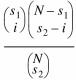Abstract
We have combined and compared three techniques for predicting functional interactions based on comparative genomics (methods based on conserved operons, protein fusions and correlated evolution) and optimized these methods to predict coregulated sets of genes in 24 complete genomes, including Saccharomyces cerevisiae, Caernorhabditis elegans and 22 prokaryotes. The method based on conserved operons was the most useful for this purpose. Upstream regions of the genes comprising these predicted regulons were then used to search for regulatory motifs in 22 prokaryotic genomes using the motif-discovery program AlignACE. Many significant upstream motifs, including five known Escherichia coli regulatory motifs, were identified in this manner. The presence of a significant regulatory motif was used to refine the members of the predicted regulons to generate a final set of predicted regulons that share significant regulatory elements.
INTRODUCTION
The availability of a rapidly growing number of complete genomes provides new opportunities to study gene regulation by comparative genomics. Microarray expression data have been used successfully in Saccharomyces cerevisiae and other organisms to predict coregulated sets of genes and to discover regulatory motifs in their upstream regions (1,2). However, for many sequenced bacterial genomes, mRNA expression data are not available. In addition, microarrays cannot be used to test all possible experimental conditions. Even in organisms for which there are no mRNA expression data, we can learn a great deal about regulation and interactions within pathways via comparative genomics. Three newly developed methods based purely on genome sequence comparisons allow us to predict functional relationships between non-homologous proteins of unknown function.
One such method is based on conserved operons (3–5). If two genes are found together in the same operon in two different organisms, but in two different operons in a third organism, then these two genes are likely to be functionally related in the third organism. Extending this general idea to the level of protein domains, we can predict functional interactions based on protein fusions (6–9). If two distinct proteins in one organism have homologs in a second organism which are fused into a single polypeptide chain, then the two proteins in the first organism are likely to be functionally related. The third method for predicting interactions between non-homologous proteins within an organism is correlated evolution, or the method of phylogenetic profiles (10). If homologs of two genes are present together or are absent from the same subset of genomes, then these genes are likely to be functionally related. The idea behind this method is that entire pathways or functional sets of genes will either be lost or passed on evolutionarily as a unit (e.g. only organisms with flagella will encode the genes for flagellar components). All three of these methods will become more powerful as the number of completely sequenced genomes increases.
In this paper, we modified and combined these three methods to predict coregulated sets of genes (regulons) in 24 complete genomes available in GenBank (22 bacterial genomes, S.cerevisiae and Caenorhabditis elegans). We then used the motif-discovery program AlignACE (1) on the upstream regions of genes making up these predicted regulons to search for potential regulatory motifs. The presence of a significant regulatory motif within one of these predicted regulons provides additional evidence that this grouping could be biologically meaningful. Therefore, the presence of a significant regulatory motif upstream of a subset of the genes comprising a predicted regulon was used to refine, or trim, the contents of the predicted regulon to include only the subset of genes with the regulatory motif in their upstream region.
MATERIALS AND METHODS
Organisms
The following 24 organisms were used in this analysis (abbreviations used in the figures are given in parentheses): Aquifex aeolicus (AA), Archaeoglobus fulgidus (AG), Aeropyrum pernix K1 (AP), Borrelia burgdorferei (BB), Bacillus subtilis (BS), Caenorhabditis elegans (CE), Chlamydia trachomatis (CT), Chlamydia pneumoniae (CP), Synechocystis sp. (CY), Escherichia coli K12 (EC), Haemophilus influenzae (HI), Helicobacter pylori (HP), Helicobacter pylori strain J99 (HY), Mycoplasma genitalium (MG), Methanococcus janaschii (MJ), Mycoplasma pneumoniae (MP), Mycobacterium tuberculosis (MT), Pyrococcus abysii (PA), Pyrococcus horokoshii (PH), Ricksettia prowazekii (RP), Saccharomyces cerevisiae (SC), Methanobacterium thermoautotrophicum (TH), Thermatoga maritima (TM) and Trepenoma pallidum (TP). The 22 prokaryotic genomes were used for motif finding.
Modifications in the three methods for predicting functional interactions
Several modifications were made to the three comparative genomics methods described above (methods based on conserved operons, protein fusions and phylogenetic profiles) to optimize and integrate these methods to design a better approach for predicting coregulated sets of genes. For each of the three methods, we calculated an N×N matrix of weighted interaction values, where N is the number of genes in the genome of interest. A value of zero at position ‘ij’ in the interaction matrix indicates that we predict no interaction between genes i and j. Greater values indicate predictions of higher confidence. Calculation of this interaction matrix for each of the three methods is described below. We then sum the matrices obtained by each of the three methods, and cluster the N genes by these matrix values in order to obtain predicted regulons. Information about obtaining our software for performing the analyses described here is available on our website, http://arep.med.harvard.edu/regulon_pred.
