37). The difference in OPL (optical path difference, OPD) between the two beams contrasts the image, reflecting local differences in the refractive index (RI) within the sample. However, the contrast in DIC images depends on the direction of displacement between the two light beams and the sample, precluding quantitative measurement of the OPL. To overcome the limitations of DIC, Shribak et al. developed an orientation-independent-DIC (OI-DIC) microscopy method (41), which allows the directions of displacement for the two light beams to be rapidly switched by 90° without mechanically rotating the sample or the prisms, generating a quantitative OPD map (SI Appendix, Fig. S1B). Based on the OPD value, it is possible to estimate the molecular density of intracellular regions in live cells. However, a previous type of OI-DIC with a wide-field epifluorescence microscope had difficulty in measuring precise thickness of target structures in the cell (42, 43).
In this study, we have combined a confocal laser scanning microscope (CLSM) with an OI-DIC module (Fig. 1B), which can provide high-resolution maps of OPD by OI-DIC and the thickness of target structures in the cell by CLSM. This OI-DIC imaging can generate a 3D volume image of the RI and molecular density (dry mass) in living cells based on the calibration data (Fig. 1 D, iii). Using this OI-DIC system, we quantified the absolute density of the molecules around chromosomes during mitosis of human HCT116 cells. We found that the molecular density surrounding mitotic chromosomes increased with the mitotic progression from prophase to anaphase, concurring with chromosome condensation. In telophase, the molecular density decreased as chromosomes began to decondense. Hypertonic treatment of the mitotic cells rapidly increased density and induced chromosome condensation, while hypotonic treatment had the opposite effects. In vitro, fiber-like condensates of native chromatin, induced by physiological concentrations of cations, were efficiently converted into chromatin liquid droplets with macromolecular crowders. Additional amounts of crowders made the chromatin droplets more solid-like structures. These results suggest that during mitosis, a transient rise in depletion attraction by proteins, RNAs, and others contributes to mitotic chromosome condensation, shedding light on a different aspect of mitotic chromosome condensation in living human cells.
Results
Development of an OI-DIC System Combined with a CLSM.
To estimate the density of total molecules in the chromosome milieu, we developed an OI-DIC module combined with a CLSM (Fig. 1B). We calculated the dry mass density of the sample (Fig. 1 D, i; for more details, see SI Appendix, Materials and Methods) based on the OPD map obtained from OI-DIC imaging (e.g., Fig. 1C and SI Appendix, Fig. S1B), the precisely measured thickness (t) of the target structure by CLSM (Fig. 1D and SI Appendix, Fig. S3C), and the known RI of the surrounding medium. Next, we evaluated whether the OI-DIC imaging and subsequent analysis could accurately estimate RIs by observing glass rods (diameter = 4 µm) in mineral oils with known refractive indices (RI = 1.54 and 1.58) and calculated the theoretical OPD (Fig. 1C). The theoretical and optically measured OPDs were almost identical (Fig. 1C), validating the accuracy of our OI-DIC imaging for estimating the RI of samples with measured OPD.
OI-DIC Imaging of Interphase and Mitotic Live Human HCT116 Cells.
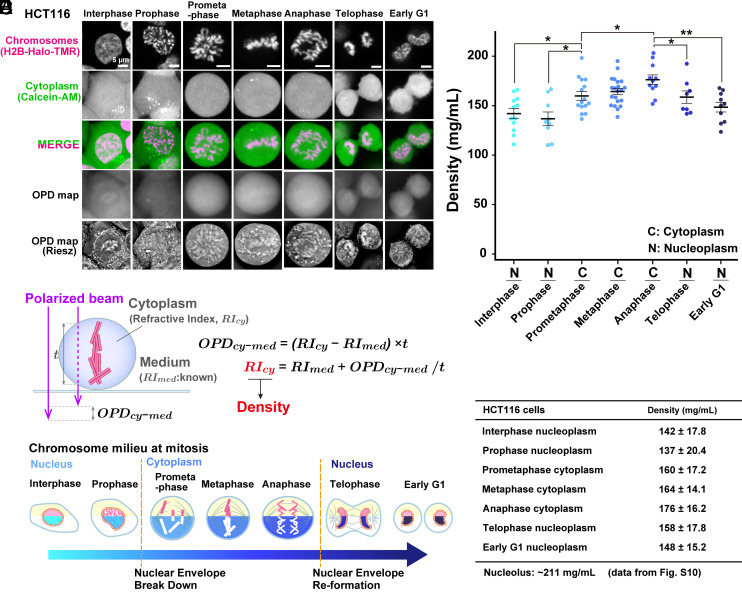
Using the procedure described above (Fig. 1D), we performed OI-DIC imaging of interphase and mitotic live human HCT116 cells stably expressing H2B-HaloTag and obtained their OPD maps (Fig. 2A). Riesz images are edge-enhanced OPD maps by the inverse Riesz transform (44, 45) and are more visually informative with the higher contrast of small details (Fig. 2 A, Bottom images). Riesz transform is equivalent to a high-pass or “sharpen” image processing filter, which attenuates low-frequency components. Note that asynchronous live cells were used for this imaging to avoid any artifactual effects, such as accumulations and aggregations of proteins and RNAs, caused by the synchronization process (For classification of mitotic cells, see SI Appendix, Materials and Methods). We clearly observed cytoplasmic organelles, nuclear envelopes, and presumably nucleoli in OPD maps of interphase HCT116 cells (Fig. 2 A, Lower Left). In the case of mitotic cells, cytoplasmic organelles, and condensed chromosomes were notable in the Riesz images (Fig. 2 A, Bottom images). We set several Region of Interest (ROIs) in each cell to avoid apparent structures in the Riesz image, presumably endoplasmic reticulum, Golgi apparatus, and mitochondria (SI Appendix, Fig. S2) (46). Cytoplasmic ROIs were set to exclude chromosomes in the entire Z-stacks (SI Appendix, Fig. S2). To obtain the thickness measurement (t) of cytoplasm (Fig. 2B and SI Appendix, Figs. S3 A and C and S4A) and chromatin (for nucleoplasm, see SI Appendix, Fig. S3 B and C; for chromosomes, see SI Appendix, Fig. S4 B and C), cells were simultaneously labeled with Calcein AM and HaloTag ligand TMR, and optical sectioning images were recorded using CLSM.
Fig. 2.
Density imaging of live human HCT116 cells using OI-DIC microscopy. (A) Confocal images, OPD maps, and Riesz images. Image intensities are not presented on the same scale for better visualization. (B) Schematic of how the RI of the cytoplasm (RIcy) is estimated. The RIcy can be calculated using the formula shown based on the OPD measured by OI-DIC, the RI of medium (RImed) measured by the refractometer, and thickness (t) values. (C) Schematic of chromatin/chromosome milieu during mitosis. The measured regions in the cells are marked in cyan, blue, or navy. (D) Total molecular densities of chromosome milieu in live HCT116 cells. Each dot represents the average of the estimated total density at several points within a single cell, where no apparent structure is obvious in the Riesz image (SI Appendix, Fig. S2). Mean ± SE are shown. Cell numbers are N = 12 (interphase), 9 (prophase), 15 (prometaphase), 21 (metaphase), 11 (anaphase), 8 (telophase), and 10 (early G1). *P < 0.05 by the two-sided unpaired t test for interphase nucleoplasm vs. prometaphase cytoplasm (P = 0.015), prophase nucleoplasm vs. prometaphase cytoplasm (P = 0.013), prometaphase cytoplasm vs. anaphase cytoplasm (P = 0.021), anaphase cytoplasm vs. telophase nucleoplasm (P = 0.044) and **P < 0.001 for anaphase cytoplasm vs. early G1 nucleoplasm (P = 6.9 × 10−4). (E) Mean ± SD of total molecular densities at each phase.
