Abstract
Extrachromosomal circular DNA (eccDNA), a pervasive yet enigmatic component of the eukaryotic genome, exists autonomously from its chromosomal counterparts. Ubiquitous in eukaryotes, eccDNA plays a critical role in the orchestration of cellular processes and the etiology of diseases, particularly cancers. However, the full scope of its influence on health and disease remains elusive, presenting a rich vein of research yet to be mined. Unraveling the complexities of eccDNA necessitates a distillation of methodologies — from biogenesis to functional analysis — a landscape we overview in this study with precision and clarity. Here, we systematically outline cutting-edge methodologies from high-throughput sequencing and bioinformatics to experimental validations, showcasing the intricate world of eccDNAs. We combed through a treasure trove of auxiliary research resources and analytical tools. Moreover, we chart a course for future inquiry, illuminating the horizon with potential groundbreaking strategies for designing eccDNA research projects and pioneering new methodological frontiers.
Keywords: Extrachromosomal circular DNA, eccDNA, ecDNA, Methodology, Approach, High-throughput sequencing, Single cell, Experimental method, Analytical tool
1. Introduction
Extrachromosomal circular DNA (eccDNA) is derived from chromosomes but is independent from chromosomal DNA (chrDNA) [1]. They ubiquitously present in eukaryotic species, including yeast, fruit flies, nematodes, and humans [2]. According to the size and copy number, extrachromosomal circular DNA are categorized into three types in human cells [3]: a) small-size eccDNA, less than 1 kb, including telomere rings, microDNA, etc., invisible to optical microscopy; b) cancer-specific large-size eccDNA (usually known as ecDNA) ranging from 1 to 3 Mb and larger, with multiple intact genes visible under light microscopy, including double microsomes and DNA lacking telomeres; c) rings and neochromosomes. It is important to note that the eccDNA and ecDNA notations may be defined and used differently in different studies. Therefore, in specific studies, eccDNA and ecDNA need to be understood and distinguished according to the context. Three key characteristics of eccDNA are: a) circular structure; b) high accessibility owing to open chromatin; c) non-Mendelian laws inheritance because of the lack of centromeres [4]. Such characteristics make eccDNA an important mechanism driving tumor heterogeneity [5], which will bring great challenges to targeted treatment to tumors. Some studies suggest that eccDNA may come from DNA damage repair, over-transcription, homologous recombination and replication stress [6], but the specific formation mechanism and function are not clear.
Presence of eccDNA plays a pivotal role in both physiological processes and disease progression, including cancer. It contributes to genome rearrangements [7,8], DNA damage and repair [9], cellular evolution and fitness [7,10], development and aging [11,12], and the regulation of immune responses [6] (Fig. 1 A). In recent years, studies have shown that eccDNA is intricately linked to cancer, where it enhances chromatin accessibility [13], accelerates oncogene amplification [[14], [15], [16], [17], [18]], drives genetic heterogeneity [5], facilitates cancer cell escape from immune surveillance [6,19], and is associated with poor cancer prognosis [4,20] (Fig. 1 B). Therefore, eccDNA holds significant potential as both a diagnostic marker and a therapeutic target. Despite its established role in cancer, the precise causal relationship between eccDNA and broader aspects of human health or disease remains to be fully elucidated and merits further investigation. Historically, the trajectory of scientific discovery has been intricately tied to technological advancements, a trend well documented across various fields of research. In order to propel the momentum of research on eccDNA and to delve deeper into its functions and potential value, there is a pressing need for a comprehensive synthesis and generalization of the diverse methodologies employed in this field. In this review, we summarize the methods for studying eccDNAs, covering high-throughput sequencing, bioinformatics analysis, experimental verifications, and auxiliary research resources and tools. In addition, we discuss designing research projects on eccDNA and outline the future perspective of emerging methods.
Fig. 1.
The pivotal role of eccDNA in various physiological processes and its implications in human health and disease. (A) The importance of eccDNA in physiological processes. This section underscores the significance of eccDNA in key biological processes, such as genome rearrangements, DNA damage and repair, and cellular evolution and fitness. It also highlights the role of eccDNA in fundamental life processes including development and aging, as well as its involvement in the regulation of immune responses. These diverse functions underscore eccDNA's influence on the cellular and molecular architecture, emphasizing its importance in maintaining cellular homeostasis and responding to environmental challenges. (B) Implications of eccDNA in human health and diseases. In this part, the focus shifts to the impact of eccDNA on various aspects of human health, particularly in the context of oncology. It delineates how eccDNA influences oncogene expression and transcription, contributing to tumor heterogeneity and the development of drug resistance. This section also explores the emerging potential of eccDNA to serve as a liquid biopsy-derived biomarker. The visual elements in the image were created by BioRender.com.
2. Methods for eccDNA study
2.1. Methods of eccDNA purification and enrichment prior to sequencing
To accurately characterize the distribution, abundance, and functionality of eccDNA, it is crucial to establish efficient and reliable purification techniques (Fig. 2A), facilitating subsequent high-performance eccDNA enrichment and sequencing.
Fig. 2.
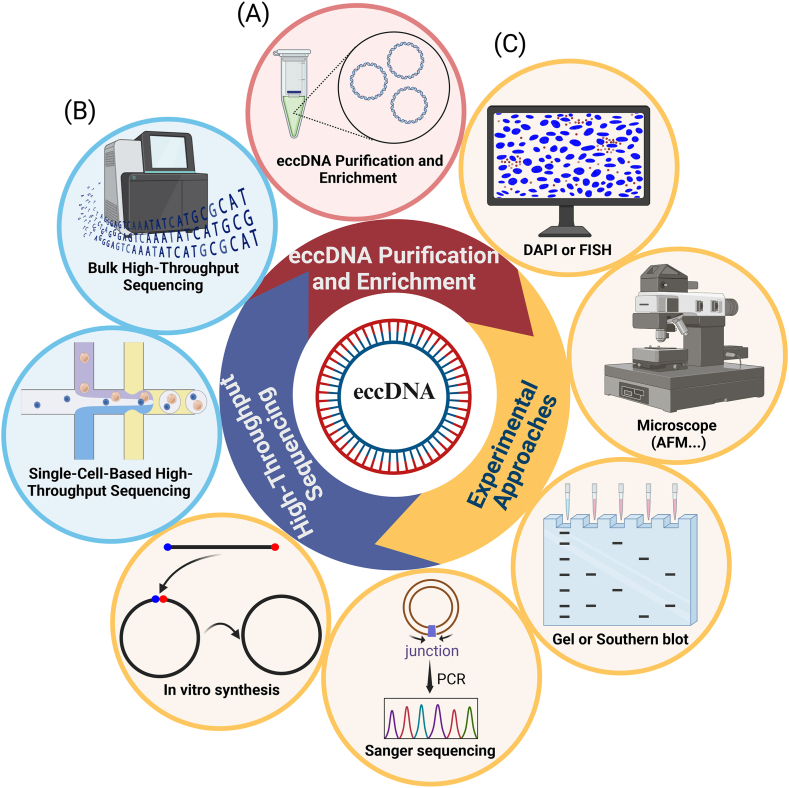
Systematic categorization of methods for investigating eccDNA. (A) Methods of eccDNA isolation and purification prior to NGS sequencing or visualization. These methods are crucial for preparing samples for subsequent high-throughput sequencing and ensuring the integrity and specificity of the eccDNA being studied. (B) High-throughput sequencing for eccDNA study. This part is further subdivided into two categories: bulk high-throughput techniques and single-cell-based high-throughput sequencing. (C) Experimental methodologies for identifying and exploring eccDNA. It includes various techniques used to confirm the presence of eccDNA in samples, as well as to investigate the biological function of eccDNA: This part focuses on the approaches including DAPI/FISH staining, microscopy, gel, southern blot, sanger sequencing and in-vito synthesis of eccDNA, employed to understand the biological roles and functional implications of eccDNA. The visual elements in the image were created by BioRender.com.
2.1.1. Non-circular purification methods
Methods in this category are generally not designed specifically for circular DNA but can be used to enrich DNA samples that may contain both linear and circular forms. These methods include general DNA extraction and purification protocols that are not selective for circular DNA. Commonly employed across multiple methodologies, the initial steps involve cell lysis and DNA purification using the genomic or plasmid DNA extraction protocols, ensuring the isolation of eccDNA. For genomic DNA, it is extracted and then subjected to enzymatic treatment to remove mitochondrial circular DNA and residual linear DNA, following the genomic DNA extraction kit guidelines [21]. For plasmid DNA, samples are treated to separate the eccDNA from chromosomal DNA, lipids, and proteins, followed by a series of enzymatic treatments to eliminate mitochondrial circular DNA and linear DNA using the Plasmid Mini AX kit protocols [[22], [23], [24], [25], [26]].
