Significance
In line with intensified efforts to combat the multifaceted impacts of malnutrition, there is a pressing need to develop diabetic-friendly, healthier rice varieties to tackle the escalating global prevalence of diabetes. In this study, we developed recombinant inbred lines with milled rice exhibiting ultralow to low glycemic index (GI) and high protein content (PC) from the cross between Samba Mahsuri and IR36 amylose extender. We performed comprehensive genomics and metabolomics complemented with modeling analyses emphasizing the importance of OsSBEIIb along with additional candidate genes whose variations allowed us to produce target rice lines with lower GI and high PC in a high-yielding background. These lines represent an important breeding resource to address food and nutritional security.
Keywords: glycemic index, starch, protein, QTLs, genomics
Abstract
To counter the rising incidence of diabetes and to meet the daily protein needs, we created low glycemic index (GI) rice varieties with protein content (PC) surpassing 14%. In the development of recombinant inbred lines using Samba Mahsuri and IR36 amylose extender (IR36ae) as parental lines, we identified quantitative trait loci and genes associated with low GI, high amylose content (AC), and high PC. By integrating genetic techniques with classification models, this comprehensive approach identified candidate genes on chromosome 2 (qGI2.1/qAC2.1 spanning the region from 18.62 Mb to 19.95 Mb), exerting influence on low GI and high amylose. Notably, the phenotypic variant with high value was associated with the recessive allele of the starch branching enzyme 2b (sbeIIb). The genome-edited sbeIIb line confirmed low GI phenotype in milled rice grains. Further, combinations of alleles created by the highly significant SNPs from the targeted associations and epistatically interacting genes showed ultralow GI phenotypes with high amylose and high protein. Metabolomics analysis of rice with varying AC, PC, and GI revealed that the superior lines of high AC and PC, and low GI were preferentially enriched in glycolytic and amino acid metabolisms, whereas the inferior lines of low AC and PC and high GI were enriched with fatty acid metabolism. The high amylose high protein recombinant inbred line (HAHP_101) was enriched in essential amino acids like lysine. Such lines may be highly relevant for food product development to address diabetes and malnutrition.
The world is facing a triple burden of malnutrition, marked by rising rates of obesity, undernutrition, and hidden hunger, particularly prevalent in poverty-stricken nations with income inequality (1). Among these challenges, diabetes is a leading contributor to global mortality, with 6.7 million deaths in 2021 alone, with one person dying every 5 s, causing at least 966 billion United States Dollar (USD) losses in health expenditures (2). Globally, 537 million adults are suffering from diabetes, and it is estimated that this will reach up to 783 million by 2045 (2). Among these incidences, type 2 diabetes accounts for 90 to 95% of diabetes (2). The Southeast Asia and Western Pacific regions are the top two regions heavily affected by diabetes, with projections indicating figures could rise to 151.5 million and 260.2 million, respectively, in the next two decades (2). Low- and middle-income countries contribute to more than 75% of diabetes incidences, with Asia accounting for 60% of the global diabetic population (2), wherein 90% of the polished rice is produced and consumed in Asia. Rice, being a staple for more than half of the global population, is consumed primarily in the polished form which is loaded with easily digestible starch (of total ~90% starch; dry weight). This means most cultivated rice varieties exhibit a high glycemic index (GI) (3). Increased consumption of high GI food and highly processed products combined with a sedentary lifestyle collectively leads to an increased risk of type 2 diabetes and other noncommunicable diseases (3). A significant number of individuals in developing countries depend heavily on staple foods such as rice for their daily calorie and protein requirements (4). The identification of rice varieties characterized by low GI with enhanced levels of protein content (PC) (particularly those high in essential amino acids; EAAs) is imperative to improve public health among low- and middle-income communities in Asia.
High-resistant starch (RS)/low-glutelin sbeIIb/Lgc1 lines were generated by CRISPR-Cas9-induced site-specific mutations in SBEIIb in an elite low-glutelin japonica rice cultivar (5). According to this study, the mutant lines showed increases of about two times in amylose content (AC) and 6% in RS with low glutelin content which is the primary rice seed storage protein. These findings point to the trade-off between increased amylose and grain PC. Furthermore, the storage PC of the grain also has an impact on slow digestibility, leading to low GI (6). Among the cereal grains, rice generally has the lowest PC, but its net protein utilization is the highest (7). Only a small number of rice PC genes have been identified to date. Thus, it is of pivotal importance to understand the genetic factors to achieve a pyramided strategy of high amylose combined with high protein lines in a high-yielding background to lower the GI and create healthier protein supplements.
Herein, we used the quantitative trait loci (QTL)-seq method using bulk segregants analysis (BSA-Seq) combined with next-generation sequencing of two pools, the high amylose and high protein (HAHP) lines, and the low amylose and low protein (LALP) lines. QTL and targeted association studies of F6 recombinant inbred lines (RILs) were used to identify the genes responsible for regulating HAHP. We further unraveled the metabolomic signatures of these lines and determined the PC and EAA composition of the pure protein isolate. The functional validation of the sbeIIb allele by gene editing confirmed the low GI phenotype. We contend that the candidate genes and pathways unraveled through this investigation leading to rice breeding lines with ultralow GI, high AC, and high PC can be deployed to develop diabetic-friendly gluten-free rice protein supplements with enriched EAAs for human health.
Results
Genetic Regions Influencing HAHP in Milled Rice Grains Identified by BSA-Seq.
