Abstract
The construction of mutant fungal strains is often limited by the poor efficiency of homologous recombination in these organisms. Higher recombination efficiencies can be obtained by increasing the length of homologous DNA flanking the transformation marker, although this is a tedious process when standard molecular biology techniques are used for the construction of gene replacement cassettes. Here, we present a two-step technology which takes advantage of an Escherichia coli strain expressing the phage λ Red(gam, bet, exo) functions and involves (i) the construction in this strain of a recombinant cosmid by in vivo recombination between a cosmid carrying a genomic region of interest and a PCR-generated transformation marker flanked by 50 bp regions of homology with the target DNA and (ii) genetic exchange in the fungus itself between the chromosomal locus and the circular or linearized recombinant cosmid. This strategy enables the rapid establishment of mutant strains carrying gene knock-outs with efficiencies >50%. It should also be appropriate for the construction of fungal strains with gene fusions or promoter replacements.
INTRODUCTION
Gene manipulation in filamentous fungi is limited by the low efficiency of homologous recombination upon DNA-mediated transformation and the high frequency of ectopic integrations of the transforming DNA molecule: a minimum of 1 kb of DNA homologous to the target site is often required to achieve efficiencies of homologous recombination ~10% (1,2). This will become a major limitation for post-genomic studies once whole genome sequences of filamentous fungi become available.
It follows that construction of knock-out mutants by gene replacement often requires several subcloning steps prior to transformation of the fungus and is therefore a lengthy process. An alternative strategy has been proposed where knock-out constructs are directly placed on a cosmid carrying a genomic region encompassing the gene of interest, thus providing large regions of homology with the genome. This can be achieved either by in vivo or in vitro transposon mutagenesis (L.Hamer and J.E.Hamer, 20th Fungal Genetics Conference, Asilomar, USA) or by recombination between the cosmid and a PCR product in Saccharomyces cerevisiae (J.A.Sweigard, 20th Fungal Genetics Conference, Asilomar, USA). However, these two strategies, which have so far not been described in detail, have drawbacks since their application would either result in random insertions or require the development of novel genomic libraries based on Escherichia coli–S.cerevisiae shuttle vectors.
Recently, it has been shown that expression of the E.coli RecE and RecT proteins or the corresponding Redα(exo) and Redβ(bet) proteins of phage λ together with the Redγ(gam) protein can drastically promote homologous recombination in E.coli. Using this approach it has been possible to achieve allelic exchanges for genes located on the E.coli genome or an episome by recombination with a PCR product containing homology arms of 50 nt (3–6).
We have developed a similar strategy where expression of Redα, Redβ and Redγ is used to promote recombination in E.coli between an amplified bi-functional transformation marker and a selected cosmid carrying an Aspergillus nidulans genomic region. The circular or linearized cosmid is then used to transform A.nidulans yielding transformants with the appropriate gene replacement at frequencies up to 60%. This methodology should be applicable to most filamentous fungi and can be used to introduce highly specific gene modifications in fungal genomes.
MATERIALS AND METHODS
Strains and culture conditions
Escherichia coli strain KS272 [F– ΔlacX74 galE galK thi rpsL ΔphoA (PvuII)] was used for propagation of the recombination vector pKOBEG and of A.nidulans cosmids. Conditional R6Kγ origin plasmids were maintained in the pir+ host DH5α λpir [F– endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacIZYA-argF)U169 deoR (φ80dlac(lacZ)M15) (λpyr+)]. Escherichia coli strains were propagated in LB medium or LBLS (1% bacto-tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.5) medium when selection for zeocin resistance was applied. The β-lactam antibiotic carbenicillin (100 µg/ml), chloramphenicol (25 µg/ml), spectinomycin (100 µg/ml) and zeocin (50 µg/ml; Invitrogen) were added to the growth medium when required. l-Arabinose or d-glucose were added as indicated to modulate expression of genes under control of the pBAD promoter (7).