Based on the OPDs, thicknesses (t) of the cytoplasm and nucleoplasm, and the known RI of the medium (1.3362 for McCoy's 5A including 10% fetal bovine serum (FBS) in Fig. 1 D, i), we calculated the RIs of cytoplasm (Fig. 2B and SI Appendix, Fig. S3A) and nucleoplasm (SI Appendix, Fig. S3B) in HCT116 cells (SI Appendix, Table S1). It is known that there is a linear correlation between the concentrations of various macromolecules (proteins, lipids, carbohydrates, and nucleic acids) and the RIs of their solutions, within the physiological concentration range (42, 47, 48). According to the calibration curve of RIs versus the density of standard solutions for protein or nucleic acid (Fig. 1 D, iii), we obtained total molecular density values in the cytoplasm and/or nucleoplasm during various cell cycle stages (Fig. 2 C and D and SI Appendix, Fig. S5). Note that we did not look at mitotic chromosomes themselves this time (Fig. 2B) since we wanted to examine macromolecular crowding around chromosomes, that is, the chromosome milieu (Fig. 2C). Mitotic stages were judged by their chromosome morphology (Fig. 2A, for details, see SI Appendix, Materials and Methods).
Macromolecular Crowding in Mitotic Chromosome Milieu Is Higher Than in Interphase Chromatin.
Our quantification (Fig. 2 D and E and SI Appendix, Table S1) shows that the density of interphase nucleoplasm is 142 mg/mL, which agrees well with our previous report (42) and is lower than that of cytoplasm (170 mg/mL, see SI Appendix, Fig. S5). Interestingly, the density around chromatin/chromosomes increased after Nuclear Envelope Break Down (NEBD). Cytoplasm density in prometaphase cells was 160 mg/mL (Fig. 2 D and E) and higher than that of interphase nucleoplasm. With mitotic progression from prophase to anaphase, density gradually increased from 137 mg/mL to 176 mg/mL (Fig. 2 D and E). Maximal cytoplasmic density in anaphase is consistent with previous reports on maximal chromosome compaction in anaphase (49, 50). Once the cytokinesis and chromosome decondensation started at telophase (Fig. 2A), the density was reduced and became similar to the interphase density at early G1 phase (Fig. 2 D and E). At early G1, the density profile returned to high for cytoplasm and low for nucleoplasm (SI Appendix, Fig. S5). These results suggest that depletion attraction in mitotic chromosome milieu is higher than in interphase nucleoplasm, increases with mitotic progression from prophase to anaphase, and starts reverting at telophase.
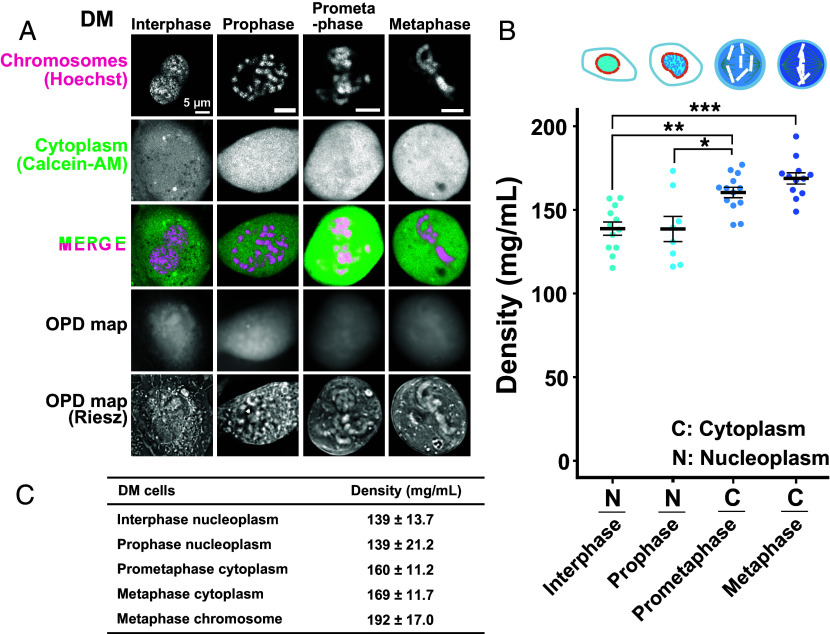
To further support our findings in HCT116 cells, we examined another cell line, Indian Muntjac DM cells (Fig. 3A). DM cells were derived from deer fibroblast cells and have very large mitotic chromosomes (51–53). Chromatin and cytoplasm were labeled with Hoechst 33342 and Calcein AM simultaneously (Fig. 3A) to obtain thickness information (t) of interphase cytoplasm (SI Appendix, Fig. S3A), interphase nucleoplasm (SI Appendix, Fig. S3B), and mitotic cytoplasm (Fig. 2B).
Fig. 3.
Density imaging of live Indian Muntjac DM cells using OI-DIC microscopy. (A) Confocal images, OPD maps, and Riesz images. Image intensities are not presented on the same scale for better visualization. (B) Total molecular densities of chromosome milieu in live DM cells. Each dot represents the average of the estimated total density at several points within a single cell, where no clear structure is apparent in the Riesz image (SI Appendix, Fig. S2). Mean ± SE are shown. Cell numbers are N = 12 (interphase), 8 (prophase), 13 (prometaphase), and 12 (metaphase). *P < 0.05 by the two-sided unpaired t test for prophase nucleoplasm vs. prometaphase cytoplasm (P = 0.024); **P < 0.001 for interphase nucleus vs prometaphase cytoplasm (P = 3.2 × 10−4); ***P < 0.0001 for interphase nucleus vs metaphase cytoplasm (P = 9.4 × 10−6). (C) Mean ± SD values of total molecular densities at each phase.
Using a similar procedure for HCT116 cells, we found that the density of cytoplasm in prometaphase DM cells, which corresponds to chromosome milieu, was 160 mg/mL (Fig. 3B and SI Appendix, Fig. S6) and higher than that of interphase nucleoplasm (139 mg/mL). The density increased from prophase to metaphase (Fig. 3B and SI Appendix, Table S2). The obtained results in DM cells were consistent with those in human HCT116 cells, suggesting that a transient rise in the density of chromosome milieu may be a general feature of mitotic cells. These findings also suggest that depletion attraction can contribute to mitotic chromosome condensation.
The Density in Mitotic Chromosomes Is 192 mg/mL.
DM cells have much larger and fewer mitotic chromosomes (7 to 9 chromosomes/cell) (51–53) than HCT116 cells (~46 chromosomes, relatively normal human karyotypes) (54). This difference enabled us to obtain the OPL of mitotic chromosomes (SI Appendix, Fig. S4B) and measure the density in mitotic chromosomes as 192 mg/mL in DM cells (SI Appendix, Fig. S7, Left and SI Appendix, Table S2). This value was significantly higher than that of mitotic cytoplasm (169 mg/mL, P = 0.0040) and comparable to the result of our previous study on the density (208 mg/mL) of the mouse pericentric heterochromatin (chromocenter) (42). In our previous work, we estimated the nucleosome concentration in mitotic chromosomes of DM cells to be about 0.52 mM (51), which corresponds to 137 mg/mL. Given that there is a considerable amount of nonhistone proteins within mitotic chromosomes (55, 56), we consider this value to be reasonable. On the other hand, because chromosome thickness is difficult to accurately measure (t3 in SI Appendix, Fig. S4B), even when using larger DM cell chromosomes, the density values of mitotic chromosomes should become more precise as measurement techniques improve in the future.
Hypertonic Treatment Raised Depletion Attraction and Induced Chromosome Hypercondensation.
