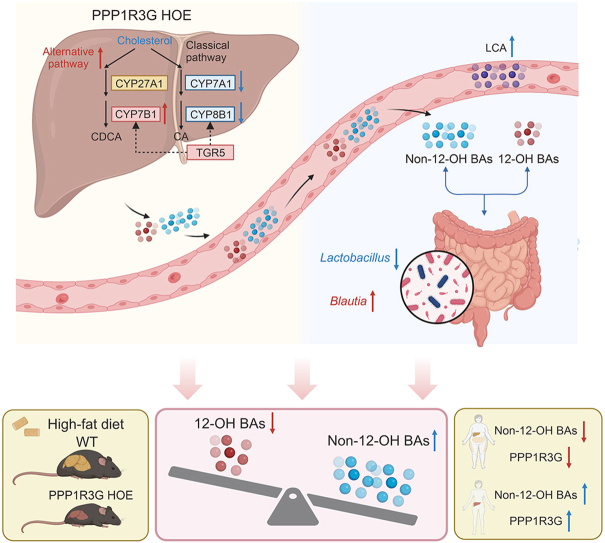
Abstract
Intestinal dysbiosis and disrupted bile acid (BA) homeostasis are associated with obesity, but the precise mechanisms remain insufficiently explored. Hepatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) plays a pivotal role in regulating glycolipid metabolism; nevertheless, its obesity-combatting potency remains unclear. In this study, a substantial reduction was observed in serum PPP1R3G levels in high-body mass index (BMI) and high-fat diet (HFD)-exposed mice, establishing a positive correlation between PPP1R3G and non-12α-hydroxylated (non-12-OH) BA content. Additionally, hepatocyte-specific overexpression of Ppp1r3g (PPP1R3G HOE) mitigated HFD-induced obesity as evidenced by reduced weight, fat mass, and an improved serum lipid profile; hepatic steatosis alleviation was confirmed by normalized liver enzymes and histology. PPP1R3G HOE considerably impacted systemic BA homeostasis, which notably increased the non-12-OH BAs ratio, particularly lithocholic acid (LCA). 16S ribosomal DNA (16S rDNA) sequencing assay indicated that PPP1R3G HOE reversed HFD-induced gut dysbiosis by reducing the Firmicutes/Bacteroidetes ratio and Lactobacillus population, and elevating the relative abundance of Blautia, which exhibited a positive correlation with serum LCA levels. A fecal microbiome transplantation test confirmed that the anti-obesity effect of hepatic PPP1R3G was gut microbiota-dependent. Mechanistically, PPP1R3G HOE markedly suppressed hepatic cholesterol 7α-hydroxylase (CYP7A1) and sterol-12α-hydroxylase (CYP8B1), and concurrently upregulated oxysterol 7-α hydroxylase and G protein-coupled BA receptor 5 (TGR5) expression under HFD conditions. Furthermore, LCA administration significantly mitigated the HFD-induced obesity phenotype and elevated non-12-OH BA levels. These findings emphasize the significance of hepatic PPP1R3G in ameliorating diet-induced adiposity and hepatic steatosis through the gut microbiota-BA axis, which may serve as potential therapeutic targets for obesity-related disorders.
Keywords: PPP1R3G, Obesity, Liver steatosis, Gut microbiota, Bile acid metabolism
Graphical abstract
Highlights
-
•
Hepatic PPP1R3G has the potential to ameliorate obesity and liver steatosis through its modulation of the liver-gut axis.
-
•
LCA treatment was able to reverse the obesity phenotype induced by HFD.
-
•
Activation of hepatic PPP1R3G holds promise as a viable therapeutic target for addressing obesity.
1. Introduction
Obesity is increasingly recognized as a global health issue and is intricately linked to type 2 diabetes mellitus (T2DM), nonalcoholic fatty liver disease, dyslipidemia, hypertension, liver cirrhosis, and cardiovascular diseases [[1], [2], [3]]. Its increasing incidence, driven by lifestyle and dietary changes, highlights the complexity of its pathogenesis [4]. Recent research has implicated bile acids (BAs) and gut microbiota in the development of obesity [5]. BAs, synthesized from cholesterol in the liver, are produced through enzymes such as cholesterol 7α-hydroxylase (CYP7A1) and sterol-12α-hydroxylase (CYP8B1), resulting in 12α-hydroxylated (12-OH) BAs. An alternative pathway generates non-12α-hydroxylated (non-12-OH) BAs [6,7]. In the intestine, bacterial enzymes convert conjugated BAs into secondary forms that are influenced by the gut microbiota, affecting host glycolipid and energy metabolism through BA co-metabolism [8,9]. Wei et al. [10] demonstrated that changes in BA profiles significantly affect the metabolic phenotype of obesity, which is likely linked to the intestinal microbiota. The ratio of non-12-OH to 12-OH BAs is related to the metabolic status of obesity. Increased 12-OH BA levels are associated with T2DM and insulin resistance [11]. Thus, the modulation of BA-gut microbiota co-metabolism offers a novel early obesity intervention model.
The dynamic interplay among BAs, the gut microbiota, and hosts presents new therapeutic avenues for metabolic disorders. Administering Parabacteroides distasonis (P. distasonis) or non-12OH BAs to mice promotes thermogenesis and mitigates body weight rebound, highlighting the critical role of the gut microbiota-BA interaction [8]. P. distasonis improves metabolism in obese mice by increasing lithocholic acid (LCA) and ursodeoxycholic acid (UDCA) levels and reducing weight gain, blood glucose, and hepatic steatosis [12]. Zheng et al. [13] showed that a high-fat diet (HFD) quickly and significantly increased BA levels within 12 h and then changed the microbiota composition within 24 h, affirming that BAs are the primary dietary regulators of gut microbiota. Transplanting the gut microbiota from obese mice fed an HFD into germ-free mice successfully replicated the obesity phenotypes [14]. The mechanisms underlying the impact of the BA-gut microbiota interplay on the metabolic status in obesity require further investigation.
Protein phosphatase 1 regulatory subunit 3G (PPP1R3G) is a regulatory subunit of protein phosphatase 1 and stimulates glycogenesis in the liver [15,16]. Hepatic PPP1R3G regulates the blood glucose equilibrium by controlling glycogen synthesis, aiding in maintaining the natural diurnal glycogen cycle [17]. Additionally, hepatic PPP1R3G is associated with altered body fat composition, as evidenced by the significantly reduced fat mass and epididymal fat pad weight in hepatocyte-specific Ppp1r3g overexpression (PPP1R3G HOE) mice compared with those in wild-type (WT) mice, as determined using nuclear magnetic resonance (NMR) [18]. PPP1R3G HOE also prevented the ethanol-induced increase in triglyceride (TG) concentration and lipid accumulation in the liver [19]. However, the underlying mechanism remains unclear. Given the significant downregulation of hepatic PPP1R3G in the HFD-induced obesity model, we postulated that manipulating PPP1R3G signaling may affect obesity and liver steatosis.
Changes in glycolipid metabolism can affect gut microbial diversity and the equilibrium of BA homeostasis [20]. This study examined the relationship between serum PPP1R3G levels and the BA profile in a clinical cohort. Additionally, we used PPP1R3G HOE mice to explore the impact of PPP1R3G on HFD-induced obesity and investigate its associated mechanisms. Our findings highlight the significant role of hepatic PPP1R3G in mitigating obesity and hepatic steatosis by regulating BAs and gut microbiota. This insight may hold promise for the treatment of metabolic disorders.
2. Materials and methods
2.1. Participants and criteria
This study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical University, China (Approval No.: XYFY2022-KL053-01). We categorized 72 participants based on body mass index (BMI) into two groups: normal weight (18.5 ≤ BMI <25) and overweight/obesity (BMI ≥ 25) [21]. The exact matching of age, sex, and other key clinical variables rendered the two groups comparable. Exclusion criteria included cancer, thyroid disorders, coronary heart disease, renal, inflammatory and infectious diseases, liver disorders, type 1 diabetes, diabetic complications, alcoholism, and medications affecting BA metabolism. We collected 5 mL of fasting blood samples, separated the serum by centrifuging at 3,500 rpm for 10 min at 4 °C, and stored it at −80 °C. Clinical parameters, including fasting blood sugar, systolic/diastolic blood pressure, lipid levels (total cholesterol (TC), TG, low-density lipoprotein (LDL), high-density lipoprotein (HDL), and liver damage markers (aspartate aminotransferase (AST), alanine aminotransferase (ALT)) were measured. Clinical parameters were measured, and the participant characteristics are displayed in Table 1.
Table 1.
The clinical characteristics of human subjects.
| Parameters | Normal-BMI | High-BMI | P-values |
|---|---|---|---|
| Total number of subjects | 36 | 36 | – |
| Number of males, n (%) | 16 (44.4) | 18 (50) | 0.81 |
| Age (years) | 37.72 ± 5.06 | 37.25 ± 4.98 | 0.53 |
| Smoking status, n (%) | 8 (22.2%) | 6 (16.7%) | 0.55 |
| Drinking status, n (%) | 8 (22.2%) | 10 (27.8%) | 0.59 |
| Systolic blood pressure (kPa) | 17.81 ± 0.26 | 18.89 ± 0.32 | 0.01 |
| Diastolic blood pressure (kPa) | 11.20 ± 0.32 | 12.11 ± 0.23 | 0.02 |
| BMI (kg/m2) | 20.86 ± 2.22 | 26.62 ± 1.57 | <0.0001 |
| Fasting blood glucose (mmol/L) | 5.34 ± 1.60 | 6.91 ± 6.02 | <0.001 |
| TC (mmol/L) | 4.01 ± 0.82 | 4.62 ± 1.29 | 0.05 |
| TG (mmol/L) | 1.14 ± 0.68 | 1.52 ± 0.60 | <0.001 |
| HDL (mmol/L) | 1.41 ± 0.38 | 1.14 ± 0.29 | <0.001 |
| LDL (mmol/L) | 2.35 ± 0.72 | 2.87 ± 1.06 | 0.04 |
| AST (U/L) | 20.00 ± 7.12 | 20.25 ± 7.64 | 0.94 |
| ALT (U/L) | 16.33 ± 15.11 | 18.75 ± 14.23 | 0.18 |
| PPP1R3G (ng/mL) | 18.81 ± 3.48 | 15.33 ± 5.22 | <0.001 |
The Mann-Whitney U test was utilized to compare the groups, and the statistically significant P-values are highlighted in bold. −: no data. BMI: body mass index; TC: total cholesterol; TG: triglyceride; HDL: high-density lipoprotein; LDL: low-density lipoprotein; AST: aspartate aminotransferase; ALT: alanine aminotransferase; PPP1R3G: protein phosphatase 1 regulatory subunit 3G.
2.2. Animals, HFD feeding, and treatment
Animal experiments shall be conducted in accordance with the protocols approved by the Animal Ethics Committee of Xuzhou Medical University (Approval No.: 202209S097) and followed the National Institutes of Health Guide for Care and Use of Laboratory Animals. We obtained specific-pathogen-free ROSA26 (gene knock-in model) and B-ALB-CreERT2 (liver-specific expression tool) mice from Beijing Biocytogen Gene Biotech Co., Ltd. and confirmed their genotypes through polymerase chain reaction (PCR) analysis of the biopsies. We used tamoxifen induction to establish mice with hepatocyte-specific overexpression of Ppp1r3g (PPP1R3G HOE) by crossbreeding ROSA26 mice and B-ALB-CreERT2 mice. DNA was extracted from the toes of mice at 21 days of age, and genotypes were identified by ordinary PCR (the genotyping primer sequences are listed in Table S1). Experiments involved male mice weaned at three weeks and housed at 3–5 per cage. At eight weeks, the mice were divided into normal diet (ND)-WT, HFD-WT, and HFD-HOE groups (HFD, 60.65% energy from fat, XTHF60; Jiangsu Synergy Pharmaceutical Co., Ltd., Fengxin, China) for 12 weeks. Energy intake and body weight were recorded weekly, averaged per cage, and calculated throughout the study period. For the fecal microbiota transplantation (FMT) experiment, 8-week-old mice were fed a 12-week HFD. At week 7, fecal microbiota from PPP1R3G HOE mice was gavaged into HFD-FMT group mice twice daily for six weeks. For the LCA experiment, male WT mice were divided into ND, HFD, and HFD + LCA (HFD with 0.5% LCA; Shanghai Jizhi Biochemical Technology Co., Ltd., Shanghai, China) groups.
