Abstract
The dynamics of a hammerhead ribozyme was analyzed by measurements of fluorescence-detected temperature jump relaxation. The ribozyme was substituted at different positions by 2-aminopurine (2-AP) as fluorescence indicator; these substitutions do not inhibit catalysis. The general shape of relaxation curves reported from different positions of the ribozyme is very similar: a fast decrease of fluorescence, mainly due to physical quenching, is followed by a slower increase of fluorescence due to conformational relaxation. In most cases at least three relaxation time constants in the time range from a few microseconds to ~200 ms are required for fitting. Although the relaxation at different positions of the ribozyme is similar in general, suggesting a global type of ribozyme dynamics, a close examination reveals differences, indicating an individual local response. For example, 2-AP in a tetraloop reports mainly the local loop dynamics known from isolated loops, whereas 2-AP located at the core, e.g. at the cleavage site or its vicinity, also reports relatively large amplitudes of slower components of the ribozyme dynamics. A variant with an A→G substitution in domain II, resulting in an inactive form, leads to the appearance of a particularly slow relaxation process (τ ≈200 ms). Addition of Mg2+ ions induces a reduction of amplitudes and in most cases a general increase of time constants. Differences between the hammerhead variants are clearly demonstrated by subtraction of relaxation curves recorded under corresponding conditions. The changes induced in the relaxation response by Mg2+ are very similar to those induced by Ca2+. The relaxation data do not provide any evidence for formation of Mg2+-inner sphere complexes in hammerhead ribozymes, because a Mg2+-specific relaxation effect was not visible. However, a Mg2+-specific effect was found for a dodeca-riboadenylate substituted with 2-AP, showing that the fluorescence of 2-AP is able to indicate inner sphere complexation. Amplitudes and time constants show that the equilibrium constant of inner sphere complexation is 1.2, corresponding to 55% inner sphere state of the Mg2+ complexes; the rate constant 6.6 × 103 s–1 for inner sphere complexation is relatively low and shows the existence of some barrier(s) on the way to inner sphere complexes.
INTRODUCTION
The hammerhead ribozyme is a small catalytic RNA with a core region containing 10 invariant nucleotides and with two substrate binding arms of varying length (1–3). It cleaves the substrate with a certain specificity 3′ to a triplet of the general formula NUH where H can be adenosine, cytidine or uridine but not guanosine. Activity requires divalent metal ions such as Mg2+ or high concentrations of monovalent ions (4). The reaction catalyzed is a phosphoryl transfer where the 2′-OH group of the nucleoside at the cleavage site attacks the phosphorus at the internucleotidic linkage in an in-line mechanism to produce a terminal nucleoside 2′,3′-cyclic phosphate. Several X-ray structures of this ribozyme have been determined, including some structures of intermediate conformations, which probably represent an approach to the transition state (5–7). The general structure is consistent with several solution studies determined mainly by FRET analysis (8–10).
Despite this information the structure of the active conformer and details of the reaction mechanism are still elusive. In particular the precise role of the metal ion is yet uncertain. The overall kinetics of the hammerhead ribozymes is dominated by the steps of substrate binding and/or product release (11), which are determined by the well known mechanism of RNA double helix formation and dissociation (12,13). The cleavage rates are Mg2+-dependent and there is good evidence that one of the roles of Mg2+ is to support the formation of the catalytically competent structure. Mg2+-dependent conformational changes have been observed by gel mobility assays and by FRET analysis (9,10). We have previously determined equilibrium constants for Mg2+-induced conformation changes using the fluorescence of 2-aminopurine (2-AP) incorporated at various positions in the ribozyme (14). We report now on the time dependence obtained by fluorescence detected temperature jump relaxation for the same ribozymes as used in the previous study to obtain insight into the internal dynamics of the ribozyme. We have demonstrated that 2-AP can be introduced without loss of catalytic function (14). Substitutions at different positions of the ribozyme facilitate an analysis of the topology of the ribozyme dynamics. The data show that there are both global and local modes of the dynamics, distributed over a broad time range.
MATERIALS AND METHODS
The hammerhead ribozymes (Fig. 1) and the dodeca-riboadenylate substituted with 2-AP, A(pA)6p(2-AP)(pA)4 [2-AP-oligo(A)], were synthesized by the solid phase method and purified as previously described (14,15). A standard buffer B containing 0.1 M NaClO4, 50 mM cacodylic acid/Tris pH 7.2 was used for most of the measurements.
