Summary
Anti‐CD20 murine or chimeric antibodies (Abs) have been used to treat non‐Hodgkin lymphomas (NHLs) and other diseases characterized by overactive or dysfunctional B cells. Anti‐CD20 Abs demonstrated to be effective in inducing regression of B‐cell lymphomas, although in many cases patients relapse following treatment. A promising approach to improve the outcome of mAb therapy is the use of anti‐CD20 antibodies to deliver cytokines to the tumour microenvironment. In particular, IL‐2‐based immunocytokines have shown enhanced antitumour activity in several preclinical studies. Here, we report on the engineering of an anti‐CD20‐human interleukin‐2 (hIL‐2) immunocytokine (2B8‐Fc‐hIL2) based on the C2B8 mAb (Rituximab) and the resulting ectopic expression in Nicotiana benthamiana. The scFv‐Fc‐engineered immunocytokine is fully assembled in plants with minor degradation products as assessed by SDS‐PAGE and gel filtration. Purification yields using protein‐A affinity chromatography were in the range of 15–20 mg/kg of fresh leaf weight (FW). Glycopeptide analysis confirmed the presence of a highly homogeneous plant‐type glycosylation. 2B8‐Fc‐hIL2 and the cognate 2B8‐Fc antibody, devoid of hIL‐2, were assayed by flow cytometry on Daudi cells revealing a CD20 binding activity comparable to that of Rituximab and were effective in eliciting antibody‐dependent cell‐mediated cytotoxicity of human PBMC versus Daudi cells, demonstrating their functional integrity. In 2B8‐Fc‐hIL2, IL‐2 accessibility and biological activity were verified by flow cytometry and cell proliferation assay. To our knowledge, this is the first example of a recombinant immunocytokine based on the therapeutic Rituximab antibody scaffold, whose expression in plants may be a valuable tool for NHLs treatment.
Keywords: molecular farming, immunocytokine, human interleukin‐2, antibody‐dependent cell‐mediated cytotoxicity, non‐Hodgkin lymphomas
Introduction
Non‐Hodgkin lymphomas (NHLs) represent a serious threat, and some 70 thousands new cases have been diagnosed in USA in 2014, only (Siegel et al., 2014). The majority of NHLs express the leucocyte antigen CD20, an integral transmembrane glycoprotein of 33–37 kDa, that represents a preferential target for immunotherapy (Calcagno et al., 2012). The mouse/human chimeric anti‐CD20 antibody Rituximab (C2B8) is the first antibody‐based drug approved for the treatment of patients with recurrent B‐cell lymphomas (Reff et al., 1994). Nevertheless, only about 48% of patients treated with Rituximab respond to the therapy, with <10% showing a complete remission of the tumour (Davis et al., 2014). For this reason, there is a need to develop novel antibodies or antibody formats with improved efficacy against B‐cell lymphomas (Eichenauer and Engert, 2014; Fowler and Oki, 2013). In recent years, recombinant antibody–cytokine fusion proteins (immunocytokines, ICs) have shown a significantly enhanced efficacy against some types of cancers (Sondel and Gillies, 2012) (Vincent et al., 2013). Different ICs have been tested using a variety of tumour‐reactive mAbs linked to different cytokines, but the most striking clinical advances have been achieved by linking immunoglobulins to human interleukin‐2 (hIL‐2) (List and Neri, 2013). IL‐2 is a cytokine produced mainly by T cells and stimulates proliferation and differentiation of T‐ and B‐lymphocytes, monocytes and natural killer cells (NKs), generally enhancing antibody‐dependent cell‐mediated cytotoxicity (ADCC) (Gubbels et al., 2011; Sondel and Gillies, 2012). Despite possible side effects, IL‐2 is the cytokine of choice for the treatment of metastatic tumours and ICs based on IL‐2 are the most advanced molecules in the clinic (Fournier et al., 2011; Gubbels et al., 2011; Pretto et al., 2014). In all cases, IL‐2‐based ICs have shown far better antitumour activity than the combination of antibody and IL‐2 administered as separate molecules (Sondel and Gillies, 2012). A few studies showed the successful application of IL‐2‐ICs for the treatment of lymphomas, produced in mammalian cells expression systems. In particular, a dual cytokine fusion (IL2 and IL12) with an anti‐CD30 scFv antibody was shown to target Hodgkin lymphoma cells and to potentiate the activation of NK and T cells in a mouse model (Jahn et al., 2012). In another approach, a combination therapy of immunocytokines based on the antibody fragment F8 (specific to the alternatively spliced extra‐domain A of fibronectin, a marker for tumour‐angiogenesis) fused with IL4 (F8‐IL4) and IL12 (IL12‐F8‐F8) inhibited tumour growth in three different immunocompetent murine cancer models (F9 teratocarcinoma, CT26 colon carcinoma and A20 lymphoma) (Hemmerle and Neri, 2014).
The production of complex heterologous proteins in plants has several advantages over traditional expression systems based on mammalian cells, such as low costs, ease of production, scalability and limited risk of contamination by human pathogens (Stoger et al., 2014; Twyman et al., 2005). The production of antibodies in plants has been achieved using different approaches, including stable transformation and transient expression using both viral or Agrobacterium‐based vectors (Komarova et al., 2010). In the past few years, transient expression systems proved to be particularly attractive for the rapid accumulation of high amounts of recombinant proteins and in particular, viral‐based vector systems have been recently devised for high yield expression of full‐size antibodies (Peyret and Lomonossoff, 2013; Sainsbury et al., 2010). Recent studies also showed the possibility to express active hIL‐2 as a single molecule or fused to proteinase inhibitors in tobacco transgenic plants (Redkiewicz et al., 2012).
We describe here the first example of a recombinant immunocytokine designed as a protein fusion utilizing the antibody scaffold derived from the biopharmaceutical product Rituximab and the human IL‐2. In particular, a dimeric bivalent antibody format based on a scFv‐Fc fused to IL‐2 (2B8‐Fc‐hIL2) has been successfully produced in N. benthamiana plants by agroinfiltration. The purified, plant‐produced 2B8‐Fc‐hIL2 revealed a CD20 binding activity comparable to that of Rituximab and a full biological activity as confirmed by cell proliferation and ADCC assay.
Results
Engineering of the anti‐CD20 human interleukin‐2 immunocytokine (2B8‐Fc‐hIL2) and transient expression in N. benthamiana
The plant‐codon‐optimized sequences encoding the variable regions of the anti‐CD20‐human (2B8) antibody, alone or fused to hIL‐2, were engineered to produce the 2B8Fc antibody or the 2B8‐Fc‐hIL2 immunocytokine in plant tissues. The corresponding nonprocessed protein products were expected to contain 493 and 631 amino acids, respectively. Both genes were cloned in a plant binary vector (pBI‐Ω) (Marusic et al., 2007) under the control of the CaMV 35S promoter and the omega (Ω) translational enhancer sequence from TMV (Gallie and Kado, 1989) (Figure 1). Hydroponically grown N. benthamiana plants were co‐infiltrated with Agrobacterium tumefaciens clones harbouring the 2B8‐Fc or 2B8‐Fc‐hIL2 construct, alone or mixed with cultures of Agrobacterium carrying the p19 silencing suppressor (Lombardi et al., 2009). Leaves from the same position on the plants (leaf number 4 and 5 from the bottom) were collected at 2, 3, 4 and 5 days postinfiltration (d.p.i.), and expression was assayed by Western blot analysis using an anti‐human γIg chain antibody (Figure 2a,b).
Figure 1.
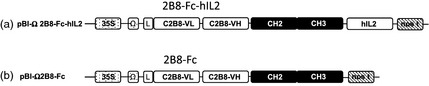
Diagram of the constructs utilized for plant agroinfiltration. (a) pBI‐Ω 2B8‐Fc‐hIL2: construct encoding the anti‐CD20 2B8‐Fc‐hIL2 immunocytokine. (b) pBI‐Ω 2B8‐Fc: construct encoding the anti‐CD20 2B8‐Fc antibody not linked to hIL‐2. The sequence encoding variable regions (VH and VL) of 2B8 monoclonal antibody, the human IgG1 Fc region (CH2‐CH3) and the human interleukin‐2 (hIL‐2) were under the control of the cauliflower mosaic virus 35S promoter (35S), Ω translational enhancer sequence and nopaline synthase terminator (nos t). (L): signal peptide sequence derived from an embryonic mouse immunoglobulin HC coding gene.
Figure 2.