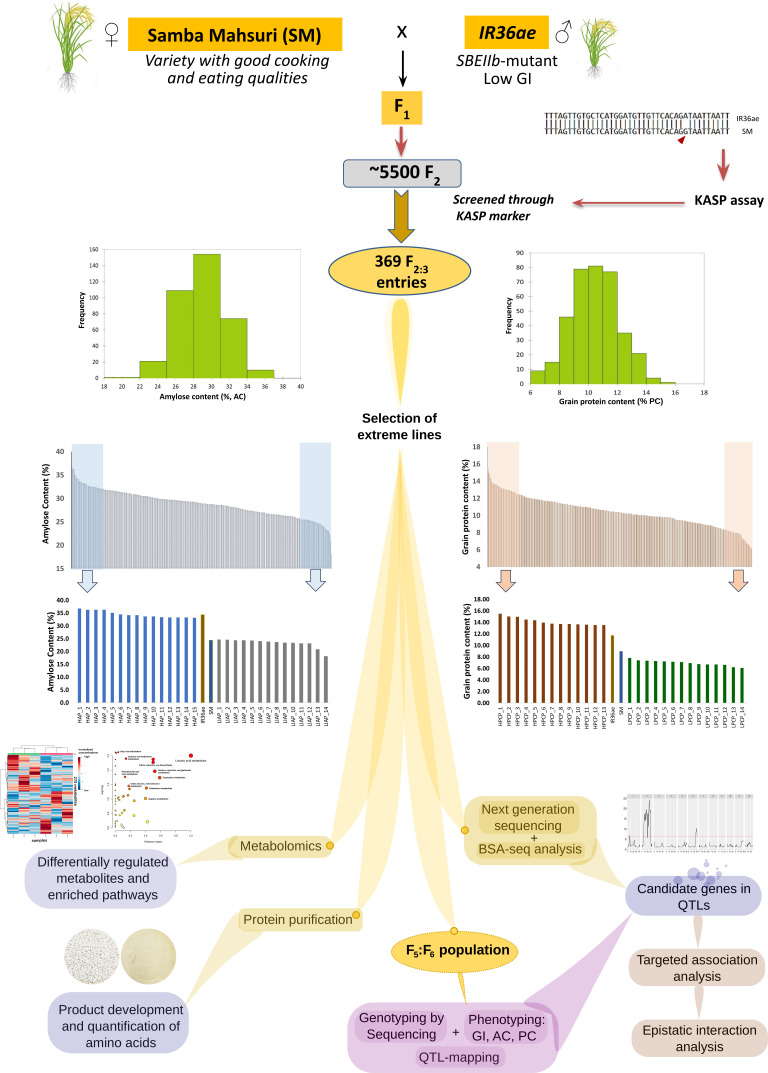
A broad variation in AC (18.1 to 36.8%) was observed in the F3 population developed from the cross of IR36 amylose extender mutant (IR36ae) with high amylose and the recipient line Samba Mahsuri with superior grain quality (Fig. 1). Amylose extender mutants such as IR36ae show higher gelatinization temperature with longer α-1,4-glucan chains and amylopectin compared to their wild-type counterparts (8). The in vitro GI values for the high and low AC bulk pools were 46 (ultralow GI) and 65 (intermediate GI), while the grain PC ranged from 6 to 15.5%. Five significant QTL peaks [qseqAC1.1 (6.02 to 6.18 Mb), qseqAC2.1 (7.74 to 8.61 Mb), qseqAC2.2 (17.15 to 19.95 Mb), qseqAC2.3 (20.72 to 21.90 Mb), and qseqAC6.1 (28.83 to 29.67 Mb)] from Chromosomes 1, 2, and 6 were detected influencing AC based on BSA-seq analysis (Fig. 2A and SI Appendix, Table S1.1). The qseqAC2.1 region showed high and moderate effect SNPs from candidate genes involved in storage protein development (LOC_Os02g15070—OsEnS-32/Glutelin precursor B6), grain size, and shape (LOC_Os02g14730—LARGE GRAIN1) and cell wall structure (LOC_Os02g14900—1,3-beta-glucan synthase), as well as transcription factors including OsEnS-34/PROLAMIN BOX BINDING FACTOR (LOC_Os02g15350) (Fig. 2A and SI Appendix, Table S2.1). A highly significant QTL, qseqAC2.2 (17.15 to 19.95 Mb), also confirmed in the QTL analysis of F6 population, comprised numerous candidate genes involved in carbohydrate, amino acid metabolism, protein synthesis, and lipid metabolism including Oryza sativa Starch Branching Enzyme IIb (OsSBEIIb) (LOC_Os02g32660), which is a known major gene modulating starch branching in rice grain and two other candidate genes regulating sucrose degradation (LOC_Os02g32730—OsNIN3/neutral/alkaline invertase 3, and LOC_Os02g33110—OsCIN1/Cell-wall invertase 1), along with genes encoding germin-like glycoproteins, ankyrin repeat family proteins, and amino acid activation (SI Appendix, Tables S1.1 and S2.1). The qseqAC2.3 also showed significant candidate genes for sucrose degradation (LOC_Os02g34560 OsNIN8/neutral/alkaline invertase 8), elongation of α-glucan chains (DP12-30) during amylopectin synthesis (LOC_Os02g51070—OsSSIIb/Soluble starch synthase IIb), and grain length regulation (LOC_Os02g51320—Positive Regulator of Grain Length 2) (SI Appendix, Tables S1.1 and S2.1).
Fig. 1.
Analytical workflow of RIL population development and downstream multigenomic analysis pipeline for identifying candidate genes and genotypes with ultralow and low GI, high AC, and high PC. The RIL population resulting from the cross between Samba Mahsuri and IR36ae carrying mutation for starch branching enzyme IIb (sbe IIb) gene was initially screened using kompetitive allele-specific PCR (KASP) markers among F2:F3 lines. Highly contrasting lines with extremely high and low AC and PC were selected for further analysis using next-generation sequencing for bulk segregant QTL (BSA-seq) analysis. RIL population advancement made until F6 generation subjected to genotyping-by-sequencing method, phenotyping, and QTL mapping in order to determine candidate genes and important lines possessing ultralow/low GI, high or intermediate AC, and high PC which were then subjected to metabolomics analysis and protein purification for further product development in diabetes-stricken communities, especially in low- and middle-income regions.
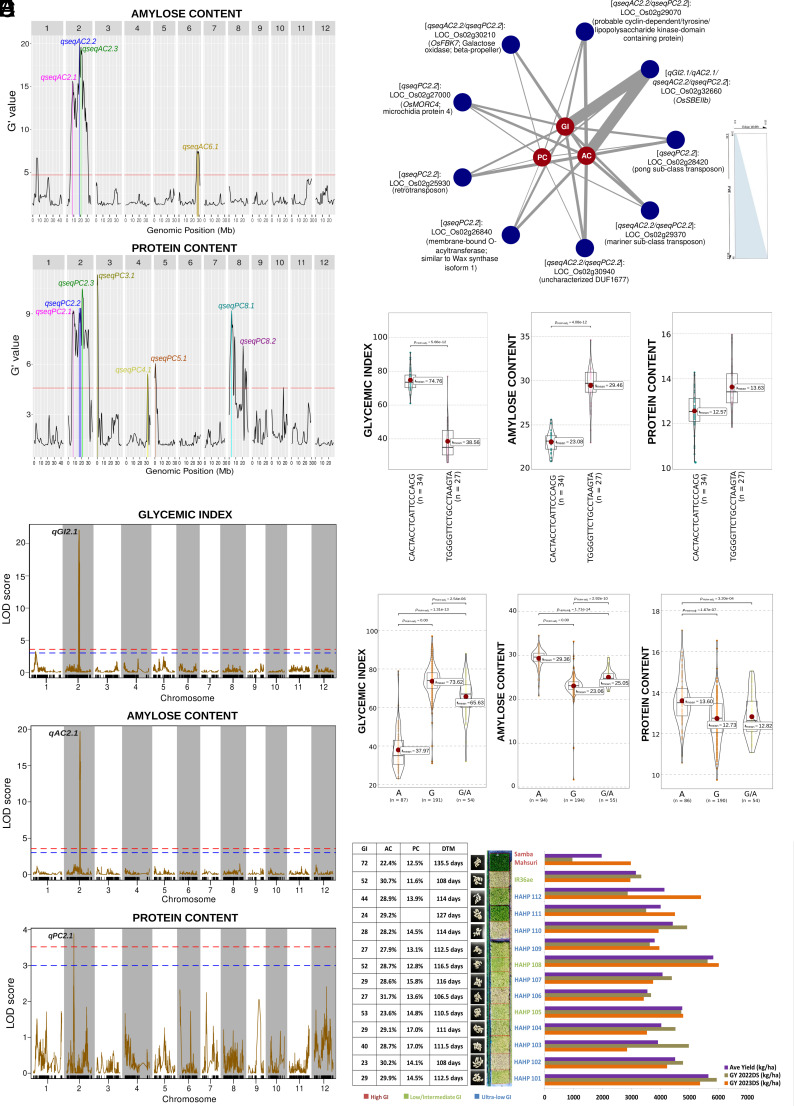
Fig. 2.