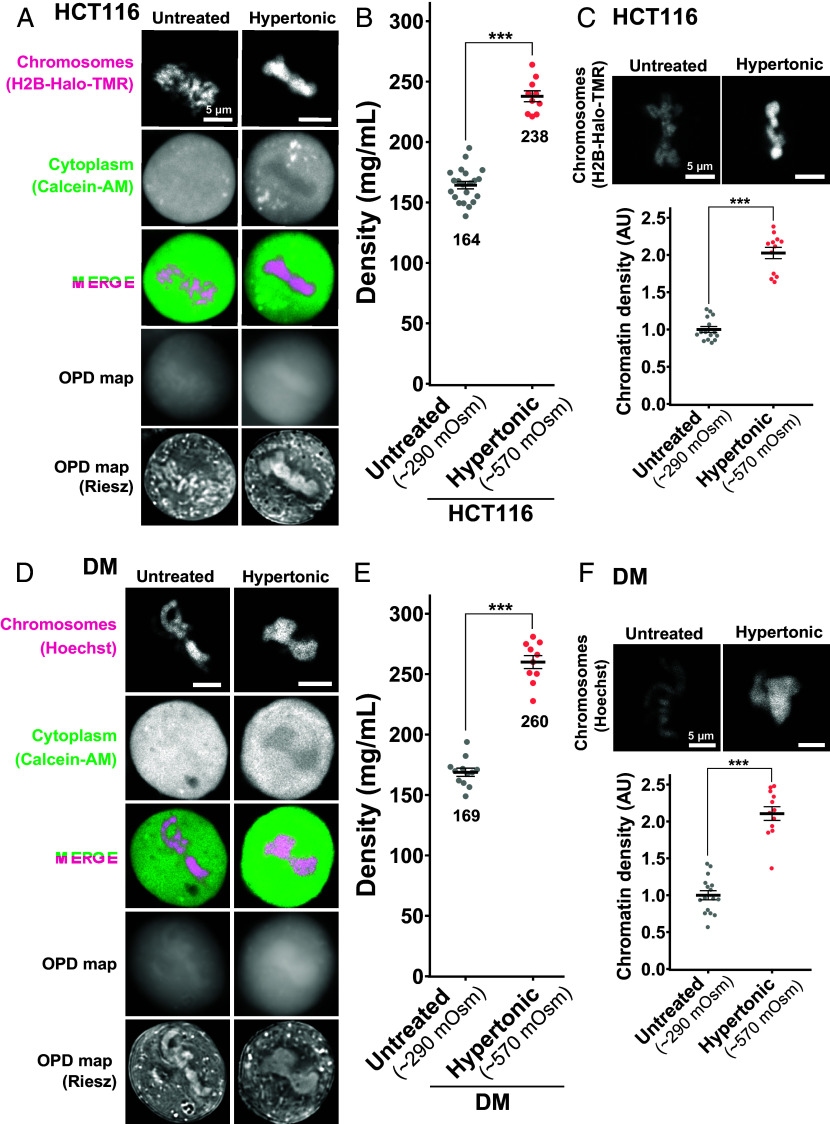
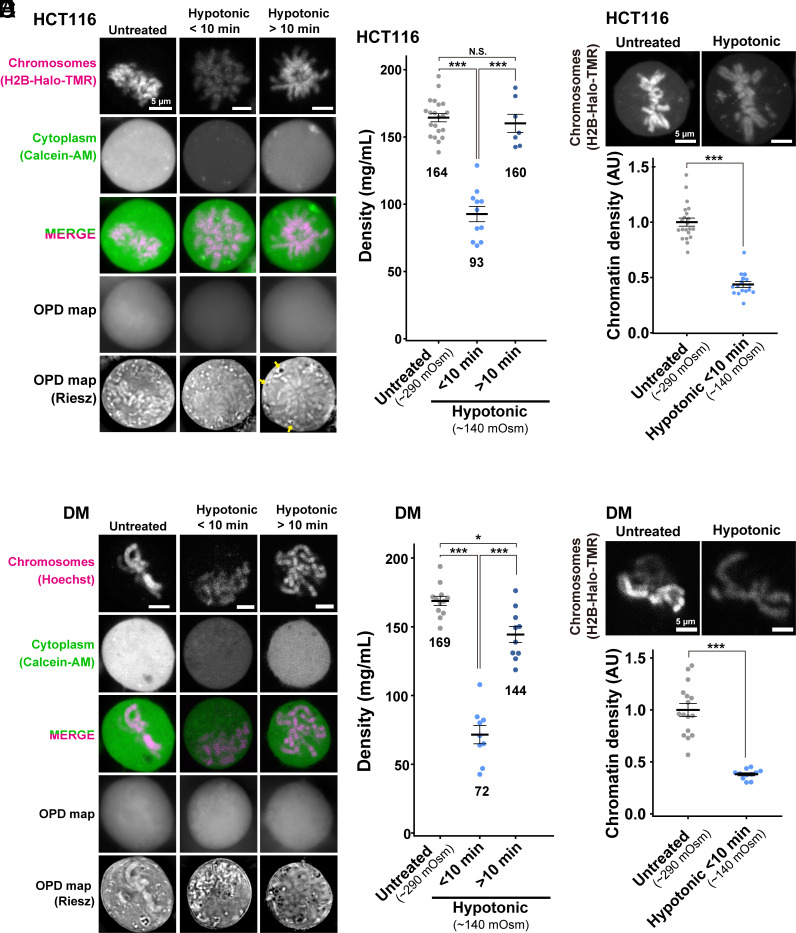
We wondered what would happen to the mitotic HCT116 cells if we changed their cytoplasmic density. To address this question, we first performed the hypertonic treatment with 570 mOsm by adding concentrated phosphate-buffered saline (Fig. 4A) as reported in refs. 57 and 58, while the physiological osmotic condition is 290 mOsm. Upon hypertonic treatment, the cytoplasmic density increased from 164 mg/mL to 238 mg/mL (Fig. 4B and SI Appendix, Table S1). Mitotic chromosomes looked clumped together (Fig. 4A). The signal intensity of mitotic chromatin labeled by H2B-HaloTag-TMR also increased (Fig. 4C), suggesting a rise in depletion attraction and induced hypercondensation of mitotic chromosomes upon hypertonic treatment.
Fig. 4.
Density imaging of hypertonic mitotic cells using OI-DIC microscopy. (A) Confocal images, OPD maps, and Riesz images in live HCT116 cells. Hypertonic treatment induced hypercondensation of chromosomes, making chromosomes clump together. Image intensities are not presented on the same scale for visualization. (B) Total densities in the cytoplasm of mitotic live HCT116 cells. Each dot represents the average of the estimated total density at several points within a single cell, mean ± SE. The average densities (mg/mL) are shown below the plots. HCT116 cell numbers are N = 12 (Untreated) and 10 (Hypertonic). ***P < 0.0001 by the two-sided unpaired unpaired t test (P = 1.0 × 10−10. Untreated data was from Fig. 2D (Metaphase). (C) In HCT116 cells treated with hypertonic solution, projections of 3 z-sections of mitotic chromosomes (top) and quantification of chromatin density (bottom) are shown. Each dot represents the mean intensity of the chromosome region within a single cell, normalized by the mean intensity of untreated cells, mean ± SE. AU, arbitrary units. HCT116 cell numbers are N = 15 (Untreated) and 15 (Hypertonic). ***P < 0.0001 by Wilcoxon rank sum test (P = 1.3 × 10−8). (D) DM cells were treated, imaged, and stained as in (A). (E) Total densities in the cytoplasm of mitotic live DM cells. DM cell numbers are N = 12 (Untreated) and 10 (Hypertonic). ***P < 0.0001 by the two-sided unpaired t test (P = 2.1 × 10−10). Untreated data was from Fig. 3B (Metaphase). (F) In DM cells treated with hypertonic solution, projections of 3 z-sections of mitotic chromosomes (top) and quantification of chromatin density (bottom) are shown. DM cell numbers are N = 16 (Untreated) and 16 (Hypertonic). ***P < 0.0001 by Wilcoxon rank sum test (P = 1.3 × 10−8).
