Summary
Several oilseed and vegetable crops of Brassica are biennials that require a prolonged winter cold for flowering, a process called vernalization. FLOWERING LOCUS C ( FLC ) is a central repressor of flowering. Here, we report that the overexpression of natural antisense transcripts (NATs) of Brassica rapa FLC (BrFLC ) greatly shortens plant growth cycles. In rapid‐, medium‐ and slow‐cycling crop types, there are four copies of the BrFLC genes, which show extensive variation in sequences and expression levels. In Bre, a biennial crop type that requires vernalization, five NATs derived from the BrFLC2 locus are rapidly induced under cold conditions, while all four BrFLC genes are gradually down‐regulated. The transgenic Bre lines overexpressing a long NAT of BrFLC2 do not require vernalization, resulting in a gradient of shortened growth cycles. Among them, a subset of lines both flower and set seeds as early as Yellow sarson, an annual crop type in which all four BrFLC genes have non‐sense mutations and are nonfunctional in flowering repression. Our results demonstrate that the growth cycles of biennial crops of Brassica can be altered by changing the expression levels of BrFLC2 NATs. Thus, BrFLC2 NATs and their transgenic lines are useful for the genetic manipulation of crop growth cycles.
Keywords: Brassica rapa, BrFLC , natural antisense transcripts, flowering time, growth cycle, vernalization
Introduction
Brassica rapa subspecies include several oilseed and vegetable crops. Brassica napus, with its 19 chromosomes, originated ~7500 years ago as the result of a hybridization between Brassica oleracea and B. rapa (Chalhoub et al., 2014). Brassica juncea originated from a hybridization between Brassica nigra and B. rapa (Nagaharu, 1935). Many Brassica species have been cultivated since prehistoric times for their seeds, edible roots, stems, leaves, buds and flowers. Many of these crops require a prolonged winter cold for flowering through a process called vernalization (Osborn et al., 1997). For crops with higher seed yields (such as oilseed rape), the earliness of flowering is an important trait. For crops with high leafy head yields (such as Chinese cabbage), lateness of flowering is a favourable trait (Mao et al., 2014; Wang et al., 2014).
FLOWERING LOCUS C (FLC) is a central repressor of flowering. It encodes a MADS‐box transcription factor that functions as a repressor of the floral transition in Arabidopsis (Michaels and Amasino, 1999; Sheldon et al., 2000). Plants with high FLC activity are late‐flowering because FLC directly represses the expression of the floral inducers FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1) (Helliwell et al., 2006). Four copies of FLC have been reported in B. rapa, and five copies were identified in B. napus (Lin et al., 2005; Kim et al., 2006, 2007). A genetic–genomics approach revealed that BrFLC2 is a major regulator of flowering time in B. rapa (Xiao et al., 2013, 2014). The overexpression of BrFLC and BnFLC in Arabidopsis delayed flowering time from 4 weeks to more than 7 weeks (Kim et al., 2007). However, the FLC cold‐sensing mechanism is not clear in B. rapa, and the genetic manipulation of vernalization traits in crops has not been successful.
Recently, natural antisense transcripts (NATs) have been identified in the FLC locus. Among them, COOLAIR (COLD‐INDUCED LONG ANTISENSE INTERGENIC RNA) plays an important role in the epigenetic silencing of FLC in Arabidopsis (Swiezewski et al., 2009; Liu et al., 2010). COOLAIR is multi‐exonic and poly‐adenylated, and its transcription covers the entire genomic DNA region of the gene. The COOLAIR antisense transcripts originate from a promoter adjacent to the FLC 3′ untranslated region and consist of two classes that terminate at proximal or distal sites (Swiezewski et al., 2007; Hornyik et al., 2010). COOLAIR has been proposed to be involved in vernalization‐mediated FLC repression and may be part of the cold‐sensing mechanism (Swiezewski et al., 2009; Liu et al., 2010; Csorba et al., 2014; Marquardt et al., 2014). Another work explains that COOLAIR plays a redundant role in regulating FLC expression and is not required for the vernalization response in Arabidopsis (Helliwell et al., 2011). Therefore, the underlying mechanism of NATs in vivo is still obscure. In addition to COOLAIR, another long intronic noncoding RNA COLD‐ASSISTED INTRONIC NONCODING RNA (COLDAIR) is required for the vernalization‐mediated epigenetic repression of FLC in Arabidopsis (Heo and Sung, 2011).
Here, we report that several NATs of BrFLC2 in B. rapa were induced during vernalization, concurrent with a rapid reduction in the activities of BrFLC genes. These NATs inhibit the activity of BrFLC genes and thereby shorten the vernalization duration and accelerate flowering. We identified B. rapa transgenic plants overexpressing BrFLC2as816, a NAT of BrFLC2, and selected 12 transgenic lines, all of which show annual properties. These data suggest that BrFLC NATs are potentially useful in the genetic manipulation of vernalization traits and growth cycles in high yield crops.
Results
BrFLC genes are triplicated and vary with crop types
Brassica rapa genomes contain four FLC copies (Lin et al., 2005; Kim et al., 2006, 2007). Using the specific primers for each copy, we isolated cDNAs of BrFLC1, BrFLC2, BrFLC3 and BrFLC5 genes from the inbred lines of Yellow sarson (YS‐143) (B. rapa var. trilocularis, rapid‐cycling crop type), Bre (B. rapa ssp. pekinensis var. bre, medium‐cycling crop type) and Wantai (B. rapa ssp. pekinensis var. wantai, slow‐cycling crop type) (Figure 1a). Interestingly, putative BrFLC1, BrFLC2, BrFLC3 and BrFLC5 in Yellow sarson show premature stop codons after the 5th, 106th, 127th and 10th AA, respectively (Figure 1b). In Bre, putative BrFLC1, BrFLC2 and BrFLC3 share a high identity (>85%) in amino acid sequences with the BrFLC reported in Chiifu‐401 (http://brassicadb.org/brad/) (Figure S1), and the putative BrFLC5 lacks an IKC domain. In Wantai, putative BrFLC1, BrFLC2 and BrFLC3 proteins have a high identity (>98%) with those in Chiifu‐401. Similar to BrFLC5 in Bre, the BrFLC5 of Wantai also lacks an IKC domain (Figure 1b).
Figure 1.
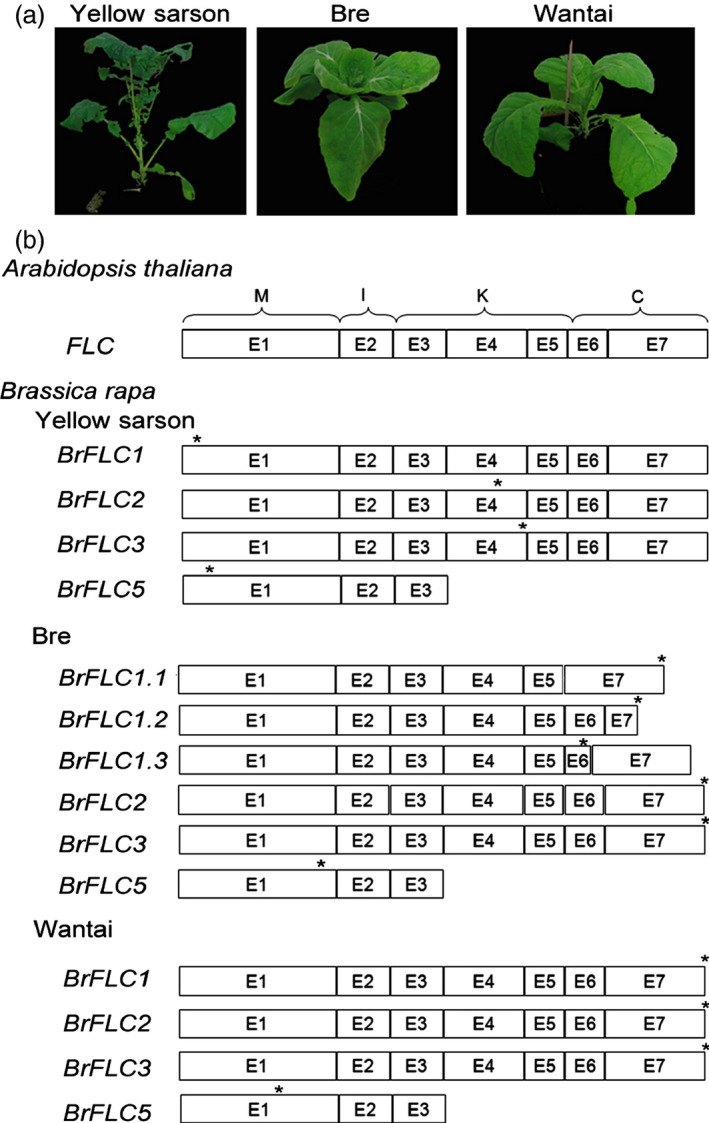
Members of BrFLC gene family in Yellow sarson (YS), Bre and Wantai. (a) Plant phenotypes of three crop types. (b) Four copies of BrFLC genes in YS, Bre and Wantai. *The stop codon. E, exon; M, MADS‐box domain for DNA binding; I, the domain for dimerization; K, the domain for protein interaction; C, the domain for the diverse functions. Arabic numbers beneath the gene structures indicate the positions of primers used for real‐time PCR.