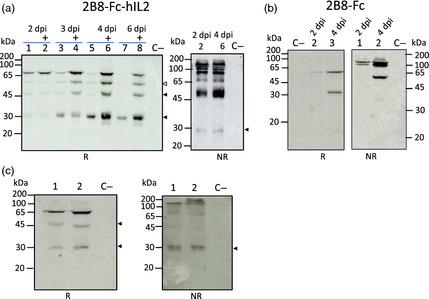
Western blot analysis of leaf extracts. N. benthamiana plants were co‐agroinfiltrated with A. tumefaciens strains harbouring the pBI‐Ω 2B8‐Fc‐hIL2 or pBI‐Ω 2B8‐Fc alone or mixed with agrobacterium strains harbouring p19 silencing suppressor from artichoke mottled crinkle virus (AMCV). Samples were separated on SDS‐12% PAGE acrylamide gel. Plant extracts were normalized for total soluble protein (TSP) content (10 μg of TSP was loaded for each sample). Extracts from noninfiltrated leaves were used as negative control (C−). Arrows indicate possible degradation fragments of the antibodies. (a) Plant extracts, expressing 2B8‐Fc‐hIL2, collected at different time points (2, 3, 4 and 6 d.p.i.) were analysed by Western blotting under reducing (R) (left side) and nonreducing (NR) conditions (right side) using an anti‐γ HRP‐conjugated antibody. (Left side and right side) Lanes 2, 4, 6 and 8 indicated by + symbol: extracts from plants co‐agroinfiltrated with 2B8‐Fc‐hIL2 and p19; lanes 1, 3, 5 and 7: extracts from plants co‐agroinfiltrated with 2B8‐Fc‐hIL2 only. (b) Western blot analysis of extracts from plants co‐agroinfiltrated with 2B8‐Fc and p19 collected at different time points (2 and 4 d.p.i.) under reducing R (left side) and nonreducing NR conditions (right side). Detection was performed with an anti‐human γ chain HRP‐conjugated antibody. (Left side) Lanes 2 and 3: extracts from sample collected at 2 and 4 d.p.i., respectively. (Right side) Lane 1 and 2: extracts from sample collected at 2 and 4 d.p.i., respectively. (c) Western blot analysis of extracts from plants co‐agroinfiltrated with 2B8‐Fc‐hIL2 and p19 collected at 4 d.p.i., under reducing (R) (left side) and nonreducing (NR) (right side) conditions using an anti‐hIL‐2 antibody. (Left side) Lanes 1 and 2: 10 and 20 μg of extracts from leaves collected at 4 d.p.i. (Right side) Lanes 1 and 2: 10 and 20 μg of extracts from leaves collected at 4 d.p.i.
Analysis under reducing conditions showed the presence of a band at approximately 65 kDa, corresponding to the monomeric 2B8‐Fc‐hIL2 immunocytokine, and three bands at lower molecular mass (approx. 50, 45, 30 kDa), most probably corresponding to degradation fragments (Figure 2a, left panel). 2B8‐Fc‐hIL2 expression was visibly improved when p19 silencing suppressor was used, starting from 3 d.p.i. and, as expected, no effects of p19 were observed at 2 d.p.i. (Figure 2a). Maximum intensity for the 65 kDa band corresponding to the intact antibody chain and for the cleavage products was detected at 4 d.p.i. (Figure 2a, lanes 5 and 6) and decreased over time (6 d.p.i.). Nevertheless, the highest relative amount of the intact antibody chain was reached at about 2 d.p.i., while increased relative levels of the degradation fragments, in particular the 30 kDa band, could be seen at later time points. Analysis under nonreducing conditions, performed at both 2 and 4 d.p.i., revealed the presence of a band at about 130 kDa, corresponding to the dimeric assembled 2B8‐Fc‐hIL2. Several bands at lower molecular mass values were also visible, probably corresponding to unassembled scFv‐Fc antibody and to degradation fragments (Figure 2a, right panel). Also in this case, highest accumulation levels of all antibody‐related bands were reached at 4 d.p.i. and the highest relative amount of intact assembled antibody was seen at 2 d.p.i.
Comparable results were obtained for the expression of 2B8‐Fc antibody with maximum protein accumulation at 4 d.p.i. (Figure 2b) and highest relative amount of intact antibody chain at 2 d.p.i. 2B8‐Fc band intensities in this Western blot analysis cannot be quantitatively compared to those of 2B8‐Fc‐hIL2 (Figure 2a) due to different exposure times used for detection. Analysis under reducing conditions at 4 d.p.i. showed the presence of the expected band at about 55 kDa corresponding to the monomeric 2B8‐Fc, together with only one band at lower molecular mass, probably corresponding to a degradation fragment (Figure 2b, left panel). Analysis under nonreducing conditions at 4 d.p.i. showed a less complex band pattern compared to 2B8‐Fc‐hIL2, revealing a band at about 110 kDa, corresponding to the dimeric assembled 2B8‐Fc and only one lower band at approximately 55 kDa, probably related to the unassembled antibody (Figure 2b right panel). Western blot analysis at 2 d.p.i under reducing and nonreducing conditions revealed the presence of only a band corresponding to the intact 2B8‐Fc chain and to the complete assembled antibody, respectively (Figure 2b).
2B8‐Fc‐hIL2 expression was also assayed using an anti‐hIL‐2 antibody to verify the presence of the fused cytokine (Figure 2c). Western blot analysis under reducing conditions showed the expected band at about 65 kDa, corresponding to monomeric 2B8‐Fc‐hIL2 and two lower bands probably related to degradation fragments (Figure 2c, left panel). Analysis under nonreducing conditions revealed a band at about 130 kDa supposedly matching with that of the dimeric assembled immunocytokine and lower bands possibly associated with partially assembled or degraded species (Figure 2c, right panel). Antibody expression levels in the extracts of agroinfiltrated leaves of the same age were determined using double antibody sandwich ELISA (DAS‐ELISA). Average levels were 28 and 41 mg/kg fresh weight (FW) for 2B8‐Fc‐hIL2 and 2B8‐Fc, which corresponded to about 0.5% and 0.7% of total soluble protein content (TSP), respectively (Table 1).
Table 1.
Expression levels of 2B8‐Fc‐hIL2 and 2B8‐Fc in agroinfiltrated N. benthamiana leaves
| Construct | % TSPa | Yieldsb (mg/kg fresh weight) | Protein‐A purification yieldc (mg/kg fresh weight) |
|---|---|---|---|
| 2B8‐Fc‐hIL2 | 0.5 | 28 ± 6 | 16 ± 6 |
| 2B8‐Fc | 0.7 | 41 ± 11 | 28 ± 4 |
% TSP values were estimated from Gleba and Giritch (2011), assuming that a yield of 5 g recombinant protein/kg FW is equivalent to 80% TSP.
Determined by quantitative ELISA as described in ‘Experimental procedures. Values are the mean of ± standard error of the mean (SEM) of triplicate samples.
Purification yield is the mean value (±standard error of the mean) obtained from three purifications of batches (40 g) of agroinfiltrated leaves.
Purification and characterization of the plant‐derived antibodies
Plant extracts were typically prepared starting from 40 g batches of agroinfiltrated leaves in the presence of the p19 silencing suppressor (harvest at 4 d.p.i.). Purification was performed using a HiTrap™ FF Protein‐A column. The average yield obtained for 2B8‐Fc‐hIL2 and 2B8‐Fc was typically in the range 0.6 and 1.1 mg of mAb, respectively. This corresponded to 16 mg/kg FW for 2B8‐Fc‐hIL2 and 28 mg/kg FW for 2B8‐Fc (Table 1). A typical SDS‐PAGE analysis of the purified antibodies under reducing and nonreducing conditions is shown in Figure 3a. Nonreducing SDS‐PAGE analysis of the purified antibodies revealed a major band of about 130 kDa for 2B8‐Fc‐hIL2 and a band with a slightly lower mass for 2B8‐Fc (Figure 3a, lanes 2 and 3, respectively), both corresponding to the dimeric assembled antibodies. Both antibodies also showed a common band with lower mass (about 100 kDa), probably corresponding to nonglycosylated products and a very faint band corresponding to the monomeric antibody chains (migrating at 65 and 55 kDa for 2B8‐Fc‐hIL2 and 2B8‐Fc, respectively). SDS‐PAGE analysis under reducing conditions of both purified antibodies showed the expected products with appropriate size plus an additional component migrating at about 30 kDa (Figure 3a, lanes 5, 6). Only in the case of 2B8‐Fc‐hIL2, a very faint additional band at higher molecular mass (<100 kDa) was also visible (Figure 3a, lane 5). To further assess assembly and integrity of the purified antibodies, size‐exclusion chromatography using a Superdex S‐200 column was performed. Analysis revealed that about 87% of the purified 2B8‐Fc‐hIL2 antibody (Figure 3b, central panel) had the expected molecular mass of approximately 130 kDa, corresponding to a major peak eluting at 2.10 mL, similar to that obtained for plant‐purified IgG used as a control (Figure 3b, top panel). A second peak eluting at 2.51 mL corresponded to lower molecular mass fragments that were also observed in nonreducing SDS‐PAGE analysis (Figure 3a, left panel and data not shown).
Figure 3.