2.1.2. Circular purification and enrichment methods
-
1
3SEP procedure
Moreover, a new three-step eccDNA purification (3SEP) procedure was developed [6,27], enhancing eccDNA purity and reproducibility by incorporating three key steps: a) modified lysis to preserve circle integrity; b) linearization of mitochondrial DNA (mtDNA) before digesting linear DNA; and c) further separation from linear DNA remnants using beads. This significantly improves the efficiency of isolating pure eccDNA.
-
2
Purification and enrichment of eccDNA according to length
For the purification of short extrachromosomal circular DNAs (such as microDNA), linear DNA is removed using an ATP-dependent exonuclease, followed by the enrichment of circular DNA through rolling circle amplification (RCA) via multiple displacement amplification [28]. In addition, a noteworthy method is clustered regularly interspaced short palindromic repeats - Cas9-assisted targeting of chromosome segments (CRISPR-CATCH) [29], which stands out for its ability to selectively enrich megabase-sized eccDNA. This strategy is effectively a fusion of in-vitro CRISPR-Cas9 cleavage and pulsed field gel electrophoresis (PFGE), resulting in high precision and efficiency in enriching eccDNA. A comparative overview of the strengths and limitations of the eccDNA purification and enrichment methods is provided (Table 1).
Table 1.
Advantages and disadvantages of eccDNA purification and enrichment methods.
| name | principles | advantages | disadvantages | applications |
|---|---|---|---|---|
| Column-based purification | Employing genomic or plasmid DNA extraction protocols to isolate eccDNA via column chromatography. | 1. Effective for isolating both small and large eccDNA fragments. 2. Offers reproducibility across different samples. |
1. Multiple steps involved can be time-consuming. 2. Risk of contamination requires careful handling. |
Suitable for large-scale eccDNA studies from cell and tissue samples, where high precision and efficiency are required. Ideal for both short and long-read sequencing applications and studies involving both high and low eccDNA abundance. |
| 3SEP procedure | Three steps are: 1) modified lysis to preserve circle integrity, 2) linearization of mtDNA before digesting linear DNA, and 3) further separation from linear DNA remnants using beads. | 1. Enhances eccDNA purity and reproducibility. 2. Significantly improves the efficiency of isolating pure eccDNA. |
1. Additional step increases the complexity of the procedure. 2. May require optimization for different sample types. |
Particularly useful for high-fidelity applications where the purity of eccDNA is crucial, such as in diagnostic and clinical research settings. Effective for both short and long eccDNA molecules. |
| CRISPR-CATCH | Combines in-vitro CRISPR-Cas9 cleavage with PFGE for selectively enriching megabase-sized eccDNA. | 1. Allows for the selective enrichment of megabase-sized eccDNA. 2. Effective in enriching large eccDNA, making it suitable for studies on large eccDNA molecules. |
1. Requires specific equipment and expertise, which might not be available in all laboratories. 2. The method is resource-intensive and may not be feasible for all research setups. |
Ideal for detailed studies on large eccDNA, particularly in cancer research where megabase-sized eccDNA is common. Suitable for long-read sequencing applications to study eccDNA dynamics and functions. |
2.2. High-throughput sequencing for eccDNA study
2.2.1. Bulk high-throughput techniques for eccDNA identification
In addition to isolation and purification, eccDNA studies also encompass high-throughput sequencing (Fig. 2B) and subsequent experimental validation (Fig. 2C). Currently, several bulk high-throughput methods exist for the systematic identification of eccDNAs, including whole-genome sequencing (WGS) [5,13,30], genome-scale enrichment and detection method for eccDNA (Circle-seq) [26,31], circular DNA enrichment sequencing (CIDER-seq) [32], assay for transposase-accessible chromatin using sequencing (ATAC-seq) [33,34] and Circulome-seq [35] (Fig. 3). Of these methods, the most commonly used in the currently published articles is WGS and circle-seq. WGS is a technique that allows for the identification of all genomic variations, including eccDNA [5,13,30], through the sequencing of an entire genome. It provides a comprehensive view of genetic information, enabling not just the identification of eccDNA, but also other types of genetic variations. However, this method may overlook some small eccDNAs. Circle-seq [26,31] is a technique that enriches and identifies eccDNA by leveraging its inherent circular structural characteristics, which are exploited molecularly to separate eccDNA from linear DNA fragments. Compared to WGS, Circle-seq can identify eccDNA more accurately, but it has higher costs and cannot identify other types of genetic variations. CIDER-seq is also a method that takes advantage of the circular structure of eccDNA for enrichment and identification [32]. It offers highly accurate identification of eccDNA and can be performed at the single-molecule level. Compared to WGS and Circle-seq, the operation of CIDER-seq is more complex and might require more technically proficient personnel. ATAC-seq is a technique primarily used for identifying open chromatin regions, and in principle, it only provides indirect evidence [33,34]. In addition, Circulome-seq is an integrated method that enriches and characterizes genomic circular DNAs by combining exonuclease digestion, CsCl/ethidium-bromide gradient centrifugation, and Tn5 transposition-based sequencing [35]. Each of these four methods has its strengths and weaknesses (Table 2), and researchers can choose the most suitable method for their studies of eccDNA based on their experimental objectives and available resources.
Fig. 3.
The evolutionary journey of various eccDNA sequencing methods. The timeline format effectively maps out key milestones and breakthroughs in this field, illustrating the transformative impact these methods have had on our understanding of eccDNA. It begins with the inception of basic techniques (such as Circle-seq and WGS) for eccDNA detection and gradually transitions to more sophisticated methods, including bulk high-throughput approaches (such as scCircle-seq and scWGS) which have allowed for the study of eccDNA in populations of cells. Further along the timeline, the focus shifts to single-cell high-throughput technologies. This distinction between bulk and single-cell methodologies highlights the increasing precision and specificity with which researchers can now study eccDNA. The visual elements in the image were created by BioRender.com.
Table 2.
High-throughput sequencing based on bulk and single cell.
| name | aliquot type | application | code availability | platform | principle | reference |
|---|---|---|---|---|---|---|
| WGS | bulk | detection of eccDNA by computational tools | AmpliconArchitect (https://github.com/jluebeck/AmpliconArchitect), AmpliconClassifier (https://github.com/jluebeck/AmpliconClassifier), AmpliconSuite-pipeline (https://github.com/jluebeck/AmpliconSuite-pipeline) | Illumina (100–150-bp paired-end reads) | by detecting copy number variation, analyzing sequencing depth, and using specific analytical tools | Turner et al. [5]; Wu et al. [13]; Luebeck et al. [30] |
| Circle-seq | bulk | genome-scale detection of eccDNAs | Circle-Map (https://github.com/iprada/Circle-Map) | Illumina HiSEq. 2000 platform | DNA is denatured, rapidly neutralized, and separated by column chromatography. It is then treated with NotI endonuclease and exonuclease to remove linear DNA, followed by circular DNA enrichment using ϕ29 DNA polymerase in RCA. | Shibata et al. [31]; Møller et al. [26] |
| CIDER-seq | bulk | full-length sequences and profiles of eccDNAs | https://github.com/devang-mehta/ciderseq2 | long-read sequencing on a Pacific Biosciences SMRT-seq instrument | Workflow is organized into three main parts: circular DNA enrichment, PacBio sequencing, and data analysis. | Mehta et al. [32] |
| ATAC-seq | bulk | identification of open chromatin regions and eccDNAs | https://github.com/pk7zuva/Circle_finder | paired-end Illumina sequencing | uses the hyperactive transposase Tn5 to cut the accessible chromatin with simultaneous ligation of adapters at cut sites | Kumar et al. [33]; Su et al. [34] |
| scEC&T-seq | single cells | detection of circular DNA and mRNA in single cells | https://github.com/henssen-lab/scEC-T-seq | Illumina PE 75 (scRNA-seq) & Illumina PE 150 or Nanopore (scCircle-seq) | isolation of DNA followed by removal of linear DNA through exonuclease digestion and enrichment of circular DNA by RCA | Chamorro González et al. [36] |
| SMOOTH-seq | single cells | detection of SVs and eccDNAs in individual cells | https://github.com/cyang235/Smooth-seq | third-generation sequencing, utilizing specifically SMRT DNA sequencing technology | Single-molecule real-time sequencing is performed on long fragments amplified through transposon insertion. This method enables accurate detection of SVs by leveraging long high-fidelity reads. | Fan et al. [37] |
| CCDA-seq | single cells | mapping of the eccDNA chromatin status at a multikilobase scale | NA | long-read nanopore sequencing | label open chromatin regions on intact circular DNA using methyltransferase, enrich circular DNA by digesting linear DNA | Chen et al. [38] |
