Summary
CRISPR/Cas9 and TALEN are currently the two systems of choice for genome editing. We have studied the efficiency of the TALEN system in rice as well as the nature and inheritability of TALEN‐induced mutations and found important features of this technology. The N287C230 TALEN backbone resulted in low mutation rates (0–6.6%), but truncations in its C‐terminal domain dramatically increased efficiency to 25%. In most transgenic T0 plants, TALEN produced a single prevalent mutation accompanied by a variety of low‐frequency mutations. For each independent T0 plant, the prevalent mutation was present in most tissues within a single tiller as well as in all tillers examined, suggesting that TALEN‐induced mutations occurred very early in the development of the shoot apical meristem. Multigenerational analysis showed that TALEN‐induced mutations were stably transmitted to the T1 and T2 populations in a normal Mendelian fashion. In our study, the vast majority of TALEN‐induced mutations (~81%) affected multiple bases and ~70% of them were deletions. Our results contrast with published reports for the CRISPR/Cas9 system in rice, in which the predominant mutations affected single bases and deletions accounted for only 3.3% of the overall mutations.
Keywords: TALEN, targeted genome editing, rice
Introduction
Feeding the growing human population is an increasingly difficult task, and an important component of the solution will require the development of new crop cultivars with increased yield potential, reduced need for fertilizer and water and increased resistance to pest and pathogens. Among the multitude of approaches that are being pursued for crop improvement, targeted genome editing technologies such as ZFNs (Zinc‐finger nucleases), TALENs (Transcription activator‐like effector nucleases) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR‐associated protein 9) are especially attractive due to their precision (Gaj et al., 2013). TALENs are artificial hybrid proteins composed of a TALE DNA‐binding domain fused to the FokI nuclease domain (Christian et al., 2010). TALEs (transcription activator‐like effectors) are proteins secreted by plant bacterial pathogens in the genus Xanthomonas after host infection and contain a DNA‐binding domain composed of a series of tandem repeats (Bogdanove et al., 2010; Mak et al., 2013). Each repeat contains a highly conserved 33–35 amino acid sequence with the exception of two residues at positions 12 and 13 which show hypervariability and are, thus, designated repeat variable diresidues or RVDs (Mak et al., 2013). In 2009, two independent groups showed that RVDs were responsible for the binding of specific nucleotides in the TALE target site following a simple code (Boch et al., 2009; Moscou and Bogdanove, 2009). For example, the RVDs His‐Asp (HD), Asn‐Ile (NI), Asn‐Gly (NG) and Asn‐Asn (NN) recognize the nucleotides C, A, T and G, respectively. According to the code of DNA‐binding specificity, TALEs can be engineered to bind any DNA sequences and fused to the endonuclease domain of FokI to create a TALEN. As FokI needs to form a dimer for DNA cleavage, two TALENs each targeting the sense and antisense strands are needed to make site‐specific DNA double stand breaks (DSBs). Localized DSBs can induce site‐specific indel mutations via the error‐prone nonhomologous end‐joining (NHEJ) repair pathway. DSBs also can be repaired by homologous recombination (HR) which can use a sister chromatid or exogenous homologous DNA template, and this can enable highly precise editing, such as gene addition or replacement, in the target sites.
In the past few years, TALEN has been successfully used for targeted genome editing in yeast, roundworms, rats, zebrafish, human embryonic stem cells, induced pluripotent stem cells, silkworm, insects and many other organisms (Gaj et al., 2013). In plants, TALEN‐induced gene mutations have been produced in many species, but only two reports are available for rice (Cermak et al., 2011; Chen et al., 2014; Christian et al., 2013; Gurushidze et al., 2014; Haun et al., 2014; Li et al., 2012; Mahfouz et al., 2011; Shan et al., 2013; Wendt et al., 2013; Zhang et al., 2013b).
In this work, we have optimized TALEN scaffolds to improve efficiency in rice. We have also performed a comprehensive study of the types and frequencies of TALEN‐induced mutations and analysed the heritability of such mutations in T0, T1 and T2 generations.