Summary of QTLs and candidate genes showing significant phenotypic variations for GI, AC, and PC of RILs based on various genomics approaches. (A) Significant QTL peaks associated with AC and PC based on bulk-segregant sequence analysis (qseq) on pooled contrasting F2-derived F3 RILs. The red horizontal line shows the G’ value significance threshold for the AC and PC BSA-seq analysis. (B) Significant QTL peaks associated with GI, AC, and PC based on composite interval mapping of F5-derived F6 RILs. Respectively, the blue and red dashed lines represent the suggestive and genome-wide Logarithm of the Odds (LOD) score significance thresholds for the QTL analysis. (C) Candidate genes (dark blue nodes) from QTL regions were filtered after targeted association analysis showing phenotypic variations for GI, AC, and PC in the F6 lines. The edge width represents the percentage of phenotypic variation explained (%PVE) by the candidate gene to the corresponding target trait (dark red nodes). (D) Combinations of alleles (shown in the x-axis of each violin plot) from nine candidate genes including OsSBEIIb showed a range of GI, AC, and PC properties in the F6 lines which can be utilized for ultralow GI and high protein rice development. (E) Phenotypic distributions of F5-derived F6 RILs grouped based on a single Single Nucleotide Polymorphism (SNP) change in OsSBEIIb. (F) GI, AC, PC, days-to-maturity (DTM), and grain yield (GY) for two different years (2022 and 2023 dry season, DS) of selected ultralow and low GI RILs for further breeding advancement and product development. The DTM was determined at 100% maturity of seeds from the day of seeding. The appearances of the selected ultralow and low GI RILs in the field were also shown, and the GI classifications are depicted in different colors as follows: red (high GI), green (low/intermediate GI), and blue (ultralow GI).
For grain PC, seven QTLs [(qseqPC2.1 (7.06 to 8.44 Mb), qseqPC2.2 (15.01 to 20.20 Mb), qseqPC2.3 (20.71 to 21.37 Mb), qseqPC3.1 (0.41 to 0.95 Mb), qseqPC4.1 (31.69 Mb), qseqPC5.1 (0.86 to 1.0 Mb), qseqPC8.1 (3.50 to 4.06 Mb), and qseqPC8.2 (20.65 to 21.31 Mb)] were identified (Fig. 2A and SI Appendix, Table S1.2). The QTL qseqPC2.1 overlapping qseqAC2.1 consists of Glutelin B6 (LOC_Os02g15070), Glutelin D (LOC_Os02g15090), and Glutelin 7 (LOC_Os02g15150), as well as some genes involved in sugar transport (LOC_Os02g13560—Tonoplast monosaccharide transporter 2) and meiotic recombination during fertilization (LOC_Os02g13810—Human enhancer of invasion 10) (SI Appendix, Tables S1.2 and S2.2). On the other hand, qseqPC2.2 (overlapping with qseqAC2.2 and qAC2.1) contained the genes including OsSBEIIb, OsNIN3, and OsCIN1 as well as candidate genes encoding glutelin (LOC_Os02g25860—Glutelin A), TRANSPARENT TESTA GLABRA 1B (LOC_Os02g32430), fasciclin-like arabinogalactan proteins, germin-like glycoproteins, and ankyrin repeat family/tetratricopeptide domain-containing proteins (TPRs), along with enzymes such as glycosyltransferases, aminotransferase, galacturonosyltransferase, nucleoside-diphosphate-sugar epimerase, digalactosyldiacylglycerol synthase, and metallo-beta-lactamase-trihelix chimera (SI Appendix, Table S1.1). Almost completely overlapping with qseqAC2.3, qseqPC2.3 also comprised OsNIN8 and PLANT GLYCOGENIN-LIKE STARCH INITIATION PROTEIN A3 (LOC_Os02g35020) with multiple high effect variants for PC (SI Appendix, Table S2.2).
Genetic Regions Influencing Ultralow GI, High Amylose, and High Protein Were Confirmed through QTL Analysis and Targeted Association Studies from the F5-Derived F6 Population.
QTL analysis of the F6 population revealed two significant QTLs: qGI2.1/qAC2.1 (spanning the region from 18.62 Mb to 19.95 Mb influencing ultralow GI and high amylose phenotypes) and qPC2.1 (interval of 10.13 Mb to 10.33 Mb influencing grain protein %) (Fig. 2B and SI Appendix, Table S3). The fine-mapped qGI2.1/qAC2.1 genetic region overlapping with qseqAC2.2 confirmed the locus identified through BSA-seq, comprising numerous candidate genes with high-effect variants significantly associated with GI and AC. The region overlapped OsSBEIIb and confirmed important candidate genes such as OsCIN1 (SI Appendix, Tables S1 and S2). On the other hand, qPC2.1 showed additional candidate genes such as LOC_Os02g17620 (isochorismatase) associated with PC.