In Bre, the BrFLC1 has three alternative transcripts, all of which encode truncated proteins without intact C‐terminal fragments (Figures 1b and S1). In Wantai, BrFLC1 encodes a full‐length protein and no alternative transcript was detected (Figure 1b). To examine whether the truncated proteins are functional in flowering repression, we transferred the BrFLC2 and BrFLC3 of Yellow sarson, two longer transcripts of BrFLC1 and BrFLC2 of Bre, and BrFLC1, BrFLC2 and BrFLC3 of Wantai into Arabidopsis ecotype C24 under the control of CaMV 35S promoter (Figure S2). The overexpression of the BrFLC2 in Bre and BrFLC1, BrFLC2 and BrFLC3 in Wantai delayed the flowering time, whereas the overexpression of BrFLC2 and BrFLC3 in Yellow sarson and two longer transcripts of BrFLC1 in Bre failed to change the flowering time. This result revealed that the four BrFLC genes in Yellow sarson and the two transcripts of BrFLC1 in Bre are nonfunctional.
We wondered whether FLC controls the growth cycle in a dose‐dependent manner in planta. If FLC quantitatively controls the growth cycle, then it could be possible to manipulate the growth cycle and to accelerate seed set by silencing FLC using a genetic engineering technique in B. rapa.
Brassica rapa crops vary in vernalization requirements
The repression of FLC on bolting and flowering in B. rapa can be relieved by vernalization. To examine the time and duration of vernalization, we observed the bolting times of 121 varieties of B. rapa using 3‐week‐old seedlings under controlled conditions. The vernalization periods of these varieties varied with cold durations lasting from 0 to 11 weeks after germination. In the absence of the cold treatment, Yellow sarson took 110 days to set seeds, thus being a rapid‐cycling cultivar without a vernalization requirement (Figure 2a,b). In the presence of the cold treatment, the genotype with the shortest cold duration threshold was Bre, while Wantai had the longest threshold. To define cold duration thresholds for vernalization, we incubated the 3‐week‐old plants of Bre and Wantai at 4 °C for various periods and then transferred them to the normal growth temperature (22 °C and 16 h light). A 20‐day cold exposure period was enough to saturate the vernalization requirement of Bre and Bre plants started flowering on the 35th day of the following normal growth temperature period (Figure 2b). In contrast, 80 days of cold exposure were required for flowering in Wantai. The flowering time of Bre and Wantai was also affected by the normal growth temperature (22 °C) periods following the cold exposure. Bre plants that were exposed to 20 days of cold conditions did not flower unless they were grown for at least another 34 days at the normal growth temperature. Wantai plants that were exposed to 80 days of cold conditions required at least another 45 days at 22 °C for bolting. This result suggests that the vernalization duration in B. rapa varied in the two biennial crop types.
Figure 2.
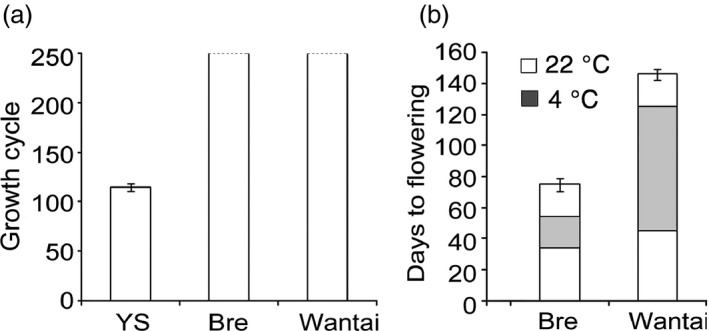
Growth cycles and flowering time of Yellow sarson (YS), Bre and Wantai. (a) Growth cycle of plants in the absence of cold treatment. The dashed lines indicate that the growth is not finished. (b) Flowering time of plants in the presence of cold treatment.
NATs of BrFLC in B. rapa are different from those of Arabidopsis FLC
COOLAIR plays an important role in epigenetic silencing of FLC in Arabidopsis (Swiezewski et al., 2009; Liu et al., 2010; Csorba et al., 2014; Marquardt et al., 2014). To examine NATs of BrFLC genes, Arabidopsis COOLAIR was aligned with the genomic sequence of four BrFLC genes in Bre. An 85‐bp region of BrFLC2 was homologous to a region of COOLAIR (Figure S3). To determine whether there were any NATs in BrFLC2 intergenic regions, we conducted reverse transcription with strand‐specific primers. A primer downstream of the 85‐bp intergenic region generated a transcript upon cold induction (Figure S4). To amplify the possible full‐length NATs, extensive rapid amplification of cDNA ends (RACE)‐PCR was performed using strand‐specific primers. In total, five types of BrFLC2 NATs were isolated (Figure 3a). Among them, three were cleaved and polyadenylated at the sequence antisense to the BrFLC2 DNA located in intron 6, and two were cleaved and polyadenylated within the sequence antisense to the BrFLC2 promoter. All five NATs were transcribed from the same sites in the intergenic regions antisense to DNA located 169 nt downstream of BrFLC2. Compared with the transcriptional start sites of AtFLC NATs, those of BrFLC2 NATs were further away from the transcriptional stop sites of BrFLC2. According to polyadenylation sites, the three NATs polyadenylated within intron 6 were classified as class I NATs, while those polyadenylated within the promoter of BrFLC2 were classified as class II NATs. The five NATs of BrFLC2 were named as BrFLC2as406, BrFLC2as599, BrFLC2as477, BrFLC2as816 and BrFLC2as755 based on their lengths (Figure 3a). There was an overlapping fragment of 330 nt between the BrFLC2 promoter and BrFLC2as816. Although extensive NAT‐specific RT‐PCR was applied, no NATs were isolated from BrFLC1, BrFLC3 or BrFLC5 genes, revealing that the NATs are specific to BrFLC2. As expected, the NATs of BrFLC2 were conserved in crop types Bre, Yellow sarson and Wantai (Table S1).
Figure 3.
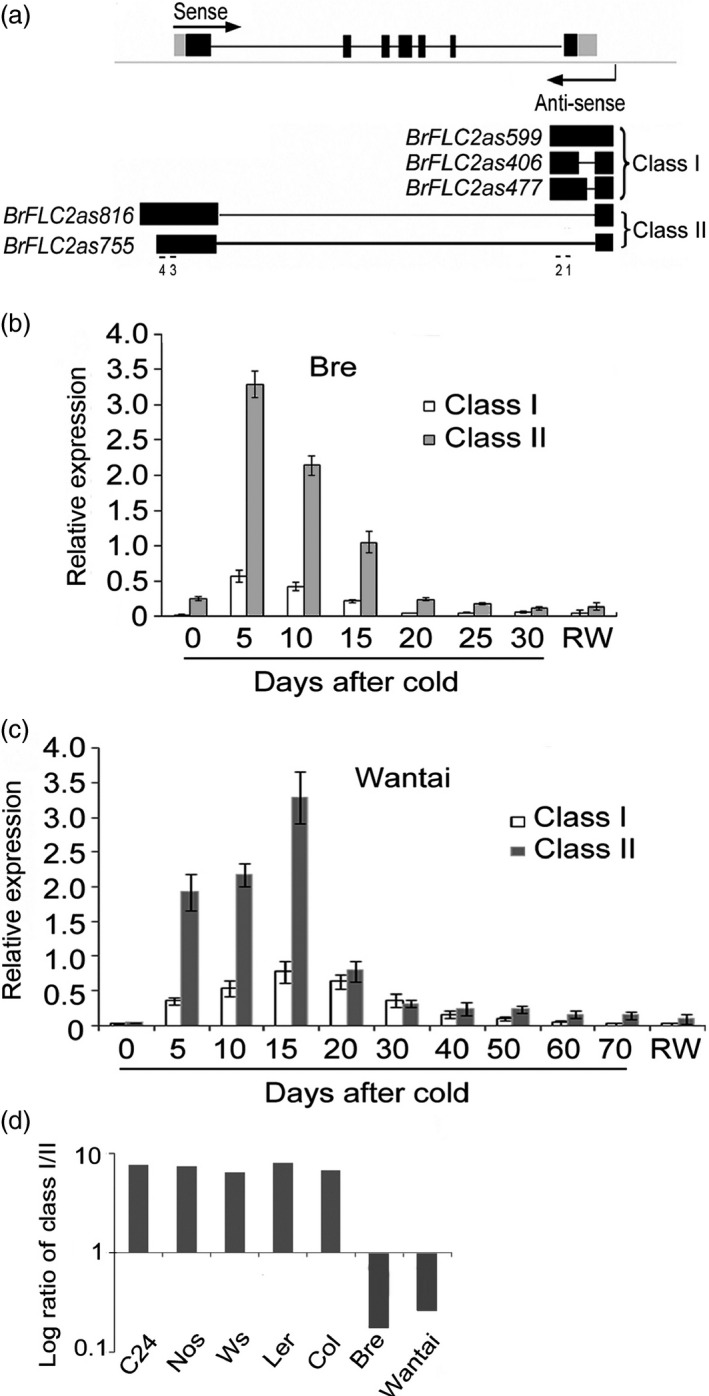
NATs (BrFLC2as) of BrFLC genes. (a) Schematical structures. Arrow heads indicate direction of transcription. Black boxes indicate exons, while grey boxes show the untranslated regions. (b) and (c) Relative expression levels of class I and class II NATs in the seedling leaves of Bre (b) and Wantai (c). (d) the COOLAIR ratios (Log) of class I to class II in Arabidopsis thaliana and B. rapa. Primers 1 and 2 were used to amplify class I NATs, while primers 3 and 4 to amplify class II NATs. RW, return to warm.