Results
Truncated TALEN backbones can significantly improve the efficiency of targeted mutagenesis in rice
As an initial approach to test the efficiency of the TALEN system in generating DSBs in rice, 10 TALEN pairs were designed targeting seven different genes (Table S1). Targeted sequences were positioned in the promoter region (predicted TATA‐box), exon region or both and transgenic lines carrying the different gene constructs were generated in the Japonica variety Nipponbare (Figure S1). The targeted genes, OsCSA, OsPMS3, OsDERF1, OsGN1a, OsTAD1, OsMST7 and OsMST8, were carefully selected for their potential agronomic value (Table S1). For example, mutations in OsCSA and OsPMS3 result in photoperiod‐sensitive male sterility, a trait highly sought after in the production of hybrid seeds, while mutations in OsGN1a and OsDERF1 could generate enhanced grain yield and drought resistance, respectively (Ashikari et al., 2005; Ding et al., 2012; Zhang et al., 2013a). The different TALEN pairs were designed using the TAL effector‐Nucleotide Targeter 2.0 (TALE‐NT) software (https://tale-nt.cac.cornell.edu/), with slightly different characteristics with the left repeats recognizing a range of nucleotide lengths ranging from 15 to 22 nt while the right recognition sequences ranged from 16 to 25 nt (Doyle et al., 2012). In addition, the spacer lengths for the different TALEN pairs ranged from 16 to 24 nt. A large number of hygromycin‐resistant putative transgenic T0 plants (between 22 and 151) were produced for each of the targets and the incorporation of each of the different transgenes verified by PCR. From each transgenic T0 plant, tissues from several leaves were collected, genomic DNA extracted and the targeted genomic region amplified by PCR. The existence and nature of the different mutations were determined by sequencing (Table 1). Six of 10 TALEN constructs showed activity in the transgenic T0 plants, but the mutation frequency was very low, with the highest efficiency observed in the OsMST7 gene, where 6.6% (10/151) of the plants contained mutations (Table 1). No direct correlation was observed between the spacer length or the lengths of the left and right repeats and the mutation efficiency.
Table 1.
Percentage of transgenic T0 plants found with TALEN‐induced mutations in targeted sequence
| Target Genes | Target Region | Left Repeats | Right Repeats | Spacer Length (bp) | No. of Plants Tested | No. of Plants with Mutations | Mutation Rate (%) |
|---|---|---|---|---|---|---|---|
| OsMST7 | Exon | 17 | 19 | 16 | 151 | 10 | 6.6 |
| OsMST8 | Exon | 15 | 16 | 19 | 75 | 4 | 5.3 |
| OsPMS3 | Promoter | 16 | 16 | 15 | 52 | 2 | 3.8 |
| OsCSA | Promoter | 16 | 16 | 15 | 44 | 1 | 2.3 |
| OsDERF1 | Promoter | 18 | 21 | 18 | 36 | 1 | 2.8 |
| OsDERF1 | Promoter | 15 | 17 | 16 | 36 | 1 | 2.8 |
| OsCSA | Promoter | 20 | 24 | 24 | 44 | 0 | 0 |
| OsGN1a | Exon | 19 | 20 | 16 | 22 | 0 | 0 |
| OsTAD1 | Exon | 19 | 21 | 16 | 35 | 0 | 0 |
| OsDERF1 | Exon | 22 | 25 | 21 | 37 | 0 | 0 |
The general TALEN architecture used to produce genomic DSBs consists of a TAL effector fused to a nuclease domain. While the most common nuclease (FokI) has been used for many years and optimized to contain just the C‐terminal 25 kDa region of the original FokI endonuclease, the optimal architecture of the TAL effector has not been established in rice. It has been reported that minimizing the length of the TAL effector can result in increased TALEN activity in animal systems but not much is known for plant systems (Bedell et al., 2012; Kim et al., 2013; Zhang et al., 2013b). Aside from the central RVD domain, which is required for the recognition of the target nucleotide sequence, TAL effectors contain relatively large N‐terminal and C‐terminal regions. We studied the importance of the C‐terminal TALE regions, by assessing the effect of truncations on the mutation efficiency of the TALEN. The original TALEN backbone, used for our initial studies, has 287 and 230 amino acids in their N‐ and C‐terminal regions, respectively, and was thus named N287C230. Two deletions of the TALEN backbone were prepared, the first one had the C‐terminal region truncated to 63 amino acids while the second one had almost the entire C‐terminal region trimmed, containing only 14 amino acids of the original protein (C63 and C14) (Figure 1). While the RVD domains for the N287C230 and N287C63 pairs were identical, we reduced the spacer length for N287C14 from 15 bp to 13 bp (Figure S1) as it has been shown that the spacer length affects the activity of different backbones (Wright et al., 2014).The results indicated that both C‐terminal deletions significantly increased mutation efficiency. Mutations induced by N287C63 were identified in 5 of 25 transgenic plants (25%), and mutations induced by N287C14 were detected in 15 of 69 transgenic plants (21.7%) representing a 658% and 571% increase in mutation efficiency over the N287C230 TALEN (Table 2).
Figure 1.

Schematic diagram of the TALEN backbones used in this study. The full length N287C230 has 287 and 230 amino acids in its N‐ and C‐termini, respectively, surrounding the RVDs region. Two truncated TALEN backbones were constructed with different amino acids lengths at the C‐terminus of 63 and 14 (N287C63 and N287C14). NLS, nuclear localization signal; N‐term, N‐terminal region; RVD, repeat variable diresidues; C‐term, C‐terminal region; FokI, flavobacterium okeanokoites (FokI) restriction endonuclease.