Targeted association analysis revealed that important candidate genes identified from the BSA-seq and QTL analyses from the RIL population showed a total of 215 SNPs from 134 candidate genes significantly associated with AC and PC (SI Appendix, Table S4.1). A single nucleotide change to homozygous A allele in SBEIIb contributed 57.2% phenotypic variance explained (PVE) to GI, 60% PVE to AC, and 8% PVE to PC (Fig. 2 C and E). Some candidate genes affecting AC and PC as well as GI and are highly expressed in early developing seeds which include those encoding galactose oxidase/beta propeller (OsFBK7: LOC_Os02g30210), probable lipopolysaccharide kinase domain-containing protein (LOC_Os02g29070), DUF1677 (LOC_Os02g30940), membrane-bound O-acyltransferase (LOC_Os02g26840; similar to Wax synthase isoform 1), OsMORC4 (LOC_Os02g27000), and some transposons (LOC_Os02g25930, LOC_Os02g28420, LOC_Os02g29370) (Fig. 2C and SI Appendix, Table S5). The allelic combinations of the significant SNPs from these candidate genes revealed distinct GI classes ranging from ultralow, intermediate, to high GI with specific groups having high to medium AC and PC. The superior set of lines exhibited ultralow GI (38.56 mean), with high AC (29.46 mean), and high PC (13.63 mean), while the inferior set of lines exhibited high GI 74.76 mean), intermediate AC (23.08 mean), and intermediate PC (12.57 mean) (Fig. 2D).
Other critical candidate genes identified for AC and PC were LOC_Os02g15070 (Glutelin B6/OsEnS-32), LOC_Os02g33110 (OsCIN1), LOC_Os02g34560 (OsNIN8), LOC_Os02g31290 (LARGE1 controlling grain size and weight), LOC_Os02g34630 (MYB transcription factor), LOC_Os02g28970 (microspore and tapetum regulator 1), LOC_Os02g33230 (nucleoside-diphosphate-sugar epimerase), and other genes encoding germin-like proteins, ankyrin repeat/TPRs, sugar and potassium transporters, cysteine protease, glycosyltransferases, fasciclin-like arabinogalactan proteins, and endosperm-specific proteins. Many of these candidate genes involved in stress response, hormone metabolism (mostly gibberellin and ethylene), yield, circadian rhythm, and flowering regulations also showed significant epistatic interactions among each other including various uncharacterized candidate genes (SI Appendix, Fig. S1 and Table S4.2). Some interesting candidate genes such as OsCIN1, OsSAP5, and OsTPR043 showed multiple epistatic interactions with AC and PC-associated genes encoding proteins such as posttranslational modification kinases, wax synthase isoform, putative bZIP protein (LOC_Os02g13810—OsHEI10/Human Enhancer of Invasion 10), signaling G-protein (LOC_Os02g25870), and tonoplast monosaccharide transporter 2 (LOC_Os02g13560), among other unknown genes linked with both AC and PC. OsHEI10 also showed numerous epistatic interactions with significant SNPs of genes encoding galactose oxidase beta-propeller (OsFBK7: LOC_Os02g30210), glycosyltransferase 2 (LOC_Os02g32750), poly ADP ribose polymerase 3 (OsEnS-38: LOC_Os02g32860), and myeloblastosis (MYB) family transcription factor (LOC_Os02g30700).
Distinct Metabolome Profiles of the Pools of High AC and PC versus Low AC and PC Lines.
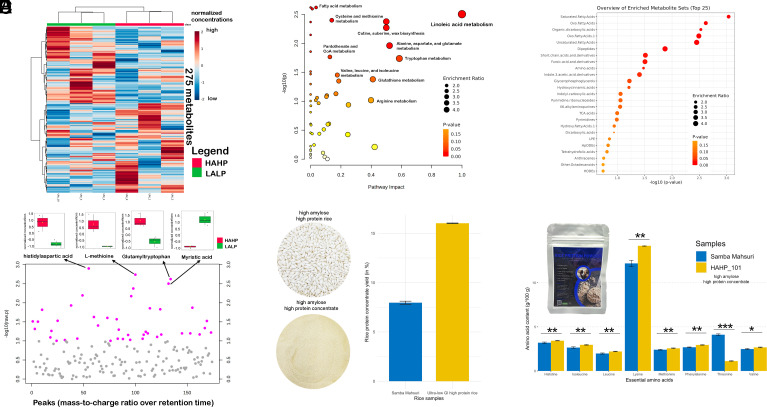
Interestingly, the major pathways distinguishing HAHP and LALP lines were related to metabolism of amino acids (Cys, Met, Val, Leu, Ile, Trp, Ala, Asp, Glu, and glutathione) and lipids (linoleic acid, cutin, suberin, and wax) based on pathway enrichment analysis (Fig. 3 A–C and SI Appendix, Figs. S2 and S3 and Table S6). Zooming into the HAHP group, the top three accumulating metabolites were dipeptides and amino acid, while a fatty acid showed low concentrations (Fig. 3D). To further support the claim of amino acid accumulation in HAHP, the line namely HAHP_101, and the recipient line Samba Mahsuri (considered as control) were selected to test the protein yield using milled grains. Intriguingly, the protein yield after alkali extraction was 15.99% for HAHP_101, whereas only 8.00% of the protein was retrieved from Samba Mahsuri (Fig. 3E). When comparing the colorimetric features of the protein concentrate, HAHP_101 showed slightly lower L* values but slightly higher a* and b* values, indicating a less light appearance compared to Samba Mahsuri (SI Appendix, Table S7). In addition, most EAAs were significantly higher in the HAHP group than in the Samba Mahsuri (Fig. 3F). Notably, lysine exhibited the highest content among the amino acids in the HAHP_101 sample.
Fig. 3.
Metabolomic analysis of HAHP and LALP groups. (A) Hierarchical clustering using 275 metabolites distinguished the HAHP (top red bar: UM_10, UM_3, UM_4) and LALP (top green bar: UM_9, UM_1, UM_2) samples. The red and green colors in the heatmap represent HAHP and LALP, respectively. (B) Pathway enrichment analysis comparing HAHP and LALP groups showed significant (P < 0.05, pathway impact > 0.1) differences in amino acid–related pathways (i.e., metabolism of Cys, Met, Val, Leu, Ile, Trp, Ala, Asp, Glu, and glutathione) and fatty acid-related pathways (i.e., linoleic acid metabolism, cutin, suberin, and wax biosynthesis). The size of the node representing each enriched pathway is directly proportional to the enrichment ratio (number of observed divided by the number of expected metabolites in a pathway), while the color intensity of the node depicts the P-value with lower values for darker orange color. (C) Top 25 types of metabolites identified from the significantly enriched pathways comparing the HAHP and LALP groups. Similarly, the size and color of the nodes represent the enrichment ratio and P-values, respectively. (D) Top four specific metabolites (P < 0.003) accumulating in the HAHP group based on mass-to-charge ratio over retention time analysis. Each dot in pink color represents a specific metabolite passing the significance threshold −log10(p) > 1.0. Boxplots comparing HAHP and LALP for these top four specific metabolites are shown above the dotplot with pairwise comparisons based on the t test. (E) Isolated protein concentrate from the milled grains of the HAHP line (HAHP_101) showed higher protein yield compared to the parental Samba Mahsuri. (F) Rice protein powder from the HAHP_101 exhibited higher levels of various EAAs compared to the parental Samba Mahsuri.