Similar findings were achieved using DM cells with hypertonic treatment (Fig. 4D). An increase in cytoplasmic density (Fig. 4E and SI Appendix, Table S2) induced hypercondensation of mitotic chromosomes and chromosome clumping (Fig. 4 D and F). In addition, the density within mitotic chromosomes increased from 192 mg/mL to 256 mg/mL (SI Appendix, Fig. S7). The cells with a higher density in the cytoplasm tended to have a higher density for their chromosomes. These results support the hypothesis that depletion attraction contributes to mitotic chromosome condensation.
Hypotonic Treatment Lowered Depletion Attraction and Induced Chromosome Decondensation.
Next, we treated HCT116 cells hypotonically (140 mOsm) by diluting the medium (Fig. 5A). We found that just after the treatment (<10 min), cytoplasmic density in prometa-/metaphase cells reduced from 164 mg/mL to 93 mg/mL (Fig. 5B and SI Appendix, Table S1). Chromatin intensity labeled by H2B-HaloTag-TMR also decreased simultaneously (Fig. 5C), suggesting a considerable decondensation of mitotic chromosomes with a reduction in depletion attraction. These findings strengthen our notion that depletion attraction is involved in mitotic chromosome condensation.
Fig. 5.
Density imaging of hypotonic mitotic cells using OI-DIC microscopy. (A) Confocal images, OPD maps, and Riesz images in live HCT116 cells. Hypotonic treatment induced a transient decondensation of chromosomes (within 10 min after treatment), and then the chromosomes seemed to re-condense. Image intensities are not presented on the same scale for visualization. (B) Total densities in the cytoplasm of mitotic live HCT116 cells. Each dot represents the average of the estimated total density at several points within a single cell, mean ± SE. The average densities (mg/mL) are shown below the plots. HCT116 cell numbers are N = 21 (Untreated), 11 (Hypotonic < 10 min), and 7 (Hypotonic > 10 min). ***P < 0.0001 by the two-sided unpaired t test for Untreated vs Hypotonic < 10 min (P = 6.4 × 10−9) and for Hypotonic < 10 min vs Hypotonic > 10 min (P = 2.8 × −6). N.S., not significant (P = 0.58). Untreated data was from Fig. 2D (Metaphase). (C) In HCT116 cells treated with hypotonic solution, projections of 3 z-sections of mitotic chromosomes (top) and quantification of chromatin density (bottom) are shown. Each dot represents the mean intensity of the chromosome region within a single cell, normalized by the mean intensity of untreated cells, mean ± SE. AU, arbitrary units. HCT116 cell numbers are N = 21 (Untreated) and 16 (Hypotonic). ***P < 0.0001 by Wilcoxon rank sum test (P = 1.6 × 10−10). (D) DM cells were treated, imaged, and stained as in (A). (E) Total densities in the cytoplasm of mitotic live DM cells. DM cell numbers are N = 12 (Untreated), 9 (Hypotonic < 10 min), and 10 (Hypotonic > 10 min). ***P < 0.0001 by the two-sided unpaired t test for Untreated vs Hypotonic < 10 min (P = 1.9 × 10−8), Hypotonic < 10 min vs Hypotonic > 10 min (P = 3.3 × 10−7), and *P < 0.05 (P = 2.5 × 10−3) for Untreated vs Hypotonic > 10 min. Untreated data was from Fig. 3B (Metaphase). (F) In DM cells treated with hypotonic solution, projections of 3 z-sections of mitotic chromosomes (top) and quantification of chromatin density (bottom) are shown. DM cell numbers are N = 16 (Untreated) and 11 (Hypotonic). ***P < 0.0001 by Wilcoxon rank sum test (P = 1.5 × 10−7).
We noticed that the cytoplasmic density recovered >10 min after the hypotonic treatment (Fig. 5B), while cellular effects caused by hypertonic treatment seemed stable after 30 min, consistent with a previous report (59). The induced decondensation of chromosomes also attenuated on the same time scale (Fig. 5A). Interestingly, we found some low-density regions or vesicles in the cytoplasm of prometaphase cells appeared with prolonged hypotonic treatment (>10 min) (shown by arrows in Fig. 5A), suggesting that the infiltrated water molecules were somehow sequestered to restore intracellular osmotic pressure.
Similar findings with hypotonic treatment were obtained using DM cells (Fig. 5D). Cytoplasmic density decreased (Fig. 5E and SI Appendix, Table S2) and induced decondensation of mitotic chromosomes (Fig. 5F) <10 min following treatment. The cytoplasmic density recovered 10 min after the hypotonic treatment (Fig. 5E), corresponding to a rise in chromosome intensity (Fig. 5D). These results strengthen our finding that depletion attraction contributes to mitotic chromosome condensation.
Macromolecular Crowders Induce Liquid Droplets of Chromatin In Vitro, and Additional Crowding Makes the Droplets Stiffer and More Solid-Like.
To finally examine how an increase in depletion attraction/macromolecular crowding during mitosis can change the physical properties of chromosomes, we performed an in vitro condensation assay using chicken native chromatin. The peak size of the native chromatin used for this assay was ~6 kb and corresponded to ~30 nucleosomes (SI Appendix, Fig. S8 A and B). Cations diminish the repulsion between negatively charged nucleosomes in the chromatin and electrostatically attract them to form chromatin condensates (22–26, 28, 58, 60, 61). Therefore, we incubated our native chromatin in a buffer containing 100 mM K+ and 0.8 mM Mg2+ to mimic physiological concentrations of cations in the cell (28) and found many fiber-like condensates of chromatin were formed (Fig. 6A, Panel 1), consistent with previous reports (28, 58, 61).
Fig. 6.