For all three regulon prediction methods, entries in the interaction matrix, aij, were set to zero if proteins i and j are homologous (BLAST E-value <10–6). This is necessary because two highly homologous proteins will have similar phylogenetic profiles, as well as links from protein fusions and conserved operons. Without setting these matrix values to zero, the final clusters will contain largely groups of homologs. Even when these homologous matrix entries are set to zero, many of the final clusters still contain a large fraction of homologous genes. Consequently, many of the motifs found by AlignACE simply correspond to homologous segments of upstream regions found upstream of homologous sets of genes. Therefore, all motifs where greater than one-third of the aligned motif instances are found upstream of homologous genes were excluded from our analysis.
Predictions based on conserved operons
To predict interactions based on conserved operons, we chose to use close homologs rather than strict orthologs to identify conserved operons. Groupings based on strict orthologs (3) result in many missed interactions. Using close homologs increases both the sensitivity (rate of true positives) and the rate of false positives (defined below). Our motif-finding method (AlignACE), however, can tolerate a reasonable rate of false positives in the input set. In addition, we included divergently transcribed genes in our operon definition. From our groups obtained using close homologs and this less strict operon definition, we were able to identify more significant regulatory motifs than using the groupings based on strict orthologs and tight operon definitions in the WIT (What Is There?) database, including more known Escherichia coli motifs (3,11).
For each of the N genes in the genome of interest, we searched for homologs in the other 23 complete genomes [BLAST (12) cutoffs were set at an E-value of 10–5]. We then searched for pairs of genes i and j in the genome of interest that had homologs contained in the same operon in two or more genomes. If there are no such pairs of homologs, then we assign this value in the N×N interaction matrix (aij) a score of zero. If there are such pairs of homologs in two or more different organisms, then we score the interaction according to the evolutionary distances between the genomes that contain these two genes within the same operon (3). Gene pairs contained in operons conserved across larger evolutionary distances will receive higher scores than those conserved between closely related genomes, as gene order in closely related genomes is more likely to be similar for reasons other than coregulation.
To calculate the matrix of evolutionary distances, we used a method similar to that described by Overbeek et al. (3,4). We aligned 16S ribosomal subunit sequences from the Ribosome Database Project (13), and then generated a matrix of evolutionary distances using the software package Phylip (14). We only add this score once per pair of genes (we do not increase the score due to the presence of multiple paralogous interactions). This matrix of scores based on evolutionary distances was clustered using a hierarchical clustering algorithm to obtain groups of interacting genes. We used an evolutionary distance cutoff score of 0.1 in the clustering process in order to exclude links due to proteins found in the same operon only in very closely related organisms (for example, M.genitalium and M.pneumoniae have an evolutionary distance score of 0.01, whereas E.coli and H.influenzae have an evolutionary distance score of 0.15).
Predictions based on protein fusions
To calculate interactions based on protein fusion events, we searched for non-overlapping BLAST hits (12) within a single polypeptide in the non-redundant database to two distinct proteins in the genome of interest. All hits in the non-redundant database with E-values below 10–5 were saved. This is a tighter cutoff than that used by Marcotte et al. (7,8) in order to reduce the rate of false positives. The rate of false positives quoted in their paper (7) was based on the loosest groupings in the MIPS (Munich Information center for Protein Sequences) database (i.e. metabolism). For our purposes (reconstruction of metabolic pathways and coregulated sets of genes, rather than functional annotation of genes) this is not a useful indication of the rate of false positives.
If there is no such protein fusion to link proteins i and j, then the aij entry in the interaction matrix is set to zero. If there are one or more genes in the non-redundant database that will link proteins i and j, then we weighted this interaction by the BLAST scores of the two non-overlapping hits. We chose the linking fusion protein with the lowest pair of BLAST scores to proteins i and j, and then we weighted the interaction using the log of the higher of the two BLAST scores (the BLAST score from i to this protein, or the BLAST score from j to this protein). This weighting scheme was set to obtain scores between 0 and 1. Interactions where the highest of the pair of BLAST E-values was <10–40 were assigned a score of 1.0. Interactions with BLAST E-values between 10–5 and 10–40 were assigned a value equal to the negative log of the BLAST value divided by 10.0. In order to reduce errors due to extremely common domains (8), we excluded proteins linked to 50 or more other proteins with scores greater than 0.1. We used a cutoff of 0.1 in the clustering process.