In Arabidopsis, a long intronic noncoding RNA, COLDAIR, is required for the vernalization‐mediated epigenetic repression of FLC (Heo and Sung, 2011). We wondered whether there was COLDAIR‐like RNA in the BrFLC genes. The first introns of BrFLC1, BrFLC2, BrFLC3 and BrFLC5 were much shorter than those in AtFLC (3.5 kb) and did not contain any fragments homologous to AtFLC COLDAIR (Figure S5). Using several pairs of strand‐specific primers, we performed tiling RT‐PCR and RACE‐PCR to detect COLDAIR homologs in the first introns of the four BrFLC genes, but failed to identify any COLDAIR transcripts. Moreover, the RNA‐seq data of Bre and Wantai (data not shown) did not show any transcripts derived from these introns. Thus, we excluded the possibility of a COLDAIR homolog existing in the BrFLC genes.
BrFLC genes are down‐regulated, while BrFLC2 NATs are up‐regulated under cold conditions
To define the relationship between BrFLC2 and the BrFLC2as, the abundance of relative transcripts was analysed using real‐time PCR. In Bre seedling leaves without a cold treatment, the transcripts of class I NATs were not detectable, while class II NATs were few (Figure 3b); after cold treatment, the transcript levels of class I and class II NATs sharply increased, reached the peaks at day 5 and then gradually declined. In Wantai, transcript levels of class I and II NATs were not detectable before cold treatment and were induced under cold conditions; however, these NATs under cold conditions were much lower than in Bre and reached the peaks much later at day 15 (Figure 3c). In both crop types, class I BrFLC2as transcript levels were much lower than class II BrFLC2as transcript levels during vernalization, which is opposite in Arabidopsis (Figure 3d). This result indicates that BrFLC2 NATs are cold‐induced in two crop types, but their peaks and durations of expression are different.
In Bre leaves, BrFLC2 expression was strong prior to cold treatment; however, BrFLC3 expression was hardly detected (Figure 4a) and BrFLC5 was not expressed, confirming that BrFLC5 was a pseudogene (data not shown). In Wantai, BrFLC2 expression prior to cold treatment was weaker than in Bre, but BrFLC1 and BrFLC3 expression levels were much stronger than in Bre (Figure 4b). During vernalization, the expression levels of BrFLC1, BrFLC2 and BrFLC3 in Bre and Wantai decreased progressively with time from 5 to 25 days, and the rate of decrease in Wantai was lower than in Bre (Figure 4a,b). This result reveals that the expression patterns of BrFLC1, BrFLC2 and BrFLC3 prior to and during cold treatment are quite different in Bre and Wantai.
Figure 4.
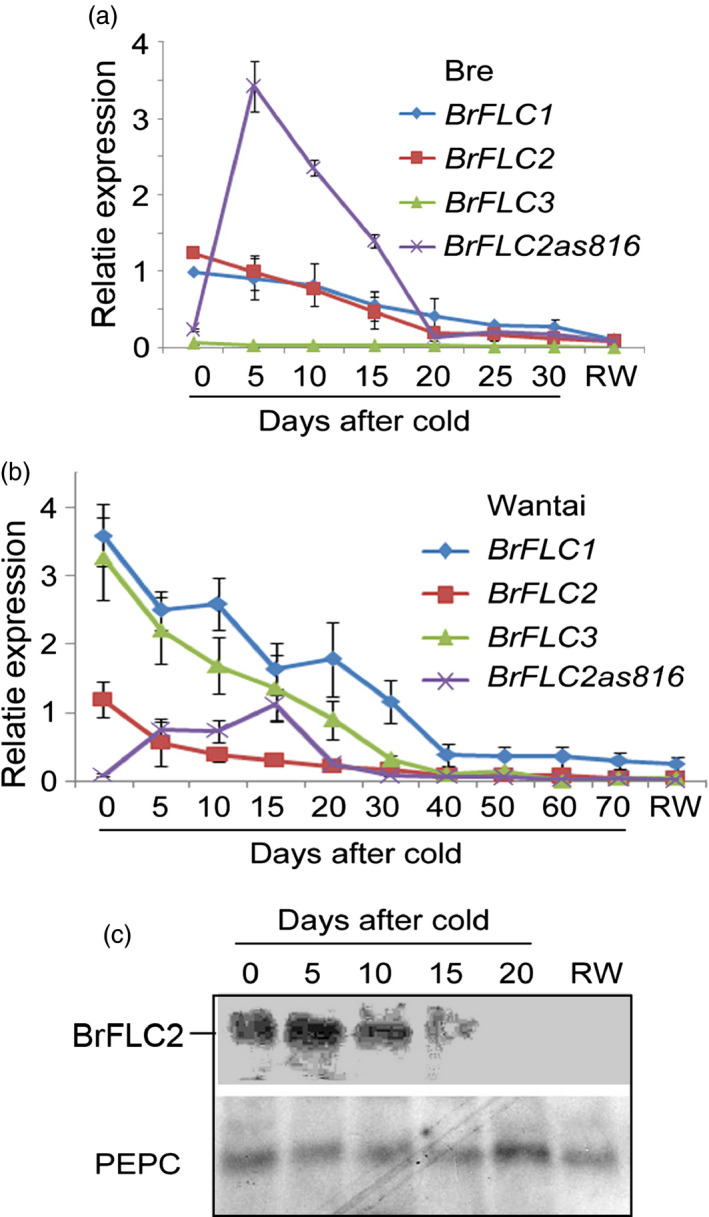
Relative expression and protein contents of BrFLC genes and BrFLC2as816. (a) Relative expression level of BrFLC genes and BrFLC2as816 in the seedling leaves of Bre. (b) Relative expression levels of BrFLC and BrFLC2as816 in the seedling leaves of Wantai. (c) BrFLC2 proteins detected by Western blotting in the seedling leaves of Bre after cold treatment. PEPC is the loading control. RW, return to warm.
We determined the BrFLC proteins in Bre leaves using Western blotting with a BrFLC antibody. During the course of vernalization, the total BrFLC protein content at day 5 after cold treatment was almost the same as before cold treatment, in contrast to the transcripts of BrFLC2, which decreased markedly. Then, the protein content decreased rapidly with time and eventually disappeared at day 20 (Figure 4c), concomitant with a decrease in BrFLC transcripts and an increase in BrFLC2 NATs. This fact indicates that there was a time lapse between the decline in BrFLC proteins and the decrease in BrFLC transcripts at the cold treatment's early stages. After 5 days, however, the production and content of BrFLC proteins in plants of B. rapa under cold conditions were highly dependent on the expression of BrFLC genes. That is, both the transcriptional and translational products of the BrFLC genes are reduced.