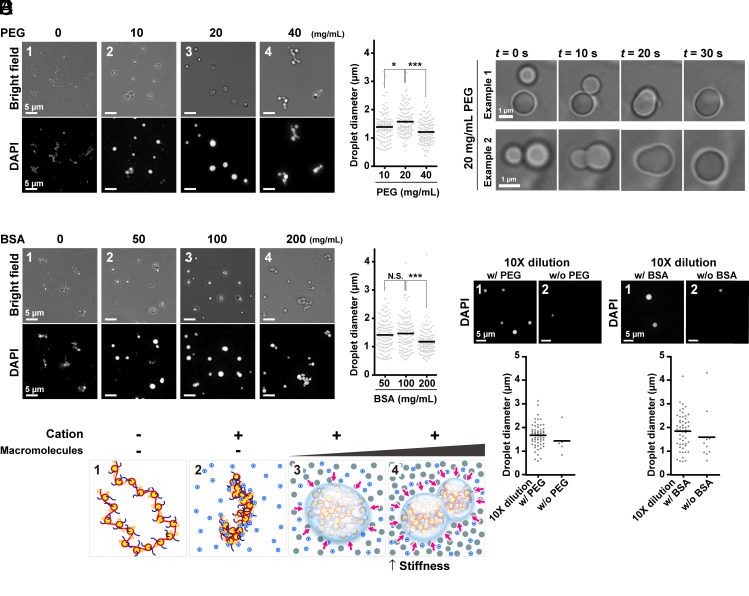
Liquid droplet formation and solidification of chicken native chromatin by macromolecular crowders. (A) Fibrous chromatin condensates were induced by 100 mM K+ and 0.8 mM Mg2+ (Panel 1, 0 mg/mL PEG). PEG converted the fibrous condensates into droplets (Panels 2 and 3). Further increases in the concentration of PEG put the droplets together without fusions (Panel 4). Magnified images are shown at the Bottom. Diameters of formed droplets are graphed on the Right. *P < 0.05 by the Wilcoxon rank-sum test for 10 mg/mL vs. 20 mg/mL PEG (P = 0.011). ***P < 0.0001 for 20 mg/mL vs. 40 mg/mL PEG (P = 7.5 × 10−9). (B) Fusions of formed droplets. Two typical examples are shown. Also see Movie S1. (C) Similar analyses to A on BSA-induced liquid droplets. N.S., not significant by the Wilcoxon rank-sum test for 50 mg/mL vs. 100 mg/mL BSA (P = 0.78). ***P < 0.0001 for 100 mg/mL vs. 200 mg/mL BSA (P = 7.0 × 10−6). (D) Schematic of conversion from fibrous chromatin condensates to liquid droplets. The chromatin is stretched by electrostatic repulsion without cations (Panel 1). Mg2+ and K+ decrease net negative charge and repulsion and induce fibrous chromatin condensates (Panel 2). Depletion attraction/macromolecular crowding converts the fibrous condensates into droplets (Panel 3). Further depletion attraction/macromolecular crowding makes the droplets stiffer (Panel 4). (E) The droplet dilution assay. After the droplets were formed with 20 mg/mL of PEG, the reaction mixtures were diluted 10-fold in a buffer with (Panel 1) or without (Panel 2) 20 mg/mL of PEG. The images depict fewer and smaller droplets in the diluted buffer without PEG, suggesting that the droplets were dissolved in the diluted buffer. (Bottom) A quantitative analysis of the droplet dilution assay. The number and diameters of the droplets in a randomly picked area (6.0 × 104 µm2) are plotted. (F) The dilution assay on formed droplets with 100 mg/mL BSA.
We simulated depletion attraction/macromolecular crowding of chromosome milieu by incubating increasing concentrations of PEG (a neutral polymer, M.W. ~8 kDa), BSA (M.W. 66 kDa), or Dextran (M.W. ~200 kDa) with our fiber-like condensates of chromatin. We first added increasing concentrations of PEG as a crowder and observed morphological changes of chromatin condensates. With 10 mg/mL (w/v) (corresponding to 1.25 mM) PEG, some spherical structures were found among fibrous condensates (Fig. 6A, Panel 2). At 20 mg/mL (2.5 mM) PEG, almost all condensates became spherical with various sizes and looked like liquid droplets formed by liquid–liquid phase separation (Fig. 6A, Panel 3) (60, 62–64). Indeed, the droplets often fused to form larger droplets (Fig. 6B and Movie S1). Upon increasing PEG to 40 mg/mL (5.0 mM), droplets were connected to one another (Fig. 6A, Panel 4). Note that their sizes were not as large as those with 20 mg/mL PEG (Fig. 6 A, Right). These were presumably stuck together and not fused into lager droplets during our observation period (up to 1 h). A higher concentration of PEG converted chromatin droplets to be stiffer and more solid-like, reducing their fluidity (Fig. 6D).
Next, we used BSA as another crowder in the same concentration of cations and native chromatin. BSA affected chromatin morphology at higher concentrations (w/v) than PEG. At 50 mg/mL (0.75 mM) BSA, chromatin droplets formed among fibers (Fig. 6C, Panel 2). Almost all condensates became droplets at 100 mg/mL (1.5 mM) BSA (Fig. 6C, Panel 3). As in the case of PEG, increasing the BSA concentration to 200 mg/mL (3.0 mM) formed clusters of droplets that appeared to stick together (Fig. 6C, Panel 4). Again, their sizes were smaller than those with 100 mg/mL (1.5 mM) BSA due to much fewer fusion events being prevented by stiffer and more solid-like properties (Fig. 6 C, Right). We also obtained a similar set of results using Dextran (M.W. ~200 kDa) (SI Appendix, Fig. S8C). These results suggest that depletion attraction/macromolecular crowding can change the physical properties of chromosomes. Addition of crowders induced the formation of chromatin liquid droplets in vitro. Further crowding increased droplet stiffness by pushing chromatin droplets from the outside. This larger depletion attraction may have prevented droplet fusions at our time scale (up to 1 h) due to the reduced fluidity (Fig. 6D).
To examine whether the chromatin droplet formation induced by depletion attraction/macromolecular crowding was reversible, we lowered the crowding effect around the droplets. After droplets were made using 20 mg/mL PEG, 100 mg/mL BSA, or 50 mg/mL Dextran, the droplet solutions were diluted 10-fold with a buffer containing 100 mM K+ and 0.8 mM Mg2+ with or without crowder (Fig. 6 E and F and SI Appendix, Fig. S8D). The number of droplets in the diluted buffer without crowder substantially decreased and more small chromatin condensates appeared as compared to the control droplets in the diluted buffer with crowder. Droplets resolved into small chromatin condensates, suggesting that droplet formation by depletion attraction/macromolecular crowding is reversible.
We measured the density of the chromatin condensates in vitro by OI-DIC (SI Appendix, Fig. S9). Interestingly, the data show that the densities of the chromatin condensates are much higher than those in mitotic chromosomes of living cells: ~448 mg/mL in PEG and ~365 mg/mL in BSA. Furthermore, the increase in crowders (from 20 to 40 mg/mL PEG or from 100 to 200 mg/mL BSA) did not further increase their densities. We consider in vitro chromatin condensates to have almost “maximum” density as proposed in ref. 65. An increase in crowders thus cannot raise the density of chromatin condensates, even though their physical properties change from liquid-like to solid-like.
Discussion
The chromosome milieu governs physical forces contributing to chromosome condensation. We developed an OI-DIC microscopy system combined with CLSM to elucidate the effects of the chromosome milieu in live mitotic cells (Fig. 1B). The subsequent analysis enabled the absolute densities in cytoplasm and/or nucleoplasm in various cell cycle stages of human HCT116 cells and Indian Muntjac DM cells to be calculated. We demonstrated that the density of cytoplasm in metaphase cells (164 mg/mL) was higher than that of interphase nucleoplasm (142 mg/mL). Density at chromosome milieu gradually increased from 137 mg/mL to 176 mg/mL (Fig. 2 D and E) as cells mitotically progressed from prophase to anaphase, concurring with chromosome condensation (49, 50). Furthermore, hypertonic treatment of the mitotic cells rapidly raised density and induced chromosome condensation (Fig. 4), while hypotonic treatment had the opposite effects (Fig. 5). Our findings suggest that a transient rise in depletion attraction (the density of proteins, RNAs, and others) during mitosis is involved in mitotic chromosome condensation, providing a unique insight into the physical bases of mitotic chromosome condensation in living human cells.
We demonstrate that the depletion attraction/macromolecular crowding converts fibrous chromatin condensates formed by cations into liquid droplets in vitro. Previous reports on chromatin liquid droplet formation have thus far only used synthetic nucleosomes such as 12-mer arrays, which have uniform properties in the length, spacing, size, and modifications (58, 60, 66). Increasing depletion attraction/macromolecular crowding in vitro made the droplets stiffer and more solid-like (Fig. 6D). While cations induced an electrostatic attraction force between chromatin, the depletion attraction/macromolecular crowding worked as external pressure from outside (Fig. 6D). Our results suggest that the depletion attraction/macromolecular crowding is another force that contributes to chromosome rigidity during mitosis (Fig. 7). The additional condensation by increased depletion attraction is particularly advantageous for the chromosome segregation and transmission processes during anaphase, when mechanical shearing stress is high. Note that the highest cytoplasmic density level was identified in anaphase (Fig. 2D) and concurred with previous reports of the highest level of chromosome compaction in anaphase (49, 50). Condensed chromatin domains in interphase have been observed in a variety of cells (5–7, 67–71) and are proposed to work as building blocks for mitotic chromosomes (8, 69). The assembled stiffer chromatin droplets that we observed in the highly crowded situations looked like mitotic chromosomes (Panels 4 in Fig. 6 A and C and SI Appendix, Fig. S8C). Hence, the depletion attraction/macromolecular crowding may facilitate the assembly of condensed chromatin domains to form mitotic chromosomes.