Predictions based on phylogenetic profiles
We used the method of phylogenetic profiles as described in Pellegrini et al. (10). The presence of a homolog in a particular organism is defined by the presence of a BLAST hit with an E-value <10–5. Euclidean distances between phylogenetic profile vectors (vectors consisting of zeros and ones) were calculated. These distances are the values used in the interaction matrix. We excluded proteins that were linked to 50 or more proteins with two or less mismatches. We used a Euclidean distance cutoff of 0.75 in the clustering process.
Calculations of sensitivity and error
The sensitivity (rate of true positives) and the rate of false positives in the predicted interactions (1.0 – positive predictive value, or errors of type II) were calculated by comparing the predicted interactions to known regulons and metabolic pathways. In E.coli 55 known regulons in our database (15), which contain a total of 444 genes, were used for comparison. In addition, 84 KEGG (Kyoto Encyclopedia of Genes and Genomes) metabolic pathway categories (16), which contain a total of 1121 genes, were used. In S.cerevisiae 82 KEGG metabolic pathway categories, which contain a total of 907 genes, as well as 171 MIPS functional group categories (17), which contain a total of 3420 genes, were used to compare to our predictions.
In S.cerevisiae the KEGG groupings were the most useful groups for our calculation of the sensitivity and the rate of false positives. The KEGG groups contain only those genes which have well-characterized function and are a part of a well-characterized pathway, whereas the MIPS functional groups attempt to classify the majority of proteins of known function. Since our goal is to reconstruct small and accurate groupings of genes in the same metabolic or regulatory pathway, the KEGG groupings are a more accurate set for comparison for our purposes.
The sensitivity is obtained by dividing the number of ‘correctly’ predicted interactions (i.e. interactions that link two genes within the same control grouping) by the total number of pairwise known interactions in the control groupings. The sensitivity is thus a measure of the ability of the predicted interactions to ‘reconstruct’ the control groupings. An interaction linking two genes of known function that are not classified together in a known control grouping is considered a false positive. The number of such false positive predictions is divided by the total number of predicted interactions linking two genes of known function to obtain the false positive rate.
Obtaining predicted regulons from the interaction matrices
We used a simple hierarchical clustering algorithm (18) to cluster the three individual interaction matrices (from conserved operons, protein fusions and phylogenetic profiles), as well as the sum of the three matrices. Cutoffs were chosen by maximizing sensitivity while retaining a reasonable rate of false positives in the predicted interactions (∼50%). The resulting clusters were compared to the known control clusters by calculating the probability P of obtaining the observed gene overlap:
This equation is similar to that used in the calculation of the specificity score (11,19). N is the total number genes in the genome of interest, s1 is the number of genes in the predicted regulon, s2 is the number of genes in the control grouping and x is number of genes that are found in both the predicted regulon and the control group (the intersection of s1 and s2).
AlignACE runs
For all bacterial genomes, upstream regions were predicted as in McGuire et al. (11). AlignACE 2.0 (19) was used for the motif searches, using default parameter values. Upstream regions from each of 22 prokaryotic genomes were aligned separately. The MAP score, site specificity score (Ssite), palindromicity and degree of similarity to known motifs were calculated as in McGuire et al. (11). These parameters were used to choose the motifs most likely to correspond to biologically significant motifs.
RESULTS
Comparisons of predicted interactions to control groupings
We compared our predicted functional interactions to known regulons and metabolic pathways in E.coli and S.cerevisiae by examining the values in the interaction matrices, as well as the final clusters obtained from this matrix and the motifs found in the upstream regions of these clusters. The sensitivity and the rate of false positives for the interaction matrix values were calculated by evaluating the subset of the predicted interactions in the interaction matrix which link two genes that are classified by KEGG (16), MIPS (17) or our E.coli regulon database (our ‘control groupings’) (15). Calculation of sensitivity and the rate of false positives is described in Materials and Methods.
The sensitivity and the rate of false positives in the predicted interactions were used as measures of the success of each method. For each parameter being optimized as we refined the three methods (i.e. cutoff values, operon definition, etc.), the sensitivity and the rate of false positives were calculated and compared. For example, Table 1 shows the effect of using three different operon definitions on the sensitivity and the rates of false positives in E.coli. In Table 1 you can see that using a broader operon definition that includes divergently transcribed genes and no distance cutoffs between adjacent genes increases both the sensitivity and the rate of false positives. By including a larger number of potentially coregulated genes in each predicted operon, we increase the number of conserved operons observed, including the number of correctly predicted interactions. However, since not all divergently transcribed genes are coregulated, we also increase our rate of false positives.