Machine Learning Classification of Ultralow, Low, and High GI and Their Metabolomic Signatures.
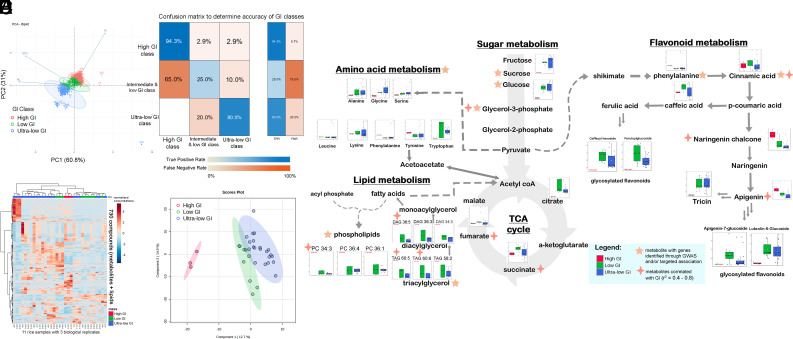
In order to develop cost-effective high-throughput breeding tools for measuring GI, we performed a machine learning strategy for GI classification of rice samples. Initially, principal component analysis (PCA) conducted on selected rice lines showed that GI classes (ultralow, low, intermediate, and high) displayed clear distinction based on AC, with minor contributions from PC (Fig. 4A). Machine learning model using an artificial neural network (ANN) following previous publications (9–11) optimized to 4-68-18-3 architecture also revealed that AC and PC in addition to two significant SNPs (snp_02_19362520 and snp_02_17959252) from OsSBEIIb and OsFBK7 are key variables able to predict the GI class to an accuracy of 74.47%. Furthermore, the model showed robust predictive power for ultralow GI and high GI lines, with true positive rates (TPRs) of 80.0% and 94.3%, respectively, showcasing the high selectivity of the aforementioned GI classes (Fig. 4B). However, challenges brought about by lower marker density from the genotyping-by-sequencing method and missing SNPs in some RILs affected the robustness of the model in accurately predicting the low/intermediate GI class (Fig. 4B). Nevertheless, the machine learning approach demonstrated important phenome-genome parameters which can aid in predicting the GI class of rice samples.
Fig. 4.
Modeling and metabolomic analysis of ultralow, low, and high GI rice lines. (A) PCA using GI, AC, and PC showed clustering of the high, low, and ultralow GI rice samples (n = 386) based on the first two PC explaining more than 90% of the total variation. (B) Confusion matrix depicting the accuracy of the ANN model for predicting the GI classes of the ultralow, low, and high GI rice lines based on AC, PC, and two significant SNPs from OsSBEIIb and OsFBK7. Shades of colors blue and orange in the boxes represent the TPR and false positive rate (FDR), respectively, for each classification based on the model. (C) Further hierarchical clustering using 730 metabolites and lipids distinguished the samples into ultralow, low, and high GI classes. (D) Validation of the clustering was demonstrated through the Partial Least Squares Discriminant Analysis (PLS-DA) model which discriminated the ultralow, low, and high GI rice samples using all the metabolites. (E) Biochemical pathway showing the differentially accumulated metabolites among the ultralow, low, and high GI rice samples, highlighting specific metabolites with candidate genes identified through Genome-Wide Association Studies (GWAS) and/or targeted association analysis and metabolites correlated with GI.
As illustrated in Fig. 4 C and D, the metabolites and lipids distinctly differentiated the low GI, ultralow GI, and high GI lines (SI Appendix, Table S8). Upon scrutinizing the primary metabolites of the rice samples, it became evident that various amino acid biosynthesis and sugar metabolism pathways differed between the ultralow GI and high GI rice samples (SI Appendix, Figs. S4 and S5). Consistent with this feature, the low and ultralow GI rice samples exhibited higher levels of tryptophan and hydroxyproline. Furthermore, sugars showed significant variation among the samples, with erythrose, glucuronic acid, maltose, and erythritol exhibiting the highest log-fold change, favoring the low and ultralow GI rice samples. Besides amino acids, phenolic compounds such as tricin, caffeoylquinic acid, feruloylglucoside, p-coumaroyglucoside, luteolin-6-glucoside, hydroxygallic acid derivative, caffeoyl hexoside, and sinapoyl glucoside were found to significantly accumulate in the low and ultralow GI rice lines based on pathway analysis (SI Appendix, Fig. S6). Conversely, the high GI line exhibited an accumulation of lipids classified as phosphatidylcholines, phosphatidylethanolamines, and their lyso-counterparts (SI Appendix, Fig. S6). Fig. 4E summarizes the metabolite compounds mapped to their respective pathways, highlighting the preferential activation of these pathways. It shows a preferential influx toward amino acid and flavonoid biosynthesis in ultralow GI and HAHP lines, and lipid accumulation in high GI lines, underscoring the potential significance of these pathways in contributing to differences in GI.
CRISPR/Cas9-Mediated OsSBEIIb Mutagenesis in Rice.