Overexpression of BrFLC2 NATs negates the requirement for vernalization in B. rapa
The genomic structure map showed that BrFLC2as816 overlapped the promoter and exon 1 region of BrFLC2 (Figure 3a). To examine whether BrFLC2as changes the growth cycles of B. rapa, we constructed BrFLC2as816 under the control of AA6 (Patent WO 2007/069894) (Figure 5a) and transformed it into Bre plants using the vernalization–infiltration method (Bai et al., 2013). In total, 12 homozygous transgenic lines were screened, named B2as (Figure 5b). Surprisingly, all of the transgenic lines in the growth room bolted prior to cold treatment, while the wild‐type plants did not bolt (Figure 5c; Table 1, Figure S6), indicating the gradient among the transgenic lines at bolting time. Among them, five bolted after 6 weeks of normal growth temperature and were designated as super‐early lines (Figures 5c and S6), with B2as‐1 being a representative line; three bolted after 8 weeks and were designated as early lines, with B2as‐4 being a representative line; and the rest bolted after 9 weeks and were designated as moderate lines, with B2as‐7 being a representative line. Overall, these lines did not require cold conditions for vernalization.
Figure 5.
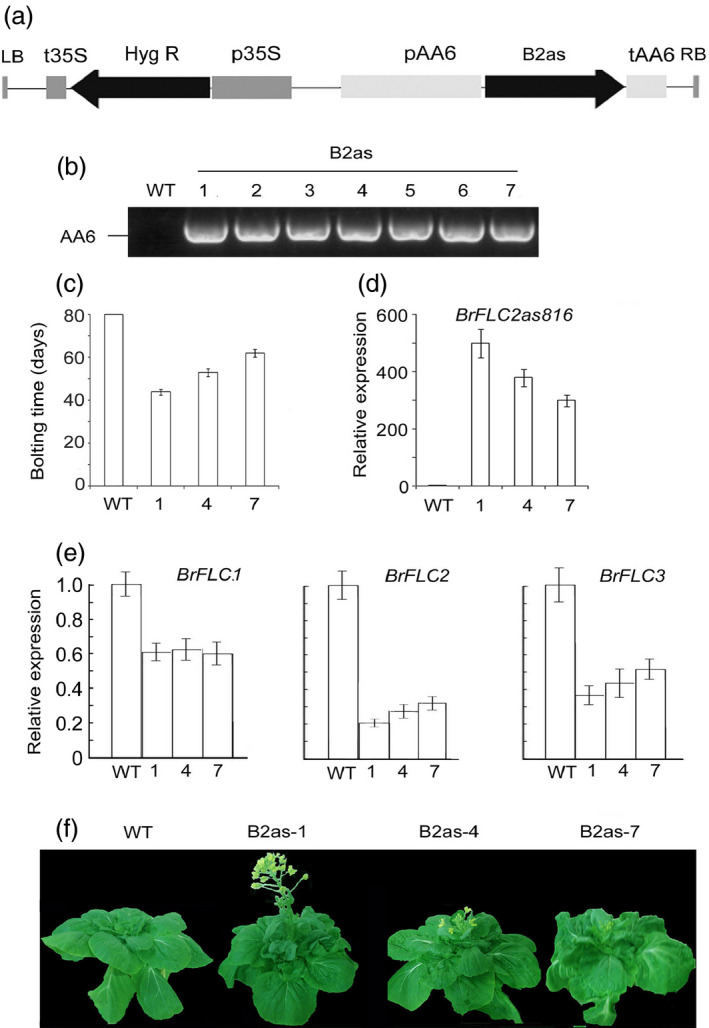
Transgenic Bre plants overexpressing BrFLC2as816 grown in growth room. (a) Schematical structures of binary vectors of BrFLC2as816. (b) PCR of AA6 promoter for identification of the transgenic lines by PCR using the primers spanning AA6 promoter and BrFLC2as816 fragment. (c) Days to bolting of BrFLC2as816‐transgenic plants after germination. Error bars indicate the standard deviation. (d) and (e) Real‐time PCR showing expression of BrFLC2as816 and BrFLC genes in BrFLC2as816‐transgenic plants. (f) Plants of the transgenic lines B2as in growth room. p35S, CaMV 35S promoter; Hyg R, hygromycin‐resistant gene; pAA, AA6 promoter; LB, left border of T‐DNA; t35S, CaMV35S terminator; tAA6, AA6 terminator.
Table 1.
Flowering time and growth cycles of transgenic lines overexpressing BrFLC2as816. In growth room, the seeds were sown in plastic pots and grown at 22 °C in SIPPE Phytotron. For each parameter, more than 20 plants were measured. Means ± standard error. DAG, days after germination; NA, not available; NF, not flowering; S, super‐early line; E, early line; M, moderate line
| Lines | Bolting (DAG) | Flowering (DAG) | Seed ripening (DAG) | Siliques per plant | Seed yield per plant (gram) | Growth cycle |
|---|---|---|---|---|---|---|
| Bre | NA | NA | NA | NA | NA | NF |
| B2as‐1 | 44 ± 1.3 | 51 ± 1.1 | 103 ± 2.6 | 189 ± 5.2 | 39.81 ± 4.35 | S |
| B2as‐3 | 47 ± 2.0 | 55 ± 2.1 | 105 ± 4.1 | 190 ± 6.7 | 43.08 ± 3.69 | S |
| B2as‐8 | 45 ± 2.2 | 54 ± 1.9 | 105 ± 2.9 | 200 ± 6.1 | 44.81 ± 4.65 | S |
| B2as‐9 | 42 ± 2.3 | 51 ± 1.5 | 103 ± 1.5 | 187 ± 7.7 | 43.08 ± 3.92 | S |
| B2as‐12 | 46 ± 1.3 | 55 ± 1.4 | 106 ± 2.1 | 197 ± 5.1 | 45.61 ± 4.63 | S |
| B2as‐4 | 55 ± 1.9 | 64 ± 1.8 | 115 ± 2.3 | 215 ± 8.1 | 49.63 ± 6.21 | E |
| B2as‐5 | 58 ± 1.2 | 69 ± 1.7 | 118 ± 1.6 | 211 ± 7.2 | 48.47 ± 5.92 | E |
| B2as‐11 | 58 ± 1.6 | 70 ± 1.8 | 118 ± 1.4 | 219 ± 5.9 | 49.01 ± 4.55 | E |
| B2as‐2 | 65 ± 1.9 | 76 ± 2.3 | 125 ± 2.1 | 248 ± 6.7 | 57.09 ± 4.96 | M |
| B2as‐6 | 69 ± 2.3 | 78 ± 1.0 | 129 ± 3.2 | 252 ± 8.2 | 55.33 ± 5.86 | M |
| B2as‐7 | 70 ± 1.4 | 80 ± 0.9 | 130 ± 2.8 | 260 ± 4.1 | 59.91 ± 4.01 | M |
| B2as‐10 | 66 ± 2.1 | 77 ± 1.2 | 130 ± 2.7 | 253 ± 6.9 | 59.19 ± 4.97 | M |
To verify the transgenic lines, we performed PCR to detect the AA6 promoter in the transformed plants of the T3 generation and real‐time PCR to quantify the transcripts of BrFLC2as816 (Figures 5b,d, and S7). BrFLC2as816 was overexpressed in the leaves of B2as‐1, B2as‐4 and B2as‐7 plants, whereas expression levels of BrFLC2, BrFLC1 and BrFLC3 decreased markedly when compared with their levels in the wild type (Figure 5e), indicating that the BrFLC genes are down‐regulated by BrFLC2as816. Among the down‐regulated BrFLC genes, BrFLC2 was the most down‐regulated. Among the three types of transgenic lines, the super‐early lines had the lowest levels of BrFLC2 on average and the moderate ones had the highest levels. We hypothesize that the early bolting of the transgenic plants is negatively correlated with the expression of BrFLC2.
Normally, plant growth starts from seed germination and finishes with seed ripening. The length of a plant's growth cycle should be designated as the number of days from seed germination to seed ripening under ideal conditions. In the growth room under ideal growth conditions, Bre plants failed to flower and thus did not produce any seed. By contrast, super‐early line B2as‐1, early line B2as‐4 and moderate line B2as‐7 set seeds at days 103, 115 and 125, respectively, indicating a gradient in time of seed ripening. This result indicates that the transgenic lines overexpressing BrFLC2 NATs are able to finish the growth cycles under ideal growth conditions without a period of cold and to provide the crop types with varying growth cycles.
To understand how BrFLC NATs affect other biological and economic traits, we examined the numbers of branches and siliques on the transgenic lines overexpressing BrFLC2as816. In the growth room, the number of rosette leaves, as well as branches and flowers in the super‐early line, was much fewer than those of the moderate lines (Table S2). Among the three types of the transgenic lines, the super‐early lines ripened earliest and the moderate lines ripened last. For seed yield per plant, the super‐early lines were the lowest and the moderate lines were the highest (Table 1). These results revealed that the seed yield is different between the transgenic lines.