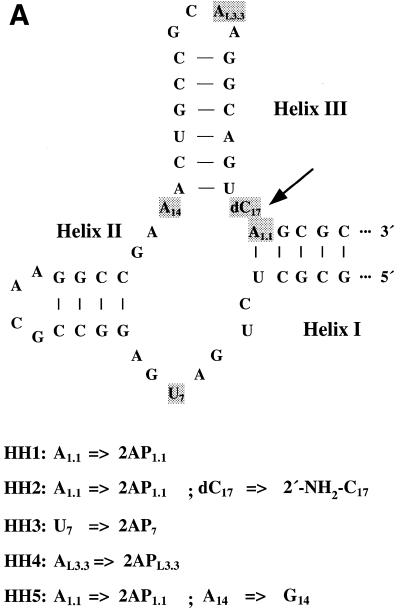
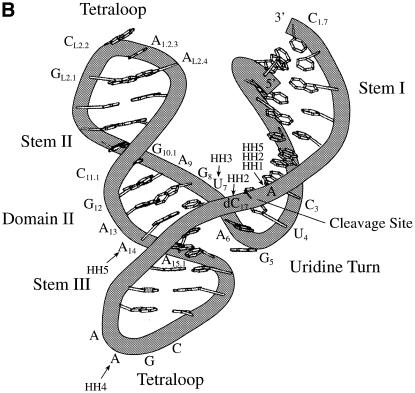
Figure 1.
(A) Sequence and secondary structure of hammerhead ribozyme variants; numbering according to Hertel et al. (20). (B) Model of the hammerhead ribozyme structure for the sequence used in the present investigation with the positions of substitutions; based on the crystal structure of Pley et al. (21), assuming that the essential parts of the structure remain unchanged.
Fluorescence intensities were measured using an SLM 8000 spectrofluorimeter, interfaced to a PC for data collection and averaging. The fluorescence was excited at 320 nm with a bandwidth of 1 nm; the emission was measured at 380 nm with a bandwidth of 16 nm. Before measurements, all solutions were centrifuged within the cuvette in a low-speed desk-top centrifuge for 2 min, in order to clear the solution from dust particles. After thermostating in the cuvette holder for 5 min, the fluorescence intensity was measured for 400 s. There was no indication of a slow process under these conditions. After correction by reference measurements, the data were fitted by least-squares fitting routines using the facilities of the Gesellschaft für wissenschaftliche Datenverarbeitung mbH (Göttingen, Germany). The model used for fitting of the titration data has been described previously (14).
Fluorescence detected temperature jump relaxation was measured with an instrument of the type described by Rigler et al. (16) with improvements by C. R. Rabl (unpublished results). A 200 W Xe/Hg arc lamp together with a Schoeffel GM250 monochromator was used as the light source. The fluorescence was excited at 313 nm and collected behind cutoff filters GG385 (2 mm) from Schott (Mainz, Germany). The transients were initially stored on a Biomation 1010 waveform recorder and finally transmitted to the facilities of the Gesellschaft für wissenschaftliche Datenverarbeitung mbH, for exponential fitting by deconvolution procedures either according to Porschke and Jung (17) or Diekmann et al. (18).
Stopped flow data were measured with fluorescence detection (excitation/emission wavelengths and cutoff filters as in T-jump experiments) using an instrument developed at the Max Planck Institut für biophysikalische Chemie. The stopped flow construction of this instrument is equivalent to that used in a recently described stopped flow field jump instrument (19).
RESULTS
Design of hammerhead constructs
The hammerhead ribozymes used in the present investigation were cis-cleaving constructs with 49 nt in which cleavage was inhibited by the presence either of 2′-deoxycytidine at position 17 for hammerheads HH1, HH3, HH4 and HH5 or of 2′-deoxy-2′-aminocytidine for HH2 (Fig. 1). The constructs were designed to analyze various parts of the molecule for any characteristic contribution to its dynamics and to metal ion binding. Thus, 2-AP reporter groups were substituted next to the 3′-side of the cleavage site (position 1.1 in HH1, HH2 and HH5), at the catalytic core close to the cleavage site (position 7 in HH3) and at the periphery in the stem III tetraloop (position L3.3 in HH4). Ribozyme HH5 was also modified at position 14, where an adenosine was exchanged by a guanosine, abolishing cleavage activity (22).