| Mobilome-seq | single cells | enrichment of eccDNA | https://github.com/njaupan | MiSeq sequencer (Illumina) | relies on linear digestion of genomic DNA followed by RCA of circular DNA | Lanciano et al. [40] |
| scCircle-seq | single cells | dissect circDNA complexity | https://github.com/BiCroLab/scCircle-seq | HiSeq X Ten (Illumina) | selectively amplifies circular DNA by RCA while linear DNA is digested by enzymes | Chen et al. [41] |
| scATAC-seq | single cells | mark active eccDNA enhancers | https://github.com/ChangLab/ecDNA-hub-code-2021 | Illumina NovaSeq 6000 | At the single-cell level, it employs the hyperactive transposase Tn5 to cut the accessible chromatin, while simultaneously ligating adapters at the cut sites. | Hung et al. [16] |
| scWGS | single cells | investigate copy-number changes | https://github.com/FrancisCrickInstitute/PEACE_melanoma_14_paper | HiSeq 4000 | Nuclei isolated from frozen tissue were stained with Hoechst and PI, sorted by ploidy via FACS, and then 48 cells from each group underwent library preparation and single-end DNA sequencing. | Spain et al. [42]; Chang et al. [43] |
| scGTP-seq | single cells | analyze eccDNA and SVs | https://github.com/fanxylab/EcDNAFinder.git | third‐generation platform | detects eccDNA candidate reads through read rearrangements, constructs breakpoint graphs for each read, concatenates consensus fragments, annotates coordinates, and filters circular DNA structures | Chang et al. [43] |
2.2.2. Single-cell-based high-throughput sequencing for eccDNA exploration
In recent years, the single cell sequencing technology has made great progress in the field of cell biology. High-throughput sequencing for eccDNA based on single cells came into being, which includes single-cell extrachromosomal circular DNA and transcriptome sequencing (scEC&T-seq) [36] and single-molecule real-time sequencing of long fragments amplified through transposon insertion (SMOOTH-seq) [37] (Fig. 3). Among these, the scEC&T-seq approach is significant as it allows for parallel analysis of eccDNA and transcriptomes at the single-cell level, providing comprehensive insights into the functional implications of eccDNA in cellular phenotypes and gene expression [36]. In addition, chromatin status of eccDNA at a single-molecule resolution is tested by a sequencing of enzyme-accessible chromatin in circular DNA (CCDA-seq) [38], while methylation status is analyzed using tagmentation and enzymatic conversion approaches [39] (Fig. 3). Active transposable elements within eccDNA are identified through Mobilome-seq [40] (Fig. 3), which has discovered transposable elements in a variety of plants and animals, containing Arabidopsis, rice, peanuts, potatoes, aspen, fruit flies. Furthermore, techniques like single‐cell Circle-seq (scCircle-seq) [41], single‐cell ATAC-seq (scATAC-seq) [16], single‐cell WGS (scWGS) [42,43] and single‐cell paralleled genome and transcriptome sequencing on a third‐generation platform (scGTP-seq) [43] have been utilized to dissect ecDNA complexity, mark active ecDNA enhancers, investigate copy-number changes, and uncover structure variation, respectively (Fig. 3). These single-cell methodologies have significantly enhanced our understanding of eccDNA, providing a more nuanced and detailed perspective on its role and dynamics in cellular biology.
2.3. Experimental approaches for eccDNA study
2.3.1. Experimental methods to verify the existence of eccDNA
While high-throughput sequencing methods provide an extensive overview of eccDNA, experimental approaches usually provide objective and direct evidence for its existence. Imaging-based methods, particularly those employing fluorescence in situ hybridization (FISH), remain the gold standard for confirming the presence of eccDNA [44]. FISH, a molecular cytogenetic technique, can detect the distribution of a known DNA fragment. The fluorescence intensity of FISH also reveals the content and location of eccDNA in different cells. By combining DNA FISH, 4′,6-diamidino-2-phenylindole (DAPI) or other staining with imaging-based methods such as scanning electron microscopy, transmission electron microscopy and 3D structure illumination microscopy, we can determine whether a known gene forms eccDNA. In addition, the combination of outward-directed PCR (inverse PCR) with Sanger sequencing, agarose gel electrophoresis and Southern blot has been utilized in published literature to verify the presence of eccDNA. A primer is designed for the junction of eccDNA, and the amplified fragment will cross the junction, while another primer can be designed for inward-directed PCR as internal reference for eccDNA circularization (Fig. 2C). In summary, these experimental methods complement high-throughput sequencing by providing crucial insights into the physical characteristics and spatial distribution of eccDNA, thereby enhancing our understanding of its biological significance.
2.3.2. Live cell imaging systems to visualize eccDNAs
Live cell imaging microscopy, a highly specific tool offering real-time dynamic monitoring of living cells and even single molecules, propels new advancements in biology and medicine. The Casilio system, developed by Cheng et al., combines CRISPR-Cas9 and Pumilio RNA-binding proteins to offer a versatile system for gene regulation and genomic labeling [45]. The CRISPR-based DNA tracking system, ecTag, derived from Casilio technology, tracks the dynamic changes of eccDNA in living cells, aiding the study of eccDNA's role in tumor heterogeneity [46]. By designing sgRNA to target eccDNA at the sequence of breakpoint sites, dCas9 and engineered Pumilio/FBF (PUF)-tethered effectors (Casilio) technology maps the breakpoint of eccDNA and recruits fluorescent molecules to the PUF binding site. Besides, clustered regularly interspaced short palindromic repeats FISH amplifier (CRISPR FISHer) imaging visualizes eccDNAs in live cells, such as HPV-human hybrid eccDNA [47]. Another study employed a Tet-operator (TetO) array insertion into eccDNA and labeled it with TetR-eGFP or TetR-A206K-eGFP for minimized GFP dimerization, enabling eccDNA detection in living cells [16]. In summary, these visualization methods, bridging experimental verification and real-time observation, provide a comprehensive view of eccDNA dynamics and interactions within live cells, offering valuable insights into eccDNA's biological implications.
2.3.3. Experimental methods to investigate the biological function of eccDNA
When a most basic scientific hypothesis is supposed, we need to prove that eccDNAs have functional phenotypes in diseases, which requires artificial perturbation of the expression level of eccDNAs. Specifically, we observe phenotype changes through two kinds of strategies, gain-of-function and loss-of-function, which might establish a causal relationship between eccDNA and functional phenotypes of diseases. On the one hand, gain-of-function can be achieved by up-regulating specific eccDNA in the cell model. A specific method involves the synthesis of circular DNA in-vitro (Fig. 2C), which can be transferred into cells for eccDNA expression using specific transfection reagents. The most common transfection agents are liposomes with high affinity to cell membranes, which are now commercially available. Some novel methods of eccDNA synthesis were developed, such as Ligase-Assisted Minicircle Accumulation (QuickLAMA) [48]. QuickLAMA can synthesize rapidly eccDNAs up to 2.6 kb using a simple PCR and ligation approach. Of note, CRISPR-C approach can create functional eccDNA by circularizing specific genomic regions in human cells, using a dual-fluorescence biosensor system [49]. This method generates eccDNAs from intergenic and genic loci and can produce eccDNAs ranging from a few hundred base pairs to large ring chromosomes. On the other hand, loss-of-function involves knockout of disease-causing eccDNAs. There is a specific operating technology: CRISPR technology, which can destroy the sequence of eccDNAs through sgRNA and Cas9 nuclease protein. Among them, model cells that overexpress Cas9 have also been commercialized, and sgRNA design strategies are very mature, so CRISPR is obviously simple and convenient.
In addition, delving into the functional studies of genes located on eccDNA, we can employ an array of methodologies analogous to those used in conventional gene function investigations. Of course, these methods must distinguish genes located on linear chromosomes from those on eccDNA. Several techniques stand out as pivotal for the precise manipulation of gene expression, enabling the dissection of gene function on eccDNA. These include RNA interference (RNAi) utilizing small interfering RNA (siRNA) and short hairpin RNA (shRNA), antisense oligonucleotides (ASO), and various gene editing tools such as CRISPR/Cas9, homologous recombination, zinc finger nuclease (ZFN), and transcription activator-like effector nuclease (TALEN). Additionally, gene overexpression strategies are also instrumental in this context. By employing these advanced molecular tools, we are able to conduct comprehensive functional analyses, shedding light on the unique roles and regulatory mechanisms of genes harbored by eccDNA, which is crucial for understanding their contribution to cellular processes and disease pathogenesis.