When the wild‐type Bre plants were grown in the field in 2011, they underwent vegetative growth during the autumn, flowered normally and set seeds in the following spring (Figure S8a,c,e). Under these conditions, the super‐early lines produced fewer rosette leaves and branches than both the Bre and the moderate lines, and flowered and set seeds during the same autumn (Table 2; Figure S8b,g,h). Their life cycles were finished before winter and thus became annual lines. However, the early lines flowered but stopped development after the commitment to the winter condition (Figure S8d,i,j), and they did not produce any seed. The moderate lines finished vegetative growth and bolted before winter (Figure S8f,i,j). Like Bre plants, the moderate lines flowered and set seeds during the following spring. Interestingly, they generated almost the same numbers of branches. Apparently, flowering and seed setting in the transgenic lines in the field were different from those in growth room. During 2012, super‐early, early and moderate lines flowered approximately at the same time (data not shown).
Table 2.
Flowering time and growing cycles of the transgenic lines overexpressing BrFLC2as816 in the field on 2011. The seedlings were transferred to the field in SIPPE Farm Station, Shanghai, China, on 9 September 2011 and grown under cultivation condition. For each parameter, more than 20 plants were measured. Means ± standard error. DAG, days after germination; NA, not available
| Lines | Bolting (DAG) | Flowering (DAG) | Seed setting (DAG) | Growing cycle |
|---|---|---|---|---|
| Bre | 123 ± 3.9 | 190 ± 3.6 | 242 ± 1.3 | Normal |
| B2as‐1 | 43 ± 1.1 | 53 ± 1.6 | 102 ± 2.0 | Short |
| B2as‐3 | 45 ± 0.9 | 53 ± 1.9 | 100 ± 1.2 | Short |
| B2as‐8 | 43 ± 2.1 | 55 ± 2.1 | 105 ± 2.1 | Short |
| B2as‐9 | 40 ± 1.2 | 52 ± 1.5 | 104 ± 1.2 | Short |
| B2as‐12 | 45 ± 1.7 | 54 ± 2.3 | 102 ± 0.9 | Short |
| B2as‐4 | 52 ± 0.7 | 68 ± 3.1 | No seed | NA |
| B2as‐5 | 54 ± 1.2 | 70 ± 1.8 | No seed | NA |
| B2as‐11 | 56 ± 1.9 | 71 ± 1.6 | No seed | NA |
| B2as‐2 | 64 ± 1.9 | 189 ± 3.2 | 230 ± 1.6 | Normal |
| B2as‐6 | 67 ± 3.2 | 193 ± 2.7 | 235 ± 1.4 | Normal |
| B2as‐7 | 68 ± 2.0 | 196 ± 3.5 | 240 ± 1.2 | Normal |
| B2as‐10 | 65 ± 1.3 | 192 ± 2.0 | 239 ± 1.5 | Normal |
BrFLC2 NATs shorten the growth cycle of Arabidopsis by repressing BrFLC genes
To demonstrate how BrFLC2as represses BrFLC genes to shorten growth cycles, we cloned three genomic fragments of BrFLC2/BrFLC2as to construct binary vectors (Figure 6a). First, a BrFLC2 genomic fragment was transferred into Arabidopsis ecotype Col, in which the expression of AtFLC was repressed owing to the interference of the fri allele (Lee and Amasino, 1995; Johanson et al., 2000). If BrFLC2 and the BrFLC2 NATs were expressed in the transgenic plants, it meant that the fri background could not block exogenous BrFLC2 expression. Thus, we were able to investigate the roles of NATs in the activities of BrFLC genes. Each line of the transgenic plants was divided into two populations: one population was grown with a 3‐week cold treatment and the other was grown without a cold treatment. The first genomic fragment, BrFLC2/BrFLC2as (2/2as), spanned the complete genomic sequence of BrFLC2, with a native promoter in the sense strand, and the DNA sequence of BrFLC2as, with a promoter in the opposite strand (Figure 6a). The resultant transgenic 2/2as plants flowered at the same time as wild‐type plants that underwent a cold treatment (3 weeks of cold exposure). However, the plants flowered later than the wild‐type plants that did not undergo a cold treatment, meaning that the cold treatment accelerated the bolting of the 2/2as plants. In the seedling leaves of 2/2as plants that were not subjected to a cold treatment, the BrFLC2as' expression levels were extremely low, but BrFLC2 was expressed. In the seedling leaves that underwent a cold treatment, BrFLC2as were expressed, but the expression level on day 20 was much higher than on day 10. During the same period, BrFLC2 expression on day 20 was lower than on day 10 (Figure 6c). This revealed that the B. rapa BrFLC2as were cold‐induced in Arabidopsis, while BrFLC2 expression was inhibited.
Figure 6.
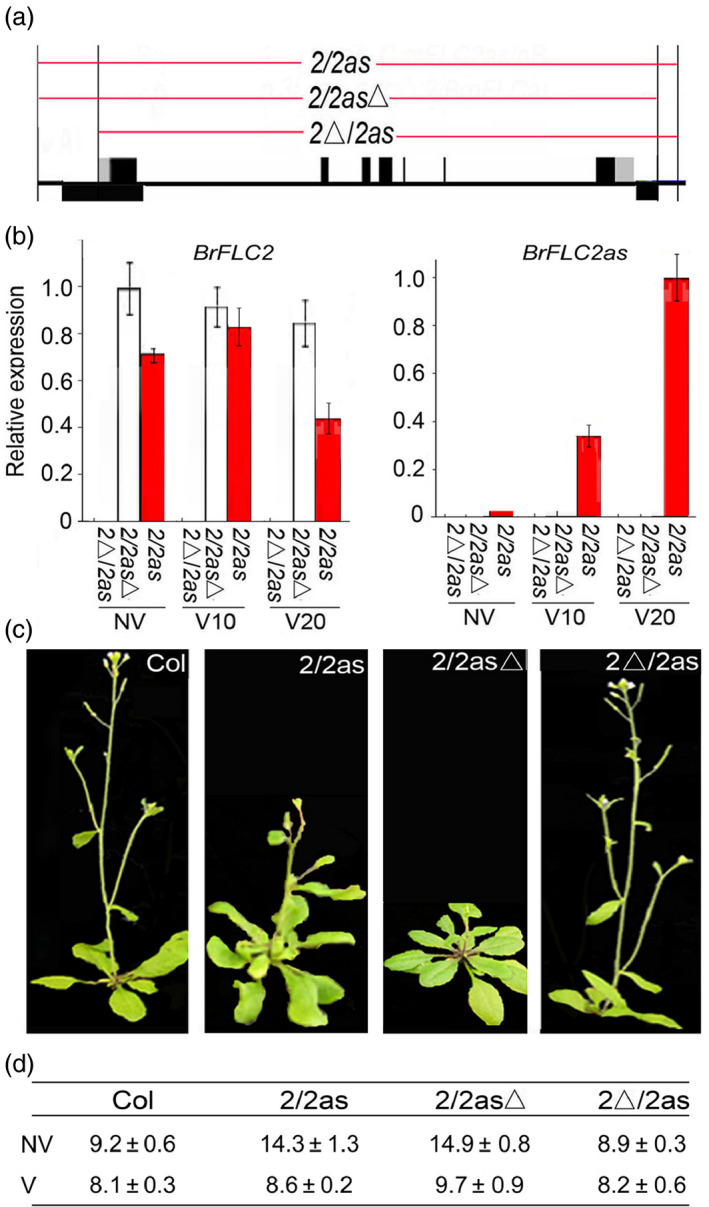
The transgenic plants of Arabidopsis (Col) overexpressing BrFLC2as816. (a) Diagrams of gene constructs of 2/2as, 2/2asΔ and 2Δ/2as. (b) Real‐time PCR showing expression of BrFLC2 and BrFLC2as in the leaves of the transgenic Arabidopsis plants. NV, nonvernalization; V10–V20 indicate vernalization for different days. Arrows indicate the direction of transcription. (c) Phenotype of transgenic plants without cold treatment. (d) The flowering time in terms of number of rosette leaves was given. 2, BrFLC2; 2as, BrFLC2as; Δ, deletion of native promoter. Flowering time is the week(s) after germination.
To verify whether BrFLC2 was repressed by BrFLC2as, we chose the second genomic fragment BrFLC2/BrFLC2asΔ (2/2asΔ) that spanned BrFLC2 with a native promoter on the sense strand and BrFLC2as without a native promoter on the opposite strand (Figure 6a). The 2/2asΔ transgenic plants flowered later than the wild type (Figure 6d). Under both conditions, BrFLC2 expression levels were at almost the same levels, while BrFLC2as expression levels were undetectable (Figure 6c). This implied that BrFLC2 expression was not inhibited when the BrFLC2as transcripts were absent.
