Abstract
Telomere length and telomerase activity directly affect the replicative capacity of primary human cells. Some have suggested that telomere length influences organismal lifespan. We compared telomere length distributions in a number of inbred and outbred established mouse strains with those of strains recently derived from wild mice. Telomere length was considerably shorter in wild-derived strains than in the established strains. We found no correlation of telomere length with lifespan, even among closely related inbred mouse strains. Thus, while telomere length plays a role in cellular lifespan in cultured human cells, it is not a major factor in determining organismal lifespan.
INTRODUCTION
Telomere length has been proposed to function as a biological clock, limiting the proliferative capacity of both cells in culture and somatic cells during organismal development (1,2). Primary human cells in culture divide for 40–60 generations; the fraction of dividing cells decreases progressively and eventually all of the cells cease dividing (3,4). In the absence of telomerase, the enzyme responsible for synthesizing telomere repeats, telomere length in primary fibroblasts decreases with increasing cell division (2). Introduction of telomerase into cultured fibroblasts allows cells to bypass senescence and proliferate indefinitely (5). Reduction in telomere length can also results in considerable defects in vivo (6–8). Although early generation telomerase knock-out mice show no significant abnormalities, late generation knock-out mice show reduced telomere length, proliferative defects in several organ systems and, ultimately, reduced lifespan (6–8).
The correlation between telomere shortening and cellular senescence has led to the hypothesis that telomere length is a significant determinant of organismal life span (9,10). Telomere length is genetically determined and differs between species (11). Within a given species telomere length is well regulated about an equilibrium length. Previous reports have suggested telomere length in the mouse is significantly longer than in human cells (12). This difference between mouse and human telomere length has been suggested to account for differences in the response to telomere shortening in these two species (13).
Established inbred mouse strains have long, hypervariable telomere lengths, ranging from 30 to 150 kb (12). In contrast, the wild-derived mouse species Mus spretus, has telomeres ranging from 8 to 10 kb (11,14–16). Because M.spretus is evolutionarily quite distinct from Mus musculus inbred strains, it was unclear whether the short telomere in M.spretus represented a unique exception to the generally long telomeres in mice.
To assess whether long telomere length is an important component of mouse chromosomes, we examined telomere length in a number of wild-derived inbred mouse strains. Wild-derived inbred mice have been inbred within the last 20–30 years, while the more established inbred strains were derived more than 60 years ago (17). Our results indicate that recently inbred mouse strains have telomere lengths considerably shorter than those found in established inbred mouse strains. This suggests that there is no requirement for long, hypervariable telomeres in mice. Additionally, mice with short telomere lengths show no significant reduction in lifespan, indicating that while telomere length may play a role in human cellular senescence it is not a natural determinant of organismal lifespan.
MATERIALS AND METHODS
Mouse strains
C57BL/6, 129/SvJ, FVB/NJ, SPRET/Ei and CAST/Ei mice were obtained from the Jackson Laboratory. Livers from Mus musculus denmark, Mus musculus castaneus, Mus hortulanus, Mus macedonicus and Mus caroli inbred mouse livers were kindly provided by the Chapman laboratory at the Roswell Park Cancer Institute. Peromyscus leucopus livers were kindly provided by Virginia Hayssen at Smith College. Swiss Webster, Black Swiss and ICR outbred mouse livers were obtained from Taconic.
Isolation of liver cells
Whole livers were harvested from 3–6-month-old mice. One gram (~50%) of each liver was homogenized in 5 ml of an ice-cold solution containing 60 mM KCl, 15 mM NaCl, 0.5 mM spermine, 0.15 mM spermidine, 15 mM Tris pH 7.5, 2 mM EDTA, 0.3 M sucrose and 16.7 mM β-mercaptoethanol. The homogenates were layered on top of 7.5 ml of a solution containing 60 mM KCl, 15 mM NaCl, 0.5 mM spermine, 0.15 mM spermidine, 15 mM Tris pH 7.5, 1 mM EDTA, 1.37 M sucrose and 14.4 mM β-mercaptoethanol in 25 × 89 mm ultracentrifuge tubes. The sucrose gradients were centrifuged at 12 000 g for 15 min at 4°C. After centrifugation, the supernatant was removed, and the pellet was washed in 10 ml Dulbecco’s phosphate buffered saline without calcium and magnesium (PBSA). After washing, the cells were pelleted by centrifugation for 5 min at 300 g and resuspended in PBSA at a density of 2 × 107 cells/ml.