Fig. 7.
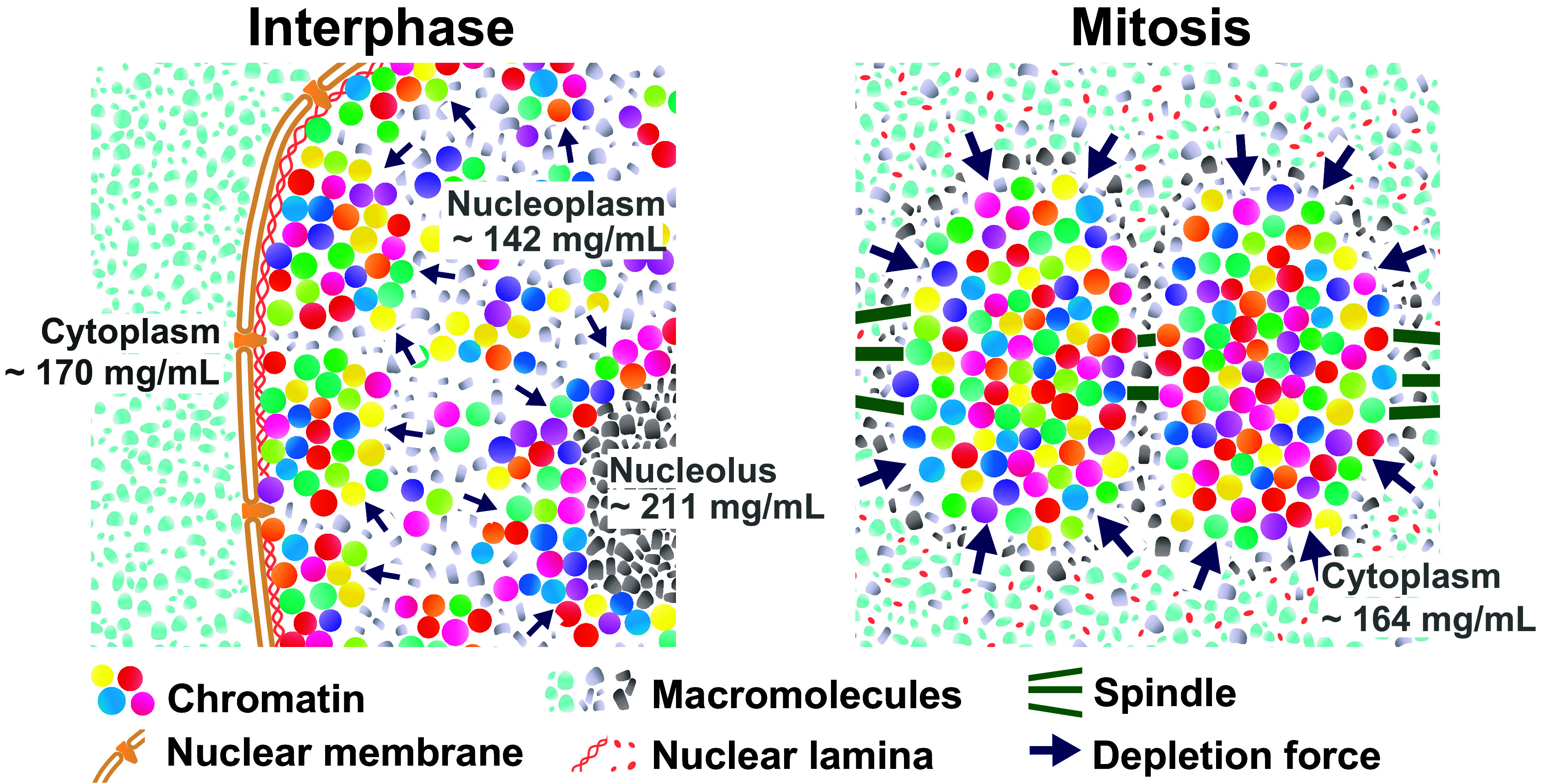
Model for upregulation of depletion attraction in mitotic cells. (Left) Schematic of depletion attraction in interphase live cells with soluble macromolecules in the cytoplasm (light-blue), nucleoplasm (gray), and nucleolus (dark gray). The cytoplasm, nucleus, and nucleolus are compartmentalized and not mixed during interphase. Molecular densities of the nucleoplasm are lower than the cytoplasm and nucleolus. (Right) After NEBD, soluble macromolecules that were localized to the cytoplasm, nucleus, and nucleolus at interphase are now mixed. Molecular density of chromosome milieu increases, making the depletion attraction stronger, and contributes to local condensation of chromosomes. These schematics are highly simplified models, and depletion attraction also works in interphase chromatin (smaller navy arrows).
What is the underlying mechanism of up-regulating macromolecule density during mitosis? Small molecules (<40 to 60 kDa) can pass through the nuclear pores by passive diffusion (72) and are rather evenly distributed within the cell. On the other hand, large macromolecules and their complexes, including RNAs, DNAs, and lipids, in eukaryotic cells are highly compartmentalized in interphase by the nuclear membrane (Fig. 7, Left and Fig. 2 A, Bottom Left). We propose that cells regulate their density localizations by compartmentalization and can up-regulate cytoplasmic density after NEBD (Fig. 7, Right). OPD maps of interphase cells show that cytoplasmic organelles (ER, Golgi apparatus, mitochondria, etc.), the nuclear membrane structure, and nucleoli are prominent high-density structures (Fig. 2 A, Bottom Left). Note that we avoided such cytoplasmic organelles for our measurements (SI Appendix, Fig. S2).
Upon NEBD, the nuclear envelope, nuclear pore complexes, nuclear lamina, and nucleoli are disassembled into small pieces (complexes or vesicles) for cell division and become a part of the “chromosome milieu.” As a result, cytoplasmic and nucleolus factors are exposed to chromosomes and fully contribute to an increase in depletion attraction/macromolecular crowding (Fig. 7, Right). For instance, nucleoli, which are enriched with proteins and RNAs, occupy ~12% of the nuclear volume and have a high-density of >200 mg/mL [SI Appendix, Fig. S10; (42)]. Ribosomes are also a potential contributor to increase depletion attraction/macromolecular crowding during mitosis. We indeed observed that in interphase and prophase, prior to NEBD, ribosome components were sequestered from chromatin and confined to the cytoplasm by the nuclear membrane (SI Appendix, Fig. S11). In contrast, in prometaphase and metaphase, after NEBD, they can come into contact with mitotic chromosomes. In addition, some of the nucleolar components also locate to the chromosome periphery, possibly causing additional depletion attraction (e.g., refs. 56, 73, and 74). Furthermore, Son et al. (75) and Zlotek-Zlotkiewicz et al. (76) found that cell volume decreased in the metaphase-anaphase transition. The reduction of cell volume can contribute to an increase in molecular density with the mitotic progression from prometaphase to anaphase (Fig. 2C). Density decreases again in telophase because the nuclear envelope reforms and active transport (77) and compartmentalization restart in telophase. Together, these fully coordinated mitotic events can lead to a transient rise in depletion attraction/macromolecular crowding observed during mitosis (Fig. 7, Right).