3. Data analysis: software and algorithms
3.1. Cell image software to visualize eccDNAs
While experimental verification methods are essential for confirming eccDNA presence, advanced visualization techniques have revolutionized our ability to observe and analyze eccDNA. ecDetect [5], a newly developed image analysis software, utilizes an unbiased, semi-automated approach to accurately quantify extrachromosomal DNA from DAPI-stained metaphase images. Similarly, ecSeg [50], a U-net-based image analysis platform, merges conventional microscopy with deep neural networks to achieve precise resolution of eccDNA and oncogene amplification at the single-cell level. This tool adeptly processes DAPI and FISH-stained metaphase images, differentiates intra-chromosomal and extrachromosomal repair mechanisms, and reveals how cell-to-cell variability impacts on tumor growth, progression, and drug resistance, currently accommodating up to two FISH probes.
3.2. Bioinformatical algorithms and software
In addressing the challenges of big data, high complexity, and wide diversity from high-throughput sequencing, the utilization of specialized bioinformatics algorithms is crucial for identifying eccDNA. Those algorithms include AmpliconArchitect (AA) [51], Circle_finder [33], Circle-Map [52], Circlehunter [53], NanoCircle [54,55], construction-based RCA for eccDNA sequence identification and location (CReSIL) [55], DeConcat [56], ecc_finder [57] ECCsplorer [58], ecDNAFinder [43], full-length ecDNA caller (Flec) [27], AmpliconReconstructor [59] and HolistIC [60]. Recently, an integrated pipeline named eccDNA-pipe has been developed to provide a comprehensive solution for the identification, analysis, and visualization of eccDNA from high-throughput sequencing data [61]. eccDNA-pipe supports data from various sequencing techniques such as whole-genome sequencing (WGS), circle-seq, and circulome-seq, obtained through both short-read and long-read sequencing. It includes tools for quality control, eccDNA identification, length distribution analysis, differential analysis of genes enriched with eccDNA, and visualization of eccDNA structures, automatically generating high-quality publication-ready plots. It is essential to recognize that different algorithms are tailored to specific sequencing contexts and data types (Table 3). When choosing an appropriate algorithm, considerations should include data characteristics, research objectives, and computational resource availability. The ongoing development and refinement of these algorithms represent a significant and evolving area in eccDNA research.
Table 3.
Bioinformatic algorithms for eccDNA identification tailored to sequencing strategies.
| name | sequencing strategy | availability | language | reference |
|---|---|---|---|---|
| AmpliconArchitect | short-read WGS | https://github.com/virajbdeshpande/AmpliconArchitect | Python, HTML and other | Deshpande et al. [51] |
| AmpliconReconstructor | short-read WGS and Bionano optical mapping | https://github.com/jluebeck/AmpliconReconstructor | Python, C++ and a Unix-based OS | Luebeck et al. [59] |
| Circle_finder | short-read ATAC-seq, WGS, WES etc. | https://github.com/pk7zuva/Circle_finder | Shell and C | Kumar et al. [33] |
| Circle-Map | short-read sequencing of eccDNA with Circle-seq enrichment | https://github.com/iprada/Circle-Map | Python | Prada-Luengo et al. [52] |
| Circlehunter | short-read ATAC-seq | https://github.com/suda-huanglab/circlehunter | Python, Shell and Awk | Yang et al. [53] |
| CReSIL | long-read WGS; long-read sequencing of eccDNA with Circle-seq enrichment or RCA | https://github.com/visanuwan/cresil | Python | Wanchai et al. [55] |
| CIDER-seq (using DeConcat algorithm) | long-read sequencing of eccDNA with random RCA or randomly primed circular DNA amplification | https://github.com/devang-mehta/ciderseq2 | Python | Mehta et al. [56] |
| ecc_finder | short-read and long-read (Oxford Nanopore) sequencing of eccDNA with RCA | https://github.com/njaupan/ecc_finder | Python | Zhang et al. [57] |
| ECCsplorer | short-read sequencing of eccDNA with RCA | https://github.com/crimBubble/ECCsplorer | Python 3 and CSS | Mann et al. [56] |
| ecDNAFinder | long-read single cell WGS | https://github.com/fanxylab/EcDNAFinder | Python, Cython and R | Chang et al. [43] |
| Flec (eccDNA_RCA_nanopore) | long-read sequencing (Oxford Nanopore) of eccDNA with RCA | https://github.com/YiZhang-lab/eccDNA_RCA_nanopore | R, Python and CSS | Wang et al. [27] |
| HolistIC | short-read WGS and Hi-C sequencing of double minute | http://www.github.com/mhayes20/HolistIC | Python | Luebeck et al. [60] |
| NanoCircle | long-read sequencing (Oxford Nanopore) of eccDNA with Circle-seq enrichment | https://github.com/RAHenriksen/NanoCircle | Python | Henriksen et al. [54], Wanchai et al. [55] |
| eccDNA-pipe | various sequencing techniques such as WGS, circle-seq, and circulome-seq | https://github.com/QuKunLab/ecc_pipe | HTML, Jupyter Notebook and other | Fang et al. [61] |
4. Auxiliary research resources and tools
While bioinformatics algorithms such as those previously mentioned are invaluable in eccDNA analysis, they often present a steep learning curve and require significant time and computational expertise, making them challenging for many researchers to utilize independently and efficiently. As a more accessible alternative, powerful and user-friendly online databases have become preferable for most researchers. Notable eccDNA databases include eccDNAdb [62], CircleBase [63], eccDNA Atlas [64], EccBase [65], eccDB [66] and the eccDNA collection database (TeCD) [67]. These databases offer publicly available eccDNA data resources, simplifying the process of eccDNA analysis and research (Table 4). In addition, several of these databases feature functional modules, like the genome browser in eccDNAdb, providing chromatin accessibility information on eccDNA, thus facilitating various aspects of eccDNA research. In summary, these databases democratize access to eccDNA data, allowing researchers to conduct comprehensive eccDNA analyses with greater ease and efficiency.
Table 4.
Information of database for eccDNA study.
| name | availability | modules | application | number of eccDNA | data source | year | reference |
|---|---|---|---|---|---|---|---|
| eccDNAdb | http://www.eccdnadb.org/ | home, browse, search, GB, statistics, submit, external links | enables users to easily determine the biological function and clinical relevance of eccDNAs in human cancers | 1270 (Homo sapiens) | analysis of WGS data from SRA & literature | 2022 | Peng et al. [62] |
| CircleBase | http://circlebase.maolab.org/ | home, search, stats, manual | targeting genes, epigenetic regulations, regulatory elements, chromatin accessibility, chromatin interactions, and genetic variants | 601,036 (Homo sapiens) | literature | 2022 | Zhao et al. [63] |
| eccDNA Atlas | http://lcbb.swjtu.edu.cn/eccDNAatlas | home, browse, search, analysis, statistics, submit | eccDNA annotation (eccDNA regions included in eccDNA Atlas or custom chromatin regions including oncogenes/lncRNAs, typical enhancers, super enhancers, CTCF-binding sites, SNPs, chromatin accessibility and eQTLs), analysis (gene expressions, functions, survival, regulatory network exploring analysis and BLAST) and genome visualization were provided for oncogenes, enhancers, SNPs, chromatin accessibility, etc. | 629,987 eccDNAs, 8221 ecDNAs from literatures and 1105 ecDNAs by AA (Homo sapiens) | literature and analysis of WGS data | 2023 | Zhong et al. [64] |
| eccBase | http://www.eccbase.net | home, browse, search, blast, submission | a total of 50 features were annotated, including basic features, molecular biology properties, and source data. | 754,391 (Homo sapiens) and 481,381 (Mus musculus) | literature curation and public algorithm/database retrieval | 2023 | Sun et al. [65] |
| eccDB | http://www.xiejjlab.bio/eccDB | home, browse, search, analysis, blast, statistics, API | eccDNAs gene expression analysis, survival analysis, GO term functional enrichment analysis, and KEGG pathway annotation, regulatory and epigenetic information annotations for eccDNAs, sequence similarity analysis | 767,981 (Homo sapiens), 372,811 (Mus musculus), 2081 (Saccharomyces cerevisiae), and 174,309 (Arabidopsis) | literature and public algorithm/database retrieval | 2023 | Yang et al. [66] |
| TeCD | http://122.224.251.240:2022 | home, search, blast, statistics, contribute | search and obtain eccDNA data, and analyze the possible potential functions of eccDNA | 193294 (Homo sapiens), 1266 (Saccharomyces cerevisiae), 591 (Arabidopsis thaliana), 434008 (Gallus gallus), 319325 (Mus musculus) | literature | 2023 | Guo et al. [67] |
5. Designing research projects on eccDNA
Embarking on a scientific research project on eccDNA demands considerable planning and the execution of numerous key steps. Here, we propose two distinct yet complementary ideas, each underpinned by rigorous scientific methodology and logic.