2.3. Euthanasia and sample collection
At 20 weeks of age, fresh fecal samples from each group were collected and stored at −80 °C for gut microbiota analysis. After an overnight fast, mice were euthanized via pentobarbital injection, and blood from ocular globes was collected, serum was separated by centrifugation at 3,000 rpm at 4 °C for 15 min. Liver and adipose tissue samples were weighed, sectioned, and processed for RNA isolation using TRIzol reagent or histopathological examination with 4% paraformaldehyde. Additionally, blood, liver, and selected intestinal tissues were quickly frozen in liquid nitrogen and stored at −80 °C for further analysis.
2.4. Biochemical indexes evaluation
In the human study, automated analytics (Beckman AU5800; Beckman Coulter, Brea, CA, USA) was utilized to quantify biochemical markers, including serum glucose, TC, TG, LDL, HDL, AST, and ALT. The same assay was used to measure serum glucose, AST, and ALT levels in mice. Liver tissues were subjected to lipid extraction using the modified Folch method [22]. The tissues were homogenized in anhydrous methanol (1:9, m/V) on ice and centrifuged at 2,500 rpm for 10 min. Commercial kits (Jiancheng Bioengineering Institute, Nanjing, China) were used to quantify serum and liver TC, TG, HDL, and LDL levels.
2.5. Enzyme-linked immunosorbent assay (ELISA)
Serum PPP1R3G levels in humans and mice and serum insulin levels in mice were quantified using ELISA kits (E8482h, E8482 m, and E0448 m; EIAab, Wuhan, China). Samples and standards (50 μL each) were loaded onto 96-well ELISA plates and incubated at 37 °C for 90 min. After incubation, the wells were sequentially treated with detection reagents A and B, followed by incubation with the substrate and stop solution. The optical density was measured at 450 nm using an ELISA microplate reader (BioTek Synergy 2, BioTek Instruments, Winooski, VT, USA).
2.6. BA extraction and analysis
The ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) system (Waters Xevo-TQS) featured a triple quadrupole mass spectrometer (Waters Corporation, Milford, MA, USA). BAs and deuterium-labeled internal standards were obtained from Sigma-Aldrich (St. Louis, MO, USA) (Table S2). The labeled standards were weighed and diluted with methanol to prepare the mixed standards. Methanol and acetonitrile (high performance liquid chromatography (HPLC) grade) were obtained from CNW Technologies (GmbH, Dusseldorf, Germany). The BAs were quantified using a modified two-step extraction method [23]. For serum, we performed protein precipitation with 50 μL of serum, 5 μL of internal standards, and 500 μL of pre-cooled methanol, followed by centrifugation. Liver tissue (200 mg) and intestinal segments (0.5–1 cm) were homogenized with 400 μL of deionized water and mixed with methanol and internal standard. Fecal samples (100 mg) were treated with the extraction solvent and internal standard and centrifuged.
2.7. Targeted BA quantification
Linearity was assessed through three sets of seven-point calibration curves (refer to Table S3). Standard curves were constructed by linear regression, correlating BA peak area ratios with concentrations, using internal standards for quantification. Multiple reaction monitoring (MRM) determined the peak area ratios of BAs to internal standards. Due to limited commercial availability, some BAs were quantified using structurally analogous internal standards. Chromatographic separation occurred on an ACQUITY UPLC BEH C18 column (1.7 μm, 100 mm × 2.1 mm; Waters Corporation) maintained at 45 °C. Detection was in negative ion mode with an electrospray ionization (ESI) source. Mobile phases comprised 10 mmol/L ammonium acetate in water (A) and 0.1% formic acid in acetonitrile (B). The elution gradient began at 35% B, increasing to 60% over 3 min, then to 100% B over 4.5 min, and returning to 35% B over 5 min, at a flow rate of 0.4 mL/min. Nitrogen was utilized as both nebulizing and cone gases. Ion source and desolvation temperatures were set at 150 and 450 °C, respectively, with a capillary voltage of 2.5 V, cone voltage of 30 V, and extraction cone of 3 V. Cone gas flowed at 50 L/h and desolvation gas at 800 L/h, with a flow rate of 0.15 mL/min. MRM was used for quantification via fragment ions. The MassLynx Workstation (version 4.1; Waters Corporation) determined the peak areas for tissue samples, which were manually inspected and corrected. Correction curves were validated, and compounds were quantified. Preprocessing involved identifying peak areas, aligning, performing baseline correction, and normalization. Quantitative results were saved for statistical analysis.
2.8. 16S ribosomal DNA (16S rDNA) sequencing
Mouse fecal samples were subjected to 16S rDNA sequencing using an Illumina MiSeq PE300 system (Illumina, San Diego, CA, USA) with 16S primers (338F and 806R). Raw reads were quality-controlled and merged using Trimmomatic and FLASH software. UPARSE software was used to conduct a cluster analysis of operational taxonomic units, followed by species classification using the Ribosomal Database Project (RDP) classifier and alignment with the SILVA (SSU123) 16S rDNA database. Principal coordinate analysis based on the unweighted_unifrac model was used to examine intergroup differences, and differentiation significance was assessed using the Adonis test.
2.9. Histopathological examinations
Histological analysis of the mouse liver and epididymal white adipose tissue (eWAT) involved immediate fixation in 4% formaldehyde buffer solution for 24 h. Subsequently, the samples were dehydrated using graded ethanol (75%, 85%, 95%, and 100%), cleared with dimethylbenzene, embedded in paraffin for hematoxylin and eosin (H&E) staining, or frozen for Oil-red-O (ORO) staining. These procedures were conducted according to established protocols. Stained slides were examined under a light microscope (Olympus, Tokyo, Japan) to assess pathological changes.
2.10. Quantitative real-time PCR (qRT-PCR)
The concentration of RNA from the liver tissue was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). The complementary DNA (cDNA) was synthesized by reverse transcription of RNA, and PCR amplifications were performed using SYBR® Premix Ex Taq™ (Takara, Tokyo, Japan). The qPCR primer sequences are listed in Table S4. The data are normalized to β-actin messenger RNA (mRNA) expression level, and the method was used to calculate the mRNA expression.
2.11. Western blotting
Liver tissue proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China) supplemented with a mixture of phosphatase and protease inhibitors (Beyotime), and protein concentration was determined by bicinchoninic acid (BCA) assay. Proteins were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes. The membrane was enclosed in 5% skim milk for 2 h and incubated with antibody targeting PPP1R3G, farnesoid X receptor (FXR), G protein-coupled BA receptor 1 (TGR5), CYP7A1, CYP27A1, CYP8B1, CYP7B1, β-tubulin, and β-actin overnight at 4 °C. Details of the antibodies used are presented in Table S5. The relative optical density of each band was analyzed using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.12. Statistical analysis
Data were analyzed using IBM SPSS Statistics 22.0 and GraphPadPrism (version 8.0.2), with quantitative data presented as mean ± standard error of the mean (SEM). BA distribution was visualized by orthogonal partial least squares discriminant analysis (OPLS-DA) using SIMCA version 13.0. Use the Student's t-test to compare the mean between the two groups against normally distributed data. Nonparametric tests, specifically the Kruskal-Wallis and Mann-Whitney U tests, were applied for non-normal data, addressing comparisons between more than two groups and two groups, respectively. One-way analysis of variance (ANOVA), augmented by Bonferroni's or Dunnett's T3 post hoc corrections, was used for multiple group comparisons. The correlation analysis included all mice from different groups. Microbiome-BA correlations were assessed using Spearman's rank correlation with false discovery rate correction and visualized using R software (version 4.2.2). P < 0.05 was considered to be statistically significant.
3. Results
3.1. PPP1R3G expression downregulated in HFD-induced obesity models and high-BMI individuals
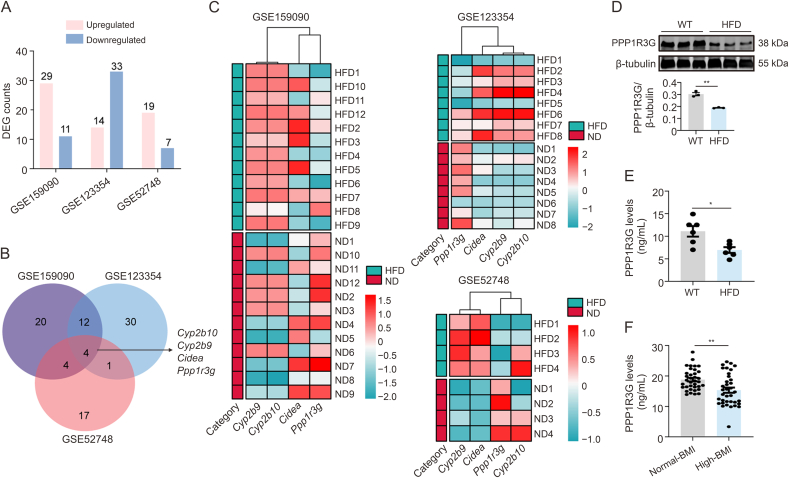
To unveil the gene expression patterns associated with obesity, we extensively analyzed the publicly available genomic datasets GSE159090, GSE123354, and GSE52748 obtained from mouse liver tissues. These datasets encompass various diet-induced obesity models and distinct dietary intervention periods and were sourced from the Gene Expression Omnibus (GEO) database. Differentially expressed gene (DEG) analysis across the three databases identified genes with altered expression, and cross-analysis highlighted consistent changes in Cyp2b10, Cyp2b9, Cidea, and Ppp1r3g (Figs. 1A and B). Ppp1r3g emerged as the exclusively downregulated gene within the intersection, implying a potential decrease in hepatic PPP1R3G levels following HFD intervention (Fig. 1C). In alignment with the database findings, Western blotting results further confirmed a notable reduction in PPP1R3G expression in the livers of HFD mice (Fig. 1D). Concurrently, serum PPP1R3G levels in WT mice fed an HFD were compared with those fed a ND (Fig. 1E). A clinical study showed a significant decrease in serum PPP1R3G levels in the high-BMI group (Fig. 1F). Collectively, these findings indicate a notable decrease in PPP1R3G expression in obesity.
Fig. 1.
Protein phosphatase 1 regulatory subunit 3G (PPP1R3G) expression downregulated in high-fat diet (HFD)-induced obesity models and high-body mass index (BMI) individuals. (A) Histogram illustrating the altered gene expression in the HFD group compared to the normal diet (ND) group, sourced from Table S1 (red: upregulated; blue: downregulated; differently expressed genes (DEGs): fold change > 1.5 and adjusted P < 0.05). (B) Intersection analysis across three databases unveiled four genes exhibiting consistent alterations. (C) Heatmaps constructed using messenger RNA (mRNA) expression levels of the four genes. (D) Representative Western blot bands and quantitative analysis of PPP1R3G in liver tissues from mice following 12-week feeding of HFD or ND (n = 3, Student's t-test). (E) Enzyme-linked immunosorbent assay (ELISA) examination of serum PPP1R3G levels in mice following 12-week feeding of HFD or ND (n = 6, Student's t-test). (F) ELISA examination of serum PPP1R3G levels in collected 36 normal-BMI individuals and 36 high-BMI individuals (Mann-Whitney U test). Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05 and ∗∗P < 0.01. WT: wild-type.