Table 1. Effect of different operon definitions on predicted interactions in E.coli.
| Operon definitiona | Number of predictionsb | False positivesc | Sensitivityd | ||
|---|---|---|---|---|---|
| KEGG (%) | Regulons (%) | KEGG (%) | Regulons (%) | ||
| Tandem genes, 300 bp cutoff | 13 602 | 32 | 44 | 6.3 | 6.2 |
| Tandem genes, no cutoff | 16 302 | 37 | 47 | 6.4 | 6.5 |
| Tandem + divergent, no cutoff | 22 117 | 45 | 51 | 6.7 | 7.0 |
aThe cutoff refers to the maximum number of base pairs allowed between adjacent genes. ‘No cutoff’ means there is no upper limit to the allowed distance between adjacent tandem or divergently oriented genes. Tandem refers to two genes on the same strand.
bThe number of pairwise interactions predicted by our method based on conserved operons, using the cutoffs described in Materials and Methods.
cPercent false positives. This value was calculated using either the KEGG metabolic pathways or our database of known E.coli regulons as a basis for comparison (see Materials and Methods).
dSensitivity was calculated by comparison to the KEGG metabolic pathways and our database of known E.coli regulons (see Materials and Methods).
Table 2 shows a comparison of the sensitivity and the rate of false positives for the three different methods, as well as their sum, in both E.coli and S.cerevisiae. As can be seen from the sensitivity value for the sum of the three methods, some of the same predictions are generated by two or more of the prediction methods, but the predictions from the three methods are also partly independent. It is therefore useful to combine the methods to generate as many predictions as possible. It is clear that the method based on conserved operons is the most powerful (it generates far more predictions with much higher sensitivity and a low rate of false positives in the predicted interactions), even for predictions in S.cerevisiae. The genes involved in these predictions in S.cerevisiae must be largely genes with bacterial homologs.
Table 2. Comparison of regulon prediction methods.
| Method | Number of predictionsa | False positivesb | Sensitivityc | ||
|---|---|---|---|---|---|
| KEGG (%) | Regulons (%) | KEGG (%) | Regulons (%) | ||
| E.coli | |||||
| Conserved operons | 22 117 | 45 | 51 | 6.7 | 7.0 |
| Protein fusions | 2 580 | 13 | 54 | 2.2 | 0.9 |
| Phylogenetic profiles | 2 329 | 51 | 53 | 0.7 | 0.6 |
| Combined method | 25 647 | 44 | 54 | 8.1 | 7.3 |
| S.cerevisiae | |||||
| Conserved operons | 12 438 | 28 | 51 | 6.4 | |
| Protein fusions | 4 205 | 37 | 55 | 0.5 | |
| Phylogenetic profiles | 1 304 | 49 | 64 | 0.1 | |
| Combined method | 16 771 | 31 | 54 | 6.6 | |
aThe number of pairwise interactions predicted by each method, using the cutoffs described in Materials and Methods.
bPercent false positives. This value was calculated using the KEGG metabolic pathways, as well as our database of known E.coli regulons or the MIPS gene groupings in S.cerevisiae (see Materials and Methods).
cSensitivity was also calculated by comparison to the KEGG metabolic pathways and our database of known E.coli regulons (see Materials and Methods).
The method based on phylogenetic profiles is the least powerful. However, we are currently using only ‘binary’ phylogenetic profiles, which simply assigns a value of ‘0’ or ‘1’, depending on whether a particular organism has a homolog to a particular protein. It is likely that this can be improved by incorporating weighting based on BLAST scores or evolutionary distances between proteins (7). However, it is likely that even with such modification, the phylogenetic profile method will not be as powerful as the other two methods.
In S.cerevisiae, the rate of false positives calculated using the MIPS functional group categories listed in Table 2 (see Materials and Methods) is not comparable with the rate of false positives published by Marcotte et al. (7) using the MIPS functional group categories, since here we are using the most specific MIPS sub-groupings (171 groupings), whereas Marcotte et al. (7) used only the 14 broadest MIPS gene divisions (i.e. metabolism). When using only these broadest groupings, the rate of false positives for totally random pairings is ∼47%. Using 171 different groupings as we have done here, the rate of false positives for random pairings is 88%. The numerical values of our false positive rates are higher than those listed in Marcotte et al. (7); however the values that we list here provide a more meaningful indicator of the rate of false positives in our predicted regulons. If we calculate rates of false positive in the same manner as Marcotte et al. (7), we obtain a false positive rate of 21% for the method based on conserved operons, 21% for protein fusions, 28% for phylogenetic profiles and 23% for the sum of the three methods. However, these values do not correlate with rates of false positives calculated using the more specific groupings; therefore we believe that these values do not provide a good estimate of the rates of false positives for the purposes of regulon prediction.