The third genomic fragment, BrFLC2Δ/BrFLC2as (2Δ/2as), spanned BrFLC2 without a promoter on the sense strand and BrFLC2as with a promoter on the opposite strand (Figure 6a). The 2Δ/2as transgenic plants flowered at the same time as the wild type.
Three BrFLC2/BrFLC2as constructs affect BrFLC2 expression and flowering time differently, with and without the native promoter. The BrFLC2as act to repress BrFLC2 and thereby to control flowering time and growth cycle. These results show that BrFLC2 NATs shorten the growth cycle of Arabidopsis by repressing BrFLC genes.
Discussion
In Arabidopsis, FLC acts as an important flowering repressor and transcription factor responding to vernalization. Recently, long noncoding RNAs from the FLC locus, such as COOLAIR and COLDAIR, have been identified and isolated in Arabidopsis (Swiezewski et al., 2009; Heo and Sung, 2011). We characterized a subset of the noncoding NATs of BrFLC2 in three crop types of B. rapa, which had different vernalization requirements. The question arises whether BrFLC2 NATs have the same roles in the three crop types. In the rapid‐cycling crop type Yellow sarson, BrFLC2 NATs did not affect the growth cycle because none of the BrFLC genes functioned in vernalization due to mutations in their genomic sequences. In the slow‐cycling crop type Wantai, BrFLC2 NAT expression levels are lower and declined more slowly during cold treatment compared with NATs in the medium‐cycling crop type Bre. However, BrFLC2 in the three crop types have NATs, while the other BrFLC genes do not. This means that the generation of the noncoding NATs is BrFLC2 specific. These findings imply that the BrFLC NATs in the mesohexaploid B. rapa are more functionally complex than those in Arabidopsis (Wang et al., 2011b). Nevertheless, it would be interesting to understand why BrFLC1 and BrFLC3 do not have any NATs even though they are highly homologous with BrFLC2.
In Arabidopsis, COLDAIR, derived from the first intron of FLC, physically interacts with the polycomb repressive complex, indicating its role in establishing and maintaining stable repressive chromatin at FLC (Heo and Sung, 2011). However, COLDAIR is not found in the first intron of BrFLC genes in B. rapa. This fact implies that the molecular mechanism underlying vernalization in B. rapa is different from that of Arabidopsis.
Generally, there are two phases in the vernalization response, the initial down‐regulation of FLC by transcriptional repression and the subsequent phase of maintaining the repressed state (Sheldon et al., 2002). Several studies indicate that the methylation of H3K37me3 inactivates the chromatin and blocks FLC expression in response to the cold (Sheldon et al., 2002; Heo and Sung, 2011). COOLAIR plays a role in the epigenetic silencing of FLC expression during vernalization (Crevillen et al., 2014; Csorba et al., 2014; Marquardt et al., 2014). We found that BrFLC NATs in B. rapa are induced by a cold treatment and highly expressed at the early stage of vernalization, concurrent with a decrease in the expression of the BrFLC genes. The transgenic lines overexpressing BrFLC2as816 are able to flower without vernalization. When exogenous BrFLC2as816 is expressed under the control of its own promoter in Arabidopsis, it inactivates the BrFLC2 gene. These results suggest that BrFLC NATs suppress the activity of the BrFLC2 gene in response to cold and accelerate vernalization. We could not estimate how much the NATs suppress the activities of BrFLC1 and BrFLC3. However, we are certain that BrFLC2 NATs contribute to the repression of all of the BrFLC genes in B. rapa. The BrFLC2 fragments that are reversely complemented to the NATs show high homology (>95%) with BrFLC1 and BrFLC3. It is reasonable to deduce that BrFLC2 NAT suppresses BrFLC1 and BrFLC3 as well. The fact is that BrFLC2as816 overexpression inhibits the expression of BrFLC1 and BrFLC3 in transgenic plants.
In Arabidopsis, the class I NATs of COOLAIR accumulate to a higher degree than class II NATs, which is consistent with proximal polyadenylation correlating with reduced levels of FLC transcription (Csorba et al., 2014). Further, a core spliceosome mutant, prp8‐6, was isolated and identified for its function in disturbing COOLAIR processing and reducing the proximal polyadenylation usage coupled with localized histone methylation reprogramming. In B. rapa, class II NATs accumulate to a higher degree than class I NATs and may be more important in vernalization. Considering the close evolutionary relationship between Arabidopsis and B. rapa, we assume that the class I NATs in B. rapa engage in the same pathway to silence BrFLC genes in localized histone methylation reprogramming as in Arabidopsis. BrFLC2as816 belongs to the class II NATs, and its overexpression suppresses BrFLC genes in B. rapa and Arabidopsis. This reveals that long class II NATs play a role in the silencing of BrFLC genes using a post‐transcriptional pathway. It remains unknown whether short class I NATs function in the repression of BrFLC genes. Anyway, class II NATs should contribute more to BrFLC repression than class I NATs because the former accumulates to a much higher degree than the latter in B. rapa. For further understanding of the function and interaction between BrFLC genes and the associated NATs, we would benefit from analysis of F2, RIL or other segregating populations derived from hybrid crosses between the different crop types.
The growth cycle of seed plants begins with seed germination and culminates with seed ripening. In a growth room where the environment is optimized for the development of B. rapa crops, the transgenic lines overexpressing BrFLC2as816 showed gradients in their growth cycles, which were closely associated with the expression levels of the BrFLC genes. Among the three types of transgenic lines, the super‐early lines had the shortest growth cycles and the moderate lines had the longest ones. This suggests that BrFLC2 NATs are involved in determining the growth cycles of B. rapa crops. Through the control of BrFLC NATs, the growth cycle of B. rapa could be adapted to different climates and regions. In this regard, the super‐early lines are useful for the genetic manipulation of crop early flowering. In particular, they are valuable for those regions where the annual oilseed crops are not cultivated.
In the field, a long winter season interrupts the growth of biennial crops. The super‐early lines finished the growth cycle before the winter season, and thus, their yields were not affected by the cold weather. The early lines flowered but failed to set seeds before winter, mainly because the reproductive organs were damaged by the cold weather. The moderate lines bolted but did not flower before winter. These lines underwent the winter season and set seeds in the same fashion as the wild type. For the yield in the field, the moderate lines need be compared with the wild type requiring vernalization. These additional data would be both desirable and of interest, in terms of better defining the roles of BrFLC genes in Brassica genus.
Thus, the three types of the transgenic lines showed the different growing cycles in the field. Growing cycles are different from growth cycles because they represent life cycles under growing conditions in the field and, as such, are greatly affected by climate and other factors in certain regions.
Both earliness of flowering and high yield are two important traits for oilseed crops. In growth rooms where the wild‐type plants are not able to flower, the moderate lines generate more branches and seeds than the other transgenic lines. In the field, they produce the same numbers of branches than the wild type. The potential for seed yield in these lines is greater than the other transgenic lines. If earliness is the priority, then the super‐early lines are the best choice. If earliness and high yield are equally important, then the moderate lines could be selected. Nevertheless, further optimization and modification of BrFLC NATs are possible for the genetic manipulation of crop flowering time and yield.
In B. rapa, NATs of BrFLC2 show extremely strong gene silencing activities because they eliminate vernalization traits from this biennial crop species by inhibiting BrFLC genes. The conversion of a biennial to an annual crop type is important for oilseed crops of B. rapa, and growth cycle gradients are useful for selecting the proper crops for growing seasons in various regions. Specifically, our transgenic lines overexpressing BrFLC2as816 are valuable because they become annual and display growth cycle gradients. In cold and chilled regions, we can grow and harvest the seeds in 1 year. In warm areas, we could use early or moderate lines for high yields, because they have many branches and a higher potential yield. As long as the high yield vigour of biennial oilseed crops is combined with rapid‐cycling traits, the productivity of B. rapa could be improved. In this way, new cultivars could be created having certain levels of early flowering using high yield cultivars. More importantly, technology that creates crops that do not require vernalization could be applied to other oilseed and grain crops, such as oilseed rape and wheat.