Aspergillus nidulans strain FGSC773 (wA3 pyroA4 pyrG89) was obtained from the Fungal Genetics Stock Centre (University of Kansas, Kansas City, USA). Growth conditions for A.nidulans strains have been described (8). Scoring of the acid trehalase phenotype was achieved by replica plating of FGSC773 transformants on minimal medium containing 1% glucose or 1% trehalose as the carbon source (8).
DNA manipulations
DNA manipulations were according to Sambrook et al. (9) and Ausubel et al. (10). Transformation of calcium–manganese-treated E.coli was as described (11). Electro-competent E.coli cells were obtained by washing exponentially growing cells three times with ice-cold distilled H2O and concentrating 300-fold with 10% glycerol. Electroporation was carried out in 0.2-cm electroporation chambers and using a BioRad GenePulser II set to the following parameters: 200 Ω, 25 µF and 2.5 kV. Shocked cells were diluted 3-fold in LB, incubated for 1 h at 30°C without aeration and subsequently plated on appropriate media.
Plasmid pTP223 (3), pBAD18 (7), pKO7, a derivative of pKO3 (12), pHP45Ω-Spc (13) and pGP704Not, a derivative of pGP704 (14), were obtained from K. Murphy, J. Beckwith, G. Church, H. Krish and D. Mazel, respectively. pUC4K and pEM7-zeo were purchased from Pharmacia and Invitrogen, respectively. pAfpyrG2, pTRE11 and pTRE12 have been described previously (8,15).
Construction of the recombination plasmid pKOBEG was as follows. The NsiI–HindIII fragment of pBAD18 carrying the pBAD promotor (7) was subcloned into PstI–HindIII-digested pUC18, yielding pUCaraC-pBAD. The λ Red region of pTP223 was amplified using oligonucleotides EBGNHe-5 (5′-CCCGCTAGCGAAAAGATGTTTCGTGAAGC-3′) and EBGh3 (5′-GGGAAGCTTATTATCGTGAGGATGCGTCA-3′). The resulting 1960 bp PCR product was digested with NheI and HindIII and subcloned into NheI–HindIII-digested pUCaraC-pBAD, yielding pUCaraC-P-EBG. The KpnI–HindIII fragment of pUCaraC-P-EBG was then subcloned into KpnI–HindIII-digested pKO7, yielding pKOBEG.
pCDA18 was obtained by subcloning the EcoRI 2.1 kb spectinomycin-resistance gene carried by pHP45Ω-Spc at the EcoRI site located in 3′ of the Aspergillus fumigatus pyrG gene in pAfpyrG2. pCDA19 is a derivative of pCDA18 where the NotI site has been replaced by EcoRV using an adaptor of the following sequence, 5′-GGCCGATATC-3′. pTRE18 was then obtained by inserting the 4 kb EcoRV of pCDA19 into EcoRI–PmlI-digested pTRE11 that had been treated with T4 DNA polymerase.
pCDA21 was obtained by subcloning the 1.9 kb EcoRV–NotI fragment of pAfpyrG2 carrying the A.fumigatus pyrG gene and the 0.5 kb NotI–EcoRI fragment of pEM7-zeo carrying a zeocin-resistance gene into EcoRV–EcoRI-digested pGP704Not.