Table 2.
Percentage of transgenic T0 plants with mutations in the OsPMS3 gene using different TALEN backbones
| Backbone | No. of Plants Tested | No. of Plants with Mutations | Mutation Rate (%) |
|---|---|---|---|
| N287C230 | 52 | 2 | 3.8 |
| N287C63 | 20 | 5 | 25.0 |
| N287C14 | 69 | 15 | 21.7 |
TALEN‐induced mutations are transmitted through the germ‐line in a Mendelian fashion
Successful germ‐line transmission of genetic mutations is essential for the application of the TALEN system to crop improvement. In a variety of species, including rice, TALEN‐induced mutations can occur in the germ‐line and be transmitted to the next generation (Li et al., 2012) although there is no detailed quantitative data determining the nature of the mutations and their inheritability over several generations. To address these issues, we extensively analysed the N287C230 TALEN‐induced mutations in eight rice T0 lines and their inheritability patterns in the T1 and T2 progenies for two different target genes, OsMST7 and OsMST8 (Table 3, Tables S2 and S3).
Table 3.
Segregation and types of TALEN‐induced mutations in target genes*

To determine the genotype of individual plants, tissues from several leaves were collected, the genomic DNA isolated and a fragment containing the TALEN‐targeted region amplified by PCR. The PCR product was then ligated to a plasmid, and multiple individual minipreps sequenced to identify any introduced mutations (T‐clone sequencing). To determine the segregation ratio in the progeny of an individual plant, multiple seedlings were grown and genomic DNA isolated. For each seedling, the targeted genomic fragment was amplified by PCR and the amplification product subjected to Sanger sequencing. The appearance of single peaks in the sequencing chromatograph denoted the homogeneity of the amplified product and was then considered to be homozygous for the predominant mutation (in red in the different tables) or wild type. The appearance of multiple peaks in a single nucleotide position indicated the existence of mixed nucleotides in that position and the plant maybe a bi‐allele, heterozygote or chimera, which is marked as ‘h’ in Table 3.
Analysis of three T0 transgenic plants targeting OsMST8 (T‐OsMST8) identified them as chimeras and revealed a very complex pattern of mutations. The predominant types of mutations in these plants were deletions; substitutions were also present, but no insertions were detected in this first analysis. Interestingly, most deletions in these plants involved multiple nucleotides, ranging from 1 to 28. The predominant mutation in the T‐OsMST8 target line 2 (T0‐2) was a 28‐nucleotide deletion (Table 3). Analysis of 159 individual T1 progeny plants revealed 46 homozygous plants harbouring the same 28‐nucleotide deletion mutation as the T0 plant, 78 ‘h’ group individuals and 35 WT plants. Eleven of the ‘h’ group T1 plants were further analysed by T‐clone sequencing, and nearly, all of them (10/11) contained the 28 nucleotide deletions (Table 3 and Table S2). Two of the ‘h’ group T1 lines harbouring the 28‐nucleotide deletions, transmitted the mutation to the next generation (T2) with this mutation being the only one producing homozygous plants (Table 3). The predominant mutation in the T‐OsMST7 target line 1 (T0‐1) was a 3‐nucleotide deletion (designated d3a), and the same mutation was by far the most abundant in the subsequent T1 and T2 generations (Table 3 and Table S2). In the remaining T‐OsMST7 and T‐OsMST8 lines analysed, the predominant TALEN‐induced mutations in the T0 generation were also found in all the T1 and T2 generations (Table 3 and Table S2). These data clearly show that mutations induced by TALEN can be efficiently transmitted to future generations confirming the TALEN system as a useful tool for rice breeding and functional genomics research.