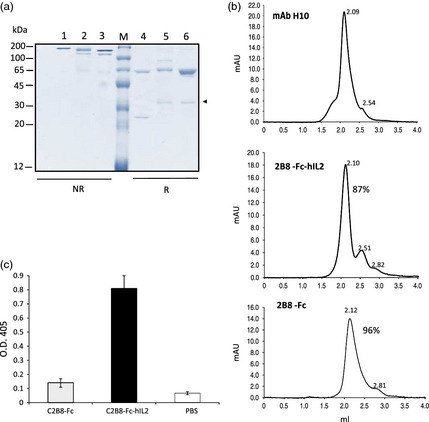
Characterization of purified antibodies from agroinfiltrated N. benthamiana. (a) Typical SDS‐PAGE analysis of purified scFv‐Fc antibodies obtained after protein‐A affinity chromatography. Nonreducing (1–3) or reducing (4–6) Coomassie‐stained SDS‐10% PAGE analysis of purified scFv‐Fc antibodies after concentration and dialysis. 1 and 4: 1 μg of the reference IgG1 Rituximab (RTX); 2, 3 and 5, 6: 1 μg of 2B8‐Fc‐hIL2 and 2B8‐Fc, respectively; M: molecular mass marker. The band indicated by an arrow is a possible degradation fragment of the antibodies. (b) Size‐exclusion chromatography of purified 2B8‐Fc‐hIL2 and 2B8‐Fc was performed on a Superdex™ 200 5/150 GL column, as described in the experimental section. Top panel represents the chromatogram obtained for the plant‐purified human mAb H10, which was used as a reference; retention volumes (mL) of the obtained peaks are reported. (c) Anti‐hIL‐2 ELISA. Wells were coated with an anti‐γ chain antibody. Ten ng of purified antibodies was used followed by anti‐hIL‐2 antibody; 2B8‐Fc and PBS were used as negative controls. Reported values are the mean of three independent experiments, and error bars represent SD of the means.
Analysis of 2B8‐Fc revealed that more than 96% of the purified antibody had the expected molecular mass (Figure 3b, bottom panel), which corresponded to a major peak eluting at 2.12 mL. To reveal the presence of hIL‐2 fused to the anti‐CD20 scFv‐Fc, the purified 2B8‐Fc‐hIL2 antibody was assayed by ELISA using an anti‐hIL‐2 antibody (Figure 3c), showing a strong immunoreactivity while the 2B8‐Fc was used as a negative control.
Structural analysis of the plant‐derived antibodies
To obtain a structural characterization of the purified, plant‐derived 2B8‐Fc‐hIL2 and 2B8‐Fc antibodies, all bands observed in SDS‐PAGE under reducing conditions (Figure 3a, lanes 5 and 6) were digested in situ with trypsin. Corresponding digests were then subjected to extensive peptide mapping experiments by combined MALDI‐TOF‐MS and nLC‐ESI‐LIT‐MS/MS analysis. A schematic representation of the results of structural characterization by mass spectrometry is reported in Figure S1. In the case of 2B8‐Fc‐hIL2, combined MS experiments of the major component (band at about 65 kDa in Figure 3a, lane 5) confirmed to be the intact antibody with a sequence coverage of 73.1% (data not shown). A unique C‐terminal peptide was observed, while multiple forms of the N‐terminal region were detected. In particular, simultaneous occurrence of peptides (17–37), (18–37) and (20–37) was revealed. Evaluation of the corresponding areas of the extracted ion chromatograms from nLC‐ESI‐LIT‐MS analysis (data not shown) provided information on peptide relative abundances, demonstrating that a major fraction of the recombinant 2B8‐Fc‐hIL2 product started at the expected Glu20 residue, with minor percentages starting at Thr18 and Gly17. Similar results for the protein N‐terminus were obtained in the case of the faint band migrating at 30 kDa also present in Figure 3a (lane 5), while a lack of various C‐terminal peptides was observed with respect to the intact 2B8‐Fc‐hIL2 (Figure S1). These results definitively demonstrated that this minor 2B8‐Fc‐hIL2 form corresponded to a recombinant antibody truncated at protein C‐terminus at the beginning of CH3 domain (YTLPPSR401).
In the case of 2B8‐Fc, combined MS experiments of the major component (band at 55 kDa in Figure 3a, lane 6) provided information on 92.0% of its sequence (data not shown), confirming to be the intact antibody. Also in this case, a unique C‐terminal peptide was observed, while the simultaneous presence of peptides (17–37), (18–37) and (20–37) was revealed at protein N‐terminus; evaluation of the corresponding areas of the extracted ion chromatograms from nLC‐ESI‐LIT‐MS analysis (data not shown) demonstrated that a major fraction of the recombinant 2B8‐Fc product started at the expected Glu20 residue, similarly to 2B8‐Fc‐hIL2. Again, peptide mapping experiments demonstrated that the faint band migrating at 30 kDa also present in Figure 3a (lane 6) corresponded to a minor 2B8‐Fc form processed at protein C‐terminus at the beginning of CH3 domain (Figure S1).
MALDI‐TOF‐MS spectra of tryptic digests from intact 2B8‐Fc‐hIL2 and 2B8‐Fc showed similar, faint MH+ signals at m/z 2766.0 and 3248.3 that, according to the known pathways of plant glycoprotein biosynthesis, were tentatively associated with peptides (339–347) and (335–347) bearing a complex‐type N‐linked glycan structure, respectively (data not shown). To provide a complete elucidation of the glycan moieties present in 2B8‐Fc‐hIL2 and 2B8‐Fc antibodies, corresponding digests were independently enriched for glycopeptides through hydrophilic interaction liquid chromatography (HILIC). Eluted fractions were then analysed by MALDI‐TOF‐MS. Spectra recorded for the two antibodies were virtually identical and showed various signals associated with glycopeptides (339–347) and (335–347). Figure 4a and b represents an enlarged portion of the MALDI‐TOF‐MS spectra recorded for 2B8‐Fc‐hIL2 and 2B8‐Fc, showing signals tentatively associated with glycopeptides (339–347) that differ from the nature of the complex‐type N‐linked glycan structures. The analysis revealed a preponderant glycopeptide species (MH+ = 2766.3), bearing a typical plant GlcNAc2Man3(Xyl)(Fuc)GlcNAc2 moiety (Fuc, fucose; GlcNAc, N‐acetylglucosamine; Man, mannose; Xyl, xylose), together with minor glycopeptides containing GlcNAc2Man3(Xyl)GlcNAc2, (MH+ = 2620.2), GlcNAc2Man3(Xyl)(Fuc)GlcNAc (MH+ = 2563.2) and GlcNAc2Man3GlcNAc2 (MH+ = 2488.1) glycan structures. It was impossible to determine whether these minor forms derived from in vivo glycan processing or originated from fragmentation reactions during MS analysis. The main glycopeptide was definitively characterized for its structure by MALDI LIFT‐TOF/TOF‐MS analysis (Figure 4c).
Figure 4.
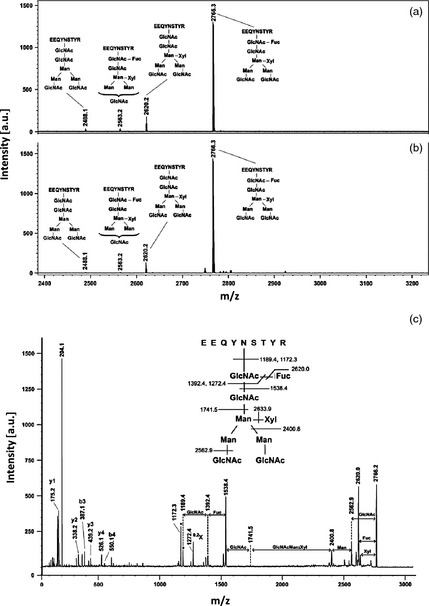
MALDI‐TOF‐MS spectrum of the peptide (339–347) bearing different complex‐type N‐linked glycan structures as obtained after enrichment by HILIC chromatography of the glycopeptide fraction from 2B8‐Fc‐hIL2 and 2B8‐Fc tryptic digests. (a) Glycopeptides from 2B8‐Fc‐hIL2; (b) Glycopeptides from 2B8‐Fc. (c) MALDI LIFT‐TOF/TOF‐MS analysis of the glycopeptide (339–347) at m/z 2766.3. Peptide fragments from y and b series are evidenced. A signal doublet corresponding to the deglycosylated peptide is marked with an asterisk. The signal corresponding to the ring fragmentation of the innermost GlcNAc is reported as 0.2X; a. u.: arbitrary units.
Functional integrity of the anti‐CD20 antibodies
CD20 binding
Human Daudi cells were used to assess the ability of 2B8‐Fc‐hIL2 and 2B8‐Fc to bind to the cell surface antigen CD20. Flow cytometry analyses showed that, when stained with Rituximab, virtually all Daudi cells were positive for CD20 expression, as expected (Figure 5a). Equimolar concentrations of 2B8‐Fc‐hIL2 and 2B8‐Fc bound to CD20 at a level comparable to Rituximab as indicated by mean fluorescence intensity values (Figure 5a,b). 2B8‐Fc‐hIL2 was also used at different dilutions, to verify the correlation between MFI values and 2B8‐Fc‐hIL2 concentration (Figure 5c). To confirm the specificity of 2B8‐Fc‐hIL2 binding to CD20, Daudi cells were co‐stained with both 2B8‐Fc‐hIL2 and Rituximab at different concentrations. In this experiment, a secondary anti‐human IgG monoclonal antibody able to recognize Rituximab but not 2B8‐Fc‐hIL2 was used. Results in Figure 5d showed that 2B8‐Fc‐hIL2 was able to partially outcompete Rituximab, confirming that 2B8‐Fc‐hIL2 specifically binds to the same epitope on the cell surface‐expressed CD20 antigen.