3.2. Serum PPP1R3G levels showed a positive correlation with non-12-OH BAs, which significantly decreased in high-BMI individuals
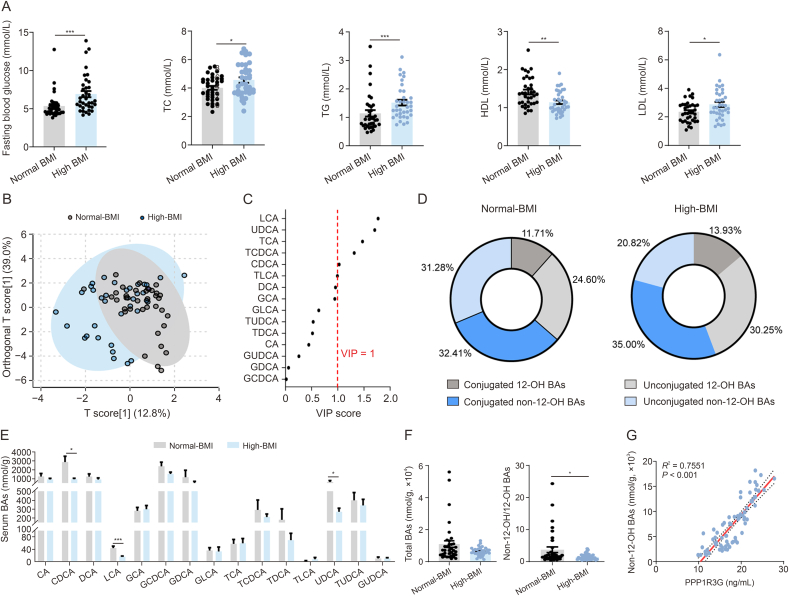
Significant differences in the clinical characteristics were observed between the normal and high-BMI groups. Specifically, the high-BMI group showed increased serum levels of fasting glucose, TC, TG, and LDL and a notable decrease in HDL (Fig. 2A). No significant differences were evident in the levels of liver damage markers, as demonstrated by the levels of AST and ALT (Fig. S1A). The OPLS-DA model distinctly separated the normal and high-BMI groups in terms of serum BA identification (Fig. 2B). The variable importance in projection (VIP) scores highlighted LCA (VIP = 1.77) and UDCA (VIP = 1.70) as pivotal variables contributing to the group differentiation (Fig. 2C and Table S6). The high-BMI group showed a lower proportion of non-12-OH BAs (Fig. 2D), with significant decreases in LCA, UDCA, and chenodeoxycholic acid (CDCA) (Fig. 2E). The total BA levels were similar between the two groups, and the high-BMI group had reduced non-12-OH/12-OH BA ratios (Fig. 2F). There was a significant positive correlation between PPP1R3G and non-12-OH BAs (Fig. 2G), but there was no correlation with fasting glucose, TC, TG, LDL, or HDL (Figs. S1B−F).
Fig. 2.
Serum biochemical markers and bile acid (BA) profiles in the normal-high-body mass index (BMI) and high-BMI groups. (A) Serum fasting glucose, total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) measurements of normal-BMI and high-BMI individuals (n = 36, Mann-Whitney U test). (B) Serum BA profiles displayed in orthogonal partial least squares discriminant analysis (OPLS-DA) scores plot for normal-BMI group (grey) and high-BMI group (blue). (C) Variable importance in projection (VIP) scores from OPLS-DA highlighting significant serum BA profile differences between the normal-BMI and the high-BMI groups (VIP value > 1 indicates discrimination importance). (D) Pie charts depict serum non-12α-hydroxylated (non-12-OH) and 12-OH BAs as proportions of total BAs, including unconjugated and conjugated forms (the proportions of BA species were calculated as: (concentrations of BA species/total BA) × 100%). (E) Analysis of serum BA composition of normal-BMI and high-BMI individuals (n = 36, Mann-Whitney U test). (F) Analysis of serum total BAs and ratios of non-12-OH/12-OH BAs of normal-BMI and high-BMI individuals (n = 36, Mann-Whitney U test). (G) Spearman correlation analysis of serum PPP1R3G and non-12-OH BA contents. Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. LCA: lithocholic acid; UDCA: ursodeoxycholic acid; TCA: taurocholic acid; TCDCA: taurochenodeoxycholic acid; CDCA: chenodeoxycholic acid; TLCA: taurolithocholic acid; DCA: deoxycholic acid; GCA: glycolcholic acid; GLCA: glycolithocholic acid; TUDCA: tauroursodeoxycholic acid; TDCA: taurodeoxycholic acid; CA: cholic acid; GUDCA: glycoursodeoxy-cholic acid; GDCA: glycodeoxycholic acid; GCDCA: glycochenodeoxycholic acid; PPP1R3G: protein phosphatase 1 regulatory subunit 3G.
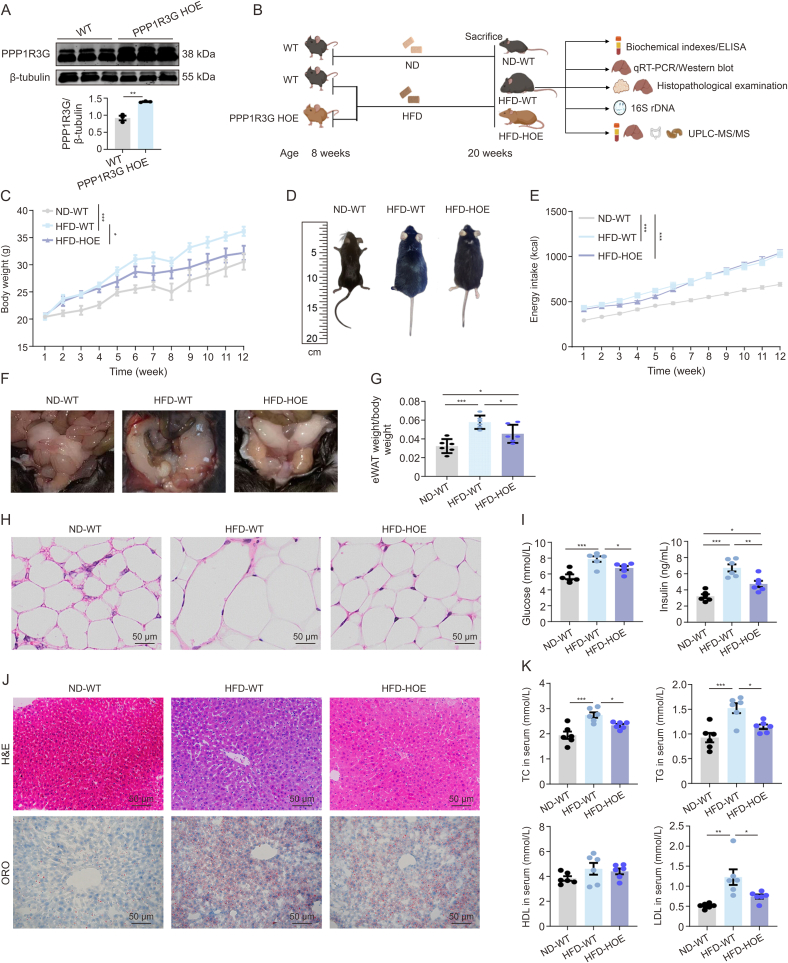
3.3. Hepatic PPP1R3G overexpression ameliorated HFD-induced obesity and liver steatosis in mice
PPP1R3G HOE mice, compared with ROSA26 controls, showed a significant increase in liver PPP1R3G expression (Fig. 3A). At eight weeks before starting the HFD, there was no significant weight difference between WT and PPP1R3G HOE mice (Fig. 3B). After 12 weeks, the HFD-WT group had gained more weight than the ND-WT group, whereas the HFD-HOE group had gained less than the HFD-WT group, suggesting restricted fat accumulation (Figs. 3C and D). Nonetheless, there was no difference in energy intake between the HFD-WT and HFD-HOE groups (Fig. 3E), indicating that the reduced body weight of the HFD-HOE mice was not due to reduced energy intake. Mice in the HFD-WT group showed increased eWAT adipocyte hypertrophy and proportion compared with ND-WT mice, with less pronounced changes in the HFD-HOE group (Figs. 3F−H). The HFD-HOE group exhibited a downward trend in fasting serum glucose levels compared with the HFD-WT group. Furthermore, HFD-WT mice had significantly higher serum insulin levels than HFD-HOE mice, where the insulin levels were slightly lower but not significantly different from those in the HFD-WT group (Fig. 3I). These results revealed that PPP1R3G HOE mitigated HFD-induced obesity, as evidenced by reduced weight and fat mass, and improved serum lipid profiles. H&E and ORO stainings revealed that HFD-exposed mice had more hepatic lipid droplets, fat vacuoles, and steatosis than pair-fed animals; however, the HFD-HOE group significantly mitigated these features (Fig. 3J). PPP1R3G HOE also reversed the HFD-induced elevation of serum AST (Fig. S2A). Comparing HFD-WT with ND-WT mice showed significant increases in serum TC, TG, and LDL levels, whereas HFD-HOE mice showed lower levels (Fig. 3K). Hepatic TC, TG, and LDL levels were significantly lower in the HFD-HOE group than in the HFD-WT group (Fig. S2B). Collectively, these findings demonstrate that PPP1R3G HOE alleviates hepatic steatosis, as shown by the normalized liver enzyme levels and histological staining in mice.
Fig. 3.
Hpatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) overexpression ameliorated high-fat diet (HFD)-induced obesity and liver steatosis in mice. (A) Representative Western Blot bands and quantitative analysis depict PPP1R3G expression in liver tissues of wild-type (WT) and PPP1R3G hepatocyte-specific overexpression (HOE) mice following normal diet (ND) (n = 3, Student's t-test). (B) Schematic of experimental design: male PPP1R3G HOE and WT littermates, aged 8 weeks, were subjected to weekly measurements of body weight, food intake, and drink consumption while being fed either a ND or an HFD until reaching 20 weeks of age. (C) The curve of the changes in body weight observed at various time points of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, two-way analysis of variance (ANOVA)). (D) Representative gross contours to indicate the body size difference of ND-WT, HFD-WT, and HFD-HOE mice. (E) The curve of energy intake at different time points of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, two-way ANOVA). (F) Illustrative depiction of epididymal white adipose tissue (eWAT) appearance in ND-WT, HFD-WT, and HFD-HOE mice. (G) The eWAT indices (eWAT weight/body weight) of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's correction). (H) Representative hematoxylin and eosin (H&E)-stained images of eWAT tissue sections from ND-WT, HFD-WT, and HFD-HOE mice (n = 3). (I) Serum fasting blood glucose and insulin measurements of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's correction). (J) Representative H&E-stained and Oil Red O (ORO)-stained images of liver tissue sections from ND-WT, HFD-WT, and HFD-HOE mice (n = 3). (K) Serum total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) measurements of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's correction). Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. ELISA: enzyme-linked immunosorbent assay; qRT-PCR: quantitative real-time PCR; 16S rDNA: 16S ribosomal DNA; UPLC-MS/MS: ultra-performance liquid chromatography-tandem mass spectrometry.