Broad spectrum of relaxation indicates existence of many conformational substates
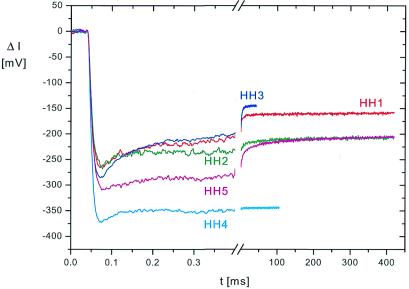
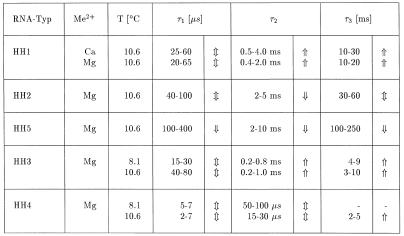
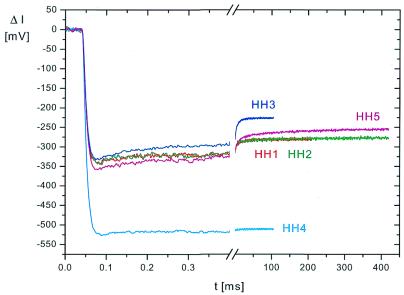
The general form of the relaxation curves induced by temperature jumps in solutions and reported by the fluorescence of 2-AP is very similar for all the hammerhead ribozyme variants: in a first part of the relaxation curves the fluorescence decreases with time constant(s) below the time resolution of the instrument (τ ≤1 µs); in a second part the fluorescence increases with time constants in the range from a few microseconds up to ~200 ms (Fig. 2). The first part is mainly due to the physical processes of thermal quenching, whereas the second part reflects conformational relaxation of the ribozymes. Exponential fitting of these relaxation curves shows the existence of at least three conformational relaxation processes in most cases. Approximate values of relaxation time constants are compiled in Table 1. An example with at least four conformational relaxation processes required for a satisfactory fit is shown in Figure 3 for HH3. Because of the limited amplitudes and the close spacing of the processes on the time scale, it is virtually impossible to deduce exact values for the time constants. Thus, the existence of a continuous spectrum of time constants cannot be excluded.
Figure 2.
Fluorescence detected relaxation curves (change of fluorescence intensity ΔI as a function of time t) for hammerhead ribozymes HH1–HH5 induced by temperature jumps from 2.0 to 10.6°C in buffer B. The data are shown with a pretrigger recording period of 40 µs, i.e. the time before application of joule heating. The time constant of the initial decrease of the fluorescence is determined by the heating time of 8.25 µs and the detector risetime of 5.4 µs (adjusted in these experiments to a relatively slow detection mode).
Table 1. Relaxation time constants τi of hammerhead ribozymes HH1–HH5 from fluorescence detected temperature jump relaxation measurements.
The arrows indicate the change of the respective time constants observed with increasing concentration of metal ions: ⇑ = increase of τ (≡ slower relaxation), ⇓ = decrease of τ (≡ faster relaxation); ⇕ = direction not clearly defined.
Figure 3.
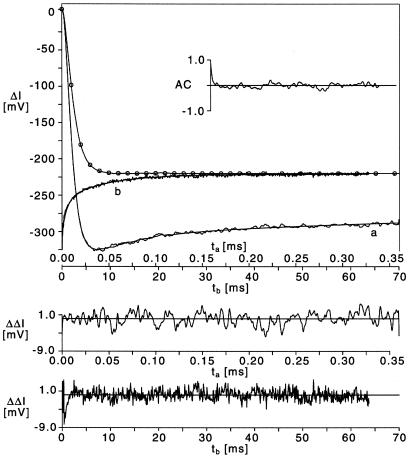
Relaxation curve for HH3 induced by a temperature jump from 2.0 to 10.6°C in buffer B with 5 mM Mg2+, shown in two different time scales (a) and (b). The line marked with circles represents the reference curve measured with a solution of tryptophan under identical experimental conditions; this reference is used for deconvolution (17). The line without noise represents a least squares fit with the time constants 0.56, 5.0, 54, 98 and 8800 µs; the corresponding relative amplitudes are –215.2, 59.7, 18.6, 20.7 and 16.3%. The two lower panels show the residuals for the fast and the slow time scale. The insert shows the autocorrelation function of the residuals.
In spite of this problem in the determination of time constants and the similarity of the general relaxation response, a more detailed comparison of the relaxation curves obtained for the hammerhead variants reveals clear differences (Fig. 2). One extreme case is that of HH4 with 2-AP in the tetraloop, which mainly exhibits a relatively fast relaxation response, which has been observed and characterized previously for isolated hairpin loops (23). The other extreme is HH5, which is not active in catalysis because of the A→G substitution at position 14. In this case the relaxation extends to relatively long times with a rather high amplitude. Thus, it is possible to identify characteristic features of given ribozyme variants.
Search for Mg2+-inner sphere complexes
Measurements of chemical relaxation can be used for an analysis of metal ion binding with respect to inner sphere complexation. Previous studies (24,25) demonstrated that inner sphere complexation of Mg2+ ions is associated with a characteristic relaxation process in the microsecond time range, which is not observed for Ca2+-ions. Thus, the hammerhead relaxation was monitored over a broad range of Mg2+- and Ca2+-ion concentrations from 0.5 to 100 mM. A sample set of relaxation curves obtained at 5 mM Mg2+ for the various hammerhead species is shown in Figure 4. Addition of Mg2+ ions decreased the relaxation amplitudes and in most cases increased the relaxation time constants. These changes reflect the increased electrostatic shielding resulting from ion binding to the ribozyme. Any additional and specific effect associated with Mg2+-binding, indicating inner sphere binding, has not been detected. However, detection would only be possible if this inner sphere effect would be associated with an amplitude at least as high as the amplitude of the conformational relaxation occurring in the same time range. Because the conformational relaxation extends over the whole time range from a few microseconds to ∼200 ms, it is difficult to get unequivocal evidence for any additional relaxation process in the same time range unless the additional process is associated with a sufficiently high amplitude.