Although the TALEN system has been successfully used in different plant species, there is very little information about the timing and tissue location where the original mutations occur. To trace the source of the mutations, different tissues such as leaves, stems and flowers from individual tillers were examined in T0 transgenic plants (Table 4 and Table S4) (tiller # in parenthesis). In total, four different tillers were analysed in the T‐OsMST8 target line 1 (T0‐1), one in the T‐OsMST8 T0‐3 line and two in the T‐OsMST7 T0‐3 and T0‐4 lines. In all studied plants, nearly all the tissues harboured the predominant mutation type. For example, in the T‐OsMST8 T0‐1 line, the predominant mutation was a 2‐bp deletion which was detected in nearly all the tissues examined (in red colour in Table 4). In the T‐OsMST7 T0‐3 line, the predominant mutation was a nucleotide substitution (named S1c) detected in all tissues examined while in the T‐OsMST7 T0‐4 line, the predominant mutation was a single base deletion (named d1a) also present in all tissues examined. As all tissues in the same tiller come from the same shoot apical meristem (SAM), the fact that they harbour the same mutation types suggests that the predominant TALEN‐induced mutations occurred in SAM cells. Those TALEN‐induced mutations occurring in later stages of organ development, which are probably reflected in the low‐frequency mutations found in different tissues, should not be transmitted to the next generation unless they give rise to the germ‐line. Of course, germ‐line cells also arise from the SAM; therefore, those TALEN‐induced mutations originating in SAM cells could be transmitted to the next generation. In addition, in nearly all examined T0 plants, which were regenerated from a transformed rice callus cell, different tillers harboured the same predominant mutation types (Table 5 and Table S5). For example, the 28‐nucleotide deletion in the T‐OsMST8 T0‐2 line was detected in all of the 8 different tillers analysed while the predominant 2‐bp deletion in the T‐OsMST8 T0‐3 line was present in all 4 tillers (Table 5). For OsMST8, all four T0‐1 tillers contained the same 3‐bp deletion (3da), all five T0‐2 tillers harboured the same single base insertion and all four T0‐5 tillers contained the same unusual mutation consisting of a 40‐bp deletion and a simultaneous di‐base substitution. In the case of the T‐OsMST7 T0‐3 line, there were two predominant mutations (s1c and d2c), both of which were present in all four tillers. These results indicate that most of the predominant TALEN‐induced mutations in different tillers of a T0 plant came from the same source, likely the SAM because cells in different tillers all originated from an early SAM. In other words, TALEN‐induced mutations are likely to occur in a very early developmental stage before the first SAM formed or during the process when the embryogenic callus cell differentiated into the SAM. Therefore, TALEN‐induced mutations could be transmitted to the next generation through germ‐line cells which originate from the SAM.
Table 4.
Extended examination of genotypes in different tissues of T0 transgenic plants

Table 5.
Extended examination of genotypes in different tillers of T0 plants

Characteristics of TALEN‐induced mutations in rice
In our study, the pattern of mutations caused by NHEJ repairs resulting from TALEN activity in rice was diverse. The combined results of eight T0 T‐OsMST7 and T‐OsMST8 TALEN target lines are shown in Figure 2. The majority of the mutations caused by NHEJ repair after the introduction of DSBs by the TALEN system were deletions (69.88%), and the deletion length varied in a wide range from 1 to 40 bps (Figure 2). Base replacement and combined mutations (at least 2 mutation types) also occurred, with frequencies of 9.78% and 16.55%, respectively (Figure 2). Base insertions were extremely infrequent (3.79%). Considering targets and mutation types, the two targets, OsMST7 and OsMST8, showed similar mutation types (Table 6).
Figure 2.

TALEN‐induced mutation types and frequency. Left insert in graph, percentage of deletions (d), insertions (i), substitutions (s) and combined (c) mutation types. Right insert, percentage of different mutation lengths. In x‐axis: d#, # of base pairs (bps) deleted from target site; i#, # of bps inserted at target site, s#, substitution mutation.
Table 6.
Mutation type and length for each target gene
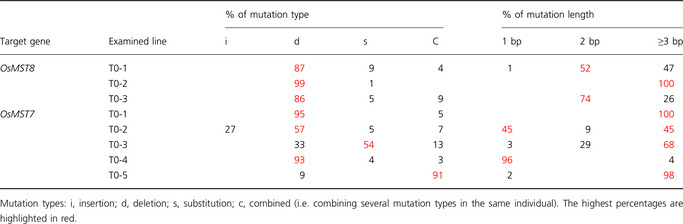
In the CRISPR/Cas9 system, mutations usually occur in the 4th base from the protospacer adjacent motif (PAM) site, but very little is known about the position of DNA cleavage by TALENs in plants. When all mutations types are examined, it is obvious that the cleavage sites are positioned in the middle of the spacer regions (Table S6). For more in depth analysis, we selected a subset of mutation types including the only observed insertion and deletions of 1 to 3 bp as it is difficult to predict the putative cleavage sites with more extended deletions. For T‐OsMST7 lines, whose target sites were separated by a 16‐bp spacer, the cleavage sites were most probably positioned either 6 nucleotides from the right TALEN target site or 5–6 nucleotides from the left TALEN target site (Table S6). In contrast, in T‐OsMST8 lines, where the spacer is 19 bp in length, the cleavage sites were positioned in the middle of the spacer region (Table S6). These results suggest that TALEN cleavage sites are dependent on the spacer length.