Allelic replacement on cosmids
Electrocompetent cells of a transformant carrying pKOBEG (CmR) and cosmid W14C08 (AmpR KmR), a derivative of pWE15 which carries an A.nidulans genomic region encompassing the treA gene (8), were prepared from a culture containing 0.2% arabinose in order to induce the expression of the red genes. Electrocompetent cells (100 µl) were electroporated with ~100 ng of the gel-purified 7 kb NotI fragment of pTRE18 or of a dialyzed PCR product obtained by amplification of pCDA21 with the following oligonucleotides: treFzeo (5′-GCTGAAGGTTCCTTTCTTGATCCTCTCTGTGGGTTCAGAAAGCGTTCAAAggaattctcagtcctgctcc-3′) and treBpyr (5′-CTCAGATTCTATGCTAAACTCGCTCATACTACTCTACATGATATTCTACAgaattcgcctcaaacaatgc-3′). These oligonucleotides have 50 bp of homology to the 5′ or 3′ non-coding region of the A.nidulans treA gene (upper case) followed by 20 bp of homology to the zeocin-resistance or A.fumigatus pyrG genes carried by pCDA21 (lower case), respectively. Amplification was performed in a 100 µl reaction containing 200 ng pCDA21, 25 nmol dNTPs, 68 pmol treFzeo, 63 pmol treBpyr, 5 U rTaq (Pharmacia) and using the following protocol: a denaturation step at 93.5°C for 5 min followed by four cycles of the following steps: denaturation at 93°C for 30 s, annealing at 58°C for 2 min, extension at 72°C for 3 min and 24 cycles of the following steps: denaturation at 93°C for 30 s, annealing at 60°C for 2 min, extension at 72°C for 3 min. A last elongation step was done at 72°C for 10 min. Shocked cells were plated on LB medium containing kanamycin and spectinomycin (NotI fragment) or LBLS medium containing kanamycin and zeocin (PCR product) and incubated at 30°C. Transformants were colony-purified once at 42°C on medium containing kanamycin and spectinomycin or zeocin and then tested for chloramphenicol resistance to test loss of pKOBEG. The resulting cosmids were further characterized by restriction enzyme digestion and PCR analysis using two sets of primers located either in the replaced region (tre#5, 5′- AGGACTCGGTCGATCGCT-3′; tre#6, 5′-CCGGATGGGAGGCGACGA-3′) or in the vicinity of the replaced region (tre#1, 5′-ATCCACTTGTTCATCGTC-3′; tre#2, 5′-AGGCAGAAAGATGATCTC-3′).
Allelic exchange in A.nidulans
Transformation of A.nidulans strain FGSC773 was according to the procedure of Osmani et al. (16) using 5 µg of circular pTRE18, cTRE19 and cTRE20 or cTRE20 DNA that had been digested with SfiI or ClaI or treated with DNase I for various times and in the presence of manganese in order to achieve partial linearization at random sites on the cosmid (10). Uridine/uracil prototrophic transformants were scored for their ability to grow on minimal medium containing trehalose as the sole carbon source. Diffuse growth on this medium was indicative of an allelic replacement at the treA locus (8). Allelic replacement at the treA locus was tested by PCR using primers tre11 (5′-ATTGGTCTTCTGGGATG-3′) and tre10 (5′-CAAAAGCCGTAAAACTTACA-3′) which generate a 434 bp fragment only in strains that have an intact treA gene or by Southern hybridization of EcoRI-digested genomic DNA. In this case, a 2.0 kb EcoRV fragment of pTRE10 (8) corresponding to the 3′-end of treA was used as a probe and detected an 8.0 kb fragment in treA+ strains and a 4.7 kb fragment in strains containing the treAΔ::zeo/pyrG allele integrated either at the treA locus or at ectopic sites. The fact that integration had not occurred at ectopic sites or in multi-copy in treA– strains is confirmed by comparing the intensity of the signals obtained using the treA probe and a probe for a single copy gene.
RESULTS
Establishment of a recombination system
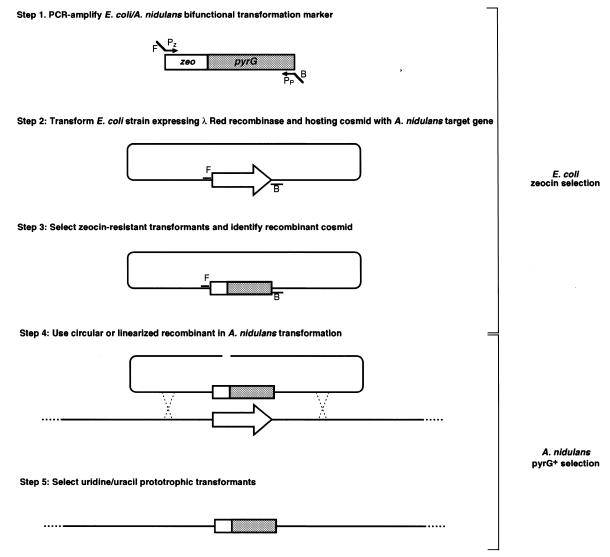
Molecular genetics of the filamentous fungus A.nidulans has been facilitated by the availability of chromosome-specific cosmid libraries and their corresponding physical maps (17,18). These libraries were constructed in the pWE15 and LORIST2 cosmid vectors which carry a ColE1 origin of replication and ampicillin and/or kanamycin resistance genes. Furthermore, DNA-mediated transformation of A.nidulans takes advantage of selective markers (e.g. pyrG which encode orotidine-5′-monophosphate decarboxylase and confers prototrophy when introduced into an A.nidulans pyrG uridine/uracil auxotroph) that are not functional in E.coli. Two plasmids were therefore designed for in vivo modification of cosmids that could be subsequently used to transform A.nidulans (Fig. 1).