Figure 4.
Fluorescence detected relaxation curves for hammerhead ribozymes HH1–HH5 induced by temperature jumps from 2.0 to 10.6°C in buffer B with 5 mM Mg2+. The data are shown with a pretrigger recording period of 40 µs, i.e. the time before application of joule heating. The time constant of the initial decrease of the fluorescence is determined by the heating time of 7.25 µs and the detector risetime of 5.4 µs (adjusted in these experiments to a relatively slow detection mode). The conditions are identical to those in Figure 2 except for the addition of 5 mM Mg2+.
Binding of metal ions to the hammerhead ribozyme HH1 was also analyzed by stopped flow experiments with fluorescence detection. Under the conditions of these experiments the fluorescence was affected by a photoreaction and, thus, the stopped flow reaction curves obtained after mixing of ribozyme with Mg2+ had to be corrected by subtraction of the change due to the photoreaction (measured after equilibration of ion binding). Addition of 10 mM Mg2+ to 1.6 µM ribozyme in the standard buffer at 10°C showed the existence of reaction effects with time constants ranging from a few milliseconds over ∼40 s up to ∼20 min (data not shown). These data demonstrate that the spectrum of reaction effects in hammerhead ribozymes extends over a wide time scale.
The effects observed upon addition of Ca2+-ions to the hammerhead ribozyme HH1 are very similar to those observed upon addition of Mg2+, both in temperature jump and stopped flow experiments (data not shown). Any clear difference in the data obtained for Mg2+ and for Ca2+ could not be identified. Thus, the Mg2+/Ca2+ comparison, which clearly indicated Mg2+-inner sphere complexation in other cases (cf. below), did not indicate inner sphere complexes in the case of hammerhead ribozymes.
Difference relaxation curves illustrate topology of relaxation response
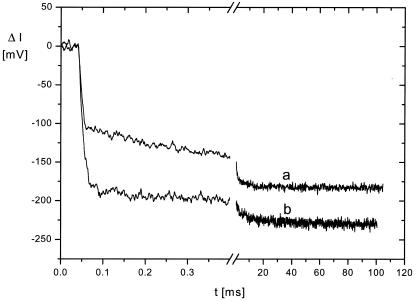
The changes of the conformational relaxation observed upon addition of Mg2+ ions provide some information on the nature of the interactions in the hammerhead ribozyme. These changes are more apparent in difference relaxation plots than in the original data. An example is given for HH2 (Fig. 5) using the data for HH1 as a reference: a difference is clearly visible in the absence of Mg2+, but disappears at 5 mM Mg2+. The ribozyme HH2 is distinguished from HH1 by a 2′-NH2-substitution of the nucleotide at the 5′-side of the cleavage site. This substitution leads to a small perturbation of the structure, which is reflected in its relaxation in the absence of Mg2+, but is not visible any more at 5 mM Mg2+. Because of electrostatic repulsion, intramolecular interactions are expected to be more labile in the absence of Mg2+. Addition of 5 mM Mg2+ reduces repulsion, increases the stability of interactions and, thus, masks the perturbation introduced by the 2′-NH2-group.
Figure 5.
Difference relaxation curves obtained by subtraction of a relaxation curve measured for HH1 from a corresponding curve measured for HH2. (a) Buffer B; (b) buffer B + 5 mM Mg2+. Experimental conditions as described in the legends to Figures 2 and 4.
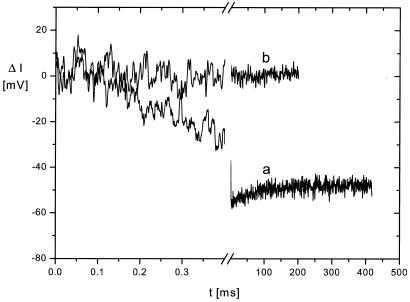
An effect in the opposite direction is found for HH4, where again HH1 is used as a reference. These ribozymes differ in the position of the reporter groups: in HH4 the 2-AP is at the periphery in the stem III tetraloop, whereas in HH1 it is in the core next to the cleavage site. The difference between these two ribozymes is clearly visible in the absence of Mg2+ and increases upon Mg2+ addition (Fig. 6). As mentioned above, the relaxation amplitudes decrease upon Mg2+ addition in all cases. However, the result shown in Figure 6 demonstrates that this decrease is much stronger for HH1 than for HH4. Thus, Mg2+ addition stabilizes the interactions in the core more strongly than those at the periphery.
Figure 6.
Difference relaxation curves obtained by subtraction of a relaxation curve measured for HH1 from a corresponding curve measured for HH4. (a) Buffer B; (b) buffer B + 5 mM Mg2+. Experimental conditions as described in the legends to Figures 2 and 4.