The results of BSA-seq, QTL mapping, and targeted association analysis pointed out OsSBEIIb as one of the key candidate genes associated with GI. To further validate these results, a single-guide RNA (sgRNA) was designed to induce mutations at the 15th exon of OsSBEIIb in the high-yielding line International Rice Research Institute (IRRI) 154. The donor splicing site of exon 15 was targeted instead of an earlier exon in order to produce different splicing variants which we expected to exhibit less severe phenotypic effects on the grains in terms of chalkiness and grain size (SI Appendix, Fig. S7A). A total of 92 T0 plants were obtained from Agrobacterium-mediated transformation, out of which 78 T0 plants confirmed transfer DNA (T-DNA) presence based on PCR screening using pUbi-F3/Cas9-97-R primers (SI Appendix, Table S9). Subsequently, T-DNA-positive T0 plants were utilized to amplify the target site with SBEIIb-TS1-F1/R1 primers (SI Appendix, Table S9). Twenty T0 individual events were then subjected to Sanger sequencing. Among the 20 T0 lines, nine mutant lines were identified comprising six heterozygous mutant lines, one biallelic mutant, and two lines exhibiting homozygous mutations. Mutation types included 1-bp insertion, 1-bp deletion, and large deletions up to 41 bp (SI Appendix, Fig. S7B). Notably, line #35 was identified as homozygous with an “A” bp insertion in both alleles, while line #46 exhibited “A” and “G” insertions on each allele (SI Appendix, Figs. S7C and S8). T1 seeds harvested from the knockout lines #35 and #46 were utilized for in vitro GI phenotyping and to estimate RS content. These T1 seeds demonstrated reduced GI values of 55 ± 0.50 and 54 ± 0.35, respectively, compared to the wild type (GI of 59 ± 0.21) and tissue culture control (GI of 58 ± 0.75). Further advanced homozygous lines showed GI of 53 ± 0.05 and 50 ± 0.30 for T2 seeds of #35 and #46, respectively. Moreover, the percentage of RS increased from 0.18 ± 0.0% to 2.50 ± 0.09% and 2.35 ± 0.00% for lines #35 and #46 in T1 seeds, respectively. While, in consequent generation, RS changed to 1.06 ± 0.08 (#35) and 4.32 ± 0.24 (#46) for T2 seeds (SI Appendix, Fig. S7E). This result confirmed that OsSBEIIb is one of the key genes associated with GI. The resulting marker-free lines will be tested in multitrial experiments and pure lines will be positioned for product development or milled rice consumption after passing all biosafety regulations.
Discussion
Addressing the triple challenge of malnutrition demands a concerted effort involving multiple interventions, particularly in staple foods consumed by millions afflicted with nutrition-related illnesses. The nutrition quality of rice together with cooking and eating quality are largely determined by the composition of starch and protein in grain endosperms. Rice with low GI and higher PC is a healthier option to support public health, especially diabetic patients, and for protein supplementation. In this study, we developed pyramided HAHP rice lines exhibiting ultralow GI. These lines were developed from the RILs from the parental lines of Samba Mahsuri (good grain quality) and IR36ae (high amylose & low GI) until the F6 generation. The present study implemented QTL-seq, QTL mapping, targeted association, and epistatic interactions analyses and identified hotspot QTL and candidate genes associated with lowering GI while increasing the PC. Through a CRISPR gene editing approach, OsSBEIIb was validated as one of the key genes for lowering GI. Metabolome analysis revealed differentially regulated metabolites and enriched pathways distinguishing the contrasting lines of HAHP and LALP having a range of GI from ultralow to intermediate classes which can be developed into functional food products.
The Genetics Underlying Low GI and High Protein Lines.
Starch digestibility is affected by numerous extrinsic and intrinsic factors including genetic makeup, composition of starch, and the extent of starch–protein interaction. High amylose crops are usually characterized by lower glycemic food load and higher RS as opposed to low amylose lines which exhibit higher starch digestibility (12). Nonetheless, starch biosynthesis is a complex process regulated by multiple genes along with numerous transcription factors and mediators which further affect grain weight, appearance, and overall quality. Fine mapping and validation of the QTLs were obtained by genotyping by sequencing (GBS) of the F5-derived F6 RILs, composite interval mapping with targeted association analysis, and epistatic interactions. Some significant genes identified in the hotspot QTLs (mostly ranging from less than 1 Mb to 2 Mb) in chromosome 2 included LOC_Os02g32660 (OsSBEIIb), LOC_Os02g15070 (Glutelin B6/OsEnS-32), LOC_Os02g33110 (Cell-wall Invertase 1), LOC_Os02g34560 (OsNIN8/neutral/alkaline invertase 8), LOC_Os02g31290 (LARGE1 controlling grain size and weight), LOC_Os02g34630 (MYB-related transcription factor), three germin-like proteins, and seven ankyrin repeat/TPRs.
By deploying high PVE-contributed SNPs for low GI, high AC, and high PC the modeling results were able to identify sbeIIb as an important locus. The G-to-A mutation at the splice junction between exon and intron 11 of SBEIIb leads to an early stop codon in IR36ae and results in a shorter SBEIIb underpinning low GI with chalky phenotype. Based on the QTL mapping results additional candidate genes influencing ultralow GI phenotype with high protein were identified. Many studies have shown that the inactivation of OsSBEIIb effectively increases the proportion of medium and long amylopectin chains (13) as well as RS (14), and decreases levels of short amylopectin chains (15), leading to overall lower starch digestibility. In the current study, we observed almost 60% variation in apparent AC and 8% variation in PC attributed to a single nucleotide change in OsSBEIIb, further supporting previous knowledge on its substantial role in AC variation and demonstrating additional effects on PC. Additional candidate genes identified in this study must be subjected to further functional validation experiments in order to unravel their importance on GI, AC, and PC. Some of the interesting candidate genes include OsCIN1 involved in cleaving and unloading sucrose in the apoplast to the developing caryopsis during early grain-filling stages, playing complementary and synergistic roles with OsCIN2/GIF1 and OsCIN3 (16–18). Both OsCIN1 and OsCIN2 were reported to be extensively subjected to selection pressure over the years for better grain filling and potential yield (19). Another important candidate gene contributing to AC and PC is OsNIN8 which encodes a neutral/alkaline invertase (Inv-N). These enzymes are known to catalyze the hydrolysis of sucrose to produce glucose and fructose (20, 21). Furthermore, previous studies on some genes encoding ankyrin repeat/TPR such as FLOURY ENDOSPERM 2 were shown to positively regulate both starch biosynthesis and storage proteins biosynthesis (22, 23) by interacting with FLO2-interacting cupin domain protein 2 (FLOC1) and a basic Helix-Loop-Helix (bHLH) transcription factor (24). Another tetratricopeptide-encoding gene, OsCEO1, was identified in a previous study as being a superior conductor of a regulatory cascade of endosperm organogenesis controlling overall rice grain quality (25). These candidate genes might also contribute to AC and PC variations in rice grains, implicating the need for further functional investigations.
Ultralow GI and High Protein Lines Share a Preferential Enrichment of Glycolysis and Amino Acid Biosynthesis.
Homozygous OsSBEIIb mutant plants exhibiting high amylose in a previous study showed broad metabolic changes and transcriptional reprogramming with upregulation of genes encoding ADP-Glucose Pyrophosphorylase (AGPase), soluble starch synthase, and other starch branching enzymes, and downregulation of genes encoding granule-bound starch synthase, debranching enzymes, pullulanase, and starch phosphorylases (26). Similarly, we observed a range of differentially abundant metabolites in the RILs with different GI classifications and high protein lines.