Figure 5.
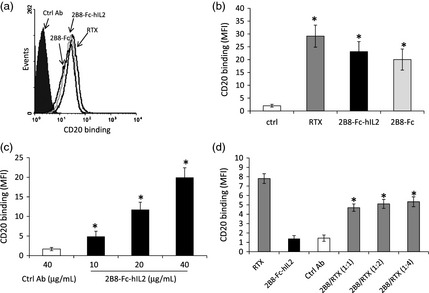
2B8‐Fc‐hIL2 and 2B8‐Fc binding to CD20 expressed on Daudi cells. (a) Daudi cells, labelled with either Rituximab (RTX), 2B8‐Fc‐hIL2, 2B8‐Fc or a control antibody (anti‐tenascin C mAb H10) were analysed by flow cytometry. Binding to CD20 was revealed with a goat anti‐human IgG polyclonal Ab, which in turn was detected with a FITC‐conjugated rabbit anti‐goat IgG polyclonal Ab. (b) mean fluorescence intensity (MFI); values represent mean SD; *, P < 0.05 for RTX, 2B8‐Fc‐hIL2 and 2B8‐Fc versus the control Ab. (c) MFI for cells stained as in (a) with different concentrations (10–40 μg/mL) of 2B8‐Fc‐hIL2. *, P < 0.05 for all concentrations of 2B8‐Fc‐hIL2 versus the control Ab and each other. (d) Competitive binding of Rituximab and 2B8‐Fc‐hIL2 to CD20. Daudi cells were stained with either Rituximab (RTX), 2B8‐Fc‐hIL2 or a control Ab (mAb H10) or with both RTX and 2B8‐Fc‐hIL2 at different ratio (2B8/RTX). Antibody binding to CD20 was revealed with a FITC‐conjugated mouse anti‐human IgG monoclonal Ab (which does not recognize the 2B8 constructs). Columns represent MFI values ± SD *, P < 0.05 for 2B8‐Fc‐hIL2/RTX versus RTX. Findings were confirmed in three independent experiments. Statistical analysis was performed using one‐way anova plus post hoc Bonferroni‐corrected two‐tailed student's t‐test.
Antibody‐dependent cell‐mediated cytotoxicity
To verify whether 2B8‐Fc‐hIL2 and 2B8‐Fc were able to elicit effector functions, an ADCC assay, using Daudi cells as target cells and human peripheral blood mononuclear cells (PBMC) as effector cells, was performed. Rituximab was used for comparison and to determine optimal conditions in preliminary experiments, including effector to target cell ratio and antibody concentration. Target cell lysis was assessed by lactate dehydrogenase (LDH) release. The results showed that 2B8‐Fc‐hIL2 and 2B8‐Fc induced ADCC, arousing a percentage of cytotoxicity comparable to that of Rituximab (Figure 6a). No significant ADCC activity was detected using a plant‐produced scFv‐Fc format antibody (F8‐Fc) directed to an irrelevant antigen. These results, together with data on CD20 binding, demonstrated that the antigen‐binding (Fab‐like) and effector‐mediating (Fc‐like) portions of 2B8‐Fc‐hIL2 and 2B8‐Fc are functional and display biological activity similar to that of Rituximab.
Figure 6.
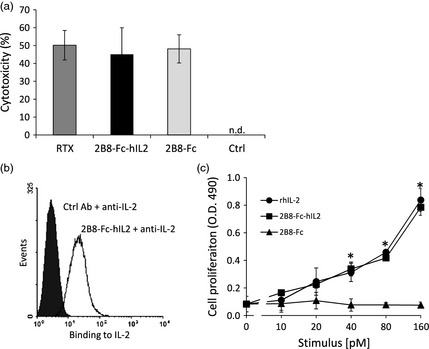
Functional integrity of 2B8‐Fc‐hIL2 and 2B8‐Fc. (a) Antibody‐dependent cell‐mediated cytotoxicity. Daudi cells were cultured with IL‐2‐activated human PBMC and either Rituximab (RTX), 2B8‐Fc‐hIL2 or 2B8‐Fc for 4 h. Percentage of cytotoxicity was assessed by LDH release. Percentage of cytotoxicity with the control Ab (F8‐Fc) was not detectable (n.d.). Results shown are means of two independent experiments ± SE. (b) Accessibility of the IL‐2 present in the 2B8‐Fc‐hIL2 immunocytokine. Daudi cells were labelled with a control Ab (mAb H10) or with 2B8‐Fc‐hIL2. Antibody binding of 2B8‐Fc‐hIL2 to CD20 was detected with a rabbit anti‐human IL‐2 and revealed with an AlexaFluor488‐conjugated goat anti‐rabbit IgG F(ab')2. (c) Biological activity of the IL‐2 present in the 2B8‐Fc‐hIL2 immunocytokine. CTLL‐2 was cultured with complete medium alone or stimulated with graded concentrations of rhIL2, 2B8‐Fc‐hIL2 or 2B8‐Fc. After 48 h, cell proliferation was assessed by a MTS to formazan reduction assay and expressed as absorbance values (O.D. 490 nm). Values represent mean ± SD of cell cultures run in triplicates. Findings were confirmed in two independent experiments. *, P < 0.05 for 2B8‐Fc‐hIL2 and rhIL2 versus 2B8‐Fc (one‐way anova plus post hoc Bonferroni‐corrected two‐tailed student's t‐test).
IL‐2 activity
The presence of IL‐2 in the 2B8‐Fc‐hIL2 immunocytokine bound on Daudi cells was assessed by flow cytometry using a FITC‐conjugated anti‐hIL‐2 mAb. The results shown in Figure 6b revealed that anti‐IL‐2 mAb could bind the immunocytokine, indicating that IL‐2 is exposed and accessible on the cell surface. Moreover, biological activity of hIL‐2 was assessed with a CTLL‐2 cell proliferation assay. As shown in Figure 6c, cell proliferation induced by 2B8‐Fc‐hIL‐2 was concentration dependent and comparable to that induced by a recombinant human IL‐2 produced in mammalian cells. Conversely, 2B8‐Fc did not induce any appreciable cell proliferation.
Discussion
Immunocytokines are a class of molecules created by linking a tumour‐reactive antibody to a cytokine to potentiate the antitumour immune response. By delivering cytokines directly to focal or disseminated diseases, ICs improve the therapeutic effect and allow the reduction of cytokine dose, limiting detrimental side effects. Currently, several ICs are undergoing clinical trials showing great promise for anticancer therapeutic applications (Sondel and Gillies, 2012). IL‐2, due to its pleiotropic effects and success in clinical use (although not free of side effects), has been included in several immunocytokines representing the most advanced group of molecules in clinical settings. Yet, so far these types of recombinant antibodies have been successfully produced using mammalian cells.
In an unconventional yet effective approach, we demonstrated here the feasibility to express an IL‐2‐based immunocytokine in plants. To this end, we engineered an anti‐CD20‐human IL‐2 immunocytokine based on the C2B8 mAb (Rituximab) and optimized the sequence for transient expression in N. benthamiana. We engineered the C2B8 monoclonal antibody to a secretory scFv‐Fc format fused to human IL‐2 through a small flexible linker. The scFv‐Fc format has several advantages: (i) it can be easily engineered for plant transient expression; (ii) it generally retains its full antigen‐binding activity; (iii) it maintains the Fc portion of human IgG1 immunoglobulins, which can activate different arms of human immune responses and can be efficiently expressed in plants at high levels (Lai et al., 2014; Van Droogenbroeck et al., 2007). Accumulation of the immunocytokine was in the range of 30 mg/kg FW; purification yields were of about 16 mg/kg of fresh leaf, quantity lower if compared to that of the scFv‐Fc format devoid of IL‐2 (2B8‐Fc). This difference can be ascribed to the higher extent of degradation found for 2B8‐Fc‐hIL2, as indicated by the presence of several specific degradation bands in Western blot analysis. Indeed, it has been extensively demonstrated that proteolysis in plant can have a major impact on the final yield of recombinant antibodies (Hehle et al., 2015; Robert et al., 2013). Three major bands were recognized on reducing SDS‐PAGE and migrated at molecular mass values of about 55, 45 and 30 kDa (see Figure 2). The relative abundance of these bands increased over time with a peak of accumulation at 4 d.p.i., when compared to that corresponding to the intact antibody chain. It can be hypothesized that the hIL‐2 fusion somehow influences antibody conformation, making the whole molecule more prone to plant proteases. Nevertheless, we demonstrated that these degradation fragments do not substantially affect quality of the protein‐A‐purified antibody. In fact, gel filtration analysis revealed that about 90% of 2B8‐Fc‐hIL2 had the expected molecular mass compared to 96% of the 2B8‐Fc, the remaining being lower molecular mass fragments. Interestingly, both purified 2B8 recombinant antibodies show the same discrete band of about 30 kDa on reducing SDS‐PAGE (Figure 3a), indicating that a similar proteolytic event on the recombinant antibody fragment occurs in plant cells. This band was also analysed by mass spectrometry, showing that it corresponds to the recombinant antibody with intact N‐terminus but degraded at protein C‐terminus. We were somehow aware of the fact that the hIL‐2 would have a negative impact on recombinant protein yield. In fact, previous literature highlighted that hIL‐2 accumulates at low levels in plant cells, due to its low molecular mass and stability (Magnuson et al., 1998; Park and Cheong, 2002). To overcome this problem, tobacco plants expressing mature hIL‐2 fused with two different proteinase inhibitors were generated. However, reported yields were in the range of 0.1% of TSP (Redkiewicz et al., 2012). In the light of these results, our approach demonstrates that the scFv‐Fc‐IL2 fusion can reach levels of IL‐2 much higher than any of those previously reported in literature.