Plug preparation
Liver cells were combined with an equal volume of 2% low melting point agarose at 45°C and cast into 100 µl plug molds. After casting, plugs were incubated in a lithium dodecyl sulfate (LDS) solution, containing 1% LDS, 100 mM EDTA pH 8.0 and 10 mM Tris pH 8.0, at 37°C for 1 h with constant agitation. The solution was then replaced with fresh solution, and the plugs were incubated overnight at 37°C. The plugs were washed twice for at least 2 h in 20% NDS (2 mM Tris, 6.8 mM N-laurylsarcosine and 127 mM EDTA brought to pH 9.5 with NaOH). Plugs were stored at 4°C in 20% NDS.
Pulsed-field gel
DNA plugs to be digested were incubated for 1 h in TE, then again for 1 h in fresh TE. The DNA plugs were then incubated for 2 h in 1× DpnII restriction enzyme buffer, with the initial enzyme buffer replaced with fresh enzyme buffer after 1 h. Each plug was then placed in 400 µl 1× DpnII buffer with 60 U DpnII and digested for 16 h at 37°C. After 16 h, an additional 30 U of enzyme was added, and the plugs were incubated for 8 h. λ DNA concatamers embedded in agarose (New England Biolabs) were used as molecular weight markers.
Plugs were run on a 1.0% Pulsed Field Grade Agarose (Bio-Rad) gel in 1× Tris acetate EDTA (TAE). Gels were run using a CHEF-DR II pulsed field gel apparatus (Bio-Rad) for 20–24 h at a constant 200 V, using ramped pulse times from 1 to 10 s. Running temperature was kept at 14°C.
In-gel hybridization
Following electrophoresis and EtBr staining, gels were dried down on filter paper for 1 h at 50°C. Gels were prehybridized for 1 h at 55°C in 20 mM NaH2PO4, 0.1% SDS, 5× Denhardt’s reagent and 5× SSC and hybridized with probe for 3 h at 55°C in 5 ml prehybridization solution. (CCCTAA)4 oligonucleotides were obtained from Operon Technologies Inc. Oligonucleotides were end-labeled with [γ-32P]ATP and then purified using NAP™5 columns (Pharmacia Biotech). After hybridization, gels were washed three times for 20 min in 3× SSC at room temperature and three times for 20 min in 3× SSC/0.1% SDS at 58°C. Image analysis was performed using a FUJIFILM BAS 1500 Bio-Imaging Analysis System including Image Reader Software version 1.3E.
Quantitative fluorescent in situ hybridization (Q-FISH) analysis
C57BL/6J and CAST/Ei mice were intra-peritoneally injected with colcemid at a concentration of 1 µg/g bodyweight. After 4 h, whole spleens were removed from the mice and macerated in 10 ml PBS. Cell suspensions were then centrifuged for 10 min at 300 g. The resulting cell pellets were resuspended in 10 ml 0.075 M KCL at 37°C and incubated for 20 min. The tubes were then centrifuged for 10 min at 300 g, and the pelleted cells were fixed in 10 ml cold 3:1 methanol:acetic acid. The cells were washed in fresh fixative at least twice before using. Cells were dropped onto clean glass slides to generate metaphase spreads.
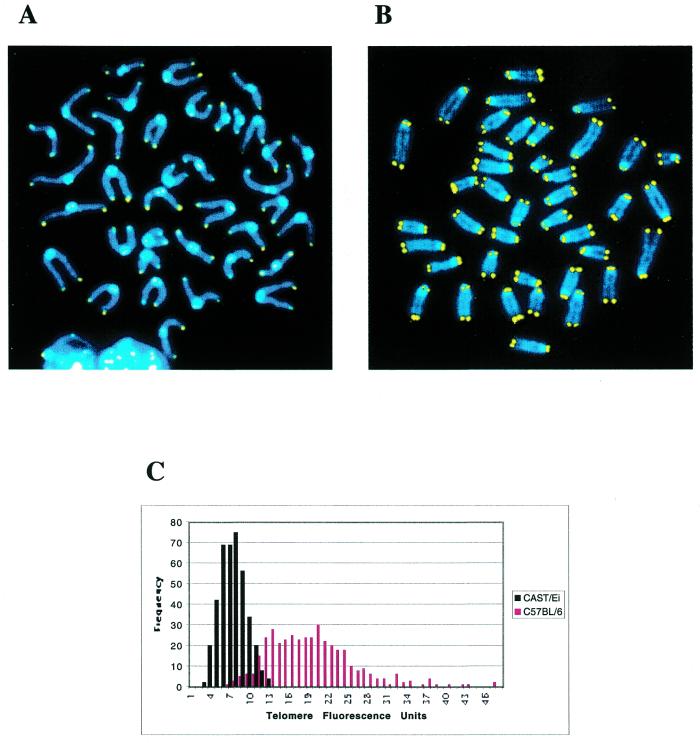
PNA FISH was performed using a Cy3-labeled (CCCTAA)3 PNA oligonucleotide (PE Biosystems) as described (18). Chromosomes were counterstained with DAPI. FluoreSpheres® fluorescent beads (Molecular Probes) were used to monitor signal intensity loss during microscope use. Signal intensities were calculated using designated Q-FISH software provided by the Lansdorp laboratory (18). Telomere signal intensities from 10 metaphases (800 telomeres) for each mouse are shown in Figure 2.