Materials and methods
Plant materials
The crop types of B. rapa used in this study include Yellow sarson (YS‐143) (B. rapa var. trilocularis, rapid‐cycling crop type), Bre (B. rapa ssp. pekinensis var. bre, medium‐cycling crop type) and Wantai (B. rapa ssp. pekinensis var. wantai, rapid‐cycling crop type). The seeds of the inbred lines of these crop types were germinated on moisture‐absorbent papers in a plant growth chamber at 22 °C for 3 days and then transplanted to peat soil in plastic pots and moved into a growth room of SIPPE Phytotron in Shanghai. In this growth room, the plants were grown at 22 °C with 16 h of light per day under a light source of warm white fluorescent tubes (colour code 990), an irradiance of 150 μmol/m2/s and a light intensity on the plant canopy of 75 μmol/m2/s. The relative humidity was 65%–70%, and the air velocity was approximately 0.9 m/s. All of the seedlings were grouped randomly and grown under identical conditions for 3 weeks. For B. rapa vernalization, 3‐week‐old plants with three leaves were transferred to 4 °C for different time periods ranging from 20 to 80 days with an interval of 5 days. Afterwards, the seedling was returned to normal growth temperature conditions for flowering time statistics. For cultivation in the field, the 4‐week‐old seedlings were acclimated for 3 days and transferred to the field in SIPPE Farm Station on 24th August 2011 and 9th September 2012.
The seeds of Arabidopsis and transgenic lines were surface‐sterilized in 70% ethanol for 1 min, followed by 20% bleaching water for 10 min, and then washed four times in sterile distilled water and plated on top of solid Murashige and Skoog medium and incubated at 22 °C for 7 days. For Arabidopsis vernalization, a 7‐day‐old plant was transferred to 4 °C for different time periods. Afterwards, the seedling was returned to normal growth temperature conditions for flowering time statistics.
Analysis of flowering time
The appearance of the first floral buds, the opening of the first flowers and the ripening of the first seeds were designated as bolting time, flowering time and ripening time, respectively. The days of bolting time, flowering time and ripening time were calculated from seed germination. More than 20 plants were measured for each cold treatment and each transgenic line. For the plants in the field, the number of rosette leaves was counted at flowering stage. In Arabidopsis, number of the rosette leaves was recorded at flowering stage, according to the method of Li et al. (2012). More than three independent transgenic lines were observed. More than 30 plants were measured for each transgenic line.
RNA extraction and real‐time PCR
Total RNA was extracted using the TRIZOL reagent (Invitrogen, Carlsbad, CA) as directed by the manufacturer. Real‐time PCR was performed as described (Wang et al., 2011a,b; Yu et al., 2012) with primers listed in Table S3. BrActin (accession code, Bra022356) was used as the internal control gene. For gene expression data analysis, we calculated differences between the C t (Cycle Threshold) values for experimental and reference genes (BrActin) as ΔΔC t and graphed the fold change of each RNA to the calibrator sample.
Rapid amplification of cDNA ends
Sequence information for BrFLC genes was retrieved from http://brassicadb.org/brad/. The RNA samples were isolated from Bre leaves using RACE ready cDNA (Takara, Tokyo, Japan), and the 5′ and 3′ ends of BrFLC2 NATs were amplified by RACE‐PCR using gene specific primers. One hundred picomoles of 5′ adaptor (rGrUrUrCrArGrArGrUrUrCrUrArCrArGrUrCrCrGrArCrGrArUrC) was directly ligated with 5 μg of total RNA. After ligation, the first‐strand cDNAs were synthesized using Superscript III Reverse Transcriptase (Invitrogen, CA) and a specific primer. The cDNA was treated with RNase H to remove the RNA strand and amplified for two rounds using two sets of primers. The 3′ and 5′ PCR products were excised from the gel and cloned into the pMD18T vector (Takara). Twenty positive colonies were sequenced from each RNA sample.
Protein analysis
Total protein samples were isolated from the leaves upon cold treatment of plants. Western blotting analyses were performed to detect BrFLC proteins. Twenty micrograms of total protein were separated on 12% denaturing polyacrylamide gels by electrophoresis and transferred to HyBond‐N+ membrane (Amersham, Buckinghamshire, UK) by electroblotting. Anti‐FLC antibody and anti‐PEPC antibody were used to hybridization.
Construction of binary vectors
The cDNA of BrFLC2as816 was isolated from Bre seedlings by PCR using KOD TOYOBO DNA polymerase (Toyobo, Osaka, Japan) and inserted into pCAMBIA1300 binary vectors (http://www.cambia.org) under the control of the AA6 promoter. The fragments of BrFLC2 genomic DNA were isolated from Bre seedlings and cloned into pCAMBIA1301 binary vectors (Table S3). The binary constructs were delivered into Agrobacterium tumefaciens strain GV3101 using a freeze–thaw method (Weigel and Glazebrook, 2006).
Genetic transformation
The Bre plants were transformed using vernalization–infiltration method (Bai et al., 2013). This method is based on vacuum infiltration method (Bechtold and Pelletier, 1998). Briefly, a pot of germinated seeds was put in growth room and incubated at 4 °C for 25 days of vernalization treatment. The seedlings in the pots were transferred to growth room at 22 °C and grew for 2 weeks. Then, the plants with small flower buds at the early bolting stage were placed upside down in vacuum desiccator that contained infiltration medium and the engineered Agrobacterium for vacuum infiltration. The Agrobacterium‐infected plants were transferred to the growth room. The seeds were harvested and screened by germination on agar medium containing hygromycin. The hygromycin‐resistant seedlings were transplanted in growth room. T2 and T3 seedlings were identified for the insertion of exogenous genes and analysed for the segregation of population.
Generation of Arabidopsis thaliana Columbia was transformed with the BrFLC genomic constructs. The inflorescences were dipped in the Agrobacterium solution containing sucrose and Silwet‐77 for 1 min as described (Clough and Bent, 1998). The infected plants were cultured for 2 days in the dark and then moved in greenhouse for seed production.
Funding
This work was supported by grants from the National Basic Research Program (‘973’ Program) of China (Grant No. 2012CB113903), National High Technology Research and Development Program (‘863’ Program) of China (Grant No. 2012AA100104) and Natural Science Foundation of China (Grant No. 31200235).
Author contributions
Y.H. designed the research. X.L. and S.Z. performed the research and analysed the data. J.B. contributed to genetic transformation of Chinese cabbage. X.L. and Y.H. wrote the article.
Supporting information
Figure S1 Alignment of putative amino acid sequences of BrFLC genes between Yellow sarson, Bre and Wantai.
Figure S2 Bolting time of the transgenic Arabidopsis lines overexpressing BrFLC genes from the crop types Yellow sarson, Bre and Wantai.
Figure S3 Homologous fragments between BrFLC2as816 and one NAT of AtFLC.
Figure S4 RT‐PCR showing BrFLC2 NATs in B. rapa.
Figure S5 Diagrammatic representation of alignment of the intron 1 fragments of BrFLC genes with that of AtFLC.
Figure S6 Bolting time of Bre plants overexpressing BrFLC2as816.
Figure S7 Real‐time PCR showing the expression of BrFLC2 in Bre plants overexpressing BrFLC2as816.
Figure S8 Bre plants overexpressing BrFLC2as816 at different stages in the field.
Table S1 Sequences of BrFLC2 antisense transcripts.
Table S2 Growth parameters of BrFLC2as816‐transgenic Bre plants in growth room and field.
Table S3 The primers used in this study.
Acknowledgements
Thank Dr. Xiaowu Wang, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, for sending the seed sample of Yellow sarson.