Comparison of clusters (predicted regulons) to the control groups
After obtaining interaction matrices as described in Materials and Methods, we then clustered these matrices to obtain ‘predicted regulons’. As a further test of our groupings, as well as of our clustering method, we compared the clusters that we obtain with the control groupings of genes of known function (KEGG pathways, MIPS categories and the known E.coli regulons in our database). For each cluster, the overlap in gene content was compared to each of the control clusters and the probability of obtaining this overlap was calculated (see Equation 1). The number of clusters with similarity to one of these control groupings with a P-value below one of three different cutoffs is listed in Table 3. We see again that the clusters based on conserved operons perform better than clusters obtained using the other two methods, and the sum of the three methods performs better than any individual method.
Table 3. Number of E.coli gene clusters with similarity to control groupsa.
| Method | Number of clusters analyzedb | Number of genes in clustersc | Regulons (P-values) | KEGG (P-values) | ||||
|---|---|---|---|---|---|---|---|---|
| 10–5 | 10–10 | 10–20 | 10–5 | 10–10 | 10–20 | |||
| WIT clusters | 229 | 546 | 28 | 8 | 0 | 43 | 19 | 6 |
| Conserved operons | 313 | 2426 | 23 | 8 | 0 | 48 | 25 | 6 |
| Protein fusions | 206 | 905 | 12 | 1 | 0 | 31 | 13 | 3 |
| Phylogenetic profiles | 249 | 2419 | 2 | 0 | 0 | 12 | 2 | 0 |
| Combined method | 372 | 2963 | 26 | 9 | 0 | 51 | 25 | 6 |
aPredicted regulons were compared to known gene groupings (known E.coli regulons and KEGG metabolic pathways). Calculation of P-values from the overlap in gene composition of the groups is described in Materials and Methods.
bTotal number of clusters (predicted regulons) analyzed.
cNumber of different genes making up the clusters analyzed.
We also compared our groups based on conserved operons to the groups constructed at the WIT database (3). The WIT clusters are composed of orthologs to genes found in close proximity in at least two organisms. These clusters contain groups of operons predicted to interact functionally in each organism. They were constructed by using a strict ortholog definition (top reciprocal FASTA hits) and a strict operon definition, and a human curator was used to define the cluster cutoffs. Because of this their clusters include far fewer genes than ours do. However, our clusters, which were automatically generated with no human curation, compare favorably to theirs in terms of reproducing control groups (Table 3). As shown below, our clusters yield far more significant upstream regulatory motifs than the WIT clusters because of the restricted gene composition of the WIT clusters (they contain 75% fewer genes than our predicted regulons).
Regulatory motifs found using AlignACE
We searched for regulatory motifs in the upstream regions of our predicted regulons by using the local alignment program AlignACE. We searched for motifs separately in each organism using predicted regulons obtained from each of the three methods for regulon prediction, as well as predicted regulons obtained by combining all three regulon prediction methods together. Motifs were found using these groups in each of the 22 prokaryotic organisms included in our analysis (see Materials and Methods). AlignACE output files and spreadsheets with parameters for all of the motifs found are available at http://arep.med.harvard.edu/ regulon_pred. Table 4 shows the number of highly significant motifs scoring above two sets of very stringent cutoff critera.
Table 4. Number of highly significant motifs obtained by AlignACE.
| Method | Number of genesa | Cutoff 1b | Cutoff 2c |
|---|---|---|---|
| WIT clusters | 546 | 7 | 0 |
| Phylogenetic profiles | 2426 | 46 | 20 |
| Protein fusions | 905 | 29 | 139 |
| Conserved operons | 2419 | 128 | 129 |
| Combined method | 2963 | 165 | 278 |
aNumber of different genes making up the clusters (predicted regulons).
bSsite <10–10, MAP >10, palindromicity >0.7, AT <80%.
cSsite <10–25, MAP >10.
We also compared the motifs we found using our predicted regulons to the motifs we obtained using the WIT groupings (3,11). In Table 4 you can see that we obtained many more motifs in our groups based on conserved operons than from the WIT groupings based on conserved operons. This is largely because their groupings only contain a quarter as many genes as ours due to the restrictive nature of their ortholog definitions.
In addition, Table 4 illustrates that, despite the fact that the groups based on phylogenetic profiles contain as many total genes (2419) as the groups based on conserved operons, these groupings yield far fewer significant motifs. It is encouraging that the groupings we believe to be better based on sensitivity and the rate of false positives (i.e. the groups obtained from the sum of the three interaction matrices and the groups based on conserved operons) yield more motifs than the groups we believe to be less useful (i.e. the groups obtained from phylogenetic profiles).