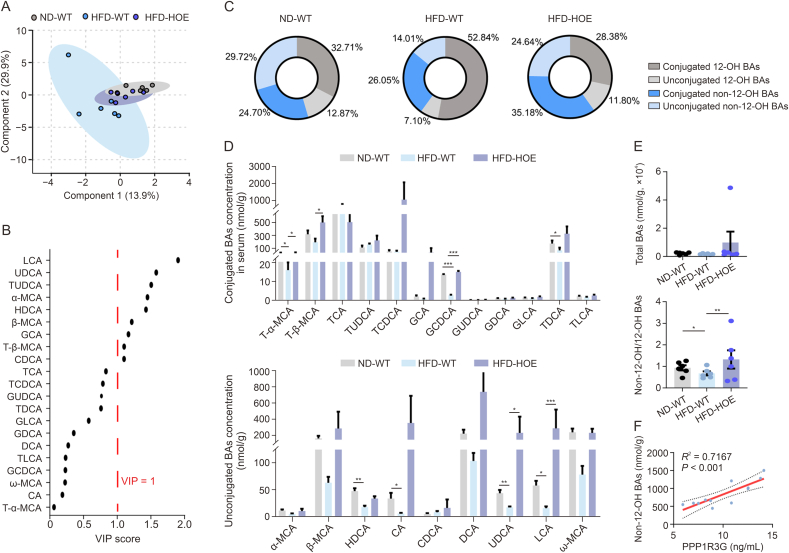
3.4. Hepatic PPP1R3G overexpression prominently upregulated the serum non-12-OH/12-OH BAs ratio during an HFD
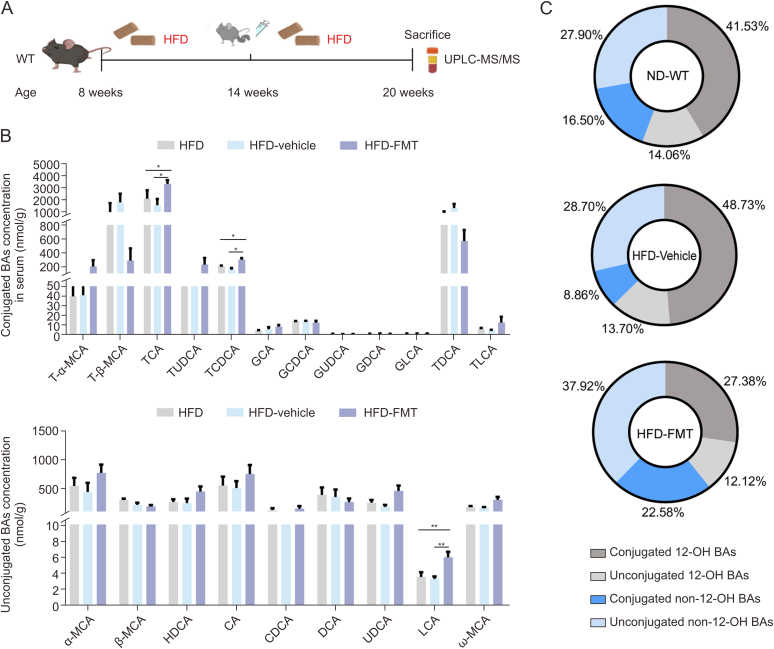
Next, we elucidated the molecular mechanisms underlying obesity and liver steatosis mediated by hepatic PPP1R3G. We evaluated the potential impact of PPP1R3G on lipid accumulation in HepG2 cells and found that its overexpression barely influenced lipid accumulation (Fig. S3). The procedures of lentivirus transfection and oleic acid induction in HepG2 cells are shown in Supplementary data. Subsequently, we conducted a thorough multisite BA profile analysis, which demonstrated a clear distinction in serum BA levels among the three groups (Fig. 4A). The VIP scores from the OPLS-DA model identified LCA (VIP = 1.90) and UDCA (VIP = 1.57), both classified as non-12-OH BAs, as the top differentiators in the HFD-WT and HFD-HOE groups (Fig. 4B and Table S7). The HFD-WT group had a lower proportion of non-12-OH BAs than the ND-WT group, while the HFD-HOE group had the highest proportion (Fig. 4C). Specific non-12-OH BAs, such as tauro-α-muricholic acid (T-α-MCA), glycochenodeoxycholic acid (GCDCA), UDCA, and LCA, decreased in the HFD-WT group but were counteracted by PPP1R3G HOE (Fig. 4D). Total BA levels were consistent across all groups, but the non-12-OH/12-OH BA ratio was higher in the HFD-HOE and ND-WT groups than in the HFD-WT group, with no significant difference between the HFD-HOE and ND-WT groups (Fig. 4E). Similar to clinical findings, a significant positive correlation was observed between PPP1R3G and non-12-OH BAs in WT mice (Fig. 4F). In summary, PPP1R3G HOE altered serum BA profiles in HFD mice by increasing the proportion of non-12-OH BA without changing total BA levels.
Fig. 4.
Hepatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) overexpression prominently elevated the serum non-12α-hydroxylated (non-12-OH)/12-OH bile acids (BAs) ratio during an high-fat diet (HFD). (A) Serum BA profiles displayed in orthogonal partial least squares discriminant analysis (OPLS-DA) scores plot for normal diet (ND)-wild-type (WT) group (grey), HFD-WT group (blue), and HFD-hepatocyte-specific overexpression (HOE) group (purple). (B) Variable importance in projection (VIP) scores from OPLS-DA highlighting significant serum BA profile differences between the HFD-WT and HFD-HOE groups (VIP > 1 indicates discrimination importance). (C) Pie charts depict serum non-12-OH and 12-OH BAs as proportions of total BAs, including unconjugated and conjugated forms of ND-WT, HFD-WT, and HFD-HOE mice. (D) Analysis of serum BA composition of ND-WT, HFD-WT, and HFD-HOE mice (n = 6; one-way analysis of variance (ANOVA) with Bonferroni's/Dunnett's correction for normal and Kruskal-Wallis for non-normal data). (E) Analysis of serum total BAs and ratios of non-12-OH/12-OH BAs of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, Kruskal-Wallis test). (F) Spearman correlation analysis of serum PPP1R3G and non-12-OH BA contents of ND-WT and HFD-WT mice. Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. LCA: lithocholic acid; UDCA: ursodeoxycholic acid; TUDCA: tauroursodeoxycholic acid; MCA: muricholic acid; HDCA: hyodeoxycholic acid; GCA: glycolcholic acid; T-β-MCA: tauro-β-MCA; CDCA: chenodeoxycholic acid; TCA: taurocholic acid; TCDCA: taurochenodeoxycholic acid; GUDCA: glycoursodeoxy-cholic acid; TDCA: taurodeoxycholic acid; GLCA: glycolithocholic acid; GDCA: glycodeoxycholic acid; DCA: deoxycholic acid; TLCA: taurolithocholic acid; GCDCA: glycochenodeoxycholic acid; CA: cholic acid.
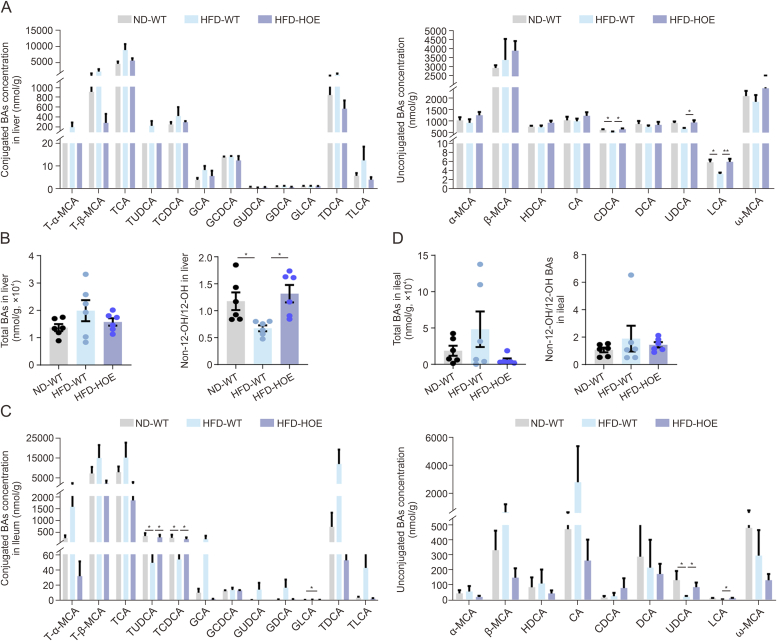
3.5. Hepatic PPP1R3G overexpression induced alterations of BA profiles within the liver, diverse intestinal segments, and fecal matter in HFD-fed mice
Hepatic BA assessment revealed that CDCA and LCA levels were notably decreased in the HFD-WT group but were significantly restored by PPP1R3G HOE (Fig. 5A). While total BA levels remained consistent, the non-12-OH/12-OH BA ratio was significantly higher in the HFD-HOE group (Fig. 5B). In the jejunum and ileum, which are key for BA reabsorption [24], BA profiles exhibited minimal variability. The levels of non-12-OH BAs, such as taurochenodeoxycholic acid (TCDCA) in the jejunum and tauroursodeoxycholic acid (TUDCA), TCDCA, and UDCA in the ileum, were significantly decreased in the HFD-WT group but increased in the HFD-HOE group (Figs. 5C and S4A). Total jejunal BA levels were reduced in the HFD group (Fig. S4B), whereas the ileal levels showed no significant group differences (Fig. 5D). Cecal BA profiles showed no notable group differences (Figs. S4C and D). We discovered distinct alterations in various individual non-12-OH BA concentrations in HFD-fed mice. The glycoursodeoxy-cholic acid (GUDCA) and LCA levels, particularly in the colon, were significantly restored by PPP1R3G HOE (Fig. S5A). Additionally, the proportion of non-12-OH BAs in the colon was higher in the HFD-HOE group than that in the HFD-WT group (Fig. S5B). Furthermore, no significant differences in the non-12-OH/12-OH BA ratios in the jejunum, ileum, or cecum were observed among the three groups (Figs. 5C, S4B, and S4D). In contrast, a significantly higher ratio in the colon was found in the HFD-HOE group than in the HFD-WT group, despite no significant discrepancy in total BA levels (Fig. S5C). Fecal BA levels and non-12-OH/12-OH BA ratios remained stable across groups (Figs. S5D and E), but the HFD-WT group showed increased total fecal BAs and a reduction in the HFD-HOE group (Fig. S5F). These findings highlight the significant influence of PPP1R3G HOE on the systemic regulation of BA homeostasis.
Fig. 5.
Hepatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) overexpression induced modifications in bile acids (BAs) profiles within the liver and ileum during an high-fat diet (HFD). (A) Analysis of hepatic BA composition of normal diet (ND)-wild-type (WT), HFD-WT, and HFD-hepatocyte-specific overexpression (HOE) mice (n = 6, one-way analysis of variance (ANOVA) with Bonferroni's/Dunnett's correction for normal and Kruskal-Wallis for non-normal data). (B) Analysis of hepatic total BAs and ratios of non-12α-hydroxylated (non-12-OH)/12-OH BAs of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's/Dunnett's correction for normal and Kruskal-Wallis for non-normal data). (C) Analysis of ileal BA composition of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's/Dunnett's correction for normal and Kruskal-Wallis for non-normal data). (D) Analysis of ileal total BAs and ratios of non-12-OH/12-OH BAs of ND-WT, HFD-WT, and HFD-HOE mice (n = 6, one-way ANOVA with Bonferroni's/Dunnett's correction for normal and Kruskal-Wallis for non-normal data). Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05 and ∗∗P < 0.01. T-α-MCA: tauro-α-muricholic acid; TCA: taurocholic acid; TUDCA: tauroursodeoxycholic acid; TCDCA: taurochenodeoxycholic acid; GCA: glycolcholic acid; GCDCA: glycochenodeoxycholic acid; GUDCA: glycoursodeoxy-cholic acid; GDCA: glycodeoxycholic acid; GLCA: glycolithocholic acid; TDCA: taurodeoxycholic acid; TLCA: taurolithocholic acid; HDCA: hyodeoxycholic acid; CA: cholic acid; CDCA: chenodeoxycholic acid; DCA: deoxycholic acid; UDCA: ursodeoxycholic acid; LCA: lithocholic acid.