Discussion
In the past decade, genome editing has emerged as an exciting technology promising to deliver crop genetic improvement with extremely high precision. Genome editing technologies work by creating specific DSBs at targeted genomic sites before making precise genetic modifications through the cellular DNA repair mechanisms, including error‐prone nonhomologous end joining (NHEJ) and homology‐directed repair (HDR) (Wyman and Kanaar, 2006). Genome editing provides a unique tool for functional genomic studies aiming to deliver critical knowledge for human health and agricultural production. The initial tools for genome editing were artificial Zinc‐Finger Nucleases but their inherent complexity precluded the broad adoption of the technology by the scientific community (Gaj et al., 2013). Nevertheless, in the last 5 years, the development of TALEN and CRISPR has seen a resurgence in interest as both technologies are highly effective for genome editing and not too technically demanding. Although there are two published reports describing the production of TALEN‐induced gene mutations in rice, a comprehensive study characterizing the nature as well as inheritability patterns of such mutations was missing. Recent work in our laboratory has analysed the patterns, specificity and heritability of CRISPR/Cas9‐induced mutations in rice, but a comparison between TALEN and CRISPR/Cas9 in the same plant system is missing (Zhang et al., 2014). In this work, we have used TALEN to engineer efficient‐targeted gene mutations in rice, analysed the patterns of TALEN‐induced mutations and studied their heritability.
When the N287C230 TALEN backbone was used for rice genome editing, TALEN‐induced mutations were detected for six of ten different constructs in T0 transgenic plants. Nevertheless, the mutation rates were very low, with the highest efficiency being 6.6%. In addition, three of the six successful TALEN constructs resulted in a single T0 plant harbouring mutations of 36, 36 and 44 lines produced. Given the low efficiency observed, it is important to produce a large number of transgenic lines to ensure the presence of TALEN‐induced mutations, suggesting that not enough lines were produced for the four unsuccessful constructs. The differences in mutation rates between the tested genes could be due to differences in the genomic target site (e.g. accessibility of the target site), differences in spacer length between the TALEN monomers and/or the necessary changes in the RVD composition of the TALENs, necessary to target different DNA sequences. Our attempts to optimize the length of the TALEN backbone clearly demonstrated that truncation of the C‐terminus dramatically improved efficiency with constructs N287C63 and N287C14, producing mutation rates of 25% and 21.7%, respectively, compared with the 3.8% obtained when using the original N287C230 backbone. Even in the optimized conditions, TALEN‐induced mutation efficiency in rice is still lower than CRISPR/Cas9, which showed an average efficiency of more than 50%, although it is high enough to be used in rice functional genomics research (Zhang et al., 2014). In order to get closer to the efficiency shown by CRISPR/Cas9, TALEN could be further improved by truncating the C‐terminal region. It has been reported that truncation of the TALEN C‐terminus can improve genome editing efficiency, and a N152/C63 backbone has shown increased mutagenesis efficiency in human genes (Bedell et al., 2012; Miller et al., 2011). The same TALEN backbone has been successfully used in Arabidopsis and tobacco showing TALEN activity (Christian et al., 2013; Zhang et al., 2013b).
The few available reports studying TALENs in plant systems focus mostly on the generation of mutations with relatively little attention devoted to the inheritability of the traits. In this work, we thoroughly analysed the inheritance characteristics with our results showing that for most of the T0 lines there was a predominant type of mutation accompanied by additional, low‐frequency mutations. We observed that the predominant mutations detected in T0 plants were also found in the corresponding T1 and T2 populations, demonstrating that TALEN‐induced mutations are highly stable and can be transmitted to the next generations in the same way as other endogenous genes. In contrast, the low‐frequency mutation types present in T0 plants were not found in the following generations suggesting that they were somatic mutations. Mutation inheritability to the next generation occurs through germ‐line cells leading us to the hypothesis that the predominant TALEN‐induced mutation occurred in the SAM cells. Consistent with this hypothesis, in the same original T0 plant, the predominant TALEN‐induced mutations were detected in different tissues from the same tiller, as well as tissues from different tillers. Therefore, TALEN‐induced mutations are likely to occur in a very early developmental stage before the first SAM formed or during the SAM formation process in the embryogenic callus. It has been reported that TALEN‐induced mutations can be detected in calli cells, which is consistent with our results (Shan et al., 2013). In rice, the CRISPR/Cas9 system generated mutations at a similar developmental stage as TALENs while in Arabidopsis, many of the CRISPR/Cas9‐induced mutations detected in T1 plants were somatic mutations only and they were not transmitted to the T2 generation (Feng et al., 2014; Zhang et al., 2014).