Mg2+-inner sphere complex reported by 2-AP-fluorescence in a substituted dodeca-riboadenylate
The relaxation effects obtained for hammerhead ribozymes in the presence of Mg2+ ions and the absence of evidence for formation of inner sphere complexes raises the question whether the fluorescence of 2-AP can be used to detect inner sphere complexes. Because oligoriboadenylic acids are known to form Mg2+-inner sphere complexes (24,25), an oligoriboadenylic acid with 2-AP-substitution was analyzed by the same approach used for the hammerhead ribozymes. Fluorescence detected temperature jump relaxation using 2-AP-oligo(A) revealed a clear relaxation effect with a time constant of ~100 µs in the presence of mM-concentrations of Mg2+ (Fig. 7), which was not observed in the presence of Ca2+ under the same conditions (data not shown). This result demonstrates formation of Mg2+-inner sphere complexes and shows that the fluorescence of 2-AP can be used to detect these complexes.
Figure 7.
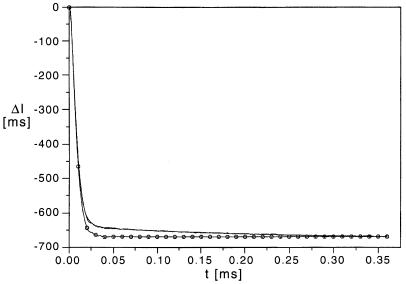
Fluorescence detected relaxation curve for 2-AP-oligo(A) in buffer B + 20 mM Mg2+ induced by a temperature jump from 2.0 to 10.6°C. The line marked with circles is the tryptophan reference curve used for deconvolution; least squares fitting provides a value of 95 µs for the chemical relaxation effect.
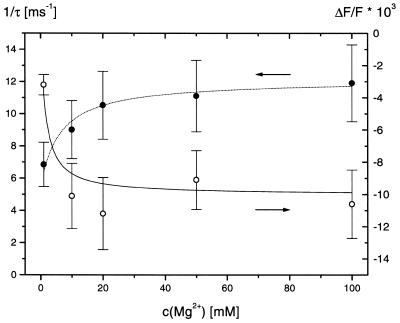
Measurements of the relaxation effect at different Mg2+ concentrations show a dependence of time constants and amplitudes (Fig. 8), which is consistent with the following mechanism
Figure 8.
Reciprocal relaxation time constants 1/τ (filled circles) and relative fluorescence amplitudes ΔF/F (open circles) obtained by temperature jump measurements for 2-AP-oligo(A) at different Mg2+ concentrations in buffer B at 10.6°C. The dotted and the continuous lines represent a combined fit of these data according the reaction model specified in equations 1 and 2. The parameters are: k12 = 1 × 1010 M–1s–1; k21 = 6.3 × 107 s–1; k23 = 6.6 × 103 s–1; k32 = 5.5 × 103 s–1; α = 0.01; β = 2.38; Δln K1 = 0; Δln K2 = –0.0222.
In a first reaction step Mg2+ combines with oligo(A) to an outer sphere complex OSC, which is converted in a second, relatively slow reaction to an inner sphere complex ISC. Because the first reaction step is known to proceed without activation barriers as a diffusion controlled reaction, we use k12 = 1 × 1010 M–1 s–1. The overall binding constant
K = K1(1 + K2) = (k12/k21)/[1 + (k23/k32)] 3
was determined by fluorescence titrations. The values obtained for Mg2+ and Ca2+ (350 and 320 M–1, respectively) are identical within the limits of accuracy (±100 M–1). Furthermore, the relative change of the fluorescence observed in titration experiments Δ [≡ (fluorescence intensity in the limit of saturation with Me2+)/(fluorescence intensity before addition of Me2+)] is related to the individual quantum yields by
Δ = [1/(K2 +1)]α + [K2/(K2 +1)]β 4
where α and β are the quantum yields of OSC and ISC relative to that of the free oligonucleotide, respectively. Again the Δ-values obtained for Mg2+ and Ca2+ (1.30 and 1.26, respectively) are identical within the limits of accuracy (±0.02).
Fitting of the data in Figure 8 provided the parameters k23 = 6.6 × 103 s–1 and k32 = 5.5 × 103 s–1. Thus, the inner sphere binding constant K2 = k23/k32 is 1.2 and the outer sphere binding constant K1 is 160 M–1. Due to mutual coupling of the enthalpy changes and the fluorescence quantum yields, these parameters cannot be determined as individual independent values. The combination of these parameters, which has been used to describe the experimental data in Figure 8, is presented in the legend to this figure.
DISCUSSION
The fluorescence of 2-AP provides a unique approach to the analysis of reaction states and their dynamics in nucleic acids at specific sites. 2-AP may be introduced at various positions within nucleic acid sequences without any serious perturbation of the structure and its fluorescence can be used to report on any process occurring in the environment. It has been shown previously that 2-AP can be introduced into hammerhead ribozymes without loss of catalytic function (14).