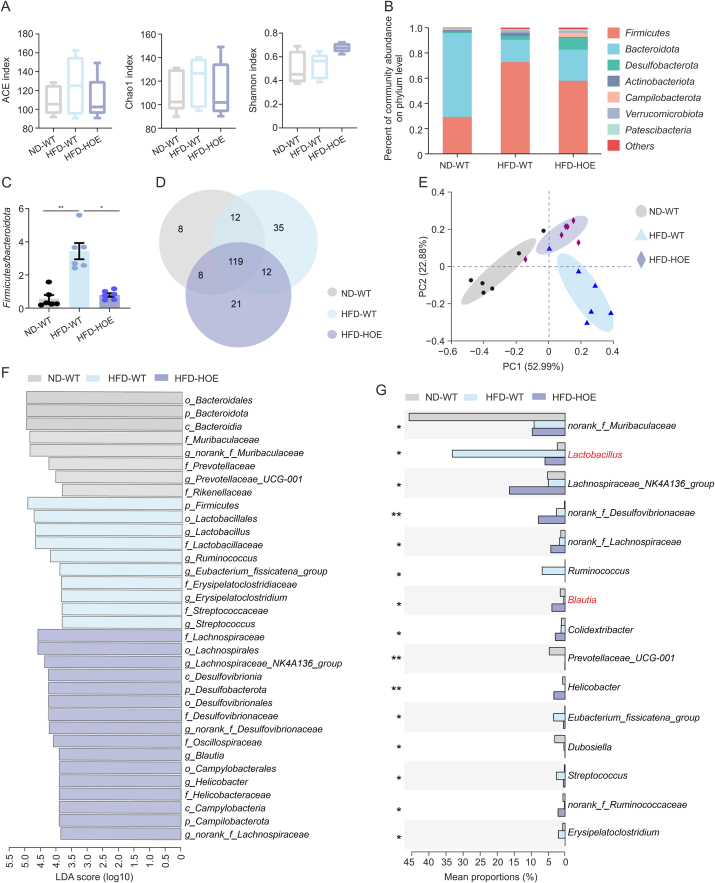
3.6. Hepatic PPP1R3G overexpression led to compositional alterations of the gut microbiome in HFD-fed mice
Given the strong link between gut microbiota, obesity, and BA composition [25,26], we investigated whether gut microbiota alterations contribute to disrupted BA profiles. PPP1R3G HOE did not affect alpha diversity, as indicated by the unaltered angiotension converting enzyme (ACE), Chao1, and Shannon index values (Fig. 6A); however, the beta-diversity analysis showed significant differences, with increased Firmicutes and decreased Bacteroidetes in the HFD-WT group compared with the ND-WT group. PPP1R3G HOE partially reversed these changes at the phylum level (Fig. 6B). The Firmicutes/Bacteroidetes ratio, positively correlated with HFD-induced obesity [27], was lower in the HFD-HOE group (Fig. 6C). At the genus level, the principal coordinate analysis revealed distinct microbiota compositions among the groups, whereas the Venn diagram analysis indicated 119 common species across all groups, with the HFD-WT group exhibiting the highest count of unique species (Figs. 6D and E). The HFD-WT group had more Lactobacillus, whereas the HFD-HOE group had higher Lachnospiraceae_NK4A136 and Blautia levels. PPP1R3G HOE significantly increased Blautia, a microbial community associated with inflammation and obesity [28], and reduced Lactobacillus, a microbial community linked to weight gain and obesity [29] (Figs. 6F and G). Overall, PPP1R3G HOE reshaped the gut microbial community structure.
Fig. 6.
Hepatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) overexpression induced modifications in gut microbiota composition during an high-fat diet (HFD). (A) Analysis of alpha diversity (including angiotension converting enzyme (ACE), Chao1, and Shannon index) based on 16S ribosomal DNA (16S rDNA) sequencing data obtained from stool samples of normal diet (ND)-wild-type (WT), HFD-WT, and HFD-hepatocyte-specific overexpression (HOE) mice (n = 6, one-way analysis of variance (ANOVA) with Bonferroni's correction). (B) Relative abundance of the seven predominant gut microbiota at the phylum level in stool from ND-WT, HFD-WT, and HFD-HOE mice. (C) Ratio of Firmicutes to Bacteroidetes in ND-WT, HFD-WT, and HFD-HOE mice (n = 6, Kruskal-Wallis test). (D) Venn diagram depicting unique and shared gut microbiota species among ND-WT, HFD-WT, and HFD-HOE mice. (E) Principal coordinate analysis conducted using Bray-Curtis dissimilarity on 16S rDNA sequencing data derived from stool samples of ND-WT, HFD-WT, and HFD-HOE mice. (F) Visualization of linear discriminant analysis (LDA) scores depicted distinctively abundant taxa among the ND-WT, HFD-WT, and HFD-HOE groups (logarithmic LDA scores >2.0; P < 0.05). (G) 15 genera exhibiting the most pronounced differences among ND-WT, HFD-WT, and HFD-HOE mice, with ∗∗P < 0.01 indicating significant intergroup differences at the corresponding genus level based on significance testing. Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05 and ∗∗P < 0.01. PC: principal component.
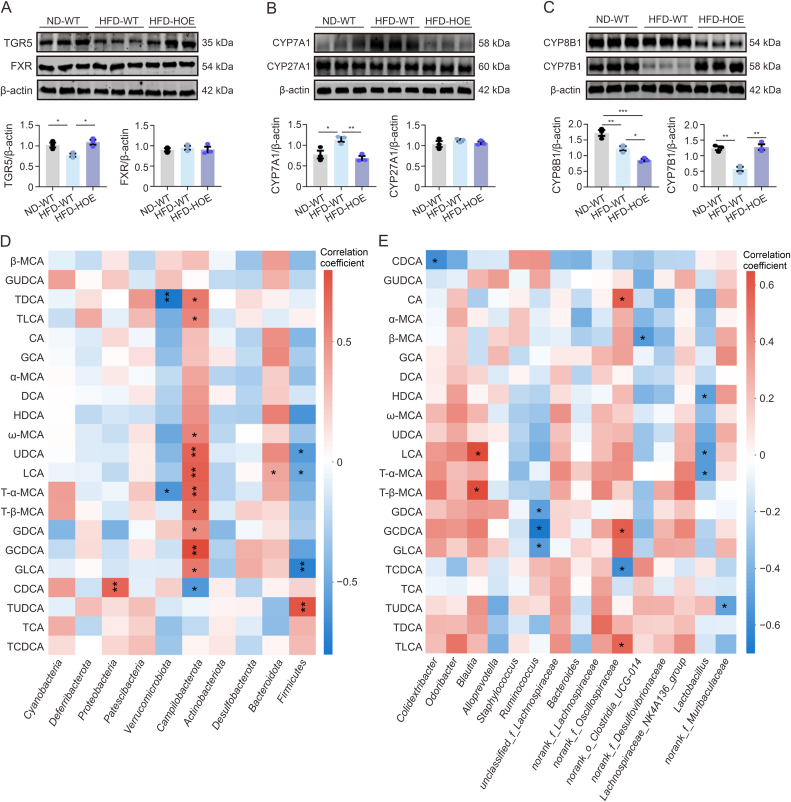
3.7. Hepatic PPP1R3G regulated relevant genes associated with BA synthesis during an HFD, and disrupted BAs were linked to the gut microbiota
To elucidate the molecular basis of the altered BA profiles, we examined the BA receptors and enzymes in the BA synthesis pathways. In the liver, HFD-WT mice showed reduced TGR5 expression, whereas HFD-HOE mice exhibited increased TGR5 levels without any changes in FXR expression (Fig. 7A), suggesting that PPP1R3G HOE mitigated abnormal TGR5 signaling. CYP7A1 expression was higher in HFD-WT mice than in ND-WT mice but lower in HFD-HOE mice, with no substantial alterations in CYP27A1 expression (Fig. 7B). Additionally, in the HFD-HOE group, CYP8B1 displayed significant downregulation compared with the ND-WT and HFD-WT groups, whereas CYP7B1 was underexpressed in the HFD-WT group relative to the ND-WT and HFD-HOE groups (Fig. 7C). Furthermore, the hepatic mRNA expression of Fgfr4 and Lrh-1, which are involved in BA regulation, and the transporters Bsep, Mdr2, and Mrp2 showed no significant differences among the three groups (Fig. S6A). Additionally, the expression of ileal BA transporters was reduced in HFD-WT mice; however, PPP1R3G-HOE did not reverse these changes (Fig. S6B).
Fig. 7.
Hepatic protein phosphatase 1 regulatory subunit 3G (PPP1R3G) regulated relevant genes in bile acid (BA) synthesis during an high-fat diet (HFD), and disrupted BAs were linked to the gut microbiota. (A) Representative Western Blot bands and quantitative analysis depict G protein-coupled BA receptor 1 (TGR5) and farnesoid X receptor (FXR) expression in liver tissues of normal diet (ND)-wild-type (WT), HFD-WT, and HFD-hepatocyte-specific overexpression (HOE) mice (n = 3, Student's t-test). (B) Representative Western Blot bands and quantitative analysis depict CYP7A1 and CYP27A1 expression in liver tissues of ND-WT, HFD-WT, and HFD-HOE mice (n = 3, Student's t-test). (C) Representative Western Blot bands and quantitative analysis depict cholesterol-12α-hydroxylase (CYP8B1) and cholesterol 7α-hydroxylase (CYP7B1) expression in liver tissues of ND-WT, HFD-WT, and HFD-HOE mice (n = 3, Student's t-test). (D, E) Correlation analysis between fecal microbiota at the phylum level (D) and genus level (E) and specific fecal BAs. The color code indicates the Spearman correlation coefficients: positive correlation (red) and negative correlation (blue). Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. β-MCA: β-muricholic acid; GUDCA: glycoursodeoxy-cholic acid; TDCA: taurodeoxycholic acid; TLCA: taurolithocholic acid; CA: cholic acid; GCA: glycolcholic acid; DCA: deoxycholic acid; HDCA: hyodeoxycholic acid; UDCA: ursodeoxycholic acid; LCA: lithocholic acid; T-α-MCA: tauro-α-MCA; GDCA: glycodeoxycholic acid; GCDCA: glycochenodeoxycholic acid; GLCA: glycolithocholic acid; CDCA: chenodeoxycholic acid; TUDCA: tauroursodeoxycholic acid; TCA: taurocholic acid; TCDCA: taurochenodeoxycholic acid.
We performed a correlation analysis between the top 15 most abundant bacteria at the genus and phylum levels and fecal BAs, revealing significant correlations, especially between non-12-OH BAs and the gut microbiota. Firmicutes negatively correlated with UDCA, LCA, and glycolithocholic acid (GLCA), whereas Bacteroidetes positively correlated with LCA at the phylum level (Fig. 7D). At the genus level, Lactobacillus negatively correlated with hyodeoxycholic acid (HDCA), LCA, and T-α-MCA, while Blautia and Ruminococcus showed positive and negative associations with certain BAs (Fig. 7E). These findings suggest that gut microbiota dysbiosis significantly affects BA composition. To further explore the impact of microbiota related to hepatic PPP1R3G on BAs, we analyzed the BA profiles of WT mice receiving PPP1R3G HOE microbiota via gavage during HFD feeding (Fig. 8A). Additionally, FMT from PPP1R3G HOE mice to WT mice during HFD notably elevated serum LCA, TCDCA, taurocholic acid (TCA), and non-12-OH BA proportions (Figs. 8B and C).
Fig. 8.