Figure 1.
A two-step methodology for allelic exchange in A.nidulans. F and B refer to the homology extensions corresponding to regions flanking the target gene (open arrow). PZ and PP refer to priming sites on the bi-functional zeo/pyrG transformation marker.
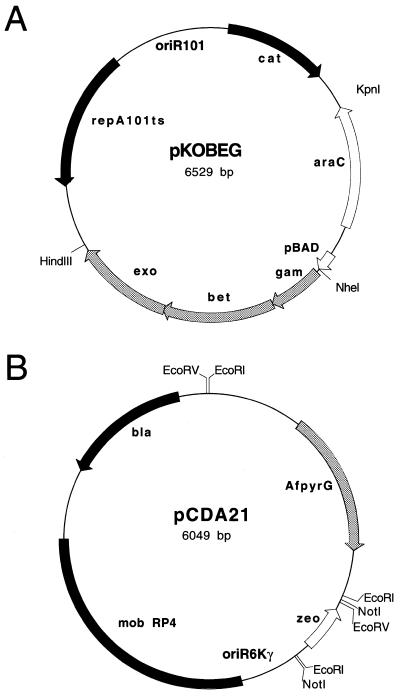
Plasmid pKOBEG (Fig. 2A) is a thermosensitive replicon that carries the λ phage redγβα operon (3) expressed under the control of the arabinose-inducible pBAD promotor (7). Because pKOBEG is a derivative of pSC101 and confers chloramphenicol resistance it can be propagated in E.coli together with most ColE1-derived plasmids. The functionality and inducibility of the Red functions encoded by pKOBEG were confirmed by inactivating the dnaK gene on the E.coli genome using a kanamycin-resistance gene flanked by 1000 bp of DNA homologous to the 5′ and 3′ regions of the dnaK gene. While none of the KmR transformants obtained from glucose-grown cells showed the appropriate gene replacement, at least 80% of the transformants obtained from arabinose-grown cells had occurred from a gene replacement at the dnaK locus (data not shown).
Figure 2.
Maps of recombination plasmid pKOBEG (A) and plasmid pCDA21 (B) carrying the zeo/pyrG bi-functional transformation marker.
Plasmid pCDA21 (Fig. 2B) contains a bi-functional transformation marker (zeo/pyrG) constructed using a zeocin resistance gene functional in E.coli and the A.fumigatus pyrG gene which can be used to select transformants of filamentous fungi that lack OMP-decarboxylase (e.g. A.nidulans pyrG, Aspergillus niger pyrA, Neurospora crassa pyr4 mutants). Because pCDA21 carries an R6Kγ origin of replication, it cannot replicate in E.coli strains that do not express the pir gene (14). Therefore, amplification products obtained from this plasmid can be introduced into E.coli pir– strains without the need for DpnI treatment or gel purification that has been used to eliminate the matrix DNA (4,5).