The first idea is inherently phenotype-driven, putting the investigation of eccDNA function and implications at the forefront. The starting point is the selection of a biologically or clinically relevant phenotype, such as cellular drug resistance. Alongside this, a proper group design is paramount, ideally incorporating both drug-resistant and non-resistant control groups. With these foundational elements in place, high-throughput sequencing techniques, such as WGS, Circle-seq or scEC&T-seq, can be harnessed to identify all extant eccDNA and the associated genes (Fig. 4 A). Subsequent to the identification phase, potential target eccDNA and genes on eccDNA, which are likely associated with the specific phenotype, are selected for further investigation. This selection is based on the sequencing results and a thorough bioinformatics analysis. An essential aspect is determining whether the eccDNA-harbored genes are transcribed. Current research on the relationship between eccDNA and eccDNA-associated transcriptomics represents a promising avenue for future investigation but remains largely unexplored, potentially due to existing technical challenges. Recent advances have described parallel sequencing of circular DNA and full-length RNA based on new platforms of single-cell resolution (scEC&T-seq) [36], facilitating the assessment of both large and small circular DNA categories. As a critical next step, the existence of the chosen eccDNA in cells is verified using FISH combined with advanced imaging technology. Functional studies of the chosen eccDNA and its genes follow, employing gain-of-function and loss-of-function experimental techniques. These studies allow for a deeper exploration of their role in phenotypes, such as drug resistance. Finally, further research is dedicated to understanding how the chosen eccDNA and its genes impact cellular drug resistance, with an ambitious aim to elucidate the underlying mechanisms (Fig. 4 A).
Fig. 4.
Two candidate designs for eccDNA research. (A) Phenotype-driven approach. This design begins with the selection of a biologically or clinically relevant phenotype, such as cellular drug resistance, and involves a group design incorporating both resistant and non-resistant cells. It includes high-throughput sequencing (e.g., WGS or Circle-seq) to identify eccDNA and associated genes, followed by a selection process for potential target eccDNA based on bioinformatics analysis. Verification of eccDNA presence is achieved through FISH and advanced imaging. Functional studies using gain-of-function and loss-of-function techniques further explore the role of eccDNA in phenotypes like drug resistance, aiming to elucidate underlying mechanisms. (B) High-throughput sequencing and bioinformatics-driven approach. This method utilizes extensive sequencing data or database-driven selection (e.g., from eccDNAdb) to identify eccDNA and genes on eccDNA. It involves a comprehensive bioinformatics analysis, including gene expression abundance, prognosis analysis, and the integration of AI models for enhanced predictive analytics and expression profiling. The existence verification of eccDNA in experimental models is optional, emphasizing flexibility in research design. The visual elements in the image were created by BioRender.com.
The second idea leans heavily on high-throughput sequencing or database-driven selection, as well as bioinformatics analysis of eccDNA. Here, large amounts of high-throughput sequencing data (like WGS or Circle-seq) are utilized to identify all present eccDNA and genes on eccDNA. Alternatively, specific eccDNA genes in certain types of disease can be directly chosen from a public database, such as the eccDNAdb (Fig. 4 B). Upon selection of the target eccDNA or genes resident on eccDNA, they undergo a bioinformatics analysis that encompasses gene expression abundance, prognosis analysis, and now, with advancements in computational research, the inclusion of artificial intelligence models (Fig. 4 B). These artificial intelligence (AI)-driven tools are increasingly being developed to refine predictive analytics, enhance the accuracy of expression profiling, and facilitate a deeper understanding of eccDNA-driven pathophysiology. Not unlike the first idea, verifying the existence of eccDNA selected from bioinformatics analysis in experimental models is often desirable, yet not strictly necessary.
These approaches are now visually represented in Fig. 4, suggesting how to integrate bioinformatics insights with experimental validation, thus enhancing the narrative of eccDNA's impact in various diseases. Though each approach provides a basic framework for eccDNA research, it is vital to bear in mind that the actual research plan should be thoughtfully customized to align with specific research objectives and resources at hand.
6. Future perspective
6.1. Technical innovations and limitations
Although great progress has been made in the methods for screening and identifying eccDNA, there is still room for improvements and ongoing challenges, especially in the study of its function and mechanism: I) The optimization of sequencing analysis algorithms. With the continuous evolution of high-throughput sequencing technology, we need more algorithms to analyze and interpret large-scale data. II) Research methods of the fundamental biology of eccDNA. Future research needs to develop new experimental methods and techniques, such as high-resolution microscopy techniques, to better study the formation, and delivery mechanisms of eccDNA. III) Research methods for three-dimensional structure and transcription regulation of eccDNA. Current studies have relied heavily on sequencing-based methods such as Hi-C and proximity ligation-assisted chromatin immunoprecipitation sequencing (PLAC-seq), but these may have certain limitations for eliminating chromosome interference. Therefore, new methods need to be developed to study the spatial organization of eccDNA and its interaction with chromosomes. Our grasp of the transcriptional regulation mechanisms of eccDNA, especially how they interact with super-enhancers to influence gene expression, is still insufficient. IV) Research methods for studying the functions of eccDNA in diseases. Future studies could optimize existing techniques of gene regulation and even develop entirely new research methods distinct from linear DNA to investigate the specific role of eccDNA in cellular functions and disease progression. These improvements and innovations will help us better understand the mechanisms and utilize the functions of eccDNA.
6.2. Biomedical applications and functional insights
EccDNA is a new research field, and the future research directions include but are not limited to the following aspects: I) Genetic stability and formation of eccDNA. The mechanisms by which eccDNA is formed and evolved, as well as how it is replicated and delivered in cells are the problems to be solved. II) Functions and mechanisms of eccDNAs in physiology and diseases. The role of eccDNAs, including both short and long eccDNAs, is multifaceted and significant in various biological processes. Short eccDNAs, with their capability to produce miRNAs and si-like RNAs, can regulate the expression of corresponding linear host genes [68], which is still under-evaluated and warrants further investigation. Although eccDNA has been found to play an important role in some cancers, its association with other diseases and its function in normal physiological processes remain unclear. Understanding the mechanisms of eccDNAs in physiology and diseases is crucial to elucidate their full impact on cellular functions and disease progression. III) Application of eccDNA in therapeutics and diagnostics. EccDNA presents novel opportunities in both therapeutic targeting and biomarker development. For therapeutic applications, the challenge lies not only in targeting disease-causing genes or drug resistance genes located on the eccDNA, but also in directly targeting the eccDNA itself. Developing strategies that target eccDNA effectively without affecting linear DNA is crucial for advancing eccDNA-based therapies. As a biomarker, the high presence of eccDNA in cancer cells offers unique opportunities for clinical diagnostics. Research should be directed towards enhancing methods to detect the presence and quantity of eccDNA, which could facilitate early diagnosis of cancer and other diseases. Additionally, the measurement of eccDNA levels could serve as a prognostic tool and assist in monitoring disease progression, helping to predict treatment efficacy and assess the long-term outcomes of therapeutic interventions. Furthermore, the potential to extract and analyze eccDNA from liquid biopsies, as illustrated in Fig. 1, opens new avenues for non-invasive diagnostic and prognostic applications, providing a novel tool for early disease detection and monitoring.
In summary, it will be needed to better understand how eccDNA forms, how eccDNA affects on genomics, epigenetics and gene transcription regulation and how eccDNA influences on the onset and progression of diseases; to explore whether and why eccDNA-driven diseases are more likely to evade treatment, how to accurately target eccDNA, and whether eccDNAs could act as biomarkers for disease diagnosis and monitoring. Concrete steps are needed to establish eccDNA as diagnostic, prognostic, and treatment-sensitive biomarkers, facilitating the transfer of expanding knowledge on circular DNA regulation into daily clinical application.
Funding statement
This work was supported by grants from the National Natural Science Foundation of China (82372617, 81972658 and 81802812 to L.P., 81803636 to X.Y.), Guangdong Basic and Applied Basic Research Foundation (2024B1515020090, 2023A1515012683, 2019A1515012114 and 2018A030313129 to L.P., 2024A1515030038 to X.Y., 2023A1515140033 (Guangdong-Dongguan Joint Fund) to Y.Z.), Guangzhou Basic and Applied Basic Research Foundation (2024A03J0845 and 2023A04J2098 to L.P.), the Science and Technology Planning Project of Guangdong Province (2023B1212060013 and 2020B1212030004) and Fundamental Research Funds for the Central Universities, Sun Yat-sen University (24ykqb004 to L.P.).
Ethics statement
This review article does not involve any ethical concerns.
Data availability statement
The authors declare that no data associated with our study has been deposited into a publicly available repository since no data was used for the research described in the review.