There are striking differences between the TALEN‐induced mutation patterns observed in our study, and the CRISPR/Cas9‐induced mutations previously reported in rice (Zhang et al., 2014). In the CRISPR/Cas9 system, most mutations (72.4%) only affected 1 bp and more than half of them were 1‐bp insertions (Zhang et al., 2014). In contrast, our results show that the vast majority of TALEN‐induced mutations in rice (82.10%) altered multiple bases with most of them (69.88%) being deletions, whereas insertions accounted for only 3.79%. While the most frequent mutation type in the CRISPR/Cas9 system was a 1‐bp insertion, over half of TALEN‐induced mutations were multiple base deletions. These characteristics give TALEN an advantage over CRISPR/Cas9 in cases where it is advantageous to modify multiple bases, such as to mutate a mature miRNA sequence. We observed that 9.78% of mutations were base substitutions in the targets although considering that Taq polymerase is not a proof‐reading enzyme, the possibility of errors during the PCR amplification might imply that the detected substitution rate may have been overestimated. Assuming that the mutations generated by the TALEN and CRISPR/Cas9 systems are caused by the same error‐prone nonhomologous end‐joining (NHEJ) repair pathway, the observed differences in their type and lengths must be due to the nature of the DSBs produced by each system. The Cas9 nuclease always produces DSBs with blunt ends, usually occurring 4 bases away from the PAM site. In contrast, the FokI nuclease used in TALEN catalyses single strand breaks, and therefore, the production of DSBs is the result of the combined action of two nucleases that are positioned in close proximity of each other by a pair of TALEN proteins. The TALEN‐induced DSBs can therefore be very variable depending on the spacer length and in most occasions will produce jagged ends. Even though our discussion is based on the analysis of TALEN and CRISPR/Cas9 results produced in rice, it is important to note that the two systems have not been compared directly in the same study with the same genomic sequences being targeted.
In summary, both TALEN and CRISPR/Cas9 are highly effective tools for targeted genome editing; however, each has specific advantages and limitations. CRISPR/Cas9 efficiency is much higher than TALEN, a big advantage in systems with difficult or cumbersome transformation systems; however, off‐target effects and the ‐NGG (two invariable Gs preceded by a variable base) PAM requirement pose an important limitation on the CRISPR/Cas9 system. The apparently different nature of the mutations generated by the CRISPR/Cas9 and TALEN systems, single versus multiple bases; insertions versus deletions, respectively, are neither an advantage nor a limitation but could determine the system of choice for a given application.
Material and methods
TALEN design and plasmid construction
The TALEN target sites were selected using the TAL effector‐Nucleotide Targeter 2.0 (TALE‐NT) software (https://tale-nt.cac.cornell.edu/) (Doyle et al., 2012). TALENs were constructed using the Golden Gate Assembly method as previously described (Cermak et al., 2011). All target sites have a T at position 0 and the RVDs HD, NI, NG and NN were chosen to specifically recognize the nucleotides C, A, T and G, respectively. TALEN components were amplified from the pTAL3 vector using primers XmaI‐SpeI‐TAL3 F and KpnI‐BstEII‐TAL3 R (Table S7) which contain XmaI/SpeI sites and KpnI‐BstEII sites, respectively. The amplified fragment was ligated into pZero/Blunt to produce an intermediate vector pTAL3(m). TAL effector repeats were later assembled into pTAL3(m) or other TALEN backbones according to the Golden Gate Assembly method (Cermak et al., 2011). Assembled TAL effector arrays and surrounding N‐terminal, C‐terminal and Fok I regions were constructed into the modified two‐gene expression vector p1301‐TALEN (Figure S2) containing the maize ubiquitin 1 promoter (Ubi1) and the cauliflower mosaic virus 35S promoter (35S). One of the paired TALEN genes was cloned into p1301‐TALEN using BstEII and SpeI sites under the control of the 35S promoter while the second paired TALEN gene was cloned into the XmaI and KpnI sites under the control of the ubi1 promoter. To generate the truncated backbones, the different N‐ or C‐terminal fragments were amplified from the pTAL3(m) vector. Standard molecular biology techniques were used to construct the different backbones.
Rice transformation and plant growth
Transgenic rice lines c.v. Nipponbare (O. sativa L. ssp. japonica) containing the different TALEN constructs were produced by Agrobacterium‐mediated transformation (Hiei et al., 1997).The transgenic lines were grown in greenhouses under standard long‐day (14 h day at 30 °C/8 h night at 28 °C) or short‐day (10 h day at 30 °C/14 h night at 28 °C) conditions or grown in the paddy field in Shanghai (30_N, 121_E), China during normal rice growing seasons.
Genomic DNA extraction and PCR‐based genotyping
Genomic DNA from transgenic plants was extracted using the CTAB (cetyltrimethyl ammonium bromide) method(Doyle, 1987). The TALEN targeted regions were amplified by PCR using specific primers (Table S7). Amplified PCR products were sequenced and those plants in which the sequencing chromatograms showed single peaks in the target regions were marked as homozygotes or wild‐type plants. For those plants with sequencing chromatograms showing multiple peaks, the amplified DNA was cloned into the pMD18‐T vector (TAKARA, Dalian, China) and multiple individual clones sequenced to identify mutations.