Amino acids usually serve as precursors for a wide array of both primary and secondary metabolites, act as intermediates in the production of end product metabolites, and contribute to the regulation of multiple metabolic pathways (27, 28). The prevalence of amino acid pathways in the superior ultralow GI group can be attributed to the close regulatory network between the aspartate family pathway and the synthesis of EAAs found in the samples. The high number of amino acids associated with the superior rice lines could indicate the effect of the level of free amino acids as precursors of protein accumulation in rice grains (29) since these ultralow GI and low GI (superior lines) contain higher PCs than the high GI lines. Dipeptides like glutamyltryptophan and histidylaspartic acid exhibited accumulations in the superior lines. While the precise roles of these dipeptides remain undefined, it is possible that dipeptides derived from aromatic amino acids act as intermediates in the formation of flavonoids (28). A previous study (30) reported that flavonoid glycosides inhibit α-glucosidase and α-amylase in both in vitro and in vivo experiments. In addition, these flavonoids are known to ameliorate diabetes by regulating numerous cellular networks such as Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB), Phosphoinositide 3-Kinase (PI3K)/Akt, Mitogen-Activated Protein Kinase (MAPK), Glycogen Synthase Kinase 3 (GSK3), and Peroxisome Proliferator-Activated Receptor Gammas (PPARγ). Both lysine degradation and biosynthesis were significant pathways associated with the high amylose/high protein group. High lysine content in the endosperm, associated with ae and mutated opaque (o2) gene expression (31), has been correlated with an increase in starch biosynthesis enzymes (32). Our findings in the high amylose/high protein group revealed a redirection of flux toward glyoxylate/dicarboxylate metabolism, suggesting that the glyoxylate cycle may contribute to the metabolic shift observed in superior ultralow GI combined with high protein rice lines, where amino acid metabolism pathways predominate. In contrast, inferior rice lines with high GI and intermediate protein exhibit a metabolic shift toward fatty acid biosynthesis, highlighting the competition between carbohydrate metabolism and lipid-associated pathways in the low amylose group (33). In particular, saturated fatty acids such as myristic acid and undecanedioic acid are accumulating in the LALP group. Generally, fatty acid content in cereal grains influences lipid stability and affects functionality properties during processing and storage (34). Vital metabolic changes for carbohydrates, protein, and fatty acids were corroborated with the findings identified in QTL-seq results in the present study. Candidate genes identified for exhibiting underlying variations could provide promising linkages to vital metabolic fluxes being regulated.
Rice Protein Enriched with EAAs.
Our current study reports a protein level of 15.99% in the ultralow GI line, representing an almost 500% increase over conventional milled rice (brown rice ≈7.4 g and white rice ≈2.7 g) (35). Additionally, the higher levels of EAAs found in the HAHP sample could significantly contribute to meeting the Recommended Dietary Allowance (RDA) for amino acids (32). For instance, 100 g of the HAHP rice provides EAAs such as isoleucine (2.96 ± 0.04 g), leucine (2.21 ± 0.03 g), lysine (14.19 ± 0.05 g), methionine (2.57 ± 0.04 g), phenylalanine (2.95 ± 0.04 g), and valine (2.67 ± 0.04 g), which can meet the RDA for individuals aged 10 and above (32). This preferential enrichment of EAAs with very high levels of protein may occur through the action of enzymes like aspartate kinase, argininosuccinate synthase, and aspartate semialdehyde (36). Notably, previous studies have demonstrated that the modulation of feedback inhibition of enzymes such as aspartate kinase and dihydrodipicolinate synthase led to a substantial increase, up to 21.7-fold, in lysine levels in rice through a transgenic strategy (37). The high PC of the rice may contribute to slower digestion and absorption rates, enhancing its ultralow GI characteristics (32). It is also important to note that the ultralow and low GI lines developed in this study have yields comparable to high-yielding rice varieties (Fig. 2F). Collectively, these findings underscore the potential of HAHP rice as a nutritional powerhouse, offering a substantial source of protein and EAAs that could play a pivotal role in meeting dietary requirements and addressing nutritional challenges (38), particularly in regions where rice is a dietary staple.
Materials and Methods
Biochemical, genetic materials, and molecular procedures are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
N.S. acknowledges funding from the Foundation for Food and Agricultural Research (CA-21-SS-0000000157), Department of Agriculture and Farmers Welfare, Government of India, and The Indian Council of Agricultural Research. We would like to acknowledge the Department of Science and Technology–Advanced Science and Technology Computing and Archiving Research Environment for providing facilities for most of our analyses. We thank Drs. Tobias Kretzschmar and Juan David Arbelaez from IRRI for providing kind support for initial KASP maker development and conducting initial segregating studies and Dr. Mallikarjuna Swamy from IRRI for providing his valuable suggestions on the breeding methodology adopted. We also thank Socorro Carandang for DNA extraction and acknowledge the support of GBS of The Elshire Group Limited. We thank Commonwealth Scientific and Industrial Research Organisation (CSIRO) Australia for offering the in vitro Glycemic Index (GI) measurement services and Integrated Service Laboratory at IRRI for protein and amylose estimation of samples and cross-cutting service team for setting up crosses, population advancement, and field support. We also acknowledge the Creative Proteomics Company for offering the untargeted metabolomics platform services. We also like to thank Jazlyn Uy for isolating the rice proteins. R.N.T. acknowledges the Academy for International Agricultural Research (ACINAR) for funding his Ph.D. ACINAR, commissioned by the German Federal Ministry for Economic Cooperation and Development (BMZ), is being carried out by German Association for Tropical and Subtropical Agricultural Research (ATSAF) (Council for Tropical and Subtropical Agricultural Research) e.V. on behalf of the Deutsche Gesellschaft für Internationale Zusammenarbeit GmbH. S.A. and A.R.F. acknowledge the European Union’s Horizon 2020 research and innovation programme, project PlantaSYST (Strategic Global Alliance for Climate-Smart Agriculture (SGA-CSA) No. 739582 under Framework Partnership Agreement (FPA) No. 664620) and the BG05M2OP001-1.003-001-C01 project, financed by the European Regional Development Fund through the Bulgarian “Science and Education for Smart Growth” Operational Programme.
Author contributions
S.B., G.S.K., and N.S. designed research; S.B., S.K., S.-R.K., A.R.R.-C., V.P., I.S.-L., J.v.S., S.A., and A.R.F. performed research; S.B., E.A.P.-U., S.K., R.N.T., G.M., R.J.Q.B., L.M.L., and A.R.R.-C. analyzed data; and E.A.P.-U., R.N.T., R.J.Q.B., A.R.F., A.K., G.S.K., and N.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: P.C., Universitat de Lleida; and G.H., Heinrich Heine Universitat Dusseldorf Mathematisch-Naturwissenschaftliche Fakultat.
Contributor Information
Gurudev S. Khush, Email: gurdev@khush.org.