Noteworthy, when the 2B8‐Fc‐hIL2 is bound to CD20 on cell membrane, the fused IL‐2 is readily accessible and its conformation can be recognized by a monoclonal antibody, as revealed by flow cytometry analyses. Moreover, proper IL‐2 conformation and functional activity was confirmed in a IL‐2‐dependent cell proliferation assay. Indeed, the biological activity of the plant‐made antibody‐hIL2 fusion was comparable to that of a recombinant human IL‐2 produced in human‐derived HEK293 cells. These results are in accordance with those obtained in a previous work in which partially purified tobacco‐derived hIL‐2 in fusion with protease inhibitors showed a similar proliferation activity to bacterial derived hIL‐2 (Redkiewicz et al., 2012).
Immunocytokines need to be characterized for their effector functions as hIL‐2 fusion could compromise the activity of the antibody. In a previous work it was shown that IL‐2 fusion to the recombinant anti‐CD20 DI‐Leu16 mAb did not significantly alter its biological activity, retaining ADCC activity similar to that of Rituximab (Gillies et al., 2005). Our results also showed that both 2B8‐Fc‐hIL2 and 2B8‐Fc are effective in eliciting ADCC of human PBMC versus Daudi cells. These findings demonstrated that the antigen binding (Fab‐like) and effector‐mediating (Fc‐like) portions of 2B8‐Fc‐hIL2 and 2B8‐Fc are functional and display biological activity equivalent to that of Rituximab. Correct antigen recognition was also confirmed by FACS competitive assay in which 2B8‐Fc‐hIL2 was able to efficiently compete with Rituximab for CD20 binding on Daudi cells indicating, as expected, that both antibodies recognize the same epitope. Even if our experiments showed that 2B8‐Fc‐fused IL2 is biologically active, it remains to determine whether this IC provides additional therapeutic effects. Biologically active IL2, being driven by 2B8 to tumour cells, could enhance local immune response and contribute to the generation of antitumour effector cells. However, future experiments with both in vitro and in vivo models are required to verify this possibility.
The 2B8‐Fc‐IL2 plant‐produced IC was also characterized for glycosylation profile, showing a largely dominant glycoform, namely Glc2Man3(Xyl)(Fuc)GlcNAc2, which occurred at Asn297. The same profile was also observed for the 2B8‐Fc molecule, indicating that hIL‐2 fusion does not alter the glycosylation processing of the 2B8‐Fc portion in the plant cell. Uniform glycosylation of monoclonal antibodies has been already demonstrated for several antibodies produced in plants (i.e. the anti‐HIV mAbs 2G12 and VRC01 and the anti‐West Nile Virus (WNV) scFv‐Fc fragment) (Lai et al., 2014; Sainsbury et al., 2010; Teh et al., 2014) and represents an advantage in terms of both product quality control and manufacturing. In several cases, it was demonstrated that plant‐produced HIV‐neutralizing antibodies with a homogeneous Glc2Man3(Xyl)(Fuc)GlcNAc2 glycoform perform as well as antibodies produced in CHO cells in terms of cell‐mediated anti‐HIV activity (Teh et al., 2014). To date, there is still no evidence that plant glycans decorating antibodies are immunogenic in humans; nevertheless, recent advances in plant glyco‐engineering highlight the potential to produce antibodies with a full ‘mammalian’ glycosylation profile (Loos and Steinkellner, 2014; Strasser et al., 2008). The removal of the fucose moiety from the plant‐produced anti‐HIV 2G12 antibody was shown to enhance binding to FcγRIIIa and to increase its potency in antibody‐dependent cell‐mediated virus inhibition (ADCVI) (Forthal et al., 2010). In a different work, a glyco‐modified mAb (MB314) lacking core fucose, recognizing the tumour‐associated carbohydrate antigen Lewis Y, was stably produced in a moss expression system. ADCC effector function of MB314 was increased up to 40‐fold, whereas complement‐dependent cytotoxicity activity was decreased 5‐fold (Kircheis et al., 2012). A study was also performed on the characterization of a novel nonfucosylated Rituximab (BLX‐300), which was produced in a Lemna aquatic plant‐based system (Gasdaska et al., 2012). Authors showed that Target cell binding and induction of apoptosis were similar for BLX‐300 and Rituximab, while an increase in ADCC was observed in BLX‐300 versus Rituximab. In the light of these evidences, future experiments will be directed in the production of the 2B8‐Fc‐hIL2 IC with a modified glycan profile to evaluate possible enhancements in the biological activity. The production of an IC with optimized glycan profile could represent an advantage of using plants as compared to traditional mammalian‐based systems.
To the best of our knowledge, this study provides the first evidence of the functional expression of a immunocytokine single‐chain variant of the biotherapeutic Rituximab produced in plants. This 2B8‐Fc‐hIL2 was at least equivalent to the commercial molecule in both binding to Daudi cells and activation of ADCC and may represent a promising plant‐derived biopharmaceutical for the treatment of NHLs.
Experimental procedures
Recombinant antibodies and fusion proteins
Anti‐CD20 2B8‐Fc‐hIL2 was synthetically constructed (GenScript) based on the v gene sequences of the murine 2B8 (US Patent No. 5,736,137). VH and VL sequences were assembled to produce a single‐chain Fv (scFv) with a 15‐residue linker fused to a human IgG1 Fc including the hinge region. The hIL‐2 sequence (GenBank: CAA25742.1) was then fused to the C‐terminus of the Fc to obtain the full‐length 2B8‐Fc‐hIL2 (Figure 1a). The 2B8‐Fc antibody was obtained from 2B8‐Fc‐hIL2 by PCR amplification excluding the hIL‐2 region (Figure 1b). Both constructs were cloned into the plant expression binary vector pBI‐Ω (Marusic et al., 2007) using the BamHI/EcoRI restriction sites, yielding plasmids pBI‐Ω2B8‐Fc‐hIL2 and pBI‐Ω2B8‐Fc. The pBI‐Ωp19 bearing the p19 silencing suppressor gene from AMCV was also used (Marusic et al., 2007). All synthetic genes were codon‐optimized for expression in N. benthamiana using the OptimumGene™ algorithm (GenScript, Piscataway, NJ).
Transient expression in N. benthamiana
Transient expression in plant leaves was performed by vacuum agroinfiltration. A. tumefaciens (LBA 4404) clones harbouring 2B8‐Fc‐hIL2, 2B8‐Fc and p19 constructs were grown separately. Bacteria were pelleted/sedimented by centrifugation at 4000 g and resuspended in infiltration buffer (10 mm MES, 10 mm MgSO4, pH 5.8). Agrobacterium suspensions harbouring different vectors were either used separately or mixed together (2B8‐Fc‐hIL2 and p19 or 2B8‐Fc and p19) to reach the final optical density (OD600) of 0.5 for each construct. Hydroponically grown 6‐week‐old N. benthamiana plants (at the 6–7 leaf stage) were infiltrated by completely submerging each plant in the Agrobacterium‐containing solution inside a desiccator. Vacuum was then applied reaching ~10 mm Hg, and then quickly released. Infiltration was confirmed visually, observing infiltrated areas as translucent. Plants were then placed in the greenhouse, and leaf sampling was performed at set time points (2–6 d.p.i). Leaves of the same age (from the ‘middle leaf’ position, typically leaf 4 and 5 from the bottom) from three individual plants were collected and stored in liquid N2. For antibody purification, batches of 40 g of agroinfiltrated leaves of all ages were typically collected at 4 d.p.i., frozen in liquid N2 and stored at −80 °C.