In the present investigation 2-AP was used to analyze the dynamics and topology of hammerhead ribozymes. Previously the fluorescence of 2-AP has been demonstrated (23) to indicate changes of base stacking interactions in its environment in an oligoriboadenylate with a time constant of <1 µs. Thus, the relaxation effects described here with time constants in the range from a few microseconds to 200 ms must be attributed to internal conformational relaxation of the hammerhead ribozyme. Most of our constructs show at least three relaxation processes which according to theory (26,27) demonstrate the existence of at least four conformational states of the ribozyme. In some cases even more relaxation effects have been observed. Because the relaxation effects are closely spaced on the time scale, it has not been possible to obtain satisfactory evidence for the existence of discrete processes and, thus, a continuous spectrum of relaxation effects reflecting a corresponding spectrum of conformational states cannot be excluded.
Although a structural assignment of individual relaxation effects is not possible, the experimental data clearly demonstrate the existence of local modes of conformational relaxation. Hammerhead ribozymes are relatively small RNA molecules and, thus, a general global mode of relaxation may have been expected. However, the experiments show particularly large amplitudes of slow relaxation processes in the catalytic core, whereas fast processes are preferentially reflected from the loop region at the periphery.
This investigation complements the earlier study (14) where fluorescence changes of the same 2-AP-containing hammerhead ribozymes were monitored as a function of Mg2+ concentration. The data indicated the presence of multiple binding sites with varying degrees of affinity towards this metal ion. The present results indicate at least three conformational changes at a constant Mg2+ concentration on the pre-steady state time scale. Thus a picture emerges of a rather flexible molecule which undergoes numerous conformational changes to reach its steady state structure. At present it is not possible to assign these changes to a particular structural change. The ribozyme constructs described here are not cleaved and thus additional changes are expected for the cleavage reaction to occur. This is evident from the X-ray structures in the ground state which all require at least one additional local conformational change to reach the transition state to be compatible with an in-line cleavage step (5–7).
RNA folding has been the subject of considerable interest (28). In particular, the folding of the domains of the Tetrahymena ribozyme, which is one of the large ribozymes, has also been studied by pre-steady state kinetics. One study employed time-resolved hydroxyl radical footprinting to follow the folding of the entire molecule (29). Another looked at the folding of the P4–P6 subdomains by stopped flow changes of covalently attached pyrene (30). There is a hierarchy of folding events which range from rates of 2 to 0.02 s–1. Thus, the rates in this system are considerably slower than those detected by temperature jump relaxation for the much smaller hammerhead ribozyme. However, except for the fastest folding of the P5a–P5c domain the processes monitored for the Tetrahymena ribozyme are associated with the formation of domain–domain interactions. The resolution of the methods used for the Tetrahymena ribozyme would not have detected the fast internal changes found for the hammerhead ribozyme.
In addition to the time range and topology of conformational relaxation, temperature measurements have been used in the present investigation to analyze the type of RNA–Mg2+ interactions. On the basis of phosphorothioate interference studies it is generally accepted that one Mg2+ should be coordinated to the pro-Rp phosphate oxygen at the cleavage site (1,31). Although this has not been shown by X-ray analysis of the ground state structures, a Mg2+ in the vicinity of this position could be identified in the X-ray structure approaching the transition state (6). Using the Rp-diastereomer of a phosphorothioate group at the cleavage site, an inner sphere complex with Hg2+ has been observed by UV–Vis spectroscopy (32). There are a number of other metal ion binding sites detected by uranyl-induced photocleavage (10), NMR spectroscopy (33), X-ray structural analysis (5) and phosphorothioate interference (22,34–36). Molecular dynamics simulations have provided evidence for two Mg2+ ions near the active site (37). A site at G5 previously identified as inhibitory in the presence of Tb(III), by displacing an essential Mg2+, has been further characterized by luminescence spectroscopy (38). These experiments indicate that Tb(III) at this position makes two to three inner sphere contacts with the RNA.
Thus, there is evidence for the formation of inner sphere complexation with metal ions other than Mg2+. It would of course be desirable to demonstrate such type of complexation also with this metal ion. Fluorescence detected temperature jump relaxation measurements are suitable to identify the presence of Mg2+-inner sphere complexation on the basis of the characteristic rate constant for inner sphere substitution (25,39,40). This approach has been successful in the analysis of a Mg2+ binding site in the anticodon loop (41) of tRNAPhe and of Mg2+-inner sphere complexes with oligoriboadenylates (24,25). The example described here for the interaction of 2-AP-oligo(A) with Mg2+ as a model system demonstrates that temperature jump relaxation detected by 2-AP fluorescence can distinguish between inner and outer sphere complexation. However, the relaxation spectra of the hammerhead ribozymes were too broad to identify the presence of Mg2+-inner sphere complexation. This does not exclude the existence of such complexes, because their characteristic signal may be hidden in the broad range of conformational relaxation processes.