While chromatin with a high concentration of macromolecules, or depletant, forms liquid droplets in vitro (Fig. 6 and SI Appendix, Fig. S8), why are the mitotic chromosomes not spherical (i.e., a kinetic effect)? We consider that long strings of interacting nucleosomes in the cell impose physical constraints and work as a kinetic barrier, which induces the kinetic effect. Consistently, when a restriction enzyme AluI was injected into living mitotic cells, which fragments chromatin to relieve the physical constraints (the barrier), the AluI-treated chromosomes lost their rod shape and became chromatin liquid droplets (18, 78) like observed in vitro (Fig. 6 and SI Appendix, Fig. S8).
It is relevant to estimate the order of magnitude of the depletion attraction in terms of kBT. We may regard a chromosome (100 Mb) as an array of chromatin domains, each about 100 kb with a radius of 100 nm with domains dispersed in interphase (8). We assume these domains are assembled into mitotic chromosomes (8). Considering proteins of about 30 kDa with a radius of 3 nm to be typical depletant molecules in the milieu and using the Asakura-Oosawa relation to estimate the depletion attraction (30, 79), the estimated free energy stabilization for chromosome condensation is 6 to 9 kBT/domain larger in mitotic chromosomes than that in interphase chromosomes, showing a considerable contribution of depletion attraction to compensate the entropy loss induced upon condensation (For details, see SI Appendix, Materials and Methods).
A transient rise in depletion attraction during mitosis can act as an additional force to form larger chromosomes in higher eukaryotic cells. Higher eukaryotic cells have a mitotic process called open mitosis. NEBD occurs during prophase, and the nuclear envelope is regenerated around the chromosomes in telophase (9, 10). Our findings suggest that the open mitosis system allows the cell to up-regulate depletion attraction during mitosis. Closed mitosis occurs in the case of lower eukaryotes (e.g., fungi and yeasts), where the nuclear envelope does not break down during mitosis (80, 81). Their chromosome milieu seems constant during mitosis. The depletion attraction may not be involved in chromosome assembly in fungi and yeasts because their genome and chromosomes are so small. We propose that eukaryotic organisms might have evolved to acquire open mitosis and an additional chromosome compaction force for their larger chromosomes. As discussed above, this force could be beneficial to make larger chromosomes more rigid to ensure faithful transmission into daughter cells.
The OI-DIC module combined with CLSM can obtain matched high-resolution OPD and confocal images to get precise absolute densities in live cells, while another density microscopy (82) may have difficulty because it requires a separate reference beam and uses two objective lenses for OPD/RI reconstruction. Our mitotic chromosome study using OI-DIC is complementary to the ones using Hi-C (83) because Hi-C reveals how genomic DNA folds in mitotic chromosomes but does not necessarily show their physical compaction. Label-free imaging such as OI-DIC and other methods, e.g., ref. 84, can dissect mitotic chromosomes from a unique viewpoint and contribute to understanding the mitotic chromosome assembly process.
Materials and Methods
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods. Briefly, HCT116 and Indian Muntjac DM cells were cultured in appropriate media with FBS. The Olympus FV3000 microscope with custom DIC assemblies enabled detailed OI-DIC imaging. Cell densities were calculated using OPD maps and confocal images. Chromatin for droplet assays was prepared from fresh chicken blood.
Supplementary Material
Appendix 01 (PDF)
Bright field movie of fusions of chromatin liquid droplets formed in the presence of 20 mg/mL of PEG (10 s/frame). Three fusion events were recorded in total.
Acknowledgments
We are grateful to Dr. K. M. Marshall for critical reading and editing of this manuscript and Dr. R. Imai for a preliminary result on density in mitotic chromosomes. We thank Dr. M. Kanemaki for providing their HCT116 cell line, Dr. H. Kimura and Dr. P. Cook for providing their DM cell line, Dr. T. Uchiumi for providing antiribosomal P protein antibody, and Dr. Y. Murayama, Dr. A. Nakano, Dr. K. Hibino, Mr. M. A. Shimazoe, and Maeshima laboratory members for their helpful discussions and support. This work was supported by JSPS grants JP21H02453 (K.M.), JP22H05606 (S. Ide), JP21H02535 (S. Ide), JP20H05936 (K.M.), JP24H00061 (K.M., M. Sasai), JP23K17398 (K.M., S. Ide, T.T.), and JP22H00406 (M. Sasai), Takeda Science Foundation (K.M.), Inoué Endowment Fund (M. Shribak), and NIGMS/NIH grant R01GM101701 (M. Shribak). S. Iida was a SOKENDAI Special Researcher (JST SPRING JPMJSP2104) and supported by a SOKENDAI Genetics Course travel fellowship and SOKENDAI Student Dispatch Program. S. Iida is currently a JSPS Fellow (JP23KJ0996).
Author contributions
S. Iida, T.T., M. Shribak, and K.M. designed research; M. Shribak developed OI-DIC microscopy; S. Iida, S. Ide, S.T., and K.M. performed research; T.G. contributed new reagents/analytic tools; S. Iida, S. Ide, S.T., M. Shribak, M. Sasai, and K.M. analyzed data; and S. Iida., M. Shribak, and K.M. wrote the paper with input from all other authors.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Michael Shribak, Email: mshribak@mbl.edu.
Kazuhiro Maeshima, Email: kmaeshim@nig.ac.jp.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Misteli T., The self-organizing genome: Principles of genome architecture and function. Cell 183, 28–45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maeshima K., Iida S., Tamura S., Physical nature of chromatin in the nucleus. Cold Spring Harb. Perspect. Biol. 13, a040675 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirny L., Dekker J., Mechanisms of chromosome folding and nuclear organization: Their interplay and open questions. Cold Spring Harb. Perspect. Biol. 14, a040147 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miron E., et al. , Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv. 6, eaba8811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nozaki T., et al. , Condensed but liquid-like domain organization of active chromatin regions in living human cells. Sci. Adv. 9, eadf1488 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakirov A. N., et al. , Fiber-like organization as a basic principle for euchromatin higher-order structure. Front. Cell Dev. Biol. 9, 784440 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeshima K., Iida S., Shimazoe M. A., Tamura S., Ide S., Is euchromatin really open in the cell? Trends Cell Biol. 34, 7–17 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Alberts B., et al. , Molecular Biology of the Cell (Norton & Company, ed. 7, 2022).