References
- Bai, J. , Wu, F. , Mao, Y. and He, Y. (2013) In planta transformation of Brassica rapa and B. napus via vernalization–infiltration methods. Protoc. Exch., doi: 10.1038/protex.2013.067. [DOI] [Google Scholar]
- Bechtold, N. and Pelletier, G. (1998) In planta Agrobacterium‐mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A.P. , Tang, H. , Wang, X. , Chiquet, J. , Belcram, H. , Tong, C. , Samans, B. , Corréa, M. , Da Silva, C. , Just, J. , Falentin, C. , Koh, C.S. , Le Clainche, I. , Bernard, M. , Bento, P. , Noel, B. , Labadie, K. , Alberti, A. , Charles, M. , Arnaud, D. , Guo, H. , Daviaud, C. , Alamery, S. , Jabbari, K. , Zhao, M. , Edger, P.P. , Chelaifa, H. , Tack, D. , Lassalle, G. , Mestiri, I. , Schnel, N. , Le Paslier, M.C. , Fan, G. , Renault, V. , Bayer, P.E. , Golicz, A.A. , Manoli, S. , Lee, T.H. , Thi, V.H.D. , Chalabi, S. , Hu, Q. , Fan, C. , Tollenaere, R. , Lu, Y. , Battail, C. , Shen, J. , Sidebottom, C.H.D. , Wang, X. , Canaguier, A. , Chauveau, A. , Bérard, A. , Deniot, G. , Guan, M. , Liu, Z. , Sun, F. , Lim, Y.P. , Lyons, E. , Town, C.D. , Bancroft, I. , Wang, X. , Meng, J. , Ma, J. , Pires, J.C. , King, G.J. , Brunel, D. , Delourme, R. , Renard, M. , Aury, J.M. , Adams, K.L. , Batley, J. , Snowdon, R.J. , Tost, J. , Edwards, D. , Zhou, Y. , Hua, W. , Sharpe, A.G. , Paterson, A.H. , Guan, C. and Wincker, P. (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Crevillen, P. , Yang, H.C. , Cui, X. , Greeff, C. , Trick, M. , Qiu, Q. , Cao, X.F. and Dean, C. (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature, 515, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba, T. , Questa, J.I. , Sun, Q. and Dean, C. (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA, 111, 16160–16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A. , Wood, C.C. , Robertson, M. , Peacock, W. and Dennis, E.S. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high‐molecular‐weight protein complex. Plant J. 46, 183–192. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A. , Robertson, M. , Finnegan, E.J. , Buzas, D.M. and Dennis, E.S. (2011) Vernalization‐repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One, 6, e21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, J.B. and Sung, S. (2011) Vernalization‐mediated epigenetic silencing by a long intronic noncoding RNA. Science, 331, 76–79. [DOI] [PubMed] [Google Scholar]
- Hornyik, C. , Terzi, L.C. and Simpson, G.G. (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell, 18, 203–213. [DOI] [PubMed] [Google Scholar]
- Johanson, U. , West, J. , Lister, C. , Michaels, S. , Amasino, R. and Dean, C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kim, J.S. , Chung, T.Y. , King, G.J. , Jin, M. , Yang, T.J. , Jin, Y.M. , Kim, H.I. and Park, B.S. (2006) A sequence‐tagged linkage map of Brassica rapa . Genetics, 174, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y. , Park, B.S. , Kwon, S.J. , Kim, J. , Lim, M.H. , Park, Y.D. , Kim, D.Y. , Suh, S.C. , Jin, Y.M. , Ahn, J.H. and Lee, Y.H. (2007) Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L.: ssp. pekinensis). Plant Cell Rep. 26, 327–336. [DOI] [PubMed] [Google Scholar]
- Lee, I. and Amasino, R.M. (1995) Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 108, 157–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Yang, X. , Wu, F. and He, Y. (2012) HYL1 controls the miR156‐mediated juvenile phase of vegetative growth. J. Exp. Bot. 63, 2787–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.I. , Wang, J.G. , Poon, S.Y. , Su, C.L. , Wang, S.S. and Chiou, T.J. (2005) Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis . Plant Physiol. 137, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Marquardt, S. , Lister, C. , Swiezewski, S. and Dean, C. (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science, 327, 94–97. [DOI] [PubMed] [Google Scholar]
- Mao, F. , Wu, F. , Yu, X. , Bai, J. , Zhong, W. and He, Y. (2014) microRNA319a‐targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt, S. , Raitskin, O. , Wu, Z. , Liu, F. , Sun, Q. and Dean, C. (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol. Cell, 54, 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D. and Amasino, R.M. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell, 11, 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaharu, U. (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 7, 389–452. [Google Scholar]
- Osborn, T.C. , Kole, C. , Parkin, I.A. , Sharpe, A.G. , Kuiper, M. , Lydiate, D.J. and Trick, M. (1997) Comparison of flowering time genes in Brassica rapa, B. napus and Arabidopsis thaliana . Genetics, 146, 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C. , Rouse, D.T. , Finnegan, E.J. , Peacock, W.J. and Dennis, E.S. (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA, 97, 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C. , Conn, A.B. , Dennis, E.S. and Peacock, W.J. (2002) Different regulatory regions are required for the vernalization‐induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell, 14, 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski, S. , Crevillen, P. , Liu, F. , Ecker, J.R. , Jerzmanowski, A. and Dean, C. (2007) Small RNA‐mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc. Natl. Acad. Sci. USA, 104, 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski, S. , Liu, F. , Magusin, A. and Dean, C. (2009) Cold‐induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature, 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Yu, X. , Wang, H. , Lu, Y. , Ruiter, M.D. , Prins, M. and He, Y. (2011a) A novel class of heat‐responsive small RNAs derived from the chloroplast genome of Chinese cabbage (Brassica rapa). BMC Genom. 12, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.W. , Wang, H.Z. , Wang, J. , Sun, R.F. , Wu, J. , Liu, S.Y. , Bai, Y.Q. , Mun, J.H. , Bancroft, I. , Cheng, F. , Huang, S. , Li, X. , Hua, W. , Wang, J. , Wang, X. , Freeling, M. , Pires, J.C. , Paterson, A.H. , Chalhoub, B. , Wang, B. , Hayward, A. , Sharpe, A.G. , Park, B.S. , Weisshaar, B. , Liu, B. , Li, B. , Liu, B. , Tong, C. , Song, C. , Duran, C. , Peng, C. , Geng, C. , Koh, C. , Lin, C. , Edwards, D. , Mu, D. , Shen, D. , Soumpourou, E. , Li, F. , Fraser, F. , Conant, G. , Lassalle, G. , King, G.J. , Bonnema, G. , Tang, H. , Wang, H. , Belcram, H. , Zhou, H. , Hirakawa, H. , Abe, H. , Guo, H. , Wang, H. , Jin, H. , Parkin, I.A.P. , Batley, J. , Kim, J.S. , Just, J. , Li, J. , Xu, J. , Deng, J. , Kim, J.A. , Li, J. , Yu, J. , Meng, J. , Wang, J. , Min, J. , Poulain, J. , Wang, J. , Hatakeyama, K. , Wu, K. , Wang, L. , Fang, L. , Trick, M. , Links, M.G. , Zhao, M. , Jin, M. , Ramchiary, N. , Drou, N. , Berkman, P.J. , Cai, Q. , Huang, Q. , Li, R. , Tabata, S. , Cheng, S. , Zhang, S. , Zhang, S. , Huang, S. , Sato, S. , Sun, S. , Kwon, S.J. , Choi, S.R. , Lee, T.H. , Fan, W. , Zhao, X. , Tan, X. , Xu, X. , Wang, Y. , Qiu, Y. , Yin, Y. , Li, Y. , Du, Y. , Liao, Y. , Lim, Y. , Narusaka, Y. , Wang, Y. , Wang, Z. , Li, Z. , Wang, Z. , Xiong, Z. and Zhang, Z . (2011b) The genome of the mesopolyploid crop species. Brassica rapa . Nat. Genet. 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wu, F. , Bai, J. and He, Y. (2014) BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. [DOI] [PubMed] [Google Scholar]
- Weigel, D. and Glazebrook, J. (2006) In planta transformation of Arabidopsis . CSH Protoc. doi: 10.1101/pdb.prot4668. [DOI] [PubMed] [Google Scholar]
- Xiao, D. , Zhao, J.J. , Hou, X.L. , Basnet, R.K. , Carpio, D.P. , Zhang, N.W. , Bucher, J. , Lin, K. , Cheng, F. , Wang, X.W. and Bonnema, G. (2013) The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co‐expression networks. J. Exp. Bot. 64, 4503–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, D. , Wang, H. , Basnet, R.K. , Zhao, J. , Lin, K. , Hou, X. and Bonnema, G. (2014) Genetic dissection of leaf development in Brassica rapa using a genetical genomics approach. Plant Physiol. 164, 1309–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Wang, H. , Lu, Y. , Ruiter, M.D. , Cariaso, M. , Prins, M. , Tunen, A.V. and He, Y. (2012) Identification of conserved and novel miRNAs that are responsive to heat stress in Brassica rapa . J. Exp. Bot. 63, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of putative amino acid sequences of BrFLC genes between Yellow sarson, Bre and Wantai.
Figure S2 Bolting time of the transgenic Arabidopsis lines overexpressing BrFLC genes from the crop types Yellow sarson, Bre and Wantai.
Figure S3 Homologous fragments between BrFLC2as816 and one NAT of AtFLC.
Figure S4 RT‐PCR showing BrFLC2 NATs in B. rapa.
Figure S5 Diagrammatic representation of alignment of the intron 1 fragments of BrFLC genes with that of AtFLC.
Figure S6 Bolting time of Bre plants overexpressing BrFLC2as816.
Figure S7 Real‐time PCR showing the expression of BrFLC2 in Bre plants overexpressing BrFLC2as816.
Figure S8 Bre plants overexpressing BrFLC2as816 at different stages in the field.
Table S1 Sequences of BrFLC2 antisense transcripts.
Table S2 Growth parameters of BrFLC2as816‐transgenic Bre plants in growth room and field.
Table S3 The primers used in this study.