Protein phosphatase 1 regulatory subunit 3G (PPP1R3G) hepatocyte-specific overexpression (HOE) microbiota gavage induced modifications in serum bile acids (BAs) profile. (A) Schematic of fecal microbiota transplantation (FMT) experimental design: wild-type (WT) littermates, aged seven weeks and fed an high-fat diet (HFD), underwent FMT from PPP1R3G HOE mice through gavage, administered every two days for a continuous duration of six weeks. (B) Analysis of serum BA composition of HFD, vehicle, and FMT mice (n = 6, one-way analysis of variance (ANOVA) with Bonferroni's for normal and Kruskal-Wallis for non-normal data). (C) Pie charts depict serum non-12α-hydroxylated (non-12-OH) and 12-OH BAs as proportions of total BAs, including unconjugated and conjugated forms of HFD, vehicle, and FMT mice. Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05 and ∗∗P < 0.01. UPLC-MS/MS: ultra-performance liquid chromatography-tandem mass spectrometry; T-α-MCA: tauro-α-muricholic acid; TCA: taurocholic acid; TUDCA: tauroursodeoxycholic acid; TCDCA: taurochenodeoxycholic acid; GCA: glycolcholic acid; GCDCA: glycochenodeoxycholic acid; GUDCA: glycoursodeoxy-cholic acid; GDCA: glycodeoxycholic acid; GLCA: glycolithocholic acid; TDCA: taurodeoxycholic acid; TLCA: taurolithocholic acid; HDCA: hyodeoxycholic acid; CA: cholic acid; CDCA: chenodeoxycholic acid; DCA: deoxycholic acid; UDCA: ursodeoxycholic acid; LCA: lithocholic acid.
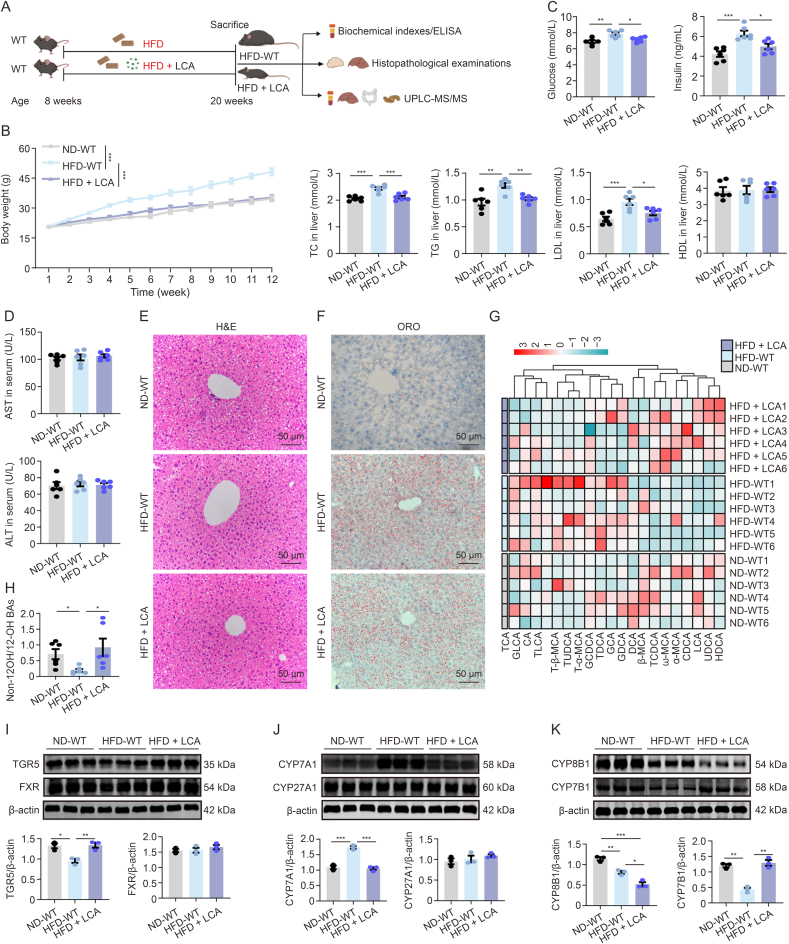
3.8. LCA treatment mitigated HFD-induced obesity and hepatic steatosis
To assess the effects of LCA modulated by the gut microbiota, we investigated mice fed an HFD and administered LCA (Fig. 9A). The HFD + LCA group showed reduced body weight gain and glucose, insulin, TC, TG, and LDL levels compared with the HFD-WT group (Figs. 9B and C). ALT and AST levels remained comparable to those in ND-WT mice, indicating the absence of hepatic damage due to the administered LCA dose (Fig. 9D). H&E staining showed fewer fat vacuoles and steatosis in the HFD + LCA group (Fig. 9E), with ORO staining confirming reduced hepatic lipid droplets compared with the HFD-WT group (Fig. 9F). The heatmap illustrating the serum BA profiles shows a significant decrease in 12-OH BAs, cholic acid (CA), TCA, taurodeoxycholic acid (TDCA), and glycodeoxycholic acid (GDCA) levels. Conversely, the quantities of non-12-OH BAs, LCA, HDCA, α-muricholic acid, ω-muricholic acid, and TCDCA levels were elevated (Fig. 9G). Furthermore, the HFD + LCA group showed a notable increase in the non-12-OH/12-OH BAs ratio (Fig. 9H). Western blotting revealed increased hepatic TGR5 and CYP7B1 protein levels in the HFD + LCA group, with decreased CYP7A1 and CYP8B1 and unchanged FXR and CYP27A1, suggesting the activation of an alternative pathway expanding non-12-OH BAs (Figs. 9I–K).
Fig. 9.
Lithocholic acid (LCA) treatment mitigated high-fat diet (HFD)-induced obesity and hepatic steatosis. (A) Schematic of LCA experimental design: wild-type (WT) littermates, aged 8 weeks, were subjected to weekly measurements of body weight while being fed normal diet (ND)-WT, HFD-WT, and HFD + LCA until reaching 20 weeks of age. (B) The curve of the changes in body weight observed at various time points of ND-WT, HFD-WT, and HFD + LCA mice (n = 6, two-way analysis of variance (ANOVA)). (C) Serum fasting blood glucose, insulin, total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) measurements of ND-WT, HFD-WT, and HFD + LCA mice (n = 6, one-way ANOVA with Bonferroni's correction). (D) Serum quantification of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) of ND-WT, HFD-WT, and HFD + LCA mice (n = 6, one-way ANOVA with Bonferroni's correction). (E) Representative hematoxylin and eosin (H&E)-stained images of liver tissue sections from ND-WT, HFD-WT, and HFD + LCA mice (n = 3). (F) Representative Oil Red O (ORO)-stained images of liver tissue sections from ND-WT, HFD-WT, and HFD + LCA mice (n = 3). (G) Heatmap illustrating the serum bile acid (BA) profiles of ND-WT, HFD-WT, and HFD + LCA mice. The gradient colors in the heatmap represent the z-scale values of serum BA concentrations. (H) Analysis of the serum ratios of non-12α-hydroxylated (non-12-OH)/12-OH BAs in ND-WT, HFD-WT, and HFD + LCA mice (n = 6, one-way ANOVA with Bonferroni's correction). (I) Representative Western blot bands and quantitative analysis depict G protein-coupled BA receptor 1 (TGR5) and farnesoid X receptor (FXR) expression in liver tissues of ND-WT, HFD-WT, and HFD + LCA mice (n = 3, Student's t-test). (J) Representative Western blot bands and quantitative analysis depict cholesterol 7α-hydroxylase (CYP7A1) and CYP27A1 expression in liver tissues of ND-WT, HFD-WT, and HFD + LCA mice (n = 3, Student's t-test). (K) Representative Western Blot bands and quantitative analysis depict cholesterol-12α-hydroxylase (CYP8B1) and CYP7B1 expression in liver tissues of ND-WT, HFD-WT, and HFD + LCA mice (n = 3, Student's t-test). Data are presented as the mean ± standard error of the mean (SEM). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. ELISA: enzyme-linked immunosorbent assay; UPLC-MS/MS: ultra-performance liquid chromatography-tandem mass spectrometry; TCA: taurocholic acid; GLCA: glycolithocholic acid; CA: cholic acid; TLCA: taurolithocholic acid; T-β-MCA: tauro-β-muricholic acid; TUDCA: tauroursodeoxycholic acid; GCDCA: glycochenodeoxycholic acid; TDCA: taurodeoxycholic acid; GCA: glycolcholic acid; GDCA: glycodeoxycholic acid; DCA: deoxycholic acid; TCDCA: taurochenodeoxycholic acid; CDCA: chenodeoxycholic acid; LCA: lithocholic acid; UDCA: ursodeoxycholic acid; HDCA: hyodeoxycholic acid.
4. Discussion
Obesity, a known risk factor for diabetes, liver disorders, cardiovascular diseases, and cancer, remains a major public health issue despite various interventions [30]. Consequently, novel pharmaceuticals and strategies are critical for combating obesity and the associated metabolic dysfunction. Studies have linked alterations in BA homeostasis to the pathogenesis of obesity, with significant changes in BA profiles observed in individuals with obesity [[31], [32], [33]]. In obese individuals with differing glucose tolerance, serum BAs, particularly GDCA and TDCA, are inversely correlated with the insulin clearance rate [32]. Our investigation revealed a marked decrease in non-12-OH BAs in individuals with high BMI compared with those with normal BMI, supporting previous findings of an intricate relationship between 12-OH BAs, lipid homeostasis, insulin sensitivity, and liver steatosis [11]. Elevated 12-OH BA levels have also been observed in obese individuals [34]. Emerging evidence underscores the strong link between non-12-OH BAs and the metabolic phenotype of obesity, suggesting that they enhance glycolipid metabolism by increasing the non-12-OH/12-OH BAs ratios [6,8]. A limitation of this study is that all subjects were recruited from a single obesity cohort, and the number of clinical serum samples was small, potentially limiting data representativeness. Future plans involve expanding the sample size and conducting cohort studies on other diseases like diabetes.
In our study, individuals with a high BMI exhibited reduced serum levels of PPP1R3G, which positively correlated with non-12-OH BAs levels. Recognized as a glycolipid metabolism regulator, the role of PPP1R3G became more compelling when genomic data from the GEO database indicated that HFD diminished liver expression in mice. A previous study reported that PPP1R3G HOE reduced fat mass and protected against ethanol-induced liver damage, underscoring its role in metabolic protection [19]. Our findings indicate that PPP1R3G HOE notably decreased fat mass and weight in HFD-fed mice, positively affecting key obesity indicators, including hepatic steatosis. However, these results are in contrast with those of Zhang et al. [35], where PPP1R3G ablation reduced glycogen deposition in the liver and decreased HFD-induced obesity. A possible explanation could be that PPP1R3G influences multiple metabolic pathways, and its manipulation could enhance glycogen synthesis with overexpression in the liver or activate alternative energy homeostasis pathways, thereby reducing fat accumulation when absent. Although our observations do not establish causality alone, they underscore the importance of further investigation into the link between PPP1R3G and obesity. Subsequent cellular studies indicated that the overexpression of PPP1R3G does not directly target hepatocytes to mitigate liver steatosis. Our murine model demonstrated that HFD reduced the serum levels of PPP1R3G, whereas PPP1R3G HOE enhanced the non-12-OH/12-OH BAs ratio in the serum and colon, contributing to obesity reduction. Additionally, HFD decreased LCA levels in the serum, liver, and colon; however, this reduction was effectively reversed by PPP1R3G HOE, which restored LCA levels. LCA significantly distinguished the groups in serum samples from humans and mice, indicating its potential as a clinical marker.