ELISA, SDS‐PAGE and Western blot analysis
For quantification of 2B8‐Fc‐hIL2 and 2B8‐Fc expression, agroinfiltrated leaves of the same age were analysed by double antibody sandwich (DAS) ELISA. Leaf tissue (100 mg) was ground in liquid N2 and homogenized in 500 μL of phosphate‐buffered saline pH 7.2 (PBS) containing a protease inhibitor cocktail (Complete™; Roche, Mannheim, Germany). After centrifugation at 20 000 g at 4 °C, for 30 min, the supernatant was recovered and quantified for TSP using the Bradford colorimetric assay as specified by the manufacturer (Bio‐Rad, Hercules, CA). The capture antibody (anti‐human γ chain, I6010; Sigma‐Aldrich, Saint Louis, MO), at a concentration of 2 μg/mL in PBS, was coated directly onto Nunc‐Immuno Maxisorp wells and incubated overnight at 4 °C. Plates were then blocked with 2% milk (w/v) in PBS at 37 °C, for 2 h. After washing, different dilutions of leaf extracts, normalized for TSP concentration, were added to the wells (100 μL) and incubated at 37 °C, for 2 h. After washing, the anti‐human γ chain HRP‐conjugated (A8419; Sigma‐Aldrich) was added at a dilution of 1 : 5000 in PBS containing 2% milk (w/v), and incubated at 37 °C, for 1 h. Enzymatic activity was measured after 30 min at 405 nm on a microtitre plate reader (TECAN‐Sunrise, Groedig, Austria) using 2,2‐azino‐di‐3‐ethylbenz‐thiazoline sulphonate (ABTS, KPL). As a positive control, the plant‐purified 2B8‐Fc‐hIL2 and 2B8‐Fc were separated by size‐exclusion chromatography and elution peaks corresponding to the complete antibodies were spectrophotometrically quantified and used in ELISA. Antibodies were spiked in wild‐type N. benthamiana extract at different dilutions (ranging from 1 to 100 ng). To assess the presence of hIL‐2, the anti‐hIL‐2 (D7A5, Cell Signaling Technology®, Danvers, MA) at a dilution of 1 : 2000 in PBS containing 3% milk (w/v) was used as secondary antibody, followed by incubation with an HRP‐conjugated anti‐rabbit antibody (0545; Sigma), at a dilution of 1 : 10 000 in PBS containing 2% milk (w/v).
Plant extracts were analysed by Western blot. Separation was performed on SDS‐12% PAGE acrylamide gel, and proteins were electrotransferred to a PVDF membrane (Millipore, Bedford, MA) using a Semi‐Dry Transfer Unit (Hoefer TE70; GE Healthcare, Freiburg, Germany). Membranes were blocked with PBS containing 4% milk (w/v), overnight, before adding anti‐human γ chain HRP‐conjugated (8419; Sigma‐Aldrich), at a dilution of 1 : 5000, or anti‐hIL‐2, at a dilution of 1 : 2000, in PBS containing 2% milk (w/v). The anti‐hIL‐2 antibody was followed by HRP‐conjugated anti‐rabbit antibody (0545; Sigma), at a dilution of 1 : 10 000 in PBS containing 2% milk (w/v). Proteins were detected by enhanced chemiluminescence (ECL, Plus; GE Healthcare). To assess the assembly of the transiently expressed antibodies, plant extracts were separated by nonreducing SDS‐10% PAGE acrylamide gel and electrotransferred proteins were detected as described above.
Protein‐A‐purified plant antibodies were separated by reducing or nonreducing SDS‐10% PAGE, and proteins were visualized by Coomassie blue staining. As a control, commercial Rituximab (RTX) was used (hcd20‐mab1; Invivogen, San Diego, CA).
Protein‐A affinity chromatography and size‐exclusion chromatography
The scFv‐Fc antibodies were purified from agroinfiltrated N. benthamiana plants by protein‐A affinity chromatography. Batches of leaves (40 g) were ground in liquid N2 to a fine powder and homogenized in 80 mL of PBS, 0.2% Tween, Complete™ Roche protease inhibitor cocktail (extraction buffer) (2 mL/g fresh tissue), using an Ultra‐Turrax homogenizer T25 (IKA, Staufen, Germany). After homogenization, the slurry was filtered through Miracloth (Sigma‐Aldrich) and clarified by centrifugation (twice at 8000 g at 4 °C, for 20 min). The resulting supernatant was loaded onto a protein‐A affinity column (1 mL HiTrap™ Protein A FF; GE Healthcare), previously equilibrated with extraction buffer, at a flow rate of 1 mL/min. The column was washed with 10 mL of PBS (10 column volumes), and the antibody was eluted with 100 mm glycine, pH 3.0 and buffered with one‐fifth volume of 1 m Tris–HCl, pH 8.0. Eluted fractions were analysed by SDS‐PAGE, followed by Coomassie staining. Antibody‐containing fractions were pooled, passed through a PD10 column (GE Healthcare) in PBS according manufacturer instructions, and concentrated in Centricon YM3 (Amicon, Bedford, MA). Antibody concentration was determined spectrophotometrically by absorbance at 280 nm (Gill and von Hippel, 1989). Purified scFv‐Fcs were analysed by size‐exclusion chromatography on a calibrated Superdex™ 200 5/150 GL column (GE Healthcare) eluted in PBS at flow rate of 0.3 mL/min, using an ÄKTA FPLC P920 instrument (GE Healthcare) thermostated at 20 °C, essentially as described before (Lombardi et al., 2010). Protein absorbance expressed as absorption units (mAU) was measured at 280 nm. Column calibration was performed using gel filtration calibration kits (Low and High Molecular Weight GE Healthcare), according to manufacturer instructions. A purified plant‐expressed human IgG1‐λ antibody (anti‐tenascin C mAbH10) was also used as a positive control (Villani et al., 2009). Peaks representing different protein fractions were collected using a fraction collector (FRAC‐920) and further analysed.
Protein digestion and glycopeptide enrichment
Bands from SDS‐PAGE under reducing conditions were excised from the gel, triturated, in‐gel reduced, S‐alkylated and digested with trypsin (Rec. Proteomics Grade; Roche) (D'Ambrosio et al., 2008). Gel particles were extracted with 25 mm NH4HCO3/acetonitrile (1 : 1 v/v), and digests were concentrated. Peptide mixtures were either desalted using μZipTipC18 pipette tips (Millipore) before MALDI‐TOF‐MS analysis or directly analysed by nanoLC‐ESI‐LIT‐MS/MS.
To isolate glycopeptides, tryptic digests from intact 2B8‐Fc‐hIL2 and 2B8‐Fc antibodies were also solved in 80% acetonitrile, 2% formic acid and loaded on NuTip Gel loader capillary HILIC columns (Glygen, Columbia, MD). After washing with 80% acetonitrile and 2% formic acid, glycopeptides were first eluted with 10 μL of 2% formic acid and then with 5 μL of 50% acetonitrile and 2% formic acid; pooled fractions were analysed by MALDI‐TOF‐MS, as described below.
Mass spectrometric analysis
Protein digests were analysed by nLC‐ESI‐LIT‐MS/MS using a LTQ XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a Proxeon nanospray source connected to an Easy‐nLC (Proxeon, Odense, Denmark) (Salzano et al., 2013). Peptide mixtures were resolved on an Easy C18 column (100 × 0.075 mm, 3 μm) (Proxeon) using a linear gradient of acetonitrile containing 0.1% formic acid in aqueous 0.1% formic acid, at a flow rate of 300 nL/min, for 60 min. Spectra were acquired in the range m/z 400–2000. Acquisition was controlled by a data‐dependent product ion scanning procedure over the three most abundant ions, enabling dynamic exclusion (repeat count 2 and exclusion duration 1 min). The mass isolation window and collision energy were set to m/z 3 and 35%, respectively.
During MALDI‐TOF‐MS analysis, whole tryptic digests or their glycopeptide fractions were mixed 1 : 1 with 2,5‐dihydroxy‐benzoic acid (10 mg/mL in 50% v/v acetonitrile, 0.1% v/v trifluoroacetic acid) and loaded on the instrument target. Spectra were acquired in the m/z range 500–5000 on a Bruker Ultraflextreme MALDI‐TOF‐TOF instrument (Bruker Daltonics, Bremen, Germany) operating in reflectron mode. Instrument settings were as follows: pulsed ion extraction = 100 ns, laser frequency = 1000 Hz and number of shots per sample = 2500–5000 (random walk, 500 shots per raster spot). Mass spectra were calibrated externally using nearest neighbour positions loaded with Peptide Calibration Standard II (Bruker Daltonics), with quadratic calibration curves. MS/MS spectra were acquired in LIFT mode. Data were elaborated using the FlexAnalysis software (Bruker Daltonics).
nLC‐ESI‐LIT‐MS/MS data were searched with MASCOT (version 2.2.06) (Matrix Science, London, UK) against a database containing the 2B8‐Fc‐hIL2 and 2B8‐Fc sequences. As searching parameters, we used a mass tolerance value of 2 Da for precursor ion and 0.8 Da for ion fragments, trypsin as proteolytic enzyme, a missed cleavage maximum value of 2, Cys carbamidomethylation as fixed modification, and Met oxidation and N‐terminal Glu/Gln cyclization as variable modifications, respectively. A MASCOT score >30 and a significant threshold (P < 0.05) were required for peptide assignment. In the case of glycopeptides, MALDI‐TOF mass signals were assigned to specific components using the GPMAW 4.23 software (Lighthouse Data, Odense, Denmark). This software generated a mass/fragment database output based on protein sequence, protease selectivity, nature of the amino acids susceptible to eventual glycosylation and the molecular mass of the modifying groups. Searching parameters were set as mentioned above; mass values were matched to protein regions using a 0.02% mass tolerance value. MALDI‐TOF‐TOF searching parameters were set with tolerances of 100 ppm and 0.5 Da for MS and MS/MS data, respectively.