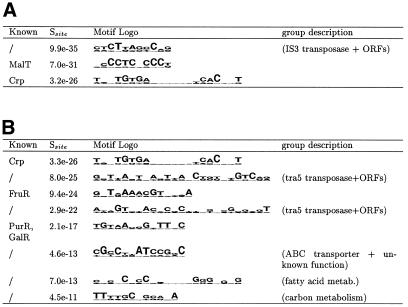
Similarities between motifs were calculated using the program CompareACE and motifs were clustered using a simple hierarchical clustering algorithm (11,19). The top-ranking clusters scoring above each of the two cutoffs listed in Table 4 are illustrated in Figures 1 and 2. Figure 1A and B shows the most significant motifs found in E.coli. It is encouraging to see that almost half of these motifs correspond to known motifs (Crp, FruR, MalT, GalR and PurR). The GalR and PurR motifs were both found in this analysis; however, because the motifs for these two homologous transcription factors are relatively similar, they are grouped together in the same category in Figure 1. Only two known motifs were found using the WIT groups in E.coli (ArgR and PurR) (11).
Figure 1.
Top motifs found in E.coli. The first column indicates if this motif is one of the 60 known E.coli motifs in our database. If the motif does not resemble a known motif, the last column (‘group description‘) describes the function of the ORFs upstream of which the motif was found. The second column contains the site specificity score (Ssite). The third column contains the motif logo (21). (A) Most specific motifs found in E.coli. All motifs with Ssite <10–25 (e–25) and MAP >10.0 were clustered (see Materials and Methods) and the most specific member of each cluster is listed here. (B) Most specific palindromic motifs found in E.coli. All motifs with palindromicity >0.7, MAP score >10.0, Ssite <10–10 and AT content <80% were clustered and the most specific member of each cluster is listed here.
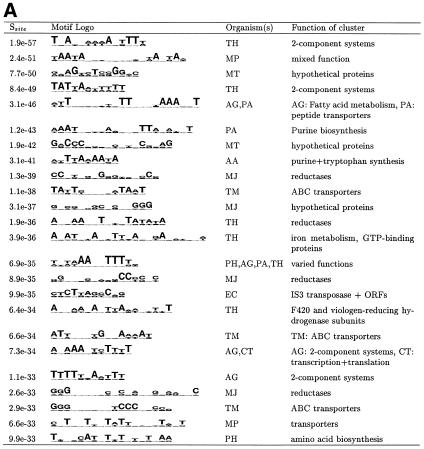
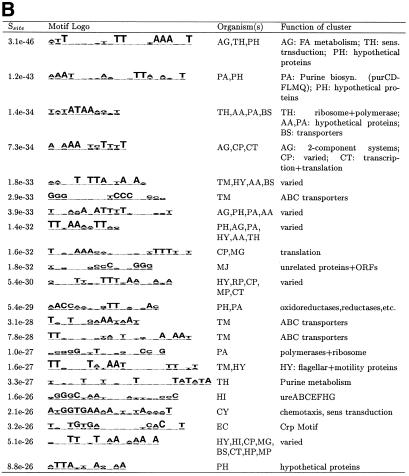
Figure 2.
Top motifs found in all 22 bacterial organisms. AlignACE was run separately on upstream regions from individual organisms. Sometimes a similar motif was found in several different organisms. The first column lists the site specificity score (Ssite), the second column contains the motif logo (21), the third column contains the two-letter abbreviation for the organism(s) in which the motif was found (see Materials and Methods) and the last column contains a broad description of the genes making up the cluster in which the motif was found. (A) Most specific motifs. All motifs with Ssite <10–25 and MAP >10.0 were clustered (see Materials and Methods). Since there are 92 motif clusters in this list, only the most specific member of each of the top 24 clusters is displayed here. (B) Most specific palindromic motifs. Motifs with palindromicity >0.7, MAP score >10.0, Ssite <10–10 and AT content <80% were clustered. Since there are 80 motif clusters in this list, only the most specific member of each of the top 22 clusters is displayed here.
The highest-scoring motifs from all 22 prokaryotic genomes are shown in Figure 2. We only show the very top motifs here as many motifs scored above our stringent cutoffs. All motifs are listed on our web site (http://arep.med.harvard.edu/regulon_pred). Several of the motifs shown in Figure 2 were found separately in more than one organism. In several cases, the same motif was found separately upstream of genes of similar function in more than one organism. For example, the same motif was found upstream of a set of oxidoreductases in both P.horokoshii (PH) and P.abysii (PA) (Fig. 2B). This could be interpreted as additional evidence that this motif is biologically relevant since it has been conserved in these two organisms. However, these two Pyrococcus genomes are closely related. Such conserved motifs found upstream of genes of similar function could also be found by grouping together upstream sequences from orthologous genes in closely related organisms (11).