Allelic replacement on cosmids
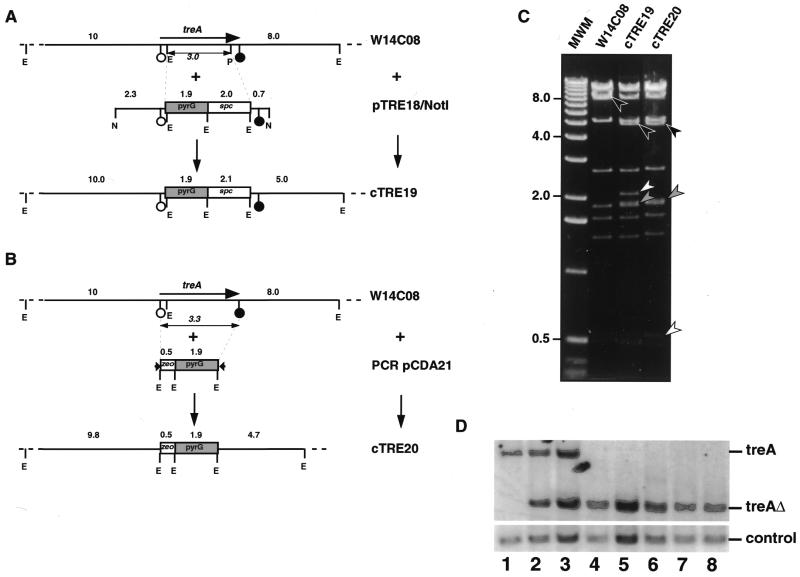
To validate the strategy outlined in Figure 1, we focused on the A.nidulans treA gene which encodes an acid trehalase required for growth of A.nidulans on trehalose: inactivation of treA can be easily scored by comparing growth of transformants on minimal medium containing glucose or trehalose as the sole carbon source (8). Cosmid W14C08 which contains the treA gene and flanking genomic region was introduced into an E.coli strain carrying pKOBEG. The resulting strain was transformed with (i) a 7 kb NotI fragment derived from plasmid pTRE18 and composed of a spc/pyrG marker flanked by 2.3 and 0.7 kb of DNA homologous to the 5′ and 3′ region of the treA gene, respectively or (ii) a PCR product obtained through amplification of the zeo/pyrG cassette of pCDA21 using oligonucleotides with 50 bp of homology to the 5′- or 3′-end of the treA coding region (regions F and B in Fig. 1) and 20 bp of homology in the zeo/pyrG cassette (regions PZ and PP in Fig. 1). Tens to hundreds of SpcR or ZeoR transformants were obtained in both cases. Characterization of a subset of these transformants by restriction enzyme analysis and PCR analysis showed that most resulted from the expected allelic exchange on cosmid W14C08. cTRE19 and cTRE20 (Fig. 3) are selected derivatives of W14C08 obtained using the NotI fragment of pTRE18 or PCR-amplified pCDA21, respectively. Sequencing of the region at the boundary between genomic DNA and the selective marker used for allelic replacement demonstrated exact gene replacement in cTRE20, confirming that 50 nt of homology are sufficient to promote recombination between the PCR product and a target gene located on an episome (5).
Figure 3.