In summary, application of the fluorescence temperature jump technique to hammerhead ribozymes containing 2-AP demonstrates the existence of many different conformational states. The time range of relaxation processes extends from a few microseconds up to 0.2 s. Evidence for a further extension of this time range up to 20 min is provided by stopped flow experiments. Thus, the time scale of conformation changes in hammerhead ribozymes is more extensive than expected. The present investigation also provides clear evidence for the existence of local relaxation modes. This approach should also be useful for the future assignment of relaxation effects to given structure changes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank U. Kutzke for her expert preparation of RNA samples. The facilities of the Gesellschaft für wissenschaftliche Datenverarbeitung mbH, Göttingen, were used for data processing. This work was financially supported by the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Birikh K.R., Heaton,P.A. and Eckstein,F. (1997) The structure, function and application of the hammerhead ribozyme. Eur. J. Biochem., 245, 1–16. [DOI] [PubMed] [Google Scholar]
- 2.Lilley D.M.J. (1999) Structure, folding and catalysis of the small nucleolytic ribozymes. Curr. Opin. Struct. Biol., 9, 330–338. [DOI] [PubMed] [Google Scholar]
- 3.Carola C. and Eckstein,F. (1999) Nucleic acid enzymes. Curr. Opin. Chem. Biol., 3, 274–283. [DOI] [PubMed] [Google Scholar]
- 4.Murray J.B., Seyhan,A.A., Walter,N.G., Burke,J.M. and Scott,W.G. (1998) The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol., 5, 587–595. [DOI] [PubMed] [Google Scholar]
- 5.Wedekind J.E. and McKay,D.B. (1998) Crystallographic structures of the hammerhead ribozyme – relationship to ribozyme folding and catalysis. Ann. Rev. Biophys. Biomol. Struct., 27, 475–502. [DOI] [PubMed] [Google Scholar]
- 6.Murray J.B., Terwey,D.P., Maloney,L., Karpeisky,A., Usman,N., Beigelman,L. and Scott,W.G. (1998) The structural basis of hammerhead ribozyme self-cleavage. Cell, 92, 665–673. [DOI] [PubMed] [Google Scholar]
- 7.Scott W.G. (1998) RNA catalysis. Curr. Opin. Struct. Biol., 8, 720–726. [DOI] [PubMed] [Google Scholar]
- 8.Tuschl T., Gohlke,C., Jovin,T.M., Westhof,E. and Eckstein,F. (1994) A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science, 266, 785–789. [DOI] [PubMed] [Google Scholar]
- 9.Bassi G.S., Murchie,A.I., Walter,F., Clegg,R.M. and Lilley,D.M.J. (1997) Ion-induced folding of the hammerhead ribozyme – a fluorescence resonance energy transfer study. EMBO J., 16, 7481–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassi G.S., Mollegaard,N.E., Murchie,A.I.H. and Lilley,D.M.J. (1999) RNA folding and misfolding of the hammerhead ribozyme. Biochemistry, 38, 3345–3354. [DOI] [PubMed] [Google Scholar]
- 11.Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porschke D. and Eigen,M. (1971) Cooperative non-enzymic base recognition III. Kinetics of the helix-coil transition of the oligoribouridylic·oligoriboadenylic acid system and of oligoriboadenylic acid alone at acid pH. J. Mol. Biol., 62, 361–381. [DOI] [PubMed] [Google Scholar]
- 13.Craig M.E., Crothers,D.M. and Doty,P. (1971) Relaxation kinetics of dimer formation by self complementary oligonucleotides. J. Mol. Biol., 62, 383–401. [DOI] [PubMed] [Google Scholar]
- 14.Menger M., Tuschl,T., Eckstein,F. and Porschke,D. (1996) Mg2+-dependent conformational changes in the hammerhead ribozyme. Biochemistry, 35, 14710–14716. [DOI] [PubMed] [Google Scholar]
- 15.Tuschl T., Ng,M.M., Pieken,W., Benseler,F. and Eckstein,F. (1993) Importance of exocyclic base functional groups of central core guanosines for hammerhead ribozyme activity. Biochemistry, 32, 11658–11668. [DOI] [PubMed] [Google Scholar]
- 16.Rigler R., Rabl,C.R. and Jovin,T.M. (1974) A temperature-jump apparatus for fluorescence measurements. Rev. Sci. Instrum., 45, 580–588. [Google Scholar]
- 17.Porschke D. and Jung,M. (1985) The conformation of single-stranded oligonucleotides and of oligonucleotide-oligopeptide complexes from their rotation relaxation in the nanosecond time range. J. Biomol. Struct. Dyn., 2, 1173–1184. [DOI] [PubMed] [Google Scholar]
- 18.Diekmann S., Hillen,W., Morgeneyer,B., Wells,R.D. and Porschke,D. (1982) Orientation relaxation of DNA restriction fragments and the internal mobility of the double helix. Biophys. Chem., 15, 263–270. [DOI] [PubMed] [Google Scholar]
- 19.Porschke D. (1998) Time resolved analysis of macromolecular structures during reactions by stopped-flow electrooptics. Biophys. J., 75, 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hertel K.J., Pardi,A., Uhlenbeck,O.C., Koizumi,M., Ohtsuka,E., Uesugi,S., Cedergren,R., Eckstein,F., Gerlach,W.L., Hodgson,R. and Symons,R.H. (1992) Numbering system for the hammerhead. Nucleic Acids Res., 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- 22.Ruffner D.E. and Uhlenbeck,O.C. (1990) Thiophosphate interference experiments locate phosphates important for the hammerhead RNA self-cleavage reaction. Nucleic Acids Res., 18, 6025–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Menger M., Eckstein,F. and Porschke,D. (2000) Dynamics of the RNA hairpin GNRA tetraloop. Biochemistry, 39, 4500–4507. [DOI] [PubMed] [Google Scholar]
- 24.Porschke D. (1979) The mode of Mg++ binding to oligonucleotides. Inner sphere complexes as markers for recognition? Nucleic Acids Res., 6, 883–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porschke D. (1995) Modes and dynamics of Mg2+-polynucleotide interactions. In Cowan,J.A. (ed.), The Biological Chemistry of Magnesium. VCH Publishers, New York, NY, pp. 85–110.