CRediT authorship contribution statement
Xiao-Qing Yuan: Writing – original draft, Visualization, Resources, Methodology, Investigation, Funding acquisition, Formal analysis, Conceptualization. Nan Zhou: Writing – review & editing, Writing – original draft, Visualization, Supervision, Software, Investigation, Conceptualization. Shi-Jian Song: Writing – review & editing, Resources, Methodology. Yi-Xia Xie: Resources, Methodology. Shui-Qin Chen: Resources, Methodology. Teng-Fei Yang: Resources, Methodology. Xian Peng: Resources. Chao-Yang Zhang: Resources, Methodology. Ying-Hua Zhu: Writing – review & editing, Validation, Supervision, Resources, Methodology, Funding acquisition. Li Peng: Writing – review & editing, Visualization, Validation, Supervision, Project administration, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the authors used ChatGPT-4.0 in order to polish language. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Nan Zhou, Email: zhnanx@live.com.
Ying-Hua Zhu, Email: zhuyinghua2003@163.com.
Li Peng, Email: pengli9@mail.sysu.edu.cn.
Abbreviations
- AA
AmpliconArchitect
- AI
artificial intelligence
- ASO
antisense oligonucleotides
- ATAC-seq
assay for transposase-accessible chromatin using sequencing
- Casilio
dCas9 and engineered Pumilio/FBF (PUF)-tethered effectors
- CCDA-seq
sequencing of enzyme-accessible chromatin in circular DNA
- CIDER-seq
circular DNA enrichment sequencing
- chrDNA
chromosomal DNA
- Circle-seq
genome-scale enrichment and detection method for eccDNA
- circSeq
Illumina-sequencing of amplified circular DNA
- CReSIL
construction-based RCA for eccDNA sequence identification and location
- CRISPR-CATCH
clustered regularly interspaced short palindromic repeats - Cas9-assisted targeting of chromosome segments
- CRISPR FISHer
clustered regularly interspaced short palindromic repeats FISH amplifier
- DAPI
4′,6-diamidino-2-phenylindole
- eccDNA/ecDNA
extrachromosomal circular DNA
- ecTag
CRISPR-based DNA tracking system to label ecDNA elements
- FISH
Fluorescence in situ hybridization
- Flec
full-length ecDNA caller
- microDNA
short extrachromosomal circular DNAs
- mtDNA
mitochondrial DNA
- PFGE
pulsed field gel electrophoresis
- PLAC-seq
proximity ligation-assisted chromatin immunoprecipitation sequencing
- PUF
Pumilio/FBF
- QuickLAMA
Ligase-Assisted Minicircle Accumulation
- RCA
rolling-circle amplicon
- RNAi
RNA interference
- sc
single‐cell
- scATAC-seq
single‐cell ATAC-seq
- scCircle-seq
single‐cell Circle-seq
- scEC&T-seq
single-cell extrachromosomal circular DNA and transcriptome sequencing
- scGTP-seq
single‐cell paralleled genome and transcriptome sequencing on a third‐generation platform
- scRNA-seq
single-cell RNA sequencing
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- scWGS
single‐cell WGS
- SMOOTH-seq
single-molecule real-time sequencing of long fragments amplified through transposon insertion
- SMRT
single-molecule real-time
- SV
structural variant
- TALEN
transcription activator-like effector nuclease
- TeCD
the eccDNA collection database
- TetO
Tet-operator
- WGS
whole-genome sequencing
- ZFN
zinc finger nuclease
- 3SEP
three-step eccDNA purification
References
- 1.Wu N., Wei L., Zhu Z., Liu Q., Li K., Mao F., Qiao J., Zhao X. Innovative insights into extrachromosomal circular DNAs in gynecologic tumors and reproduction. Protein Cell. 2024;15:6–20. doi: 10.1093/procel/pwad032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhaak R.G.W., Bafna V., Mischel P.S. Extrachromosomal oncogene amplification in tumour pathogenesis and evolution. Nat. Rev. Cancer. 2019;19:283–288. doi: 10.1038/s41568-019-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koche R.P., Rodriguez-Fos E., Helmsauer K., Burkert M., MacArthur I.C., Maag J., Chamorro R., Munoz-Perez N., Puiggròs M., Dorado Garcia H., Bei Y., Röefzaad C., Bardinet V., Szymansky A., Winkler A., Thole T., Timme N., Kasack K., Fuchs S., Klironomos F., Thiessen N., Blanc E., Schmelz K., Künkele A., Hundsdörfer P., Rosswog C., Theissen J., Beule D., Deubzer H., Sauer S., Toedling J., Fischer M., Hertwig F., Schwarz R.F., Eggert A., Torrents D., Schulte J.H., Henssen A.G. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 2020;52:29–34. doi: 10.1038/s41588-019-0547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H., Nguyen N.P., Turner K., Wu S., Gujar A.D., Luebeck J., Liu J., Deshpande V., Rajkumar U., Namburi S., Amin S.B., Yi E., Menghi F., Schulte J.H., Henssen A.G., Chang H.Y., Beck C.R., Mischel P.S., Bafna V., Verhaak R.G.W. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner K.M., Deshpande V., Beyter D., Koga T., Rusert J., Lee C., Li B., Arden K., Ren B., Nathanson D.A., Kornblum H.I., Taylor M.D., Kaushal S., Cavenee W.K., Wechsler-Reya R., Furnari F.B., Vandenberg S.R., Rao P.N., Wahl G.M., Bafna V., Mischel P.S. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Wang M., Djekidel M.N., Chen H., Liu D., Alt F.W., Zhang Y. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature. 2021;599:308–314. doi: 10.1038/s41586-021-04009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt S., Arrey G., Regenberg B. Did circular DNA shape the evolution of mammalian genomes? Trends Biochem. Sci. 2023;48:317–320. doi: 10.1016/j.tibs.2022.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Yerlici V.T., Lu M.W., Hoge C.R., Miller R.V., Neme R., Khurana J.S., Bracht J.R., Landweber L.F. Programmed genome rearrangements in Oxytricha produce transcriptionally active extrachromosomal circular DNA. Nucleic Acids Res. 2019;47:9741–9760. doi: 10.1093/nar/gkz725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen T., Malapati P., Shibata Y., Wilson B., Eki R., Benamar M., Abbas T., Dutta A. MicroDNA levels are dependent on MMEJ, repressed by c-NHEJ pathway, and stimulated by DNA damage. Nucleic Acids Res. 2021;49:11787–11799. doi: 10.1093/nar/gkab984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng H., Mirouze M., Bucher E. Extrachromosomal circular DNA: a neglected nucleic acid molecule in plants. Curr. Opin. Plant Biol. 2022;69 doi: 10.1016/j.pbi.2022.102263. [DOI] [PubMed] [Google Scholar]
- 11.Cohen S., Menut S., Méchali M. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol. Cell Biol. 1999;19:6682–6689. doi: 10.1128/mcb.19.10.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull R.M., Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr. Genet. 2020;66:889–894. doi: 10.1007/s00294-020-01069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S., Turner K.M., Nguyen N., Raviram R., Erb M., Santini J., Luebeck J., Rajkumar U., Diao Y., Li B., Zhang W., Jameson N., Corces M.R., Granja J.M., Chen X., Coruh C., Abnousi A., Houston J., Ye Z., Hu R., Yu M., Kim H., Law J.A., Verhaak R.G.W., Hu M., Furnari F.B., Chang H.Y., Ren B., Bafna V., Mischel P.S. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoshani O., Brunner S.F., Yaeger R., Ly P., Nechemia-Arbely Y., Kim D.H., Fang R., Castillon G.A., Yu M., Li J.S.Z., Sun Y., Ellisman M.H., Ren B., Campbell P.J., Cleveland D.W. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591:137–141. doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.J., Jung Y.L., Cheong T.C., Espejo Valle-Inclan J., Chu C., Gulhan D.C., Ljungström V., Jin H., Viswanadham V.V., Watson E.V., Cortés-Ciriano I., Elledge S.J., Chiarle R., Pellman D., Park P.J. ERα-associated translocations underlie oncogene amplifications in breast cancer. Nature. 2023;618:1024–1032. doi: 10.1038/s41586-023-06057-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung K.L., Yost K.E., Xie L., Shi Q., Helmsauer K., Luebeck J., Schöpflin R., Lange J.T., Chamorro González R., Weiser N.E., Chen C., Valieva M.E., Wong I.T., Wu S., Dehkordi S.R., Duffy C.V., Kraft K., Tang J., Belk J.A., Rose J.C., Corces M.R., Granja J.M., Li R., Rajkumar U., Friedlein J., Bagchi A., Satpathy A.T., Tjian R., Mundlos S., Bafna V., Henssen A.G., Mischel P.S., Liu Z., Chang H.Y. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature. 2021;600:731–736. doi: 10.1038/s41586-021-04116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosswog C., Bartenhagen C., Welte A., Kahlert Y., Hemstedt N., Lorenz W., Cartolano M., Ackermann S., Perner S., Vogel W., Altmüller J., Nürnberg P., Hertwig F., Göhring G., Lilienweiss E., Stütz A.M., Korbel J.O., Thomas R.K., Peifer M., Fischer M. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat. Genet. 2021;53:1673–1685. doi: 10.1038/s41588-021-00951-7. [DOI] [PubMed] [Google Scholar]