Nese Sreenivasulu, Email: n.sreenivasulu@irri.org.
Data, Materials, and Software Availability
All study data are included in the article and SI Appendix.
Supporting Information
References
- 1.Blankenship J. L., Rudert C., Aguayo V. M., Triple trouble: Understanding the burden of child undernutrition, micronutrient deficiencies, and overweight in East Asia and the Pacific. Matern. Child Nutr. 16, e12950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Diabetes Federation, IDF Diabetes Atlas (International Diabetes Federation, Brussels, Belgium, ed. 10, 2021). [Google Scholar]
- 3.Mohan V., et al. , Effect of brown rice, white rice, and brown rice with legumes on blood glucose and insulin responses in overweight Asian Indians: A randomized controlled trial. Diabetes Technol. Ther. 16, 317–325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henchion M., Hayes M., Mullen A. M., Fenelon M., Tiwari B., Future protein supply and demand: Strategies and factors influencing a sustainable equilibrium. Foods 6, 53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo D., et al. , Evaluation of the quality of a high-resistant starch and low-glutelin rice (Oryza sativa L.) generated through CRISPR/Cas9-mediated targeted mutagenesis. J. Agric. Food Chem. 68, 9733–9742 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Badoni S., Parween S., Henry R. J., Sreenivasulu N., Systems seed biology to understand and manipulate rice grain quality and nutrition. Crit. Rev. Biotechnol. 43, 716–733 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Peng B., et al. , OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 5, 4847 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yano M., Okuno K., Kawakami J., Satoh H., Omura T., High amylose mutants of rice, Oryza sativa L. Theor. Appl. Genet. 69, 253–257 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Buenafe R. J., Tiozon R. Jr., Boyd L. A., Sartagoda K. J., Sreenivasulu N., Mathematical modeling to predict rice’s phenolic and mineral content through multispectral imaging. Food Chem. Adv. 1, 100141 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenafe R. J. Q., Kumanduri V., Sreenivasulu N., Deploying viscosity and starch polymer properties to predict cooking and eating quality models: A novel breeding tool to predict texture. Carbohydr. Polym. 260, 117766 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenafe R. J., Rathnam A., Añonuevo J. J., Sundar S., Sreenivasulu N., Application of classification models in screening superior rice grain quality in male sterile and pollen parents. J. Food Compost. Anal. 104, 104137 (2021). [Google Scholar]
- 12.Emide D., et al. , Molecular insights into the role of amylose/amylopectin ratio on gluten protein organization. Food Chem. 404, 134675 (2023). [DOI] [PubMed] [Google Scholar]
- 13.Butardo V. M., et al. , Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 62, 4927–4941 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang R., et al. , A single amino acid mutation of OsSBEIIb contributes to resistant starch accumulation in rice. Breed. Sci. 66, 481–489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishi A., Nakamura Y., Tanaka N., Satoh H., Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127, 459–472 (2001). [PMC free article] [PubMed] [Google Scholar]
- 16.Wang E., et al. , Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Hirose T., Takano M., Terao T., Cell wall invertase in developing rice caryopsis: Molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant Cell Physiol. 43, 452–459 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Li G., et al. , Phloem unloading in developing rice caryopses and its contribution to non-structural carbohydrate translocation from stems and grain yield formation. Plant Cell Physiol. 63, 1510–1525 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Wang E., et al. , Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication. BMC Evol. Biol. 10, 1–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao S.-G., et al. , Analysis of the rice SHORT-ROOT5 gene revealed functional diversification of plant neutral/alkaline invertase family. Plant Sci. 176, 627–634 (2009). [Google Scholar]
- 21.Jia L., et al. , OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.). Planta 228, 51–59 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki T., et al. , Coordinated regulation of the genes participating in starch biosynthesis by the rice floury-2 locus. Plant Physiol. 110, 89–96 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.She K.-C., et al. , A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22, 3280–3294 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki R., et al. , A novel FLOURY ENDOSPERM2 (FLO2)-interacting protein, is involved in maintaining fertility and seed quality in rice. Plant Biotechnol. 37, 47–55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.She K.-C., et al. , A novel superior factor widely controlling the rice grain quality. Nat. Prec. 6.2, 1–17 (2009). [Google Scholar]
- 26.Baysal C., et al. , Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. U.S.A. 117, 26503–26512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama R., de Oliveira M. V., Kleven B., Maeda H. A., The entry reaction of the plant shikimate pathway is subjected to highly complex metabolite-mediated regulation. Plant Cell 33, 671–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiozon R. J. N., et al. , Metabolomics and machine learning technique revealed that germination enhances the multi-nutritional properties of pigmented rice. Commun. Biol. 6, 1000 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trovato M., Funck D., Forlani G., Okumoto S., Amir R., Amino acids in plants: Regulation and functions in development and stress defense. Front. Plant Sci. 12, 772810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadeghi M., Miroliaei M., Ghanadian M., Inhibitory effect of flavonoid glycosides on digestive enzymes: In silico, in vitro, and in vivo studies. Int. J. Biol. Macromol. 217, 714–730 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Wang W., et al. , The Zea mays mutants opaque2 and opaque 16 disclose lysine change in waxy maize as revealed by RNA-Seq. Sci. Rep. 9, 12265 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia M., et al. , Identification and characterization of lysine-rich proteins and starch biosynthesis genes in the opaque2mutant by transcriptional and proteomic analysis. BMC Plant Biol. 13, 1–14 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Z., Wang Y.-S., Chen H.-H., Li Q.-Q., Wang Q., The gelatinization and retrogradation properties of wheat starch with the addition of stearic acid and sodium alginate. Food Hydrocoll. 81, 77–86 (2018). [Google Scholar]
- 34.Lu X., et al. , Pasting, rheology, and fine structure of starch for waxy rice powder with high-temperature baking. Int. J. Biol. Macromol. 146, 620–626 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Juliano B. O., Tuaño A. P. P., Gross Structure and Composition of the Rice Grain, Rice J.-S. Bao., Ed. (AACC International Press, 2019), pp. 31–53. [Google Scholar]
- 36.Han M., et al. , L-Aspartate: An essential metabolite for plant growth and stress acclimation. Molecules 26, 1887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Q. Q., et al. , Lysine biofortification in rice by modulating feedback inhibition of aspartate kinase and dihydrodipicolinate synthase. Plant Biotechnol. J. 19, 490–501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eggum B. O., Juliano B. O., Perez C. M., Khush G. S., Starch, energy, and protein utilization by rats in milled rice of IR36-based amylose extender mutant. Cereal Chem. 70, 275–279 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and SI Appendix.