Changes in the BA composition are closely linked to the progression and development of obesity. Hepatic BA synthesis occurs via two primary routes: the classical pathway driven by CYP7A1 and CYP8B1 for 12-OH BAs and the alternative route initiated by CYP27A1 and CYP7B1 for non-12-OH BAs [6]. Variation in the non-12-OH BA ratio is regulated by the hepatic expression of CYP8B1 and CYP7B1 [36]. Our study found that HFD intervention decreased CYP7A1 expression in the livers of PPP1R3G HOE mice. The transition from the classical to the alternative BA synthesis pathway has been shown to protect against obesity in liver-specific transgenic Cyp7a1 mice [37]. We also observed a notable reduction in hepatic CYP8B1 expression in the HFD-HOE mice, which increased the relative proportion of non-12-OH BAs. This corresponds with earlier findings that Cyp8b1 knockdown decreases BA hydrophobicity and confers resistance to metabolic disorders such as HFD-induced obesity and hepatic steatosis in mice by reducing lipid absorption and increasing the ratio of conjugated non-12-OH/12-OH BAs, thereby enhancing lipid metabolism [38,39]. CYP7B1 expression was higher in the HFD-HOE group than in the HFD-WT group. Reduced hepatic CYP7B1 expression has been reported in obese individuals with T2DM [40]. Previous studies have suggested that activating the alternative synthesis pathway enhances glycolipid metabolism [6], indicating a potential link between glycolipid regulation by PPP1R3G and BA synthesis. However, reports have also indicated that adding CA, CDCA, or deoxycholic acid (DCA) to rat chow leads to downregulating CYP7B1 and CYP7A1-specific activities [41]. Administering various doses of CA, CDCA, DCA, and LCA to mice for one week decreased Cyp7a1 and Cyp8b1 expression in the liver [42]. Similarly, in vitro, hepatocytes treated with these BAs exhibited suppressed CYP7A1 and CYP8B1 [43]. Although the direct regulation of CYP7B1 and CYP8B1 by PPP1R3G has not been explicitly demonstrated, an indirect relationship through BA regulation may exist. Although PPP1R3G-HOE might affect certain aspects of hepatic BA regulation, it did not markedly change the expression of crucial genes involved in BA transport in the liver and ileal tissues under HFD conditions. Previous studies demonstrated that HDCA enhances hepatic lipid metabolism in nonalcoholic fatty liver disease mice by suppressing CYP7A1 expression, thus ameliorating fatty liver symptoms [44]. Gavage with a mixture of LCA and UDCA reduced hyperlipidemia, and administration of bacteria capable of converting and elevating the levels of UDCA, LCA, and succinic acid in mice improved obesity, hyperglycemia, and fatty liver degeneration induced by HFD [12]. In our study, the LCA intervention significantly improved obesity, hepatic steatosis, and serum biochemical parameters. LCA intervention also downregulated hepatic CYP7A1 and CYP8B1 and increased CYP7B1 expression under HFD, with an augmented proportion of non-12-OH BAs in the HFD + LCA group, aligning with the PPP1R3G HOE effects. This aligns with previous reports that feeding mice with LCA decreases Cyp7a1 and Cyp8b1 levels. Moreover, LCA feeding did not alter the mRNA or protein levels of Ntcp or Bsep [42]. These outcomes provide evidence of the hepatic safety of the administered LCA dose, which is an essential consideration in therapeutic strategies. In addition, the safety and effectiveness of LCA in broader populations remains to be determined in future clinical studies.
LCA, a gut-derived metabolic product of CDCA, is a potent natural agonist for TGR5 [45]. The binding of BAs to TGR5 on L cells stimulates glucagon-like peptide-1 release and enhances metabolic stability [46]. Similarly, the binding of BAs to TGR5 in adipose tissue increases energy expenditure, thus exerting beneficial metabolic effects in obese mice [47]. Additionally, the selective transport of LCA from the gut to the liver activates the vitamin D receptor, resulting in the production of cholic acid-7-sulfate and improvement in diabetic phenotypes following sleeve gastrectomy [48]. This indicates an alternative pathway for the metabolic effects of LCA independent of direct TGR5 activation. A growing body of literature has highlighted the critical role of TGR5 signaling in hepatic metabolic regulation, particularly in managing BA profiles. Ablation or inactivation of TGR5 signaling increases susceptibility to cholestatic liver injury in mice, likely because it maintains a healthy BA profile [[49], [50], [51]]. TGR5 agonists have shown promise in improving hepatic steatosis and reducing weight gain in mice fed an HFD [52]. Conversely, mice with TGR5 ablation have decreased total BA pool size [53,54]. A previous study has found that whole-body Tgr5−/− mice exhibited an increased ratio of 12-OH BAs/non-12-OH BAs [55]. This finding emphasizes the crucial role of TGR5 in maintaining the BA equilibrium. However, the underlying mechanism remains unclear. Given the central role of the liver in systemic metabolic regulation, TGR5 expressed in hepatocytes is a pivotal regulator of whole-body glucose homeostasis [56]. Activation of hepatic TGR5 in mice reduces CYP7A1 expression, thereby altering the overall BA pool composition [57]. Moreover, increased hepatic concentrations of conjugated 12-OH BAs contribute significantly to liver fibrosis via TGR5-mediated signaling, suggesting that antagonizing TGR5 may effectively prevent or reverse liver fibrosis [58]. Administration of the TGR5-specific agonist compound 18 improved glucose tolerance independent of body weight, glucagon-like peptide-1 secretion, and glucose-stimulated insulin secretion. This effect, which is absent in hepatocyte-specific TGR5 knockout mice, indicates the distinct role of hepatic TGR5 in glucose regulation [56]. Furthermore, treating HFD-fed mice with the TGR5-specific agonist INT-777 reduced liver steatosis, indicating that TGR5 signaling in the liver helps decrease TG accumulation [54,57]. Therefore, we speculated that altered liver TGR5 levels in PPP1R3G HOE mice likely affected HFD-induced obesity and liver steatosis by regulating BA profiles and overall metabolism. Future studies should use mice with specific knockouts to provide more definitive evidence. Additionally, future studies should use TGR5 inhibitors or knockout models alongside LCA administration and monitor changes in energy metabolism and weight without TGR5 signaling to reveal the independent role of LCA regulation.
The complex interplay between BA metabolism and the gut microbiota is crucial for regulating nutrient metabolism, energy balance, and overall health [22,59,60]. A study in 2007 identified metabolic differences between obesity-prone and obesity-resistant animals that were largely attributed to microbial metabolites [61]. Subsequent studies found that subtle changes in gut microbiota-mediated BA metabolism can lead to increased weight under the same diet, promoting an obesity-prone phenotype. Germ-free mice were resistant to HFD-induced obesity and exhibited significantly higher non-12-OH BA levels than their conventionally raised counterparts [62]. We also observed a dynamic interaction between BAs and intestinal microbes following HFD treatment. FMT of PPP1R3G HOE mice notably increased serum LCA and non-12-OH BA levels. Gut microbiota plays an important role in obesity. Lactobacillus is known for its health benefits, and its abundance often correlates with obesity [63]. Lactobacillus reuteri J1, for instance, mitigated obesity by inhibiting the FXR pathway and altering white adipose tissue, which is influenced by changes in UDCA and LCA [22]. Conversely, a decreased presence of Blautia, associated with disrupted glycolipid homeostasis and less visceral fat, was observed in obese individuals and HFD-fed mice [[64], [65], [66]]. Our study showed that PPP1R3G HOE increased Blautia levels and reduced Lactobacillus abundance, aiding obesity reduction. Additionally, the HFD-HOE group showed increased Lachnospiraceae_NK4A136_group levels, which are linked to BA metabolism and anti-obesity effects [67]. Importantly, Lactobacillus and Blautia are strongly linked to specific non-12-OH BAs. These findings suggest that PPP1R3G HOE affects the gut microbiota composition, potentially contributing to altered BA profiles. The metabolism of BAs by the gut microbiome influences the expression of many key enzymes involved in the de novo synthesis of BAs, including CYP7A1, CYP7B1, CYP8B1, and CYP27A1 [68]. Determining the primary function of hepatic PPP1R3G and its direct impact on the role of the gut microbiota in obesity prevention remains challenging. The shifts in non12-OH BAs might stem from the broader metabolic impact of PPP1R3G or alterations in the gut microbiome-BA interaction, which require further investigation to clarify the underlying mechanisms. Addressing microbiome dysbiosis remains challenging; however, our study highlights the potential of enhancing BA homeostasis as an anti-obesity treatment strategy alongside microbiome manipulation.
5. Conclusions
Our study emphasizes the capacity of hepatic PPP1R3G to mitigate obesity and liver steatosis by modulating the liver-gut axis. We established that the protective effects of hepatic PPP1R3G against obesity stem from its role in altering BA synthesis, modifying gut microbiota composition, and enhancing the levels of circulating non-12-OH BAs. Thus, our research reveals a new mechanism and therapeutic target for combating obesity relevant to other conditions affected by the gut microbiome and BA metabolism disturbances.
CRediT author statement
Chu Zhang: Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - Original draft preparation, Reviewing and Editing, Visualization, Funding acquisition; Gui Wang: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing - Original draft preparation; Xin Yin: Validation, Formal analysis, Investigation, Data curation, Funding acquisition; Lingshan Gou: Writing - Original draft preparation, Funding acquisition; Mengyuan Guo: Resources, Investigation; Feng Suo: Methodology, Investigation, Funding acquisition; Tao Zhuang and Zhenya Yuan: Methodology; Yanan Liu: Investigation, Funding acquisition; Maosheng Gu: Resources, Supervision, Funding acquisition; Ruiqin Yao: Resources, Writing - Reviewing and Editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the Natural Science Foundation for Young Scientists of China (Grant No.: 82201545), the Natural Science Foundation of Jiangsu Province, China (Grant No.: BK20221221), the Practice Innovation Program of Jiangsu Province, China (Grant No.: KYCX21_2641), the Medical Science Foundation of Jiangsu Province, China (Grant No.: H2019007), the Key Medical Talents Training Project of Xuzhou, China (Grant No.: XWRCHT20220060), the Xuzhou “Pengcheng Talent” Youth Medical Reserve Talent Project, China (Grant No.: XWRCHT20220014), and the Science and Technology Projects of Xuzhou, China (Grant No.: KC21061). We acknowledge the Prof. Chanmin Liu and Prof. Xingqi Wang from Jiangsu Normal University for providing us with transgenic mice. Graphical abstract was created with BioRender.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2024.100976.
Contributor Information
Maosheng Gu, Email: gumaosheng2007@126.com.