Flow cytometry analyses
CD20‐expressing Daudi human cells, purchased by ATCC (American Type Culture Collection, code CCL213), were stained with either RTX, 2B8‐Fc‐hIL2 or 2B8‐Fc; plant‐produced anti‐tenascin C mAb H10 was used as a control (Villani et al., 2009). Their binding to CD20 was revealed with a goat anti‐human IgG polyclonal Ab (Life Technologies‐Invitrogen, H10700, Carlsbad, CA), which in turn was detected with a fluorescein isothiocyanate (FITC)‐conjugated rabbit anti‐goat IgG polyclonal Ab (Sigma‐Aldrich F7367). In the competitive binding experiments, CD20‐bound RTX was detected with a FITC‐conjugated mouse anti‐human IgG monoclonal Ab (555786, Clone G18‐145; BD Bioscience, San Diego, CA). Noteworthy, in our preliminary experiments (data not shown), monoclonal antibody G18‐145 was assessed to bind the RTX complete IgG1 but not the recombinant 2B8‐Fc‐hIL2 antibody. Thus, when RTX and 2B8‐Fc‐hIL2 are competing for binding, the fluorescent signal (FITC) due to the secondary antibody is specifically associated to the CD20‐bound RTX. IL‐2 present in 2B8‐Fc‐hIL2 was detected with a rabbit anti‐human IL‐2 (clone D7A5; Cell Signaling) and revealed with an AlexaFluor488‐conjugated goat anti‐rabbit IgG F(ab')2 (Life Technologies‐Invitrogen, H10700, Carlsbad, CA). Optimal concentrations of the Abs and reagents were assessed in preliminary experiments. Fluorescence signals were collected in log mode using a FACSCalibur (BD Biosciences). Analyses of cell populations were performed on events gated according to forward scatter/side scatter light parameters.
Antibody‐dependent cell‐mediated cytotoxicity
Antibody‐dependent cell‐mediated cytotoxicity was assessed by the LDH release assay, using IL‐2‐activated (20 ng/mL, cat H7041; Sigma‐Aldrich) human PBMCs as effector cells (E) and Daudi cells as target cells (T) at an E : T ratio of 25 : 1. Briefly, target cells (104/well) were incubated at 37 °C in a humidified, 5% CO2 atmosphere, into 96‐well U‐bottomed culture plates with commercial anti‐hCD20‐hIgG1 mAb (RTX) at 10 μg/mL or the plant‐produced recombinant antibodies at equimolar concentrations. As control of the assay, the irrelevant plant‐produced scFv‐Fc recombinant antibody (F8‐Fc) was used (Capodicasa et al., 2011). After 30 min, PBMCs were added (2.5 × 105/well); they were derived from peripheral blood of healthy donors by discontinuous gradient centrifugation (Histopaque1077; Sigma‐Aldrich). LDH release was assessed after 4 h of incubation, according to the manufacturer instructions (Cat. G1780; Promega, Madison, WI). Optimal conditions for ADCC and LDH detection were assessed in preliminary experiments. The percentage of cytotoxic lyses was calculated according to the formula: 100 × (experimental release‐spontaneous release)/(maximal release‐spontaneous release).
IL‐2‐dependent cell proliferation
CTLL‐2 cell line, as obtained from ATCC (code TIB‐214), was maintained in complete RPMI 1640 medium (Euroclone ECB9006L, Milano, Italy) supplemented with 10% FBS (Hyclone SH3007103), 2 mm glutamine, penicillin (100 U/mL), streptomycin (100 mg/mL) and 5 ng/mL rhIL‐2 (Sigma‐Aldrich H7041) at 37 °C, in a humidified 5% CO2 atmosphere. Cells were left to proliferate and divided in subcultures according to the concentrations indicated by the producer.
For the bioassay, the cells were washed twice and left in complete medium without hIL‐2 for 2 h. After an additional wash, cells were seeded in 96‐well plates (2.5 × 105/mL) and cultured with complete medium alone or stimulated with graded concentrations of rhIL‐2, 2B8‐Fc‐hIL2 or 2B8‐Fc for 48 h. Then, Cell Titer 96TM Aqueous one solution (Promega G3580) was added (20 μL/well) to each well according the producer's instructions. Six hours later, absorbance at 490 nm was read using an ELISA microtiter plate reader (TECAN‐Sunrise).
Statistical analyses
One‐way anova was used to compare groups. When significant differences were observed, a Bonferroni‐corrected two‐tailed Student's t‐test for unpaired samples was used. Data represent mean values ± standard deviation (SD). A chi‐squared test was applied to compare groups for proportions. Significance levels are indicated in the figure legends; P values <0.05 were considered statistically significant.
Supporting information
Figure S1 Structural analysis of the plant‐derived antibodies.
Acknowledgements
A special grant from the Italian Ministry of Research (MIUR): ‘Development of new biopharmaceuticals for anti‐cancer therapy’ in the framework of CNR‐ENEA Joint Programme on Research and Innovation, supported the work performed at ENEA Laboratories. Authors also acknowledge the Italian Ministry of Foreign Affairs, ‘Direzione Generale per la Promozione del Sistema Paese, Unita' per la cooperazione scientifica e tecnologica bilaterale e multilaterale’ for the support. We are grateful to Dr. Cristina Capodicasa for providing the scFv‐Fc recombinant antibody (F8‐Fc).
References
- Calcagno, A. , Rostagno, R. and Di Perri, G. (2012) Anti‐CD20 antibody therapy for B‐cell lymphomas. N. Engl. J. Med. 367, 877–878. [DOI] [PubMed] [Google Scholar]
- Capodicasa, C. , Chiani, P. , Bromuro, C. , De Bernardis, F. , Catellani, M. , Palma, A.S. , Liu, Y. , Feizi, T. , Cassone, A. , Benvenuto, E. , et al. (2011) Plant production of anti‐β‐glucan antibodies for immunotherapy of fungal infections in humans. Plant Biotechnol. J. 9, 776–787. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio, C. , Arena, S. , Salzano, A.M. , Renzone, G. , Ledda, L. and Scaloni, A. (2008) A proteomic characterization of water buffalo milk fractions describing PTM of major species and the identification of minor components involved in nutrient delivery and defense against pathogens. Proteomics, 8, 3657–3666. [DOI] [PubMed] [Google Scholar]
- Davis, B.T.A. , Grillo‐lo, A.J. , White, C.A. , Mclaughlin, P. , Czuczman, M.S. , Link, B.K. , Maloney, D.G. , Weaver, R.L. , Rosenberg, J. and Levy, R. (2014) Rituximab anti‐CD20 monoclonal antibody therapy in non‐Hodgkin' s lymphoma : safety and efficacy of re‐treatment. J. Clin. Oncol. 18, 3135–3143. [DOI] [PubMed] [Google Scholar]
- Eichenauer, D.A. and Engert, A. (2014) Antibodies and antibody‐drug conjugates in the treatment of Hodgkin lymphoma. Eur. J. Haematol. 93, 1–8. [DOI] [PubMed] [Google Scholar]
- Forthal, D.N. , Gach, J.S. , Landucci, G. , Jez, J. , Strasser, R. , Kunert, R. and Steinkellner, H. (2010) Fc‐glycosylation influences Fcγ receptor binding and cell‐mediated anti‐HIV activity of monoclonal antibody 2G12. J. Immunol. 185, 6876–6882. [DOI] [PubMed] [Google Scholar]
- Fournier, P. , Aigner, M. and Schirrmacher, V. (2011) Targeting of IL‐2 and GM‐CSF immunocytokines to a tumor vaccine leads to increased anti‐tumor activity. Int. J. Oncol. 38, 1719–1729. [DOI] [PubMed] [Google Scholar]
- Fowler, N. and Oki, Y. (2013) Developing novel strategies to target B‐cell malignancies. In Am. Soc. Clin. Oncol. Educ. Book (ed.), pp. 366–372. Publisher: ASCO University, Alexandria, VA, USA. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R. and Kado, C.I. (1989) A translational enhancer derived from tobacco mosaic virus is functionally equivalent to a Shine‐Dalgarno sequence. Proc. Natl Acad. Sci. USA, 86, 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasdaska, J.R. , Sherwood, S. , Regan, J.T. and Dickey, L.F. (2012) An afucosylated anti‐CD20 monoclonal antibody with greater antibody‐dependent cellular cytotoxicity and B‐cell depletion and lower complement‐dependent cytotoxicity than rituximab. Mol. Immunol. 50, 134–141. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Gillies, S.D. , Lan, Y. , Williams, S. , Carr, F. , Forman, S. , Raubitschek, A. and Lo, K.‐M. (2005) An anti‐CD20‐IL‐2 immunocytokine is highly efficacious in a SCID mouse model of established human B lymphoma. Blood, 105, 3972–3978. [DOI] [PubMed] [Google Scholar]
- Gleba, Y.Y. and Giritch, A. (2011). Plant viral vectors for protein expression. In Recent Advances in Plant Virology ( Caranta, X. et al., eds), pp. 387–412. Norfolk: Caister Academic Press. [Google Scholar]