- 10.Pollard T. D., Earnshaw W. C., Lippincott-Schwartz J., Johnson G., Cell Biology (Elsevier, ed. 4, 2022). [Google Scholar]
- 11.Hirano T., Condensin-based chromosome organization from bacteria to vertebrates. Cell 164, 847–857 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Paulson J. R., Hudson D. F., Cisneros-Soberanis F., Earnshaw W. C., Mitotic chromosomes. Semin. Cell Dev. Biol. 117, 7–29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlmann F., SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Batty P., Gerlich D. W., Mitotic chromosome mechanics: How cells segregate their genome. Trends Cell Biol. 29, 717–726 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Nielsen C. F., Zhang T., Barisic M., Kalitsis P., Hudson D. F., Topoisomerase IIalpha is essential for maintenance of mitotic chromosome structure. Proc. Natl. Acad. Sci. U.S.A. 117, 12131–12142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidson I. F., Peters J. M., Genome folding through loop extrusion by SMC complexes. Nat. Rev. Mol. Cell Biol. 22, 445–464 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Meijering A. E. C., et al. , Nonlinear mechanics of human mitotic chromosomes. Nature 605, 545–550 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider M. W. G., et al. , A mitotic chromatin phase transition prevents perforation by microtubules. Nature 609, 183–190 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsden M. P., Laemmli U. K., Metaphase chromosome structure: Evidence for a radial loop model. Cell 17, 849–858 (1979). [DOI] [PubMed] [Google Scholar]
- 20.Earnshaw W. C., Laemmli U. K., Architecture of metaphase chromosomes and chromosome scaffolds. J. Cell Biol. 96, 84–93 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson D. F., Vagnarelli P., Gassmann R., Earnshaw W. C., Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell 5, 323–336 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Widom J., Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J. Mol. Biol. 190, 411–424 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Dorigo B., Schalch T., Bystricky K., Richmond T. J., Chromatin fiber folding: Requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Allahverdi A., et al. , The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 39, 1680–1691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pepenella S., Murphy K. J., Hayes J. J., A distinct switch in interactions of the histone H4 tail domain upon salt-dependent folding of nucleosome arrays. J. Biol. Chem. 289, 27342–27351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeshima K., et al. , Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 35, 1115–1132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gan H. H., Schlick T., Chromatin ionic atmosphere analyzed by a mesoscale electrostatic approach. Biophys. J. 99, 2587–2596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeshima K., et al. , A transient rise in free Mg(2+) ions released from ATP-Mg hydrolysis contributes to mitotic chromosome condensation. Curr. Biol. 28, 444–451.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Asakura S., Oosawa F., On interaction betweentwo bodies immersed in a solution of macromolecules. J. Chem. Phys. 22, 1255–1256 (1954). [Google Scholar]
- 30.Marenduzzo D., Finan K., Cook P. R., The depletion attraction: An underappreciated force driving cellular organization. J. Cell Biol. 175, 681–686 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hancock R., The crowded nucleus. Int. Rev. Cell Mol. Biol. 307, 15–26 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Delarue M., et al. , mTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hancock R., Packing of the polynucleosome chain in interphase chromosomes: Evidence for a contribution of crowding and entropic forces. Semin Cell Dev. Biol. 18, 668–675 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Hancock R., Structure of metaphase chromosomes: A role for effects of macromolecular crowding. PLoS One 7, e36045 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinchenko A., Berezhnoy N. V., Chen Q., Nordenskiold L., Compaction of single-molecule megabase-long chromatin under the influence of macromolecular crowding. Biophys. J. 114, 2326–2335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zinchenko A., Chen Q., Berezhnoy N. V., Wang S., Nordenskiold L., Compaction and self-association of megabase-sized chromatin are induced by anionic protein crowding. Soft Matter 16, 4366–4372 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Allen R. D., David G. B., Nomarski G., The zeiss-Nomarski differential interference equipment for transmitted-light microscopy. Z. Wiss. Mikrosk. 69, 193–221 (1969). [PubMed] [Google Scholar]
- 38.Salmon E. D., Tran P., High-resolution video-enhanced differential interference contrast (VE-DIC) light microscopy. Methods Cell Biol. 56, 153–184 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Inoué S., The role of microtubule assembly dynamics in mitotic force generation and functional organization of living cells. J. Struct. Biol. 118, 87–93 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Oldenbourg R., Polarized light microscopy: Principles and practice. Cold Spring Harb. Protoc. 2013, pdb.top078600 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Shribak M., Quantitative orientation-independent differential interference contrast microscope with fast switching shear direction and bias modulation. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 30, 769–782 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai R., et al. , Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy. Mol. Biol. Cell 28, 3349–3359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagashima R., et al. , Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol. 218, 1511–1530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larkin K. G., Fletcher P. A., Isotropic scalar image visualization of vector differential image data using the inverse Riesz transform. Biomed. Opt. Express 5, 907–920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shribak M., Larkin K. G., Biggs D., Mapping optical path length and image enhancement using quantitative orientation-independent differential interference contrast microscopy. J. Biomed. Opt. 22, 16006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim T., et al. , RNA-mediated demixing transition of low-density condensates. Nat. Commun. 14, 2425 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barer R., Joseph S., Refractometry of living cells: Part 1. Basic principles. J. Cell Sci. 95, 399–423 (1954). [Google Scholar]
- 48.Liu X., Oh S., Peshkin L., Kirschner M. W., Computationally enhanced quantitative phase microscopy reveals autonomous oscillations in mammalian cell growth. Proc. Natl. Acad. Sci. U.S.A. 117, 27388–27399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mora-Bermudez F., Gerlich D., Ellenberg J., Maximal chromosome compaction occurs by axial shortening in anaphase and depends on Aurora kinase. Nat. Cell Biol. 9, 822–831 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Liang Z., et al. , Chromosomes progress to metaphase in multiple discrete steps via global compaction/expansion (stress) cycles. Cell 161, 1124–1137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hihara S., et al. , Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2, 1645–1656 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Manders E. M., Kimura H., Cook P. R., Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 144, 813–821 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoencamp C., et al. , 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Sci. 372, 984–989 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iida S., et al. , Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells. Sci. Adv. 8, eabn5626 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohta S., et al. , The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell 142, 810–821 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booth D. G., et al. , 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol. Cell 64, 790–802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albiez H., et al. , Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 14, 707–733 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Strickfaden H., et al. , Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772–1784.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 59.Burgess A., Rasouli M., Rogers S., Stressing mitosis to death. Front. Oncol. 4, 140 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson B. A., et al. , Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Itoh Y., et al. , 1,6-hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci. Alliance 4, e202001005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hyman A. A., Weber C. A., Julicher F., Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Gorisch S. M., Wachsmuth M., Toth K. F., Lichter P., Rippe K., Histone acetylation increases chromatin accessibility. J. Cell Sci. 118, 5825–5834 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Gibson B. A., et al. , In diverse conditions, intrinsic chromatin condensates have liquid-like material properties. Proc. Natl. Acad. Sci. U.S.A. 120, e2218085120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belmont A. S., Bruce K., Visualization of G1 chromosomes: A folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127, 287–302 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kireeva N., Lakonishok M., Kireev I., Hirano T., Belmont A. S., Visualization of early chromosome condensation: A hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166, 775–785 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nozaki T., et al. , Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell 67, 282–293.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Xu J., et al. , Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells. Cell Rep. 24, 873–882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olins D. E., Olins A. L., Epichromatin and chromomeres: A “fuzzy” perspective. Open Biol. 8, 180058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma J., Goryaynov A., Sarma A., Yang W., Self-regulated viscous channel in the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 109, 7326–7331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Hooser A. A., Yuh P., Heald R., The perichromosomal layer. Chromosoma 114, 377–388 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Stenstrom L., et al. , Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 16, e9469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Son S., et al. , Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J. Cell Biol. 211, 757–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zlotek-Zlotkiewicz E., Monnier S., Cappello G., Le Berre M., Piel M., Optical volume and mass measurements show that mammalian cells swell during mitosis. J. Cell Biol. 211, 765–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haraguchi T., et al. , Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 113, 779–794 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Maeshima K., A phase transition for chromosome transmission when cells divide. Nature 609, 35–36 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Marenduzzo D., Micheletti C., Cook P. R., Entropy-driven genome organization. Biophys. J. 90, 3712–3721 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sazer S., Lynch M., Needleman D., Deciphering the evolutionary history of open and closed mitosis. Curr. Biol. 24, R1099–R1103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang D., Oliferenko S., Remodeling the nuclear membrane during closed mitosis. Curr. Opin. Cell Biol. 25, 142–148 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Kim T.-K., et al. , Mitotic chromosomes in live cells characterized using high-speed and label-free optical diffraction tomography. Cells 8, 1368 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibcus J. H., et al. , A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le Gratiet A., Marongiu R., Diaspro A., Circular intensity differential scattering for label-free chromatin characterization: A review for optical microscopy. Polymers (Basel) 12, 2428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Bright field movie of fusions of chromatin liquid droplets formed in the presence of 20 mg/mL of PEG (10 s/frame). Three fusion events were recorded in total.
Data Availability Statement
All study data are included in the article and/or supporting information.