Acknowledgements
We would like to thank Dr. Daniel Voytas for supplying the Golden Gate TALEN and TAL Effectors. The work was supported by the Chinese Academy of Sciences. The authors declare that they have no conflict of interests with respect to this work.
Supporting information
Figure S1 Targeted gene's loci structures and targeted sequences.
Figure S2 Schematic diagram of TALEN vector p1301‐TALEN designed for Agrobacterium‐mediated rice transformation.
Table S1 Rice genes targeted using the TALEN system.
Table S2 Segregation and types of TALEN‐induced mutations in target genes.
Table S3 Sequencing results of T0 and T1 plants.
Table S4 Sequencing results of different tissues of T0 transgenic plants.
Table S5 Sequencing results of different tillers of T0 transgenic plants.
Table S6 Summary of all TALEN induced mutation types in OsMST7 and OsMST8 target sites.
Table S7 List of PCR primers and their applications.
Hui Zhang and Feng Gou contributed equally to this work.
J‐K.Z. and H.Z. conceived the study. H.Z., F.G., J.Z., W.L. Q.L and Y.M. performed the experiments. J‐K.Z., H.Z. and J.R.B. analysed the data. J‐K.Z., H.Z. and J.R.B. wrote the manuscript.
References
- Ashikari, M. , Sakakibara, H. , Lin, S. , Yamamoto, T. , Takashi, T. , Nishimura, A. , Angeles, E.R. , Qian, Q. , Kitano, H. and Matsuoka, M. (2005) Cytokinin oxidase regulates rice grain production. Science, 309, 741–745. [DOI] [PubMed] [Google Scholar]
- Bedell, V.M. , Wang, Y. , Campbell, J.M. , Poshusta, T.L. , Starker, C.G. , KrugII, R.G. , Tan, W. , Penheiter, S.G. , Ma, A.C. and Leung, A.Y. (2012) In vivo genome editing using a high‐efficiency TALEN system. Nature, 491, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Scholze, H. , Schornack, S. , Landgraf, A. , Hahn, S. , Kay, S. , Lahaye, T. , Nickstadt, A. and Bonas, U. (2009) Breaking the code of DNA binding specificity of TAL‐type III effectors. Science, 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A.J. , Schornack, S. and Lahaye, T. (2010) TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. [DOI] [PubMed] [Google Scholar]
- Cermak, T. , Doyle, E.L. , Christian, M. , Wang, L. , Zhang, Y. , Schmidt, C. , Baller, J.A. , Somia, N.V. , Bogdanove, A.J. and Voytas, D.F. (2011) Efficient design and assembly of custom TALEN and other TAL effector‐based constructs for DNA targeting. Nucleic Acids Res. 39, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Shan, Q. and Gao, C. (2014) An efficient TALEN mutagenesis system in rice. Methods, 69, 2–8. [DOI] [PubMed] [Google Scholar]
- Christian, M. , Cermak, T. , Doyle, E.L. , Schmidt, C. , Zhang, F. , Hummel, A. , Bogdanove, A.J. and Voytas, D.F. (2010) Targeting DNA double‐strand breaks with TAL effector nucleases. Genetics, 186, 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, M. , Qi, Y. , Zhang, Y. and Voytas, D.F. (2013) Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 (Bethesda), 3, 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J. , Lu, Q. , Ouyang, Y. , Mao, H. , Zhang, P. , Yao, J. , Xu, C. , Li, X. , Xiao, J. and Zhang, Q. (2012) A long noncoding RNA regulates photoperiod‐sensitive male sterility, an essential component of hybrid rice. Proc. Natl Acad. Sci. USA, 109, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J.J. (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Doyle, E.L. , Booher, N.J. , Standage, D.S. , Voytas, D.F. , Brendel, V.P. , Vandyk, J.K. and Bogdanove, A.J. (2012) TAL Effector‐Nucleotide Targeter (TALE‐NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 40, W117–W122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Z. , Mao, Y. , Xu, N. , Zhang, B. , Wei, P. , Yang, D.L. , Wang, Z. , Zhang, Z. , Zheng, R. , Yang, L. , Zeng, L. , Liu, X. and Zhu, J.K. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas‐induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111, 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj, T. , Gersbach, C.A. and Barbas, C.F. 3rd . (2013) ZFN, TALEN, and CRISPR/Cas‐based methods for genome engineering. Trends Biotechnol. 31, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurushidze, M. , Hensel, G. , Hiekel, S. , Schedel, S. , Valkov, V. and Kumlehn, J. (2014) True‐breeding targeted gene knock‐out in barley using designer TALE‐nuclease in haploid cells. PLoS ONE, 9, e92046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun, W. , Coffman, A. , Clasen, B.M. , Demorest, Z.L. , Lowy, A. , Ray, E. , Retterath, A. , Stoddard, T. , Juillerat, A. , Cedrone, F. , Mathis, L. , Voytas, D.F. and Zhang, F. (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. [DOI] [PubMed] [Google Scholar]