- 26.Eigen M. and De Maeyer,L. (1963) Relaxation methods. In Weissberger,A. (ed.), Technique of Organic Chemistry. Wiley, New York, NY, pp. 895–1054.
- 27.Bernasconi C.F. (1976) Relaxation Kinetics. Academic Press, New York, NY.
- 28.Treiber D.K. and Williamson,J.R. (1999) Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol., 9, 339–345. [DOI] [PubMed] [Google Scholar]
- 29.Sclavi B., Sullivan,M., Chance,M.R., Brenowitz,M. and Woodson,S.A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science, 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- 30.Silverman S.K. and Cech,T.R. (1999) RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry, 38, 14224–14237. [DOI] [PubMed] [Google Scholar]
- 31.Scott E.C. and Uhlenbeck,O.C. (1999) A re-investigation of the thio effect at the hammerhead cleavage site. Nucleic Acids Res., 27, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunningham L.A., Li,J. and Lu,Y. (1998) Spectroscopic evidence for inner-sphere coordination of metal ions to the active site of a hammerhead ribozyme. J. Am. Chem. Soc., 120, 4518–4519. [Google Scholar]
- 33.Hansen M.R., Simorre,J.P., Hanson,P., Mokler,V., Bellon,L., Beigelman,L. and Pardi,A. (1999) Identification and characterization of a novel high affinity metal-binding site in the hammerhead ribozyme. RNA, 5, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoll R., Bald,R. and Fürste,J.P. (1997) Complete identification of nonbridging phosphate oxygens involved in hammerhead cleavage. RNA, 3, 132–140. [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S.L., Karbstein,K., Peracchi,A., Beigelman,L. and Herschlag,D. (1999) Identification of the hammerhead ribozyme metal ion binding site responsible for rescue of the deleterious effect of the cleavage site phosphorothioate. Biochemistry, 38, 14363–14378. [DOI] [PubMed] [Google Scholar]
- 36.Murray J.B. and Scott,W.G. (2000) Does a single metal ion bridge the A-9 and scissile phosphate groups in the catalytically active hammerhead ribozyme structure? J. Mol. Biol., 296, 33–41. [DOI] [PubMed] [Google Scholar]
- 37.Hermann T., Auffinger,P., Scott,W.G. and Westhof,E. (1997) Evidence for a hydroxide ion bridging two magnesium ions at the active site of the hammerhead ribozyme. Nucleic Acids Res., 25, 3421–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feig A.L., Panek,M., Horrocks,W.D. and Uhlenbeck,O.C. (1999) Probing the binding of Tb(III) and Eu(III) to the hammerhead ribozyme using luminescence spectroscopy. Chem. Biol., 6, 801–810. [DOI] [PubMed] [Google Scholar]
- 39.Diebler H., Eigen,M. and Hammes,G.G. (1960) Relaxations-spektrometrische Untersuchungen schneller Reaktionen von ATP in wässriger Lösung. Z. Naturf., 15b, 554–560. [Google Scholar]
- 40.Diebler H., Eigen,M., Ilgenfritz,G., Maass,G. and Winkler,R. (1969) Kinetics and mechanism of reactions of main group metal ions with biological carriers. Pure Appl. Chem., 20, 93–115. [Google Scholar]
- 41.Labuda D. and Porschke,D. (1982) Magnesium ion inner sphere complex in the anticodon loop of phenylalanine transfer ribonucleic acid. Biochemistry, 21, 49–53. [DOI] [PubMed] [Google Scholar]