- 18.Morton A.R., Dogan-Artun N., Faber Z.J., MacLeod G., Bartels C.F., Piazza M.S., Allan K.C., Mack S.C., Wang X., Gimple R.C., Wu Q., Rubin B.P., Shetty S., Angers S., Dirks P.B., Sallari R.C., Lupien M., Rich J.N., Scacheri P.C. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–1341.e1313. doi: 10.1016/j.cell.2019.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu T., Wu C., Zhao X., Wang G., Ning W., Tao Z., Chen F., Liu X.S. Extrachromosomal DNA formation enables tumor immune escape potentially through regulating antigen presentation gene expression. Sci. Rep. 2022;12:3590. doi: 10.1038/s41598-022-07530-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pongor L.S., Schultz C.W., Rinaldi L., Wangsa D., Redon C.E., Takahashi N., Fialkoff G., Desai P., Zhang Y., Burkett S., Hermoni N., Vilk N., Gutin J., Gergely R., Zhao Y., Nichols S., Vilimas R., Sciuto L., Graham C., Caravaca J.M., Turan S., Tsai-Wei S., Rajapakse V.N., Kumar R., Upadhyay D., Kumar S., Kim Y.S., Roper N., Tran B., Hewitt S.M., Kleiner D.E., Aladjem M.I., Friedman N., Hager G.L., Pommier Y., Ried T., Thomas A. Extrachromosomal DNA amplification contributes to small cell lung cancer heterogeneity and is associated with worse outcomes. Cancer Discov. 2023;13:928–949. doi: 10.1158/2159-8290.CD-22-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou S., Chen S., Rao G., Zhang G., Ma M., Peng B., Du X., Huang W., Lin W., Tian Y., Fu X. Extrachromosomal circular MiR-17-92 amplicon promotes HCC. Hepatology. 2024;79:79–95. doi: 10.1097/HEP.0000000000000435. [DOI] [PubMed] [Google Scholar]
- 22.Møller H.D., Mohiyuddin M., Prada-Luengo I., Sailani M.R., Halling J.F., Plomgaard P., Maretty L., Hansen A.J., Snyder M.P., Pilegaard H., Lam H.Y.K., Regenberg B. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat. Commun. 2018;9:1069. doi: 10.1038/s41467-018-03369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cen Y., Fang Y., Ren Y., Hong S., Lu W., Xu J. Global characterization of extrachromosomal circular DNAs in advanced high grade serous ovarian cancer. Cell Death Dis. 2022;13:342. doi: 10.1038/s41419-022-04807-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin C., Chen Y., Zhang F., Liu B., Xie C., Song Y. Encoding gene RAB3B exists in linear chromosomal and circular extrachromosomal DNA and contributes to cisplatin resistance of hypopharyngeal squamous cell carcinoma via inducing autophagy. Cell Death Dis. 2022;13:171. doi: 10.1038/s41419-022-04627-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Møller H.D., Ramos-Madrigal J., Prada-Luengo I., Gilbert M.T.P., Regenberg B. Near-random distribution of chromosome-derived circular DNA in the condensed genome of pigeons and the larger, more repeat-rich human genome. Genome Biol Evol. 2020;12:3762–3777. doi: 10.1093/gbe/evz281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Møller H.D., Parsons L., Jørgensen T.S., Botstein D., Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E3114–E3122. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Wang M., Zhang Y. Purification, full-length sequencing and genomic origin mapping of eccDNA. Nat. Protoc. 2023;18:683–699. doi: 10.1038/s41596-022-00783-7. [DOI] [PubMed] [Google Scholar]
- 28.Luo X., Zhang L., Cui J., An Q., Li H., Zhang Z., Sun G., Huang W., Li Y., Li C., Jia W., Zou L., Zhao G., Xiao F. Small extrachromosomal circular DNAs as biomarkers for multi-cancer diagnosis and monitoring. Clin. Transl. Med. 2023;13 doi: 10.1002/ctm2.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung K.L., Luebeck J., Dehkordi S.R., Colón C.I., Li R., Wong I.T., Coruh C., Dharanipragada P., Lomeli S.H., Weiser N.E., Moriceau G., Zhang X., Bailey C., Houlahan K.E., Yang W., González R.C., Swanton C., Curtis C., Jamal-Hanjani M., Henssen A.G., Law J.A., Greenleaf W.J., Lo R.S., Mischel P.S., Bafna V., Chang H.Y. Targeted profiling of human extrachromosomal DNA by CRISPR-CATCH. Nat. Genet. 2022;54:1746–1754. doi: 10.1038/s41588-022-01190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luebeck J., Ng A.W.T., Galipeau P.C., Li X., Sanchez C.A., Katz-Summercorn A.C., Kim H., Jammula S., He Y., Lippman S.M., Verhaak R.G.W., Maley C.C., Alexandrov L.B., Reid B.J., Fitzgerald R.C., Paulson T.G., Chang H.Y., Wu S., Bafna V., Mischel P.S. Extrachromosomal DNA in the cancerous transformation of Barrett's oesophagus. Nature. 2023;616:798–805. doi: 10.1038/s41586-023-05937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata Y., Kumar P., Layer R., Willcox S., Gagan J.R., Griffith J.D., Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta D., Cornet L., Hirsch-Hoffmann M., Zaidi S.S., Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat. Protoc. 2020;15:1673–1689. doi: 10.1038/s41596-020-0301-0. [DOI] [PubMed] [Google Scholar]
- 33.Kumar P., Kiran S., Saha S., Su Z., Paulsen T., Chatrath A., Shibata Y., Shibata E., Dutta A. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z., Saha S., Paulsen T., Kumar P., Dutta A. ATAC-Seq-based identification of extrachromosomal circular DNA in mammalian cells and its validation using inverse PCR and FISH. Bio Protoc. 2021;11 doi: 10.21769/BioProtoc.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoura M.J., Gabdank I., Hansen L., Merker J., Gotlib J., Levene S.D., Fire A.Z. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 (Bethesda). 2017;7:3295–3303. doi: 10.1534/g3.117.300141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamorro González R., Conrad T., Stöber M.C., Xu R., Giurgiu M., Rodriguez-Fos E., Kasack K., Brückner L., van Leen E., Helmsauer K., Dorado Garcia H., Stefanova M.E., Hung K.L., Bei Y., Schmelz K., Lodrini M., Mundlos S., Chang H.Y., Deubzer H.E., Sauer S., Eggert A., Schulte J.H., Schwarz R.F., Haase K., Koche R.P., Henssen A.G. Parallel sequencing of extrachromosomal circular DNAs and transcriptomes in single cancer cells. Nat. Genet. 2023;55:880–890. doi: 10.1038/s41588-023-01386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan X., Yang C., Li W., Bai X., Zhou X., Xie H., Wen L., Tang F. SMOOTH-seq: single-cell genome sequencing of human cells on a third-generation sequencing platform. Genome Biol. 2021;22:195. doi: 10.1186/s13059-021-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W., Weng Z., Xie Z., Xie Y., Zhang C., Chen Z., Ruan F., Wang J., Sun Y., Fang Y., Guo M., Tong Y., Li Y., Tang C. Sequencing of methylase-accessible regions in integral circular extrachromosomal DNA reveals differences in chromatin structure. Epigenet. Chromatin. 2021;14:40. doi: 10.1186/s13072-021-00416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sin S.T.K., Ji L., Deng J., Jiang P., Cheng S.H., Heung M.M.S., Lau C.S.L., Leung T.Y., Chan K.C.A., Chiu R.W.K., Lo Y.M.D. Characteristics of fetal extrachromosomal circular DNA in maternal plasma: methylation status and clearance. Clin. Chem. 2021;67:788–796. doi: 10.1093/clinchem/hvaa326. [DOI] [PubMed] [Google Scholar]
- 40.Lanciano S., Zhang P., Llauro C., Mirouze M. Identification of extrachromosomal circular forms of active transposable elements using mobilome-seq. Methods Mol. Biol. 2021;2250:87–93. doi: 10.1007/978-1-0716-1134-0_7. [DOI] [PubMed] [Google Scholar]