Figure 2.
Q-FISH analysis of CAST/Ei and C57BL/6J telomeres. (A) CAST/Ei metaphase spread hybridized with a Cy3-labeled (CCCTAA)3 PNA probe. Chromosomes are counterstained with DAPI. (B) C57BL/6J metaphase spread hybridized with a Cy3-labeled (CCCTAA)3 PNA probe. (C) Frequency distributions of CAST/Ei (black) and C57BL/6J (red) telomere fluorescence intensities. Fluorescent signals from both p-arm and q-arm telomeres were used for this analysis.
RESULTS
Mus musculus castaneus mice have short telomeres
To examine whether long telomeres are a universal feature of M.musculus mice, we examined the telomere restriction fragment (TRF) size of four different inbred M.musculus strains. TRF lengths of previously examined mouse strains vary between 30 and 200 kb and cannot be resolved via conventional gel electrophoresis (12). Liver cells isolated from three independent C57BL/6J, 129S/SvJ, FVB/NJ and CAST/Ei mice were embedded in agarose plugs, digested with DpnII and resolved on a pulsed-field gel. The gel was analyzed by denaturing in-gel hybridization with a 32P-end-labeled telomere repeat oligonucleotide, (CCCTAA)4 (Fig. 1). As has been shown previously, C57BL/6J and 129/SvJ mice both exhibited long heterogeneous TRF distributions (12). Likewise, FVB/NJ mice also exhibited long heterogeneous TRF lengths, ranging from ~30 to 120 kb. Interestingly, CAST/Ei mice showed a considerably shorter TRF distribution, with an average length range of ~18–20 kb.
Figure 1.
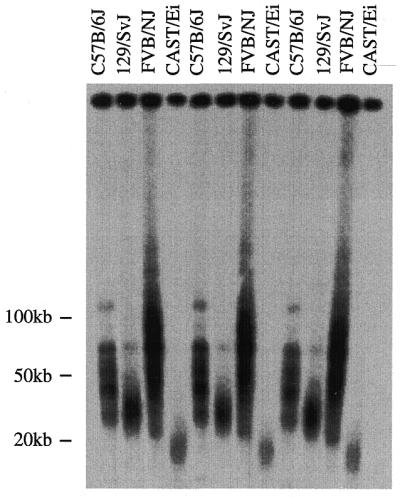
CAST/Ei mice have short, homogeneous telomere length distributions. End-labeled (CCCTAA)3 in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. TRF distributions from three different C57B/6J, 129/SvJ, FVB/NJ and CAST/Ei mice are shown.
Because variations in TRF length can potentially include differences in the location of the terminal restriction site, we also performed Q-FISH (18) on C57BL/6J and CAST/Ei metaphase spreads. This method uses directly labeled peptide nucleic acid (PNA) telomere repeat oligonucleotides to quantitate the number of repeats on individual metaphase chromosome ends. Results from this experiment for C57BL/6J and CAST/Ei mice are represented as frequency distributions of telomere signal intensities. As was the case for TRF distributions, CAST/Ei telomere signal intensities were considerably lower and more homogeneous than C57BL/6J telomere signal intensities (Fig. 2). This result confirms that long, hypervariable telomere lengths are not a conserved or required feature of M.musculus chromosomes.
Because it has been suggested that telomere length might, in part, determine organismal lifespan, we compared the telomere length to the lifespan of these two strains. Interestingly there was no significant difference in maximal lifespan between C57BL/6J (767 ± 147 days) and CAST/Ei (681 ± 168 days) mice (D.Harrison, personal communication), arguing against a significant role for telomere length in determining lifespan.