Allelic exchange on cosmid W14C08. (A and B) Replacement using a 7.0 kb NotI fragment derived from pTRE18, yielding cosmid cTRE19 (A) or PCR-amplified pCDA21, yielding cosmid cTRE20 (B). Solid line, A.nidulans DNA; open box, spectinomycin-resistance gene (A) or zeocin-resistance gene (B); gray box, A.fumigatus pyrG gene; E, EcoRI; P, PmlI; N, NotI; arrow, A.nidulans treA coding region; open circle, treA start codon; solid circle, treA stop codon; double-arrow, deletion in the treA coding region resulting from the allelic exchange; arrowheads, 50 bp homology extensions present in the composite primers used to amplify the zeo/pyrG bi-functional transformation marker. The size of the EcoRI restriction fragments in the cosmids, pTRE18/NotI fragment and PCR product are indicated in kb. (C) Ethidium bromide-stained gel of EcoRI-digested W14C08, cTRE19 and cTRE20. The 8.0 kb fragment of W14C08 and the resulting 5.0 and 4.7 kb fragments in cTRE19 and cTRE20, respectively, are indicated by solid arrowheads. The 2.1 and 0.5 kb fragments corresponding to the spc (cTRE19) and zeo (cTRE20) genes, respectively, are indicated by open arrowheads. The 1.9 kb fragment corresponding to the A.fumigatus pyrG gene is indicated by a grey arrowhead. MWM, molecular weight markers, 1 kb ladder (Gibco BRL). Representative molecular weights are indicated in kb. (D) Southern hybridization of EcoRI-digested genomic DNA of A.nidulans FGSC773 (wild-type, lane 1) and of seven transformants: two treA+ transformants with ectopic integrations of pTRE20 (lanes 2 and 3) and five treA– transformants with an appropriate gene replacement at the treA locus (lanes 4–8). Genomic DNA was transferred onto nylon membranes and probed for treA (top) or a control single-copy gene (bottom). An 8.0 kb fragment is detected when treA is intact while a 4.7 kb fragment is detected when the treAΔ::(zeo/pyrG) allele is present. Similar intensities of the hybridization signals obtained using the treA and control probes demonstrates that the treAΔ::(zeo/pyrG) allele is present in a single copy.
Allelic replacement in A.nidulans
Allelic replacement in A.nidulans is obtained using linear replacement cassettes with DNA homologous to the target locus flanking a selectable transformation marker. However, we reasoned that allelic replacement at the treA locus might occur upon transformation with circular cTRE19 or cTRE20 because of the large region of homology to the flanking regions of the treA locus that are located on both sides of the pyrG gene in these cosmids. Hence, protoplasts of A.nidulans strain FGSC773 were transformed using circular cTRE19 and cTRE20 as well as pTRE12 and pTRE18, which carry standard replacement cassettes for the treA gene cloned into pBLUESCRIPT SN+ (8,19). Approximately 25 prototrophic transformants obtained with each plasmid were tested for their ability to grow on minimal trehalose medium. The results in Table 1 show that while no treA– transformants could be obtained upon transformation of FGSC773 by pTRE12 or pTRE18, up to 29% of the transformants obtained using cTRE20 had a treA– phenotype although the frequency of allelic replacement using cTRE20 was variable. Allelic replacement in five treA– transformants obtained with cTRE20 was confirmed by PCR (data not shown) and Southern analysis (Fig. 3), thus suggesting that in contrast to plasmids carrying standard allelic replacement cassettes, circular cosmids can efficiently recombine with the A.nidulans genome to induce allelic replacement. Furthermore, Southern analysis revealed that gene replacement events were not accompanied by the integration of ectopic copies of the transforming DNA (Fig. 3).
Table 1. Allelic replacement at the treA locus using circular or linearized recombinant cosmids.
| Plasmid/cosmid | Experimenta | Homology to treA locusb | treA–/totalc |
|---|---|---|---|
| Circular | |||
| pTRE12 | 1 | 2.3/1.2 | 0/25 |
| pTRE18 | 1 | 2.3/1.2 | 0/25 |
| cTRE19 | 1 | ND | 2/24 |
| cTRE20 | 1 | ND | 5/24 |
| cTRE20 | 2 | ND | 8/24 |
| cTRE20 | 3 | ND | 0/24 |
| Linearized | |||
| pTRE12/NotI | Ref. 10 | 2.3/0.7 | 4/75 |
| cTRE20/ClaI | 2 | ND | 13/25 |
| cTRE20/SfiI | 2 | ND | 2/3 |
| cTRE20/DNase I (1 min) | 3 | ND | 1/25 |
| cTRE20/DNase I (2 min) | 3 | ND | 3/25 |
| cTRE20/DNase I (5 min) | 3 | ND | 6/25 |
| cTRE20/DNase I (10 min) | 3 | ND | 2/20 |
| cTRE20/DNase I (15 min) | 3 | ND | 8/24 |
aThree independent experiments were performed.
bLength in kb of the DNA homologous with the treA locus located on each side of the selectable marker in pTRE12 and pTRE18. These values are not known but >5 kb for cTRE19 and cTRE20. ND, not determined.
cNumber of transformants with a treA– phenotype relative to the total number of transformants tested. The treA phenotype was scored on minimal trehalose medium.