- Hiei, Y. , Komari, T. and Kubo, T. (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol. Biol. 35, 205–218. [PubMed] [Google Scholar]
- Kim, Y. , Kweon, J. , Kim, A. , Chon, J.K. , Yoo, J.Y. , Kim, H.J. , Kim, S. , Lee, C. , Jeong, E. , Chung, E. , Kim, D. , Lee, M.S. , Go, E.M. , Song, H.J. , Kim, H. , Cho, N. , Bang, D. and Kim, J.S. (2013) A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 31, 251–258. [DOI] [PubMed] [Google Scholar]
- Li, T. , Liu, B. , Spalding, M.H. , Weeks, D.P. and Yang, B. (2012) High‐efficiency TALEN‐based gene editing produces disease‐resistant rice. Nat. Biotechnol. 30, 390–392. [DOI] [PubMed] [Google Scholar]
- Mahfouz, M.M. , Li, L. , Shamimuzzaman, M. , Wibowo, A. , Fang, X. and Zhu, J.K. (2011) De novo‐engineered transcription activator‐like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double‐strand breaks. Proc Natl Acad Sci USA 108, 2623–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, A.N. , Bradley, P. , Bogdanove, A.J. and Stoddard, B.L. (2013) TAL effectors: function, structure, engineering and applications. Curr. Opin. Struct. Biol. 23, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.C. , Tan, S. , Qiao, G. , Barlow, K.A. , Wang, J. , Xia, D.F. , Meng, X. , Paschon, D.E. , Leung, E. , Hinkley, S.J. , Dulay, G.P. , Hua, K.L. , Ankoudinova, I. , Cost, G.J. , Urnov, F.D. , Zhang, H.S. , Holmes, M.C. , Zhang, L. , Gregory, P.D. and Rebar, E.J. (2011) A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148. [DOI] [PubMed] [Google Scholar]
- Moscou, M.J. and Bogdanove, A.J. (2009) A simple cipher governs DNA recognition by TAL effectors. Science, 326, 1501. [DOI] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Chen, K. , Liang, Z. , Li, J. , Zhang, Y. , Zhang, K. , Liu, J. , Voytas, D.F. and Zheng, X. (2013) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant, 6, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt, T. , Holm, P.B. , Starker, C.G. , Christian, M. , Voytas, D.F. , Brinch‐Pedersen, H. and Holme, I.B. (2013) TAL effector nucleases induce mutations at a pre‐selected location in the genome of primary barley transformants. Plant Mol. Biol. 83, 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, D.A. , Li, T. , Yang, B. and Spalding, M.H. (2014) TALEN‐mediated genome editing: prospects and perspectives. Biochem. J. 462, 15–24. [DOI] [PubMed] [Google Scholar]
- Wyman, C. and Kanaar, R. (2006) DNA double‐strand break repair: all's well that ends well. Annu. Rev. Genet. 40, 363–383. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Xu, C. , He, Y. , Zong, J. , Yang, X. , Si, H. , Sun, Z. , Hu, J. , Liang, W. and Zhang, D. (2013a) Mutation in CSA creates a new photoperiod‐sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl Acad. Sci. U.S.A. 110, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Zhang, F. , Li, X. , Baller, J.A. , Qi, Y. , Starker, C.G. , Bogdanove, A.J. and Voytas, D.F. (2013b) Transcription activator‐like effector nucleases enable efficient plant genome engineering. Plant Physiol. 161, 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Zhang, J. , Wei, P. , Zhang, B. , Gou, F. , Feng, Z. , Mao, Y. , Yang, L. , Xu, N. and Zhu, J.K. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Targeted gene's loci structures and targeted sequences.
Figure S2 Schematic diagram of TALEN vector p1301‐TALEN designed for Agrobacterium‐mediated rice transformation.
Table S1 Rice genes targeted using the TALEN system.
Table S2 Segregation and types of TALEN‐induced mutations in target genes.
Table S3 Sequencing results of T0 and T1 plants.
Table S4 Sequencing results of different tissues of T0 transgenic plants.
Table S5 Sequencing results of different tillers of T0 transgenic plants.
Table S6 Summary of all TALEN induced mutation types in OsMST7 and OsMST8 target sites.
Table S7 List of PCR primers and their applications.