Wild-derived mice have short telomeres
The presence of short telomeres in two wild-derived inbred mouse strains, M.musculus castaneus and M.spretus, suggested that short telomeres may be a common feature of recently inbred mouse strains. To further investigate telomere length in wild-derived mice, we used pulsed-field gels to examine the TRF distributions in several recently inbred strains: M.musculus denmark, M.caroli, M.hortulanus and M.macedonicus. All of these wild-derived strains examined, including an independent colony of M.musculus castaneus, showed average TRF lengths of <25 kb, with most TRFs <20 kb (Fig. 3). Analysis of at least two independent mice from each strain revealed similar results (data not shown). We next examined the TRF distribution of a non-Mus wild-derived mouse strain, P.leucopus. These mice show unusual longevity for a mouse species, averaging between 5 and 7 years (19). TRF lengths from these mice were also quite short, averaging ~12 kb (Fig. 3). The TRF distributions from these wild-derived strains were similar to those seen for normal human fibroblast cell lines, including IMR90 and WI38. Finally, analysis of telomere length from several trapped M.musculus wild mice showed TRF distributions similar to M.musculus castaneus (data not shown). These results suggest that, although established inbred mouse strains have long, hypervariable TRF distributions, long telomeres are not commonly present in wild-derived mice. The increase in TRF length that apparently occurred in established inbred strains could have arisen as a result of the infrequent appearance and retention of long telomeres within a small, isolated colony of mice. Alternatively, gradual telomere elongation could occur as a consequence of extensive brother–sister matings during the inbreeding process. To differentiate between these possibilities, we examined the TRF distributions in several outbred mouse strains: ICR, Black Swiss and Swiss Webster. These are established strains that are bred in small colonies, but they are specifically not inbred (20). Pulsed-field gel analysis of these established outbred mouse telomeres revealed long, hypervariable distributions in all three strains (Fig. 4). Thus, it appears that TRF elongation occurs with extensive breeding of an isolated colony of mice not specifically with inbreeding.
Figure 3.
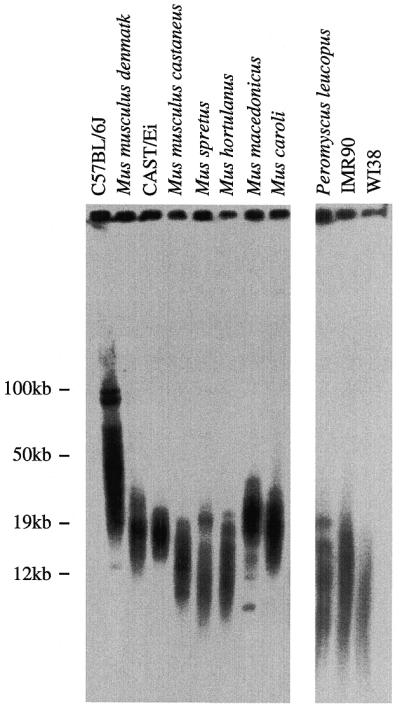
Wild-derived mouse strains have short telomeres. End-labeled (CCCTAA)3 in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. TRF distributions from C57BL/6J, M.musculus denmark, CAST/Ei, M.musculus castaneus, M.spretus, M.hortulanus, M.macedonicus, M.caroli and P.leucopus mice and two human fibroblast cell lines (IMR90 and WI38).
Figure 4.
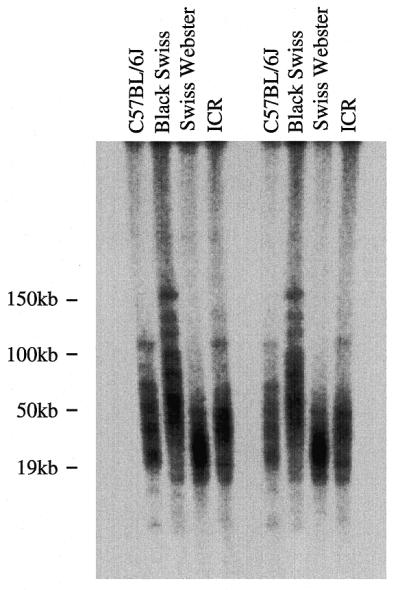
Established outbred mouse strains have long telomeres. End-labeled (CCCTAA)3 in-gel hybridization to DpnII digests of agarose-embedded genomic DNA. TRF distributions from two different C57BL/6J, Black Swiss, Swiss Webster and ICR mice.
DISCUSSION
The discovery of long, hypervariable TRFs in a number of inbred mouse strains led to the hypothesis that long telomeres are characteristic of M.musculus chromosomes (21,22). Interestingly, our data suggests that, not only are long telomeres not conserved among mouse species, but that most mouse species studied have short telomeres. These differences in telomere length do not likely represent strain-specific requirements for telomere length, because closely related strains of M.musculus show very different TRF distributions. Additionally, we found no correlation between telomere length and longevity in closely related mouse strains. In fact, the strain with the shortest telomeres of those we studied, P.leucopus, had the longest lifespan.