In other cases, the same motif is found separately in different organisms upstream of groups of genes with seemingly different functions, or genes of known function in one organism and genes of unknown function in another organism. However, the genomes in which a similar motif has been found are often closely related evolutionary (Fig. 2B). A similar DNA motif could regulate different cellular processes in different organisms via a similar DNA-binding protein, or there could be errors in annotation of gene function in these organisms. Alternatively, the fact that these two motifs were grouped together could simply be an artifact of our motif clustering process (see Materials and Methods). In some cases, the clusters used to find the motifs are composed of genes of varied function.
In many cases, two or more different motifs were found upstream of overlapping sets of genes. For example, within a single predicted regulon derived from conserved operons in T.maritima, three different and significant motifs were found upstream of partially overlapping subsets of genes. Most of the genes comprising this predicted regulon are involved in two-component systems. These three motifs (motifs 1, 2 and 3) were found in 15, 23 and 11 upstream regions from this predicted regulon, respectively. The predicted regulon contains a total of 42 genes. Motifs 1 and 2 were both found upstream of five genes; motifs 2 and 3 were both found upstream of five genes; and motifs 2 and 3 were both found upstream of three genes within this group. Only one gene (TM0852, a gene of unknown function) has all three motifs within its upstream region. From the annotated functions for these genes, it is not clear how the functions of the genes relate to their classification within the three overlapping sub-groups. A complex network involving regulation of overlapping regulons could coordinate the intricate regulation of these two-component systems in T.maritima. Such mechanisms appear to be common in other organisms as well, as we have observed many such overlapping patterns of genes containing motifs in their upstream regions.
The presence of a significant regulatory motif in the upstream regions of a subset of genes comprising one of our predicted regulons was used to prune the contents of the predicted regulon. We have created a list of 338 refined regulons in 22 prokaryotic genomes, available at http://arep.med.harvard.edu/regulon_pred. Members of these regulons share significant upstream regulatory motifs scoring above our very stringent cutoffs in addition to being linked by at least one of the three regulon prediction methods.
DISCUSSION AND CONCLUSIONS
In the analysis described here, a number of motifs were found independently in the upstream regions of several different organisms. As we have done previously (11), we could search for conserved motifs by pooling together upstream sequence from orthologous genes in groups of closely related organisms. We found that this improves our ability to discover conserved motifs (11). However, as the groups that we have constructed in this analysis are relatively large, we were able to find a great number of motifs within sequence from individual organisms.
A large number of motifs were found in our AlignACE calculations. We must select those motifs that are both biologically relevant and statistically significant. Of the measures we used to assess our motifs, we found that the specificity score is the most useful measure for selecting biologically relevant motifs (11,19). The specificity score is a measure of how specific a motif is to the sequence in which it was found. To eliminate motifs which are not statistically significant, we have applied a MAP score cutoff. The MAP score is a measure of the over-representation of the motif within the sequence input to the Gibbs sampling algorithm (20). The MAP score and specificity score cutoffs that we have chosen here are very stringent (11). We have also eliminated those motifs with AT content >80% (11). Many specific and very AT-rich motifs were found in our analysis, yet all known E.coli motifs have AT content <80% (11).
An additional approach for assessing the significance of the motifs found in our analysis is to align randomly selected groups of upstream regions with the AlignACE algorithm and to measure the frequency of finding significant motifs. Hughes et al. (19) have performed such an analysis in the S.cerevisiae genome. They found that, although a small number of specific motifs with high MAP scores are found in the upstream regions of randomly selected ORFs, many more such motifs are in the upstream regions of groups of related genes. Using the number of high-scoring motifs found in the random runs as an estimate of the background noise, they calculated false positive rates for various cutoffs in MAP score and group specificity score (19). Their highest cutoffs (analogous to the cutoffs that we have used here) had low rates of false positives (<20%). Performing such an analysis on prokaryotic sequences with the cutoffs that we have used here could be used to calculate the rate of false positives in the motifs that we have obtained.
The presence of a significant regulatory motif upstream of the genes comprising a predicted regulon lends additional evidence to the hypothesis that this is a biologically significant regulon. In addition, if a subset of the genes within a predicted regulon contain a significant regulatory motif in their upstream regions, this information can be used to revise the contents of this predicted regulon. We have used our motif analysis together with our regulon prediction methods to generate a final set of predicted regulons.