In order to increase the frequency of allelic replacement, transformation of A.nidulans FGSC773 with linearized cTRE20 was tested. First, cTRE20 was treated with two restriction enzymes that do not cleave the zeo/pyrG marker and were known to generate large DNA fragments encompassing the mutated treA allele: ClaI has several cleavage sites in W14C08, including two located at both ends of a 10 kb fragment encompassing the treA gene; SfiI cleaves W14C08 (and hence cTRE20) at only one site in the vector backbone. The results in Table 1 show that using ClaI-treated cTRE20, allelic replacements at a frequency >50% could be obtained. A higher replacement frequency (66%) was observed with SfiI-treated cTRE20 but might be considered not significant because of the small number of transformants obtained in this experiment. These replacement frequencies were significantly higher than those observed when using NotI-digested pTRE12 (5.3%; 8) or with circular cTRE20 (see above). Second, cTRE20 was treated with DNase I to an extent where only linearization of the cosmid would occur (data not shown). The results in Table 1 show that the frequency of allelic replacement increased steadily with the extent of DNase I treatment.
DISCUSSION
In this report, we have presented a rapid, simple and efficient methodology to generate allelic replacements in the filamentous fungus A.nidulans. This methodology takes advantage of the Redα and Redβ recombination functions of phage λ to promote in E.coli the in vivo exchange of a wild-type allele located on a cosmid by a PCR-generated mutant allele. The resulting cosmid with the mutant allele is then used to transform A.nidulans and obtain allelic exchange with the chromosomal wild-type locus (Fig. 1). The method is rapid because it does not involve a single subcloning step and only requires limited sequence data on the target gene to be able to generate mutant cosmid. Indeed, we have confirmed the previously published results of Muyrers et al. (5) who showed that 50 bp of homology between a episomally-located gene and a PCR product is sufficient to sustain recombination when Redα and Redβ (or E.coli RecE and RecT) are produced in combination with Redγ which inhibits the RecBCD exonuclease V. The method is simple because the vectors that carry the recombination functions and the zeo/pyrG bi-functional marker have been optimized to be compatible with ColE1-derived cosmids and to avoid purification and DpnI treatment of the PCR product that is used to generate the mutant cosmid. Finally, the method is efficient because allelic replacements on cosmids are obtained at high frequencies and, most interestingly, allelic replacements in A.nidulans are obtained at frequencies that are unprecedented in this fungus. In particular, we have shown that allelic replacement at the A.nidulans treA locus could be obtained at frequencies up to 30% when using a circular recombinant cosmid while allelic exchange is never observed when a vector carrying a standard replacement cassette with 1–2 kb of homology to the target locus is used in a circular form. Furthermore, by linearizing the recombinant cosmid we have been able to increase the replacement frequency to above 50% which is significantly higher than the replacement efficiency we had observed at this locus with a standard replacement cassette (5%; 10) which was close to the 5–10% replacement efficiency commonly observed in A.nidulans and other filamentous fungi when using this type of methodology. Although linearization is not always possible because of the lack of knowledge of the genomic region flanking the target gene, the design of new cosmid libraries with a very rare cleavage site (e.g. for a meganuclease) and the increasing knowledge of genome sequence should facilitate this step. Nevertheless, we have experienced that in most instances the use of a circular cosmid is sufficient to identify the appropriate allelic replacement within a limited population of transformants (M.-K.Chaveroche and C.d’Enfent, unpublished data).