The presence of long, hypervariable TRF distributions specifically in established inbred and outbred mouse strains and not in recently derived inbred lines suggests that telomere elongation occurs as a function of extensive breeding in an isolated colony. The mechanism for this elongation is currently unclear. One possibility is that a selection for shorter telomeres exists among wild mice, and upon placement in a non-selective breeding environment, long telomeres can be maintained. The de novo appearance of single long telomeres in individual mice could, after extensive breeding, promote the elongation of the majority of telomeres in a given colony. Alternatively there may be a mechanism for the general elongation of short telomeres. In crosses between mice with long and short telomeres, a specific elongation of the short telomeres in the offspring occurs (23). It may be that, unlike yeast (24), there is no specific mechanism to shorten long telomeres once they are introduced into a mouse.
Comparisons between mouse and human telomere length dynamics and the consequences of telomere dysfunction have been problematic given the significant differences in telomere length. For example, it is unclear whether the ability of mice to survive multiple generations in the absence of telomerase is due to the extremely long telomeres in C57BL/6 mice or a fundamental difference in the mouse and human response to telomere dysfunction. The discovery of M.musculus mice with telomere lengths similar to those in human cells should significantly enhance our ability to develop mouse models for telomere dysfunction that more closely resemble the human telomere length distribution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr David Harrison for helpful discussions and Margaret Strong for critical reading of the manuscript. We also thank Virginia Hayssen and the Chapman laboratory for generously providing mouse tissue. This work was supported by NIH grant CA16519 to C.W.G.
REFERENCES
- 1.Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 3.Hayflick L. (1965) Exp. Cell Res., 37, 614–636. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein S. (1990) Science, 249, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 6.Blasco M.A., Lee,H.W., Hande,M.P., Samper,E., Lansdorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 7.Lee H.W., Blasco,M.A., Gottlieb,G.J., Horner,J.W.,II, Greider,C.W. and DePinho,R.A. (1998) Nature, 392, 569–574. [DOI] [PubMed] [Google Scholar]
- 8.Rudolph K.L., Chang,S., Lee,H.W., Blasco,M., Gottlieb,G.J., Greider,C. and DePinho,R.A. (1999) Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- 9.Harley C.B., Vaziri,H., Counter,C.M. and Allsopp,R.C. (1992) Exp. Gerontol., 27, 375–382. [DOI] [PubMed] [Google Scholar]
- 10.Harley C.B. (1991) Mutat. Res., 256, 271–282. [DOI] [PubMed] [Google Scholar]
- 11.Kipling D. (1995) The Telomere. Oxford University Press, New York, NY, pp. 31–77.
- 12.Kipling D. and Cooke,H.J. (1990) Nature, 347, 400–402. [DOI] [PubMed] [Google Scholar]
- 13.de Lange T. and Jacks,T. (1999) Cell, 98, 273–275. [DOI] [PubMed] [Google Scholar]
- 14.Starling J.A., Maule,J., Hastie,N.D. and Allshire,R.C. (1990) Nucleic Acids Res., 18, 6881–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prowse K.R. and Greider,C.W. (1995) Proc. Natl Acad. Sci. USA, 92, 4818–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coviello-McLaughlin G.M. and Prowse,K.R. (1997) Nucleic Acids Res., 25, 3051–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck J.A., Lloyd,S., Hafezparast,M., Lennon-Pierce,M., Eppig,J.T., Festing,M.F. and Fisher,E.M. (2000) Nature Genet., 24, 23–25. [DOI] [PubMed] [Google Scholar]
- 18.Lansdorp P.M., Verwoerd,N.P., van de Rijke,F.M., Dragowska,V., Little,M.T., Dirks,R.W., Raap,A.K. and Tanke,H.J. (1996) Hum. Mol. Genet., 5, 685–691. [DOI] [PubMed] [Google Scholar]
- 19.Sacher G.A. and Hart,R.W. (1978) Birth Defects Orig. Artic. Ser., 14, 71–96. [PubMed] [Google Scholar]
- 20.Green M.C. (1981) Genetic Variants and Strains of the Laboratory Mouse. Gustav Fischer Verlag, New York, NY.
- 21.Kipling D. (1995) The Telomere. Oxford University Press, New York, NY, pp. 130–145.
- 22.Wright W.E. and Shay,J.W. (2000) Nature Med., 6, 849–851. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L., Hathcock,K.S., Hande,P., Lansdorp,P.M., Seldin,M.F. and Hodes,R.J. (1998) Proc. Natl Acad. Sci. USA, 95, 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcand S., Brevet,V. and Gilson,E. (1999) EMBO J., 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]