Ruiqin Yao, Email: wenxi_yao@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Breitfeld J., Kehr S., Müller L., et al. Developmentally driven changes in adipogenesis in different fat depots are related to obesity. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmody R.N., Bisanz J.E. Roles of the gut microbiome in weight management. Nat. Rev. Microbiol. 2023;21:535–550. doi: 10.1038/s41579-023-00888-0. [DOI] [PubMed] [Google Scholar]
- 3.Middeldorp M.E., Kamsani S.H., Sanders P. Obesity and atrial fibrillation: Prevalence, pathogenesis, and prognosis. Prog. Cardiovasc. Dis. 2023;78:34–42. doi: 10.1016/j.pcad.2023.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Di Ciaula A., Bonfrate L., Khalil M., et al. Contribution of the microbiome for better phenotyping of people living with obesity. Rev. Endocr. Metab. Disord. 2023;24:839–870. doi: 10.1007/s11154-023-09798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L., Liu Y., Wang S., et al. Cholestasis: Exploring the triangular relationship of gut microbiota-bile acid-cholestasis and the potential probiotic strategies. Gut Microbes. 2023;15 doi: 10.1080/19490976.2023.2181930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia W., Wei M., Rajani C., et al. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell. 2021;12:411–425. doi: 10.1007/s13238-020-00804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogelson K.A., Dorrestein P.C., Zarrinpar A., et al. The gut microbial bile acid modulation and its relevance to digestive health and diseases. Gastroenterology. 2023;164:1069–1085. doi: 10.1053/j.gastro.2023.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M., Wang S., Li Y., et al. Gut microbiota-bile acid crosstalk contributes to the rebound weight gain after calorie restriction in mice. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-29589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erawijantari P.P., Mizutani S., Shiroma H., et al. Influence of gastrectomy for gastric cancer treatment on faecal microbiome and metabolome profiles. Gut. 2020;69:1404–1415. doi: 10.1136/gutjnl-2019-319188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei M., Huang F., Zhao L., et al. A dysregulated bile acid-gut microbiota axis contributes to obesity susceptibility. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haeusler R.A., Astiarraga B., Camastra S., et al. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013;62:4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Liao M., Zhou N., et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26:222–235.e5. doi: 10.1016/j.celrep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X., Huang F., Zhao A., et al. Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 2017;15 doi: 10.1186/s12915-017-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang L. Pioglitazone trial for NASH: Results show promise. Gastroenterology. 2007;132:836–838. doi: 10.1053/j.gastro.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Ceulemans H., Stalmans W., Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. BioEssays. 2002;24:371–381. doi: 10.1002/bies.10069. [DOI] [PubMed] [Google Scholar]
- 16.Luo X., Zhang Y., Ruan X., et al. Fasting-induced protein phosphatase 1 regulatory subunit contributes to postprandial blood glucose homeostasis via regulation of hepatic glycogenesis. Diabetes. 2011;60:1435–1445. doi: 10.2337/db10-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.U.S. Udoh. Valcin J.A., Swain T.M., et al. Genetic deletion of the circadian clock transcription factor BMAL1 and chronic alcohol consumption differentially alter hepatic glycogen in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314:G431–G447. doi: 10.1152/ajpgi.00281.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Xu D., Huang H., et al. Regulation of glucose homeostasis and lipid metabolism by PPP1R3G-mediated hepatic glycogenesis. Mol. Endocrinol. 2014;28:116–126. doi: 10.1210/me.2013-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu J., Zhang Y., Xu D., et al. Ethanol-induced hepatic steatosis is modulated by glycogen level in the liver. J. Lipid Res. 2015;56:1329–1339. doi: 10.1194/jlr.M056978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y., Yang X., Zhang J., et al. Ketogenic diets induced glucose intolerance and lipid accumulation in mice with alterations in gut microbiota and metabolites. mBio. 2021;12 doi: 10.1128/mBio.03601-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Obesity Preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ. Tech. Rep. Ser. 2000;894:1–253. i-xii. [PubMed] [Google Scholar]
- 22.Zhang C., Fang R., Lu X., et al. Lactobacillus reuteri J1 prevents obesity by altering the gut microbiota and regulating bile acid metabolism in obese mice. Food Funct. 2022;13:6688–6701. doi: 10.1039/d1fo04387k. [DOI] [PubMed] [Google Scholar]
- 23.Xie G., Wang Y., Wang X., et al. Profiling of serum bile acids in a healthy Chinese population using UPLC-MS/MS. J. Proteome Res. 2015;14:850–859. doi: 10.1021/pr500920q. [DOI] [PubMed] [Google Scholar]
- 24.Ma J., Hong Y., Zheng N., et al. Gut microbiota remodeling reverses aging-associated inflammation and dysregulation of systemic bile acid homeostasis in mice sex-specifically. Gut Microbes. 2020;11:1450–1474. doi: 10.1080/19490976.2020.1763770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X., Li J., Lee A., et al. Satiety induced by bile acids is mediated via vagal afferent pathways. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albillos A., de Gottardi A., Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J. Hepatol. 2020;72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe Y., Fujisaka S., Ikeda K., et al. Gut microbiota, determined by dietary nutrients, drive modification of the plasma lipid profile and insulin resistance. iScience. 2021;24 doi: 10.1016/j.isci.2021.102445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., Mao B., Gu J., et al. Blautia - a new functional genus with potential probiotic properties? Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1875796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Million M., Thuny F., Angelakis E., et al. Lactobacillus reuteri and Escherichia coli in the human gut microbiota may predict weight gain associated with vancomycin treatment. Nutr. Diabetes. 2013;3 doi: 10.1038/nutd.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lingvay I., Agarwal S. A revolution in obesity treatment. Nat. Med. 2023;29:2406–2408. doi: 10.1038/s41591-023-02538-7. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., van den Munckhof I.C.L., Schraa K., et al. Genetic and microbial associations to plasma and fecal bile acids in obesity relate to plasma lipids and liver fat content. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108212. [DOI] [PubMed] [Google Scholar]
- 32.Fu Z., Wu Q., Guo W., et al. Impaired insulin clearance as the initial regulator of obesity-associated hyperinsulinemia: Novel insight into the underlying mechanism based on serum bile acid profiles. Diabetes Care. 2022;45:425–435. doi: 10.2337/dc21-1023. [DOI] [PubMed] [Google Scholar]
- 33.Hyogo H., Roy S., Paigen B., et al. Leptin promotes biliary cholesterol elimination during weight loss in ob/ob mice by regulating the enterohepatic circulation of bile salts. J. Biol. Chem. 2002;277:34117–34124. doi: 10.1074/jbc.M203912200. [DOI] [PubMed] [Google Scholar]
- 34.Haeusler R.A., Camastra S., Nannipieri M., et al. Increased bile acid synthesis and impaired bile acid transport in human obesity. J. Clin. Endocrinol. Metab. 2016;101:1935–1944. doi: 10.1210/jc.2015-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Gu J., Wang L., et al. Ablation of PPP1R3G reduces glycogen deposition and mitigates high-fat diet induced obesity. Mol. Cell. Endocrinol. 2017;439:133–140. doi: 10.1016/j.mce.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 36.Trauner M., Fuchs C.D., Halilbasic E., et al. New therapeutic concepts in bile acid transport and signaling for management of cholestasis. Hepatology. 2017;65:1393–1404. doi: 10.1002/hep.28991. [DOI] [PubMed] [Google Scholar]
- 37.Li T., Owsley E., Matozel M., et al. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology. 2010;52:678–690. doi: 10.1002/hep.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertaggia E., Jensen K.K., Castro-Perez J., et al. Cyp8b1 ablation prevents Western diet-induced weight gain and hepatic steatosis because of impaired fat absorption. Am. J. Physiol. Endocrinol. Metab. 2017;313:E121–E133. doi: 10.1152/ajpendo.00409.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P., Ruan X., Yang L., et al. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21:455–467. doi: 10.1016/j.cmet.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian F., Huang S., Xu W., et al. Compound K attenuates hyperglycemia by enhancing glucagon-like peptide-1 secretion through activating TGR5 via the remodeling of gut microbiota and bile acid metabolism. J. Ginseng Res. 2022;46:780–789. doi: 10.1016/j.jgr.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren S., Marques D., Redford K., et al. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in the rat. Metabolism. 2003;52:636–642. doi: 10.1053/meta.2003.50106. [DOI] [PubMed] [Google Scholar]
- 42.Song P., Rockwell C.E., Cui J.Y., et al. Individual bile acids have differential effects on bile acid signaling in mice. Toxicol. Appl. Pharmacol. 2015;283:57–64. doi: 10.1016/j.taap.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J., Lu H., Lu Y.-F., et al. Potency of individual bile acids to regulate bile acid synthesis and transport genes in primary human hepatocyte cultures. Toxicol. Sci. 2014;141:538–546. doi: 10.1093/toxsci/kfu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuang J., Wang J., Li Y., et al. Hyodeoxycholic acid alleviates non-alcoholic fatty liver disease through modulating the gut-liver axis. Cell Metab. 2023;35:1752–1766.e8. doi: 10.1016/j.cmet.2023.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Navarro S.L., Levy L., Curtis K.R., et al. Effect of a flaxseed lignan intervention on circulating bile acids in a placebo-controlled randomized, crossover trial. Nutrients. 2020;12 doi: 10.3390/nu12061837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pols T.W., Noriega L.G., Nomura M., et al. The bile acid membrane receptor TGR5: A valuable metabolic target. Dig. Dis. 2011;29:37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L., Yang Q., Zhang E., et al. Notoginsenoside Ft1 acts as a TGR5 agonist but FXR antagonist to alleviate high fat diet-induced obesity and insulin resistance in mice. Acta Pharm. Sin. B. 2021;11:1541–1554. doi: 10.1016/j.apsb.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaudhari S.N., Luo J.N., Harris D.A., et al. A microbial metabolite remodels the gut-liver axis following bariatric surgery. Cell Host Microbe. 2021;29:408–424.e7. doi: 10.1016/j.chom.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merlen G., Kahale N., Ursic-Bedoya J., et al. TGR5-dependent hepatoprotection through the regulation of biliary epithelium barrier function. Gut. 2020;69:146–157. doi: 10.1136/gutjnl-2018-316975. [DOI] [PubMed] [Google Scholar]
- 50.Reich M., Deutschmann K., Sommerfeld A., et al. TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut. 2016;65:487–501. doi: 10.1136/gutjnl-2015-309458. [DOI] [PubMed] [Google Scholar]
- 51.Péan N., Doignon I., Garcin I., et al. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58:1451–1460. doi: 10.1002/hep.26463. [DOI] [PubMed] [Google Scholar]
- 52.Poupon R. Bile acid mimetic-activated TGR5 receptor in metabolic-related liver disorder: The good and the bad. Gastroenterology. 2010;138:1207–1209. doi: 10.1053/j.gastro.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 53.Maruyama T., Tanaka K., Suzuki J., et al. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J. Endocrinol. 2006;191:197–205. doi: 10.1677/joe.1.06546. [DOI] [PubMed] [Google Scholar]
- 54.Vassileva G., Golovko A., Markowitz L., et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem. J. 2006;398:423–430. doi: 10.1042/BJ20060537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donepudi A.C., Boehme S., Li F., et al. G-protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis in mice. Hepatology. 2017;65:813–827. doi: 10.1002/hep.28707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holter M.M., Chirikjian M.K., Briere D.A., et al. Compound 18 improves glucose tolerance in a hepatocyte TGR5-dependent manner in mice. Nutrients. 2020;12 doi: 10.3390/nu12072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas C., Gioiello A., Noriega L., et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie G., Jiang R., Wang X., et al. Conjugated secondary 12α-hydroxylated bile acids promote liver fibrogenesis. EBioMedicine. 2021;66 doi: 10.1016/j.ebiom.2021.103290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duparc T., Plovier H., Marrachelli V.G., et al. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut. 2017;66:620–632. doi: 10.1136/gutjnl-2015-310904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mori H., Svegliati Baroni G., Marzioni M., et al. Farnesoid X receptor, bile acid metabolism, and gut microbiota. Metabolites. 2022;12 doi: 10.3390/metabo12070647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Ni Y., Su M., et al. Pharmacometabonomic phenotyping reveals different responses to xenobiotic intervention in rats. J. Proteome Res. 2007;6:1364–1370. doi: 10.1021/pr060513q. [DOI] [PubMed] [Google Scholar]
- 62.Sayin S.I., Wahlström A., Felin J., et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Zhou M., Johnston L.J., Wu C., et al. Gut microbiota and its metabolites: Bridge of dietary nutrients and obesity-related diseases. Crit. Rev. Food Sci. Nutr. 2023;63:3236–3253. doi: 10.1080/10408398.2021.1986466. [DOI] [PubMed] [Google Scholar]
- 64.Benátez-Páez A., Gómez Del Pugar E.M., López-Almela I., et al. Depletion of Blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. mSystems. 2020;5 doi: 10.1128/mSystems.00857-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B., Kong Q., Li X., et al. A high-fat diet increases gut microbiota biodiversity and energy expenditure due to nutrient difference. Nutrients. 2020;12 doi: 10.3390/nu12103197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong X., Xu J., Lian F., et al. Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: A multicenter, randomized, open label clinical trial. mBio. 2018;9 doi: 10.1128/mBio.02392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang P., Li D., Ke W., et al. Resveratrol-induced gut microbiota reduces obesity in high-fat diet-fed mice. Int. J. Obes. (Lond.) 2020;44:213–225. doi: 10.1038/s41366-019-0332-1. [DOI] [PubMed] [Google Scholar]
- 68.Wahlström A., Sayin S.I., Marschall H.U., et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.