- 41.Chen J.P., Diekmann C., Wu H., Chen C., Della Chiara G., Berrino E., Georgiadis K.L., Bouwman B.A.M., Virdi M., Harbers L., Bellomo S.E., Marchiò C., Bienko M., Crosetto N. scCircle-seq unveils the diversity and complexity of extrachromosomal circular DNAs in single cells. Nat. Commun. 2024;15:1768. doi: 10.1038/s41467-024-45972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spain L., Coulton A., Lobon I., Rowan A., Schnidrig D., Shepherd S.T.C., Shum B., Byrne F., Goicoechea M., Piperni E., Au L., Edmonds K., Carlyle E., Hunter N., Renn A., Messiou C., Hughes P., Nobbs J., Foijer F., van den Bos H., Wardenaar R., Spierings D.C.J., Spencer C., Schmitt A.M., Tippu Z., Lingard K., Grostate L., Peat K., Kelly K., Sarker S., Vaughan S., Mangwende M., Terry L., Kelly D., Biano J., Murra A., Korteweg J., Lewis C., O'Flaherty M., Cattin A.L., Emmerich M., Gerard C.L., Pallikonda H.A., Lynch J., Mason R., Rogiers A., Xu H., Huebner A., McGranahan N., Al Bakir M., Murai J., Naceur-Lombardelli C., Borg E., Mitchison M., Moore D.A., Falzon M., Proctor I., Stamp G.W.H., Nye E.L., Young K., Furness A.J.S., Pickering L., Stewart R., Mahadeva U., Green A., Larkin J., Litchfield K., Swanton C., Jamal-Hanjani M., Turajlic S. Late-stage metastatic melanoma emerges through a diversity of evolutionary pathways. Cancer Discov. 2023;13:1364–1385. doi: 10.1158/2159-8290.CD-22-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L., Deng E., Wang J., Zhou W., Ao J., Liu R., Su D., Fan X. Single-cell third-generation sequencing-based multi-omics uncovers gene expression changes governed by ecDNA and structural variants in cancer cells. Clin. Transl. Med. 2023;13 doi: 10.1002/ctm2.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu S., Bafna V., Chang H.Y., Mischel P.S. Extrachromosomal DNA: an emerging hallmark in human cancer. Annu. Rev. Pathol. 2022;17:367–386. doi: 10.1146/annurev-pathmechdis-051821-114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng A.W., Jillette N., Lee P., Plaskon D., Fujiwara Y., Wang W., Taghbalout A., Wang H. Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016;26:254–257. doi: 10.1038/cr.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi E., Gujar A.D., Guthrie M., Kim H., Zhao D., Johnson K.C., Amin S.B., Costa M.L., Yu Q., Das S., Jillette N., Clow P.A., Cheng A.W., Verhaak R.G.W. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Discov. 2022;12:468–483. doi: 10.1158/2159-8290.CD-21-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian R., Huang Z., Li L., Yuan J., Zhang Q., Meng L., Lang B., Hong Y., Zhong C., Tian X., Cui Z., Jin Z., Liu J., Huang Z., Wang Y., Chen Y., Hu Z. HPV integration generates a cellular super-enhancer which functions as ecDNA to regulate genome-wide transcription. Nucleic Acids Res. 2023;51:4237–4251. doi: 10.1093/nar/gkad105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuo S., Li X., Yang Y., Zhou J., He Q. A quick method to synthesize extrachromosomal circular DNA in vitro. Molecules. 2023;28:4236. doi: 10.3390/molecules28104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Møller H.D., Lin L., Xiang X., Petersen T.S., Huang J., Yang L., Kjeldsen E., Jensen U.B., Zhang X., Liu X., Xu X., Wang J., Yang H., Church G.M., Bolund L., Regenberg B., Luo Y. CRISPR-C: circularization of genes and chromosome by CRISPR in human cells. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rajkumar U., Turner K., Luebeck J., Deshpande V., Chandraker M., Mischel P., Bafna V. EcSeg: semantic segmentation of metaphase images containing extrachromosomal DNA. iScience. 2019;21:428–435. doi: 10.1016/j.isci.2019.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deshpande V., Luebeck J., Nguyen N.D., Bakhtiari M., Turner K.M., Schwab R., Carter H., Mischel P.S., Bafna V. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun. 2019;10:392. doi: 10.1038/s41467-018-08200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prada-Luengo I., Krogh A., Maretty L., Regenberg B. Sensitive detection of circular DNAs at single-nucleotide resolution using guided realignment of partially aligned reads. BMC Bioinf. 2019;20:663. doi: 10.1186/s12859-019-3160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M., Zhang S., Jiang R., Chen S., Huang M. Circlehunter: a tool to identify extrachromosomal circular DNA from ATAC-Seq data. Oncogenesis. 2023;12:28. doi: 10.1038/s41389-023-00476-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henriksen R.A., Jenjaroenpun P., Sjøstrøm I.B., Jensen K.R., Prada-Luengo I., Wongsurawat T., Nookaew I., Regenberg B. Circular DNA in the human germline and its association with recombination. Mol. Cell. 2022;82:209–217.e207. doi: 10.1016/j.molcel.2021.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wanchai V., Jenjaroenpun P., Leangapichart T., Arrey G., Burnham C.M., Tümmler M.C., Delgado-Calle J., Regenberg B., Nookaew I. CReSIL: accurate identification of extrachromosomal circular DNA from long-read sequences. Briefings Bioinf. 2022;23 doi: 10.1093/bib/bbac422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehta D., Hirsch-Hoffmann M., Were M., Patrignani A., Zaidi S.S., Were H., Gruissem W., Vanderschuren H. A new full-length circular DNA sequencing method for viral-sized genomes reveals that RNAi transgenic plants provoke a shift in geminivirus populations in the field. Nucleic Acids Res. 2019;47:e9. doi: 10.1093/nar/gky914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang P., Peng H., Llauro C., Bucher E., Mirouze M. ecc_finder: a robust and accurate tool for detecting extrachromosomal circular DNA from sequencing data. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.743742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mann L., Seibt K.M., Weber B., Heitkam T. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinf. 2022;23:40. doi: 10.1186/s12859-021-04545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luebeck J., Coruh C., Dehkordi S.R., Lange J.T., Turner K.M., Deshpande V., Pai D.A., Zhang C., Rajkumar U., Law J.A., Mischel P.S., Bafna V. AmpliconReconstructor integrates NGS and optical mapping to resolve the complex structures of focal amplifications. Nat. Commun. 2020;11:4374. doi: 10.1038/s41467-020-18099-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayes M., Nguyen A., Islam R., Butler C., Tran E., Mullins D., Hicks C. HolistIC: leveraging Hi-C and whole genome shotgun sequencing for double minute chromosome discovery. Bioinformatics. 2022;38:1208–1215. doi: 10.1093/bioinformatics/btab816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang M., Fang J., Luo S., Liu K., Yu Q., Yang J., Zhou Y., Li Z., Sun R., Guo C., Qu K. eccDNA-pipe: an integrated pipeline for identification, analysis and visualization of extrachromosomal circular DNA from high-throughput sequencing data. Briefings Bioinf. 2024;25 doi: 10.1093/bib/bbae034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peng L., Zhou N., Zhang C.Y., Li G.C., Yuan X.Q. eccDNAdb: a database of extrachromosomal circular DNA profiles in human cancers. Oncogene. 2022;41:2696–2705. doi: 10.1038/s41388-022-02286-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao X., Shi L., Ruan S., Bi W., Chen Y., Chen L., Liu Y., Li M., Qiao J., Mao F. CircleBase: an integrated resource and analysis platform for human eccDNAs. Nucleic Acids Res. 2022;50:D72–d82. doi: 10.1093/nar/gkab1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong T., Wang W., Liu H., Zeng M., Zhao X., Guo Z. eccDNA Atlas: a comprehensive resource of eccDNA catalog. Briefings Bioinf. 2023;24 doi: 10.1093/bib/bbad037. [DOI] [PubMed] [Google Scholar]
- 65.Sun H., Lu X., Zou L. EccBase: a high-quality database for exploration and characterization of extrachromosomal circular DNAs in cancer. Comput. Struct. Biotechnol. J. 2023;21:2591–2601. doi: 10.1016/j.csbj.2023.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang M., Qiu B., He G.Y., Zhou J.Y., Yu H.J., Zhang Y.Y., Li Y.S., Li T.S., Guo J.C., Li X.C., Xie J.J. eccDB: a comprehensive repository for eccDNA-mediated chromatin contacts in multi-species. Bioinformatics. 2023;39 doi: 10.1093/bioinformatics/btad173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo J., Zhang Z., Li Q., Chang X., Liu X. TeCD: the eccDNA Collection Database for extrachromosomal circular DNA. BMC Genom. 2023;24:47. doi: 10.1186/s12864-023-09135-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paulsen T., Shibata Y., Kumar P., Dillon L., Dutta A. Small extrachromosomal circular DNAs, microDNA, produce short regulatory RNAs that suppress gene expression independent of canonical promoters. Nucleic Acids Res. 2019;47:4586–4596. doi: 10.1093/nar/gkz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that no data associated with our study has been deposited into a publicly available repository since no data was used for the research described in the review.