- Gubbels, J.A.A. , Gadbaw, B. , Buhtoiarov, I.N. , Horibata, S. , Kapur, A.K. , Patel, D. , Hank, J.A. , Gillies, S.D. , Sondel, P.M. and Patankar, M.S. , et al. (2011). Ab‐IL2 fusion proteins mediate NK cell immune synapse formation by polarizing CD25 to the target cell‐effector cell interface. Cancer Immunol. Immunother. 60, 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehle, V.K. , Lombardi, R. , van Dolleweerd, C.J. , Paul, M.J. , Di Micco, P. , Morea, V. , Benvenuto, E. , Donini, M. and Ma, J.K.‐C. (2015) Site‐specific proteolytic degradation of IgG monoclonal antibodies expressed in tobacco plants. Plant Biotechnol. J. 13, 235–245. [DOI] [PubMed] [Google Scholar]
- Hemmerle, T. and Neri, D. (2014) The antibody‐based targeted delivery of interleukin‐4 and 12 to the tumor neovasculature eradicates tumors in three mouse models of cancer. Int. J. Cancer, 134, 467–477. [DOI] [PubMed] [Google Scholar]
- Jahn, T. , Zuther, M. , Friedrichs, B. , Heuser, C. , Guhlke, S. , Abken, H. and Hombach, A.A. . (2012) An IL12‐IL2‐antibody fusion protein targeting Hodgkin's lymphoma cells potentiates activation of NK and T cells for an anti‐tumor attack. PLoS ONE, 7, e44482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircheis, R. , Halanek, N. , Koller, I. , Jost, W. , Schuster, M. , Gorr, G. , Hajszan, K. and Nechansky, A. (2012) Correlation of ADCC activity with cytokine release induced by the stably expressed, glyco‐engineered humanized Lewis Y‐specific monoclonal antibody MB314. MAbs, 4, 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova, T.V. , Baschieri, S. , Donini, M. , Marusic, C. , Benvenuto, E. and Dorokhov, Y.L. (2010) Transient expression systems for plant‐derived biopharmaceuticals. Expert Rev. Vaccines, 9, 859–876. [DOI] [PubMed] [Google Scholar]
- Lai, H. , He, J. , Hurtado, J. , Stahnke, J. , Fuchs, A. , Mehlhop, E. , Gorlatov, S. , Loos, A. , Diamond, M.S. and Chen, Q. (2014) Structural and functional characterization of an anti‐West Nile virus monoclonal antibody and its single‐chain variant produced in glycoengineered plants. Plant Biotechnol. J. 12, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List, T. and Neri, D. (2013) Immunocytokines: a review of molecules in clinical development for cancer therapy. Clin. Pharmacol. 5, 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, R. , Circelli, P. , Villani, M.E. , Buriani, G. , Nardi, L. , Coppola, V. , Bianco, L. , Benvenuto, E. , Donini, M. and Marusic, C. (2009) High‐level HIV‐1 Nef transient expression in Nicotiana benthamiana using the P19 gene silencing suppressor protein of Artichoke Mottled Crinckle Virus. BMC Biotechnol. 9, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi, R. , Villani, M.E. , Di Carli, M. , Brunetti, P. , Benvenuto, E. and Donini, M. (2010) Optimisation of the purification process of a tumour‐targeting antibody produced in N. benthamiana using vacuum‐agroinfiltration. Transgenic Res. 19, 1083–1097. [DOI] [PubMed] [Google Scholar]
- Loos, A. and Steinkellner, H. (2014) Plant glyco‐biotechnology on the way to synthetic biology. Front. Plant Sci. 5, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson, N.S. , Linzmaier, P.M. , Reeves, R. , An, G. , HayGlass, K. and Lee, J.M. (1998) Secretion of biologically active human interleukin‐2 and interleukin‐4 from genetically modified tobacco cells in suspension culture. Protein Expr. Purif. 13, 45–52. [DOI] [PubMed] [Google Scholar]
- Marusic, C. , Nuttall, J. , Buriani, G. , Lico, C. , Lombardi, R. , Baschieri, S. , Benvenuto, E. and Frigerio, L. (2007) Expression, intracellular targeting and purification of HIV Nef variants in tobacco cells. BMC Biotechnol. 7, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. and Cheong, H. (2002) Expression and production of recombinant human interleukin‐2 in potato plants. Protein Expr. Purif. 25, 160–165. [DOI] [PubMed] [Google Scholar]
- Peyret, H. and Lomonossoff, G.P. (2013) The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83, 51–58. [DOI] [PubMed] [Google Scholar]
- Pretto, F. , Elia, G. , Castioni, N. and Neri, D. (2014) Preclinical evaluation of IL2‐based immunocytokines supports their use in combination with dacarbazine, paclitaxel and TNF‐based immunotherapy. Cancer Immunol. Immunother. 63, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkiewicz, P. , Więsyk, A. , Góra‐Sochacka, A. and Sirko, A. (2012) Transgenic tobacco plants as production platform for biologically active human interleukin 2 and its fusion with proteinase inhibitors. Plant Biotechnol. J. 10, 806–814. [DOI] [PubMed] [Google Scholar]
- Reff, M.E. , Carner, K. , Chambers, K.S. , Chinn, P.C. , Leonard, J.E. , Raab, R. , Newman, R.A. , Hanna, N. and Anderson, D.R. (1994) Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood, 83, 435–445. [PubMed] [Google Scholar]
- Robert, S. , Khalf, M. , Goulet, M.‐C. , D'Aoust, M.‐A. , Sainsbury, F. and Michaud, D. (2013) Protection of recombinant mammalian antibodies from development‐dependent proteolysis in leaves of Nicotiana benthamiana . PLoS ONE, 8, e70203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury, F. , Sack, M. , Stadlmann, J. , Quendler, H. , Fischer, R. and Lomonossoff, G.P. (2010) Rapid transient production in plants by replicating and non‐replicating vectors yields high quality functional anti‐HIV antibody. PLoS ONE, 5, e13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzano, A.M. , Novi, G. , Arioli, S. , Corona, S. , Mora, D. and Scaloni, A. (2013) Mono‐dimensional blue native‐PAGE and bi‐dimensional blue native/urea‐PAGE or/SDS‐PAGE combined with nLC‐ESI‐LIT‐MS/MS unveil membrane protein heteromeric and homomeric complexes in Streptococcus thermophilus . J. Proteomics, 94, 240–261. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Ma, J. , Zou, Z. and Jemal, A. (2014) Cancer statistics, 2014. CA: Cancer J. Clin., 64, 9–29. [DOI] [PubMed] [Google Scholar]
- Sondel, P.M. and Gillies, S.D. (2012) Current and potential uses of immunocytokines as cancer immunotherapy. Antibodies, 1, 149–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger, E. , Fischer, R. , Moloney, M. and Ma, J.K.‐C. (2014) Plant molecular pharming for the treatment of chronic and infectious diseases. Annu. Rev. Plant Biol. 65, 743–768. [DOI] [PubMed] [Google Scholar]
- Strasser, R. , Stadlmann, J. , Schähs, M. , Stiegler, G. , Quendler, H. , Mach, L. , Glössl, J. , Weterings, K. , Pabst, M. and Steinkellner, H. (2008) Generation of glyco‐engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human‐like N‐glycan structure. Plant Biotechnol. J. 6, 392–402. [DOI] [PubMed] [Google Scholar]
- Teh, A.Y.‐H. , Maresch, D. , Klein, K. and Ma, J.K.‐C. (2014) Characterization of VRC01, a potent and broadly neutralizing anti‐HIV mAb, produced in transiently and stably transformed tobacco. Plant Biotechnol. J. 12, 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyman, R.M. , Schillberg, S. and Fischer, R. (2005) Transgenic plants in the biopharmaceutical market. Expert Opin. Emerg. Drugs, 10, 185–218. [DOI] [PubMed] [Google Scholar]
- Van Droogenbroeck, B. , Cao, J. , Stadlmann, J. , Altmann, F. , Colanesi, S. , Hillmer, S. , Robinson, D.G. , Van Lerberge, E. , Terryn, N. , Van Montagu, M. , et al. (2007) Aberrant localization and underglycosylation of highly accumulating single‐chain Fv‐Fc antibodies in transgenic Arabidopsis seeds. Proc. Natl Acad. Sci. USA, 104, 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani, M.E. , Morgun, B. , Brunetti, P. , Marusic, C. , Lombardi, R. , Pisoni, I. , Bacci, C. , Desiderio, A. , Benvenuto, E. and Donini, M. (2009) Plant pharming of a full‐sized, tumour‐targeting antibody using different expression strategies. Plant Biotechnol. J. 7, 59–72. [DOI] [PubMed] [Google Scholar]
- Vincent, M. , Bessard, A. , Cochonneau, D. , Teppaz, G. , Solé, V. , Maillasson, M. , Birklé, S. , Garrigue‐Antar, L. , Quéméner, A. and Jacques, Y. (2013) Tumor targeting of the IL‐15 superagonist RLI by an anti‐GD2 antibody strongly enhances its antitumor potency. Int. J. Cancer, 133, 757–765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Structural analysis of the plant‐derived antibodies.