Of the three regulon prediction methods that we have compared in this paper, the most powerful method is based on conserved operons. The method based on protein fusions is essentially a special case of the method based on conserved operons (the two genes involved are fused into a single polypeptide in some organism, rather than being located close to one another in the genome and transcribed onto the same piece of mRNA). Even in S.cerevisiae, the method based on conserved operons yields the largest number of predictions and the highest rate of true positives with the lowest rate of false positives. Loosening the operon definition to include divergently transcribed genes increases the recovery of true positives by 10–20%. It is not expected that this percentage could be any higher because only 10–20% of the genes contained in the same known E.coli regulons and KEGG metabolic pathways are divergently transcribed. Using our groups based on conserved operons, we were able to find many more significant upstream regulatory motifs than using the groups from the WIT database (3). This is because our groups were constructed using close homologs rather than strict orthologs; therefore, our groups include a total of four times more genes than the WIT groupings.
The least powerful of the three methods is the method based on conserved phylogenetic profiles. The idea behind this method is that the genes comprising entire pathways are either lost or passed on evolutionarily as a unit. However, this is often not the case. Homologs to an enzyme in a pathway may be present in an organism that no longer contains the pathway if this enzyme has become adapted for another cellular purpose. On the other hand, a homolog to an enzyme in a pathway may not be present in an organism that does contain the pathway if another non-homologous enzyme has evolved to fill in the missing function. The frequency of such events severely limits the usefulness of this method for regulon prediction.
Summing the three methods together yields the most useful groups, as there are independent predictions from all three methods. An additional improvement would be to optimize a weighted sum of the three methods. Predicted regulons obtained by these three methods can be used to find both known and new upstream regulatory motifs using local alignment programs such as AlignACE. We believe that groups of genes predicted to be coregulated by comparative genomics, and which also share a significant upstream regulatory motif, are likely to be coregulated. Experimental testing of some of these predicted regulons and their predicted cis-regulatory motifs is needed. All of the methods described here for predicted regulons and regulatory motifs will rapidly become more powerful as the number of completely sequenced genomes increases.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Jason Hughes, Jason Johnson, Jeremy Edwards, and Tzachi Pilpel for help and discussions. A.M.M. is a Howard Hughes Medical Institute predoctoral fellow. This work was supported by the office of Naval Research (grant no. N00014-97-1-0865), the Department of Energy (grant no. DE-FG02-87-ER60565) and a grant from Hoechst Marion Roussel.
REFERENCES
- 1.Roth F.P., Hughes,J.D., Estep,P.W. and Church,G.M. (1998) Nature Biotechnol., 16, 939–945. [DOI] [PubMed] [Google Scholar]
- 2.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 3.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1999) Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Overbeek R., Fonstein,M., D’Souza,M., Pusch,G.D. and Maltsev,N. (1998) In silico biology, 1, 0009 (http://www.bioinfo.de/isb/1998/01/0009). [PubMed] [Google Scholar]
- 5.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X. and Smith,T.F. (1998) Microbial Compar. Genomics, 3, 133–140. [DOI] [PubMed] [Google Scholar]
- 7.Marcotte E.M., Pellegrini,M., Thompson,M.J., Yeates,T.O. and Eisenberg,D. (1999) Nature, 402, 83–86. [DOI] [PubMed] [Google Scholar]
- 8.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 9.Enright A.J., Iliopoulos,I., Kyrpides,N.C. and Ouzounis,C.A. (1999) Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 10.Pellegrini M., Marcotte,E.M., Thompson,M.J., Eisenberg,D. and Yeates,T.O. (1999) Proc. Natl Acad. Sci. USA, 96, 4285–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGuire A.M., Hughes,J.D. and Church,G.M. (2000) Genome Res., 10, 744–757. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maidak B.L., Cole,J.R., Parker,C.T., Jr., Garrity,G.M., Larsen,N., Li,B., Lilburn,T.G., McCaughey,M.J., Olsen,G.J., Overbeek,R., Pramanik,S., Schmidt,T.M., Tiedje,J.M. and Woese,C.R. (1999) Nucleic Acids Res., 27, 171–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein J. (1985) Evolution, 39, 783–791. [DOI] [PubMed] [Google Scholar]
- 15.Robison K., McGuire,A.M. and Church,G.M. (1998) J. Mol. Biol., 284, 241–254. [DOI] [PubMed] [Google Scholar]
- 16.Ogata H., Goto,S., Sato,K., Fujibuchi,W., Bono,H. and Kanehisa,M. (1999) Nucleic Acids Res., 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A., Podkolodny,N.L. and Kolchanov,N.A. (1998) Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartigan J.A. (1975) Clustering Algorithms. Wiley, New York.
- 19.Hughes J.D., Estep,P.W., Tavazoie,S. and Church,G.M. (2000) J. Mol. Biol., 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 20.Liu S.J., Neuwald,A.F. and Lawrence,C.E. (1995) J. Am. Stat. Assoc., 90, 1156–1170. [Google Scholar]
- 21.Schneider T.D. and Stephens,R.M. (1990) Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]