Although pCDA21 has been developed for the generation of allelic replacements in A.nidulans, it can be used to create similar events in filamentous fungi where OMP-decarboxylase mutants are available (e.g. A.niger pyrA and N.crassa pyr-4 mutants). In this plasmid, we have used the A.fumigatus gene to prevent recombination in A.nidulans between the selectable marker and the endogenous pyrG89 allele. A similar cassette has been designed that uses the A.nidulans pyrG gene and can therefore be used to generate allelic replacements in A.fumigatus (M.-K.Chaveroche and C.d’Enfent, unpublished data). Furthermore, new cassettes are being developed using dominant selectable markers, in particular hygromycin resistance, which has proven to be functional in a wide variety of filamentous fungi without the need for an auxotrophic mutant (20).
In this study, we have applied this two-step replacement methodology to exchange the entire open reading frame of the A.nidulans treA gene with a selectable marker. Because the site of recombination of the selectable marker at the target locus in the cosmid is precisely defined by the sequence of the oligonucleotides that are used for its amplification, other types of allelic exchanges and hence mutations can be generated using this methodology. In particular it should be possible to generate conditional mutants by promoter replacement or gene fusions provided that allelic replacement cassettes with a regulatable promoter or a reporter gene in addition to the selectable marker are available.
Acknowledgments
ACKNOWLEDGEMENTS
We thank K. Murphy, J. Beckwith, G. Church, H. Krish and D. Mazel for providing the different plasmids that were used in the course of this study.
REFERENCES
- 1.van den Hondel C.A.M.J.J. and Punt,P.J. (1991) In Peberdy,J.F., Caten,C.E., Ogden,J.E. and Bennett,J.W. (eds), Applied Molecular Genetics of Fungi. Cambridge University Press, Cambridge, UK, pp. 1–28.
- 2.d’Enfert C., Weidner,G. and Brakhage,A.A. (1999) In Brakhage,A.A., Jahn,B. and Schmidt,A. (eds), Aspergillus fumigatus: Biology, Clinics and Molecular Approaches to Pathogenicity. S. Karger AG, Basel, Switzerland, Vol. 2, pp. 149–166.
- 3.Murphy K.C. (1998) J. Bacteriol., 180, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Buchholz,F., Muyrers,J.P.P. and Stewart,A.F. (1998) Nature Genet., 20, 123–128. [DOI] [PubMed] [Google Scholar]
- 5.Muyrers J.P.P., Zhang,Y., Testa,G. and Stewart,A.F. (1999) Nucleic Acids Res., 27, 1555–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko K.A. and Wanner,B.L. (2000) Proc. Natl Acad. Sci. USA, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman L.M., Belin,D., Carson,M.J. and Beckwith,J. (1995) J. Bacteriol., 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.d’Enfert C. and Fontaine,T. (1997) Mol. Microbiol., 24, 203–216. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1992) Short Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 11.Hanahan D., Jessee,J. and Bloom,F.R. (1991) Methods Enzymol., 204, 63–113. [DOI] [PubMed] [Google Scholar]
- 12.Link A.J., Phillips,D. and Church,G.M. (1997) J. Bacteriol., 179, 6228–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prentki P. and Krisch,H.M. (1984) Gene, 29, 303–313. [DOI] [PubMed] [Google Scholar]
- 14.Miller V.L. and Mekalanos,J.J. (1988) J. Bacteriol., 170, 2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weidner G., d’Enfert,C., Koch,A., Mol,P.C. and Brakhage,A.A. (1998) Curr. Genet., 33, 378–385. [DOI] [PubMed] [Google Scholar]
- 16.Osmani S.A., May,G.S. and Morris,R.N. (1987) J. Cell Biol., 104, 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prade R.A., Griffith,J., Kochut,K., Arnold,J. and Timberlake,W.E. (1997) Proc. Natl Acad. Sci. USA, 94, 14564–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brody H., Griffith,J., Cuticchia,A.J., Arnold,J. and Timberlake,W.E. (1991) Nucleic Acids Res., 19, 3105–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.d’Enfert C., Diaquin,M., Delit,A., Wuscher,N., Debeaupuis,J.-P., Huerre,M. and Latgé,J.-P. (1996) Infect. Immunol., 64, 4401–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Punt P.J. and van den Hondel,C.A. (1992) Methods Enzymol., 216, 447–457. [DOI] [PubMed] [Google Scholar]